Abstract
The current standard of adjuvant therapies for patients with early stage non-small-cell lung cancers largely depends on the stage of disease. Liquid biopsy for circulating tumor DNA (ctDNA) has the potential to detect minimal residual disease, depict genomic evolution, guide precision medicine to individual patients and revolutionize the management of early stage lung cancers. In light of the seminal work published by Abbosh and colleagues, we discuss the potential paradigm changing clinical implications of ctDNA, the biological and technological challenges to consider, and the future of ctDNA driven therapeutic studies.
Keywords: Liquid biopsy, circulating tumor DNA (ctDNA), lung cancer, genetic sequencing
Despite diagnostic and therapeutic advances in thoracic oncology, lung cancers remain the leading cause of cancer-related deaths by far (1). For early stage non-small cell lung cancers that are potentially curable, current diagnostic tools are unable to provide precise prognostic information on micrometastases or the predictive benefit from adjuvant therapy in an individual patient. Adjuvant cisplatin-based chemotherapy is routinely recommended for resected stage II and IIIA non-small cell lung cancers (2). This generalized approach to chemotherapy is today’s best practice based on the overall survival benefit of 5% demonstrated in randomized controlled trials, but admittedly this modest benefit is associated with potential over-treatment or under-treatment in many patients (3). Recent technological advances in genetic sequencing and the use of “liquid biopsy” for plasma circulating tumor DNA (ctDNA) analysis have raised the hope of capturing tumor evolution to guide precision therapy for individual patients and radically improve their outcomes (4,5).
In a recent edition of Nature, Abbosh and colleagues published a study of ctDNA analysis in 100 patients with early stage non-small cell lung cancers as part of the TRACERx (NCT01888601) and PEACE post-mortem studies to depict clonal evolution and the potential for ctDNA-driven therapeutic studies (6). Plasma was collected in each patient pre- and post-operatively, including at follow up every 3 months for the first 2 years and every 6 months thereafter. Resected lung tumors underwent multi-region exome sequencing, and multiplex-PCR assay panels targeting tumor derived single nucleotide variants (SNVs) were synthesized for each patient to perform amplicon-based next generation sequencing (NGS) of pre- and post-operative ctDNA. Based on the ctDNA positive threshold of detecting two tumor derived SNVs, the clinical sensitivity for pre-operative detection was 97% (30/31) for lung squamous cell carcinomas and 19% (11/58) for lung adenocarcinomas. Multivariable analysis showed that non-adenocarcinoma histology, lymphovascular invasion and high Ki67 proliferation index are independent predictors of ctDNA detection. Other biological factors such as PET FDG avidity, clonal SNVs and tumor volume also correlated with ctDNA release. Volumetric analysis from CT showed a linear correlation between tumor volume and mean clonal plasma variant allele frequency (VAF). Through linear modeling predictions, a primary tumor burden of 10 cm3 (approximately 2.68 cm in diameter if spherical) equates to a plasma VAF of 0.1% (95% confidence interval, 0.05–0.17%), and approximately 326 million malignant cells. Persistent detection of post-operative ctDNA predicted relapse in 93% (13/14) with a median lead time of 70 days prior to radiologic confirmation. Serial ctDNA tracking revealed the emergence of tumor subclonal mutations as detected in plasma, corresponding to phylogenetic analysis on multiregion tissue sequencing including one patient who underwent post-mortem examination. Such examples of tumor heterogeneity also conferred adjuvant chemotherapy resistance.
These biological findings raise several potentially paradigm changing clinical implications for patients with early stage lung cancers. The concept of minimal residual disease (MRD) which originated from childhood leukemia management is now being investigated across solid tumors in the adjuvant setting, to precisely define patients who are at high risk of relapse and are in need of adjuvant therapies to eradicate micrometastatic disease (7-9). Analysis of post-operative ctDNA was shown to reliably prognosticate for relapse in patients with resected stage II colorectal cancers (79% vs. 10%, P<0.001), which forms the basis of clinical trial designs investigating precision adjuvant chemotherapy only for those who have evidence of MRD on ctDNA (10). Abbosh and colleagues showed that the MRD concept may be applied to patients with early stage lung cancers, not only using ctDNA analysis in prognosticating relapse to guide adjuvant chemotherapy decision making, but also in directing subsequent targeted and immunotherapies to patients who have persistent ctDNA positivity despite chemotherapy, in order to intercept and prevent cancer relapse. To prove the superiority of this precision approach, a potential future clinical trial design may be to randomize patients with stage I to IIIA non-small cell lung cancers to a standard of care arm of adjuvant therapy based on stage, and an experimental arm of ctDNA guided adjuvant therapy, with disease-free survival and overall survival as primary endpoints. In the experimental arm, it is conceivable that some patients with resected stage II lung cancers who are ctDNA negative may forego chemotherapy, and some with stage IA lung cancers who are ctDNA positive may require aggressive adjuvant therapies until ctDNA becomes negative. Should such a study demonstrate that ctDNA guided adjuvant therapy could improve survival, this would transform the standard of care of these patients and radically change how we approach thoracic oncology.
