Abstract
A series of novel naphthalimide aminothiazoles were developed for the first time and evaluated for their antimicrobial activity. Some prepared compounds possessed good inhibitory activity against the tested bacteria and fungi. Noticeably, the piperazine derivative 4d displayed superior antibacterial activity against MRSA and Escherichia coli with MIC values of 4 and 8 μg/mL, respectively, to reference drugs. The most active compound 4d showed low toxicity against mammalian cells with no obvious triggering of the development of bacterial resistance, and it also possessed rapid bactericidal efficacy and efficient membrane permeability. Preliminarily investigations revealed that compound 4d could not only bind with gyrase–DNA complex through hydrogen bonds but could effectively intercalate into MRSA DNA to form 4d–DNA supramolecular complex, which might be responsible for the powerful bioactivity. Further transportation behavior evaluation indicated that molecule 4d could be effectively stored and carried by human serum albumin, and the hydrophobic interactions and hydrogen bonds played important roles in the binding process.
Keywords: Naphthalimide, aminothiazole, antibacterial, MRSA DNA, human serum albumin
The emergence and spread of multidrug-resistant bacteria resulting from the overuse and misuse of antibiotics have been a major obstacle to the successful treatment of infectious diseases.1−3 Methicillin-resistant Staphylococcus aureus (MRSA) is currently a major nosocomial pathogen, which arises from the acquisition of staphylococcal cassette chromosome mec (SCCmec) through horizontal gene transfer in methicillin-susceptible Staphylococcus aureus. An altered penicillin-binding protein PBP2a expressed by SCCmec has fairly lower binding affinity for nearly all available β-lactam antibiotics, making them invalid against MRSA.3 Despite numerous efforts devoted to ensure sterility and limit the spread of MRSA, the morbidity and mortality rates still remain high per year. This deeply highlights the urgent and unmet medical need for developing a new type of potential antimicrobial agents against MRSA.4
Naphthalimides are highly versatile functional compounds, which have received an increasing attention in the development of DNA-binding agents,5 and established great therapeutic importance in medicinal chemistry.6,7 Especially as anticancer agents, some naphthalimides such as amonafide, mitonafide, and aristolochic acid effectively inhibit the growth of various human and murine cancer cell lines. This type of compound exerts its anticancer function by intercalating into DNA to directly inhibit the normal replication and transcription of DNA, and/or into topoisomerase (TOP) II to stabilize supramolecular complex of TOP and DNA and to prevent the correction of the topology structure of DNA.8 This special interaction model of naphthalimides with DNA has been recently applied to the rational discovery of potential antibacterial and antifungal agents, and some great achievements have been acquired.9,10
Aminothiazole is a beneficial structural fragment for bioactivity and prevalently present in a broad range of therapeutic agents11 due to its unique structure with both electron-donating groups (NH2, −S−) and an electron-accepting group (C=N), which could readily interact with DNA and many other biomacromolecules.12,13 A lot of drugs containing aminothiazole fragment have been launched into the market,14,15 such as the antibacterial cephalosporins and sulfathiazole, the antifungal drug abafungin. The successful development of various clinical aminothiazole drugs has provoked extensive studies to construct more bioactive molecules with aminothiazole fragment in antimicrobial field.16−18
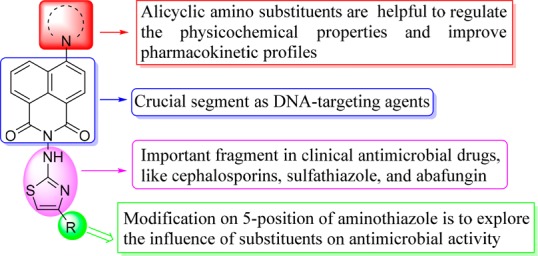
In view of this, we introduced aminothiazole moiety into the N-position of the naphthalimide backbone to generate novel naphthalimide–aminothiazole hybrids and investigated their effect on antimicrobial activity.19 It is known that the bioactivities of naphthalimides are significantly affected by functional groups on the naphthalimide ring especially at the 3- or 4-position,6 which are beneficial for targeting biomolecules. Alicyclic amines such as piperazine, morpholine, and pyrrolidine are important structural fragments in regulating the physicochemical properties and improving pharmacokinetic profiles,20 and they are prevalently present in numerous clinical antimicrobial agents. Rationally, various alicyclic amine moieties were incorporated into the 4-position of naphthalimide skeleton to study their influence on bioactive profiles (Figure 1).9
Figure 1.
Design of novel naphthalimide aminothiazoles.
The antimicrobial screening was executed among naphthalimide aminothiazoles 3–5 to investigate their biological activities. Bacterial resistance study, bactericidal kinetic assay, and cytotoxicity were explored to further assess the potentiality of the bioactive compounds as new antimicrobial agents. Additionally, the antimicrobial mechanism was also investigated through membrane permeabilization assay, molecular docking study with gyrase–DNA complex, and interaction with MRSA DNA. The transportation behavior of human serum albumin (HSA) to a highly active compound was also performed.
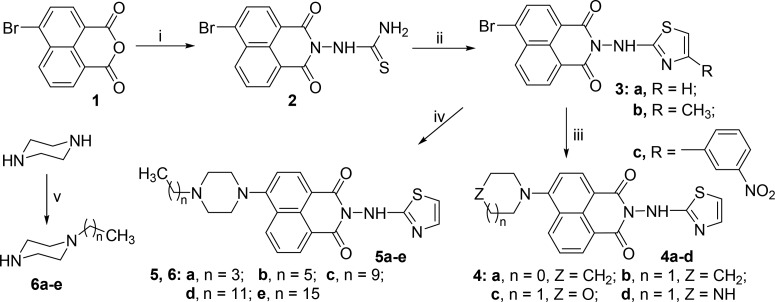
The target naphthalimide aminothiazoles 3–5 were synthesized via multistep reactions starting from commercial 4-bromo-1,8-naphthalic anhydride according to Scheme 1. Condensation of 4-bromo-1,8-naphthalic anhydride and thiosemicarbazide afforded compound 2 in 60.1% yield, which was further cyclized with the α-halogenated carbonyl compounds in ethanol at 80 °C via the typical Hantsch thiazole synthesis to give aminothiazole derivatives 3a–c in yields of 21.1–40.2%.17 The N-alkylation of piperazine with alkyl halides generated monosubstituted alkyl piperazines 6a–e with yields of 35.1–80.0%. Compound 3a was further treated with alicyclic amines in 2-methoxyethanol at 125 °C under nitrogen atmosphere to achieve the desired compounds 4a–d in yields of 30.0–46.2%. Under the same reaction conditions, the substitution of bromo atom in compound 3a by piperazinyl derivatives 6a–e produced the target naphthalimide aminothiazoles 5a–e with yields ranging from 26.0% to 40.6%.9 All the new compounds were confirmed by 1H NMR, 13C NMR, IR, and HRMS spectra. Purity was determined with the help of the quantitative nuclear magnetic resonance (QNMR) method using 1,3,5-trioxane as the internal standard. All the target compounds were found to be >95% pure, and the spectral data are in the Supporting Information.
Scheme 1. Synthetic Route of Naphthalimide Aminothiazoles 3–5.
Reagents and conditions: (i) thiosemicarbazide, DMF, 100 °C, 8 h; (ii) α-halogenated carbonyl compounds, ethanol, 80 °C, 7 h; (iii) alicyclic amines, 2-methoxyethanol, reflux, N2, 7 h; (iv) N-alkylated piperazines, 2-methoxyethanol, reflux, N2, 7 h; (v) alkyl halides, ethanol, 80 °C, 8 h.
The antimicrobial data evaluated using a two-fold serial dilution technique recommended by Clinical and Laboratory Standards Institute (CLSI)21 are depicted in Table S1. Some prepared compounds could effectively inhibit the growth of the tested strains in vitro. Preliminary active screening showed that aminothiazole derivative 3a possessed slightly better antibacterial efficacy than 4-substituted aminothiazoles 3b–c. It suggested that the modification on the 4-position of the aminothiazole ring might be unfavorable for the antibacterial activity. Among the series of compounds 4a–d, morpholine-modified naphthalimide 4c gave the strongest anti-A. baumanii potency (MIC = 8 μg/mL), two-fold more potent than chloromycin (MIC = 16 μg/mL) and equivalent to norfloxacin (MIC = 8 μg/mL). Notably, piperazinyl derivative 4d could effectively inhibit the growth of all the tested bacteria, except for A. baumanii. Especially against Gram-positive bacteria, this compound demonstrated efficient bioactivity against S. aureus 29213 (MIC = 2 μg/mL), S. aureus 25923 (MIC = 4 μg/mL), and MRSA (MIC = 4 μg/mL), which was more active than chloromycin (MICs = 4, 8, and 16 μg/mL, respectively). Besides, it was also effective in inhibiting Gram-negative bacteria including E. coli, E. coli 25922, P. aeruginosa, and P. aeruginosa 27853 at low concentrations (MICs = 4–16 μg/mL), particularly against E. coli 25922 strain, which was four- and two-fold more potent than chloromycin (MIC = 16 μg/mL) and norfloxacin (MIC = 8 μg/mL), respectively. These indicated that target molecule 4d had immense potentiality to be a lead compound in the development of more effective board-spectrum antimicrobial agents. Compounds 4a and 4b with pyrrolidinyl and piperidinyl substituents gave unfavorable antibacterial profile against the tested strains. Among six-membered alicyclic amine derivatives 4b–d, it was noteworthy that the higher active compound 4d and the lower active ones 4b–c were attributable solely to the different substitution on the 4-position of six-membered alicyclic amines, which revealed that the NH group had quite a positive influence on bioactivity compared to O or CH2 moiety. So it would be interesting and rational to continue our work to modify the N-position on piperazine ring of derivative 4d.
Further studies were focused on the effect of the alkyl chain in the piperazine ring of compound 4d on antimicrobial activity because a large amount of work had revealed the importance of the alkyl chain in affecting antimicrobial efficacy.9 Therefore, the alkyl groups were introduced into compound 4d to produce derivatives 5a–e with conspicuous loss of inhibitory efficacies against the tested pathogens, which suggested the necessity of a NH group for antimicrobial activity probably due to its participation in formation of hydrogen bonds so as to helpfully interact with biomolecules in biological system. Besides, the lengths of alkyl chains have different effects on biological activity in which analogue 5b with the hexyl group gave better antibacterial activity in comparison with other alkyl derivatives. Moreover, when the alkyl substituents were extended to decyl, dodecyl, and hexadecyl groups, compounds 5c–e displayed weak or no obvious activity in inhibiting the growth of the tested bacterial strains. This fact pointed out that only a suitable length of alkyl chain in piperazine ring was necessary for good antibacterial activity. The antifungal activities (Table S2) were also discussed and are in the Supporting Information.
Many of the clinically approved drugs have suffered defeat from the resistant clinical strains due to the decreased potency or increased susceptibility to the degradation mechanisms.22 Thereby, it would be important to evaluate the ability of compound 4d to suppress the development of resistance. The resistant pathogen MRSA was employed as the tested strains. Norfloxacin and chloromycin were used as positive control. As shown in Figure 2, this result indicated that MRSA was much more difficult to develop resistance against compound 4d than the clinical drugs, and such structure might be a promising candidate for an anti-MRSA agent with considerable therapeutic efficacy and low drug-resistance.23
Figure 2.
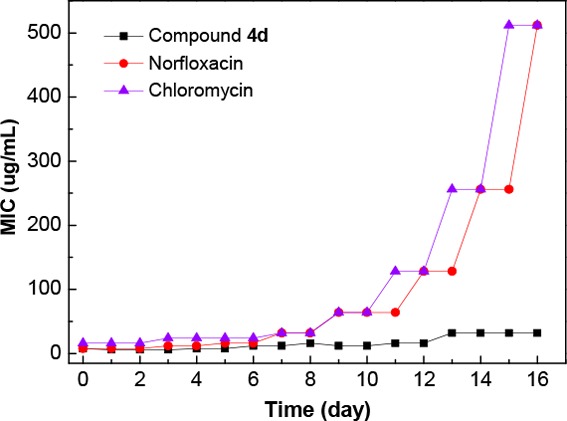
Propensity to induce bacterial resistance of compound 4d against MRSA.
To understand the antibacterial nature of the piperazinyl derivative 4d, time–kill kinetics assay was investigated against MRSA, which would provide information about the bactericidal activity of compound 4d against the tested bacteria.24 As can be seen from Figure S1, compound 4d could cause more than 3-Log (CFU/mL) reduction in the number of viable bacteria within an hour at a concentration of 4 × MIC. It was suggested that this compound showed rapid bactericidal activity against MRSA.
The selectivity toward mammalian cells over bacterial cells has been one of the major concerns in the development of clinically available antibacterial agents for biomedical applications.25,26 The active molecule 4d was further evaluated for its cytotoxicity against normal rat macrophage cells (RAW 264.7) using the MTT assay. Results in Figure S2 showed that the cell viability of compound 4d was at least 80.1% within a concentration of 128 μg/mL. This indicated that compound 4d exhibited relatively low toxicity toward mammalian cells within the MIC values against most of the test bacteria.
Membrane-targeting antimicrobials provide potential solutions for the problem of bacterial resistance, for the reason that the membrane-active nature gives low propensity for the development of drug resistance unless there are large-scale changes in the membrane compositions of bacteria. Therefore, bacterial membrane has been considered as a particularly valuable antibacterial target to overcome resistance.27 Compound 4d was further assessed for the ability to permeabilize the cytoplasmic membrane of bacteria using propidium iodide (PI) dye.28 As can be seen from Figure S3, the rapid fluorescence intensity enhancements of the mixtures of compound 4d and PI-treated MRSA or Escherichia coli appeared and became steady after 60 min, while the two control groups almost kept constant, which proved that the tested compound 4d was efficient in permeabilizing the membranes of both Gram-positive (MRSA) and Gram-negative (Escherichia coli) bacteria.
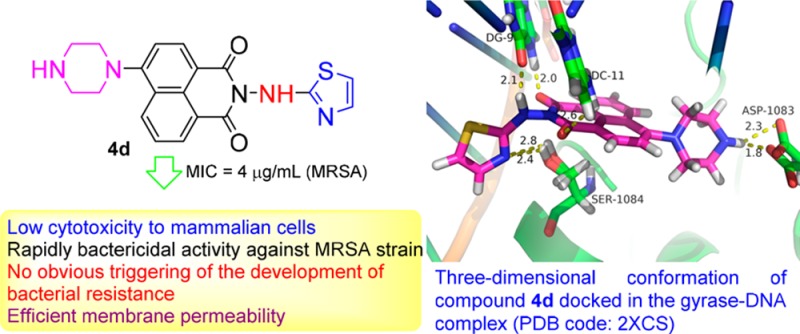
Molecular docking investigation was performed to rationalize the observed antibacterial activity and explore the possible action mechanism of hybrid 4d.29 Crystal structure of gyrase–DNA complex was selected as a representative target in the Protein Data Bank (PDB code: 2XCS). The docking mode with the lowest binding energy (−11.95 kcal mol–1) was shown in Figures 3 and S4. Molecule 4d could interact with gyrase–DNA complex through the formation of hydrogen bonds between the two carbonyl groups of the naphthalimide backbone and base pairs of DNA (DG-11 and DG-9). The amino group of aminothiazole was also adjacent to DG-9 base pairs with distance of 2.1 Å. Besides, derivative 4d could form two hydrogen bonds with residue SER-1084 of gyrase–DNA complex through the nitrogen atom of the thiazole ring. Furthermore, the NH group of the piperazine ring in the 4-position of the naphthalimide fragment was in close proximity to the residue ASP-1083, which demonstrated the necessity of the NH group for the increased bioactivity and also rationalized the obtained antimicrobial result that introduction of alkyl groups at the piperazine ring of compounds 5a–e could lead to the decreased antimicrobial potency. Compound 4d was also disclosed to be involved in hydrophobic interactions at the active site with the hydrophobic residues. All of these cooperative bindings might be helpful to stabilize the 4d–enzyme–DNA supramolecular complex, which might be responsible for the good inhibitory efficacy of compound 4d against the tested strains.
Figure 3.
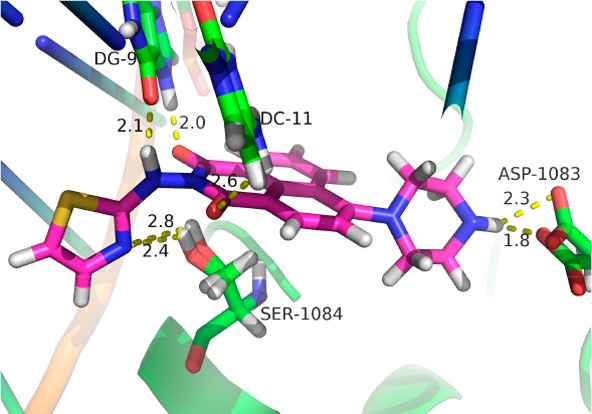
Three-dimensional conformation of compound 4d docked in the gyrase–DNA complex (PDB code: 2XCS).
The research of drug–DNA interactions is of great importance and interest in rational design and construction of novel and effective DNA–targeting drugs.30 The interaction between the highly active compound 4d and MRSA DNA was explored to further study the preliminary mechanism of antimicrobial action at the molecular level by UV–vis spectroscopy. The results revealed that compound 4d could effectively intercalate into MRSA DNA to form 4d–DNA complex, which might block DNA replication and thus exert powerful antimicrobial activity (Supporting Information).
Further binding behavior between compound 4d and HSA was carried out to preliminarily evaluate their transportation and pharmacokinetic properties. HSA is the principal extracellular protein of the circulatory system.31 Thorough investigations involving the interactions between drugs and HSA could not only provide the absorption, transportation, distribution, metabolism, and excretion properties of drugs but also be instructive to design, modify, and screen new drug molecules.32,33 The transportation behavior disclosed that the association of molecule 4d with HSA was spontaneous, and the hydrophobic interactions and hydrogen bonds played important roles in the binding process. The calculated parameters indicated that compound 4d could be effectively stored and carried by HSA (Supporting Information).
In conclusion, a series of novel naphthalimide–aminothiazole hybrids were successfully developed via a convenient synthetic strategy. The structure–activity relationships were summarized in Figure 4. Piperazinyl derivative 4d exhibited broad antibacterial spectrum. Particularly, it could effectively inhibit the growth of MRSA and Escherichia coli with MIC values of 4 and 8 μg/mL, respectively. Furthermore, compound 4d was membrane active and had low toxicity against mammalian cells, and it could rapidly kill MRSA and did not obviously trigger bacterial resistance. Preliminary research revealed that compound 4d could not only bind with gyrase–DNA complex through hydrogen bonds but also form a steady 4d–DNA complex with MRSA DNA by intercalation, which might be responsible for the powerful bioactivity. The binding behavior revealed that the association of HSA with molecule 4d was spontaneous, and the hydrophobic interactions and hydrogen bonds played significant roles in the transportation of 4d. Thereby naphthalimide aminothiazole 4d might be a potential multitargeting anti-MRSA candidate. Further researches such as accurate antimicrobial action mechanism, metabolic stability, and synergism with clinical drugs are now underway in our group.
Figure 4.
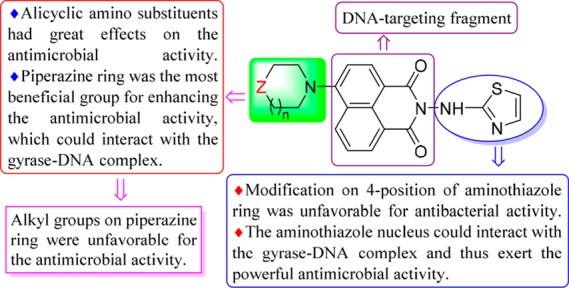
Preliminary summary of the structure–activity relationships for naphthalimide aminothiazoles.
Acknowledgments
Grateful appreciation for the help of Dr. Gao Wei-Wei in polishing language of the revised manuscript.
Glossary
ABBREVIATIONS
- DMF
dimethylformamide
- MIC
minimal inhibitory concentration that completely inhibited the growth of microbes
- HSA
human serum albumin
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.7b00452.
Experimental procedures, compounds characterization, antibacterial and antifungal activities data, bactericidal kinetic assay, cytotoxicity, bacterial membrane permeabilization, molecular docking data, and interactions with MRSA DNA and HSA of compound 4d (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
This work was partially supported by National Natural Science Foundation of China [(No. 21672173, 21372186), the Research Fund for International Young Scientists from International (Regional) Cooperation and Exchange Program (No. 81650110529)], and Chongqing Special Foundation for Postdoctoral Research Proposal (No. Xm2017184, Xm2017185).
The authors declare no competing financial interest.
Supplementary Material
References
- Brown E. D.; Wright G. D. Antibacterial Drug Discovery in the Resistance Era. Nature 2016, 529, 336–343. 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Peng X. M.; Damu G. L. V.; Geng R. X.; Zhou C. H. Comprehensive Review in Current Developments of Imidazole-Based Medicinal Chemistry. Med. Res. Rev. 2014, 34, 340–437. 10.1002/med.21290. [DOI] [PubMed] [Google Scholar]
- Lin S. M.; Koh J. J.; Aung T. T.; Sin W. L. W.; Lim F. H.; Wang L.; Lakshminarayanan R.; Zhou L.; Tan D. T. H.; Cao D.; Beuerman R. W.; Ren L.; Liu S. P. Semisynthetic Flavone-Derived Antimicrobials with Therapeutic Potential against Methicillin-Resistant Staphylococcus aureus (MRSA). J. Med. Chem. 2017, 60, 6152–6165. 10.1021/acs.jmedchem.7b00380. [DOI] [PubMed] [Google Scholar]
- Peng X. M.; Damu G. L. V.; Zhou C. H. Current Developments of Coumarin Compounds in Medicinal Chemistry. Curr. Pharm. Des. 2013, 19, 3884–3930. 10.2174/1381612811319210013. [DOI] [PubMed] [Google Scholar]
- Gong H. H.; Addla D.; Lv J. S.; Zhou C. H. Heterocyclic Naphthalimides as New Skeleton Structure of Compounds with Increasingly Expanding Relational Medicinal Applications. Curr. Top. Med. Chem. 2016, 16, 3303–3364. 10.2174/1568026616666160506145943. [DOI] [PubMed] [Google Scholar]
- Banerjee S.; Veale E. B.; Phelan C. M.; Murphy S. A.; Tocci G. M.; Gillespie L. J.; Frimannsson D. O.; Kelly J. M.; Gunnlaugsson T. Recent Advances in the Development of 1,8-Naphthalimide Based DNA Targeting Binders, Anticancer and Fluorescent Cellular Imaging Agents. Chem. Soc. Rev. 2013, 42, 1601–1618. 10.1039/c2cs35467e. [DOI] [PubMed] [Google Scholar]
- Lv J. S.; Peng X. M.; Kishore B.; Zhou C. H. 1,2,3-Triazole-Derived Naphthalimides as a Novel Type of Potential Antimicrobial Agents: Synthesis, Antimicrobial Activity, Interaction with Calf Thymus DNA and Human Serum Albumin. Bioorg. Med. Chem. Lett. 2014, 24, 308–313. 10.1016/j.bmcl.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Dai F. J.; Li Q.; Wang Y. X.; Ge C. C.; Feng C. Y.; Xie S. Q.; He H. Y.; Xu X. J.; Wang C. J. Design, Synthesis, and Biological Evaluation of Mitochondria-Targeted Flavone–Naphthalimide Polyamine Conjugates with Antimetastatic Activity. J. Med. Chem. 2017, 60, 2071–2083. 10.1021/acs.jmedchem.6b01846. [DOI] [PubMed] [Google Scholar]
- Gong H. H.; Baathulaa K.; Lv J. S.; Cai G. X.; Zhou C. H. Synthesis and Biological Evaluation of Schiff Base-linked Imidazolyl Naphthalimides as Novel Potential Anti-MRSA Agents. MedChemComm 2016, 7, 924–931. 10.1039/C5MD00574D. [DOI] [Google Scholar]
- Damu G. L. V.; Wang Q. P.; Zhang H. Z.; Zhang Y. Y.; Lv J. S.; Zhou C. H. A Series of Naphthalimide Azoles: Design, Synthesis and Bioactive Evaluation as Potential Antimicrobial Agents. Sci. China: Chem. 2013, 56, 952–969. 10.1007/s11426-013-4873-1. [DOI] [Google Scholar]
- Cui S. F.; Wang Y.; Lv J. S.; Damu G. L. V.; Zhou C. H. Recent Advances in Application of Thiazole Compounds. Zhongguo Kexue: Huaxue 2012, 42, 1105–1131. 10.1360/032011-788. [DOI] [Google Scholar]
- Gao W. W.; Zhou C. H. Antimicrobial 2-Aminothiazolyl Quinolones: What Is their Potential in the Clinic?. Future Med. Chem. 2017, 9, 1461–1464. 10.4155/fmc-2017-0108. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Avula S. R.; Gao W. W.; Addla D.; Tangadanchu V. K. R.; Zhang L.; Lin J. M.; Zhou C. H. Multi-targeting Exploration of New 2-Aminothiazolyl Quinolones: Synthesis, Antimicrobial Evaluation, Interaction with DNA, Combination with Topoisomerase IV and Penetrability into Cells. Eur. J. Med. Chem. 2016, 124, 935–945. 10.1016/j.ejmech.2016.10.011. [DOI] [PubMed] [Google Scholar]
- Das D.; Sikdar P.; Bairagi M. Recent Developments of 2-Aminothiazoles in Medicinal Chemistry. Eur. J. Med. Chem. 2016, 109, 89–98. 10.1016/j.ejmech.2015.12.022. [DOI] [PubMed] [Google Scholar]
- Peng L. P.; Nagarajan S.; Rasheed S.; Zhou C. H. Synthesis and Biological Evaluation of a New Class of Quinazolinone Azoles as Potential Antimicrobial Agents and their Interactions with Calf Thymus DNA and Human Serum Albumin. MedChemComm 2015, 6, 222–229. 10.1039/C4MD00281D. [DOI] [Google Scholar]
- Mondal S.; Mandal S. M.; Mondal T. K.; Sinha C. Structural Characterization of New Schiff Bases of Sulfamethoxazole and Sulfathiazole, their Antibacterial Activity and Docking Computation with DHPS Protein Structure. Spectrochim. Acta, Part A 2015, 150, 268–279. 10.1016/j.saa.2015.05.049. [DOI] [PubMed] [Google Scholar]
- Cui S. F.; Addla D.; Zhou C. H. Novel 3-Aminothiazolquinolones: Design, Synthesis, Bioactive Evaluation, SARs, and Preliminary Antibacterial Mechanism. J. Med. Chem. 2016, 59, 4488–4510. 10.1021/acs.jmedchem.5b01678. [DOI] [PubMed] [Google Scholar]
- Addla D.; Wen S. Q.; Gao W. W.; Maddili S. K.; Zhang L.; Zhou C. H. Design, Synthesis, and Biological Evaluation of Novel Carbazole Aminothiazoles as Potential DNA-targeting Antimicrobial Agents. MedChemComm 2016, 7, 1988–1994. 10.1039/C6MD00357E. [DOI] [Google Scholar]
- Marco-Contelles J.; Soriano E. The Medicinal Chemistry of Hybrid-based Drugs Targeting Multiple Sites of Action. Curr. Top. Med. Chem. 2011, 11, 2714–2715. 10.2174/156802611798184382. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Addla D.; Ponmani J.; Wang A.; Xie D.; Wang Y. N.; Zhang S. L.; Geng R. X.; Cai G. X.; Li S.; Zhou C. H. Discovery of Membrane Active Benzimidazole Quinolones-Based Topoisomerase Inhibitors as Potential DNA-binding Antimicrobial Agents. Eur. J. Med. Chem. 2016, 111, 160–182. 10.1016/j.ejmech.2016.01.052. [DOI] [PubMed] [Google Scholar]
- Wang X. L.; Wan K.; Zhou C. H. Synthesis of Novel Sulfanilamide-derived 1,2,3-Triazoles and their Evaluation for Antibacterial and Antifungal Activities. Eur. J. Med. Chem. 2010, 45, 4631–4639. 10.1016/j.ejmech.2010.07.031. [DOI] [PubMed] [Google Scholar]
- Li Z. Z.; Gopala L.; Tangadanchu V. K. R.; Gao W. W.; Zhou C. H. Discovery of Novel Nitroimidazole Enols as Pseudomonas aeruginosa DNA Cleavage Agents. Bioorg. Med. Chem. 2017, 25, 6511–6522. 10.1016/j.bmc.2017.10.028. [DOI] [PubMed] [Google Scholar]
- Konai M. M.; Ghosh C.; Yarlagadda V.; Samaddar S.; Haldar J. Membrane Active Phenylalanine Conjugated Lipophilic Norspermidine Derivatives with Selective Antibacterial Activity. J. Med. Chem. 2014, 57, 9409–9423. 10.1021/jm5013566. [DOI] [PubMed] [Google Scholar]
- Fang X. F.; Li D.; Tangadanchu V. K. R.; Gopala L.; Gao W. W.; Zhou C. H. Novel Potentially Antifungal Hybrids of 5-Flucytosine and Fluconazole: Design, Synthesis and Bioactive Evaluation. Bioorg. Med. Chem. Lett. 2017, 27, 4964–4969. 10.1016/j.bmcl.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Cui S. F.; Peng L. P.; Zhang H. Z.; Rasheed S.; Vijaya Kumar K.; Zhou C. H. Novel Hybrids of Metronidazole and Quinolones: Synthesis, Bioactive Evaluation, Cytotoxicity, Preliminary Antimicrobial Mechanism and Effect of Metal Ions on their Transportation by Human Serum Albumin. Eur. J. Med. Chem. 2014, 86, 318–334. 10.1016/j.ejmech.2014.08.063. [DOI] [PubMed] [Google Scholar]
- Ma Z.; Yang J.; Han J. Z.; Gao L.; Liu H. X.; Lu Z. X.; Zhao H. Z.; Bie X. M. Insights into the Antimicrobial Activity and Cytotoxicity of Engineered α-Helical Peptide Amphiphiles. J. Med. Chem. 2016, 59, 10946–10962. 10.1021/acs.jmedchem.6b00922. [DOI] [PubMed] [Google Scholar]
- Jeyakkumar P.; Zhang L.; Avula S. R.; Zhou C. H. Design, Synthesis and Biological Evaluation of Berberine-Benzimidazole Hybrids as New Type of Potentially DNA-targeting Antimicrobial Agents. Eur. J. Med. Chem. 2016, 122, 205–215. 10.1016/j.ejmech.2016.06.031. [DOI] [PubMed] [Google Scholar]
- Gao W. W.; Rasheed S.; Tangadanchu V. K. R.; Sun Y.; Peng X. M.; Cheng Y.; Zhang F. X.; Lin J. M.; Zhou C. H. Design, Synthesis and Biological Evaluation of Amino Organophosphorus Imidazoles as a New Type of Potential Antimicrobial Agents. Sci. China: Chem. 2017, 60, 769–785. 10.1007/s11426-016-9009-6. [DOI] [Google Scholar]
- Zhang H. Z.; He S. C.; Peng Y. J.; Zhang H. J.; Gopala L.; Tangadanchu V. K. R.; Gan L. L.; Zhou C. H. Design, Synthesis and Antimicrobial Evaluation of Novel Benzimidazole-Incorporated Sulfonamide Analogues. Eur. J. Med. Chem. 2017, 136, 165–183. 10.1016/j.ejmech.2017.04.077. [DOI] [PubMed] [Google Scholar]
- Peng X. M.; Peng L. P.; Li S.; Avula S. R.; Kannekanti V. K.; Zhang S. L.; Tam K. Y.; Zhou C. H. Quinazolinone Azolyl Ethanols: Potential Lead Antimicrobial Agents with Dual Action Modes Targeting MRSA DNA. Future Med. Chem. 2016, 8, 1927–1940. 10.4155/fmc-2016-0002. [DOI] [PubMed] [Google Scholar]
- Hu Y. J.; Liu Y.; Xiao X. H. Investigation of the Interaction between Berberine and Human Serum Albumin. Biomacromolecules 2009, 10, 517–521. 10.1021/bm801120k. [DOI] [PubMed] [Google Scholar]
- Yin B. T.; Yan C. Y.; Peng X. M.; Zhang S. L.; Rasheed S.; Geng R. X.; Zhou C. H. Synthesis and Biological Evaluation of α-Triazolyl Chalcones as a New Type of Potential Antimicrobial Agents and their Interaction with Calf Thymus DNA and Human Serum Albumin. Eur. J. Med. Chem. 2014, 71, 148–159. 10.1016/j.ejmech.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Dai L. L.; Zhang H. Z.; Nagarajan S.; Rasheed S.; Zhou C. H. Synthesis of Tetrazole Compounds as a Novel Type of Potential Antimicrobial Agents and their Synergistic Effects with Clinical Drugs and Interactions with Calf Thymus DNA. MedChemComm 2015, 6, 147–154. 10.1039/C4MD00266K. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.