However, for ctDNA guided interventions to become a reality for patients, there are significant biological and technological challenges that need to be overcome. In the study by Abbosh and colleagues, the striking difference in pre-operative ctDNA detection between lung squamous cell carcinoma and adenocarcinoma (97% vs. 19%, P=0.001) is concerning, particularly as adenocarcinoma is now increasingly the dominant lung cancer histology (11). The sensitivity in detecting lung adenocarcinoma in pre-operative ctDNA is consistently low regardless of stage or mutation status, thus making the ctDNA assay used inadequate for determination of MRD in the lung adenocarcinoma post-operative setting. While biological factors such as tumor necrosis and proliferation may account for differences in ctDNA shedding, whether technological improvements in the limit of detection in ctDNA may overcome this issue in lung adenocarcinomas is unknown. Recently, ultrasensitive capture based NGS assays with broader coverage and digital error suppression have been shown to lead to improved detection of ctDNA at very low concentrations from early stage and MRD settings (12,13).
As most ctDNA assays currently used for the detection of MRD requires a priori knowledge of tissue sequencing results, substantial effort is required to design individualized ctDNA analysis for each patient, such as developing individual primers for patient specific SNVs in the study by Abbosh and colleagues (6,9,10). This presents a logistical challenge in clinical practice, as results from this relatively labor intense process need to be ready in a timely manner to inform treatment. Furthermore, in patients who undergo definitive radiation therapy and have very little biopsy tissue available for sequencing, this approach may not be feasible. While de novo mutation calling from ctDNA alone by deep sequencing of a large gene panel (508 genes, 2.1 MB) has shown good concordance with tissue sequencing in the metastatic disease setting, its performance in the early stage lung cancer or MRD setting is yet to be demonstrated (14). Current commercially available ctDNA assays are not designed for the detection of MRD, mostly lacking sensitivity.
De novo mutation calling from ctDNA may also be necessary in the neoadjuvant therapy setting when tissue availability is often limited. Neoadjuvant targeted therapies or immunotherapies may be precisely selected and guided by analysis and monitoring of ctDNA, to achieve best pathologic response at definitive therapy. Other plasma markers such as cell-free RNA, methylation, exosomes, nucleosomes and circulating tumor cells may add additional biological information to improve sensitivity and specificity. The refinement of the ctDNA assay used to guide patient care is critical, as past experiences from biomarker directed chemotherapy clinical trials have taught us that inadequate assays may even cause harm to patients (15).
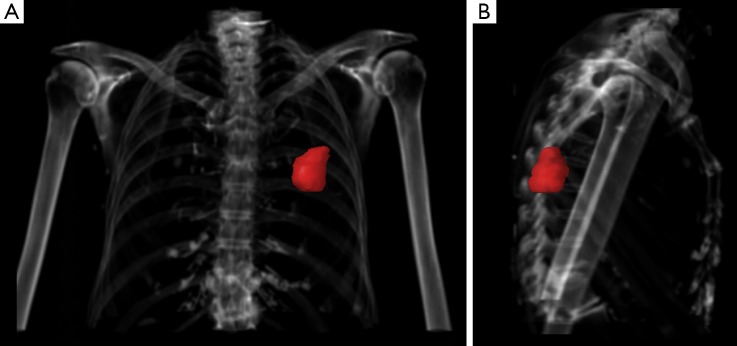
Abbosh and colleagues have used volumetric analyses to correlate with ctDNA shedding and concluded that 0.1% VAF of ctDNA equates to approximately 10 cm3 of lung tumor which is 2.68 cm in diameter if spherical (6). This finding requires validation as it has direct implications for the future use of ctDNA in early detection and screening. Volumetric analyses are no stranger to radiation oncologists who perform this every day in routine practice (Figure 1), and independent validation of ctDNA VAF from a large cohort of patients with early stage lung cancers undergoing radiotherapy will be required in order to determine the biological and technological limits of ctDNA detection.
Figure 1.
PA (A) and lateral (B) views of chest at radiation simulation with gross tumor volume in red, measured at 20.15 cm3. A biological question would be how much ctDNA shedding occurs from this lung adenocarcinoma, and technologically what is the best ctDNA assay to reliably detect it? ctDNA, circulating tumor DNA.
In conclusion, the use of liquid biopsy for ctDNA has potential to guide precision medicine and transform the management of early stage lung cancers. To get there, technological refinements of ctDNA assays are needed along with large cohort validation especially for the ctDNA detection of lung adenocarcinomas. Multidisciplinary close collaboration between thoracic surgeons, radiation oncologists, medical oncologists, radiologists, pathologists, geneticists and computational biologists are crucial to answer the abovementioned biological and technological questions. Ultimately, using the right assays, ctDNA driven clinical trials are needed to revolutionize the care of patients with early stage lung cancers.
Acknowledgements
Funding: All authors are supported by the Comprehensive Cancer Center Core Grant (P30 CA008748) at Memorial Sloan Kettering Cancer Center from the National Institutes of Health, USA.
Provenance: This is a Guest Editorial commissioned by Section Editor Xue-Feng Leng (Department of Cardiothoracic Surgery, the Affiliated Hospital of Chengdu University, Chengdu, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Kris MG, Gaspar LE, Chaft JE, et al. Adjuvant Systemic Therapy and Adjuvant Radiation Therapy for Stage I to IIIA Completely Resected Non-Small-Cell Lung Cancers: American Society of Clinical Oncology/Cancer Care Ontario Clinical Practice Guideline Update. J Clin Oncol 2017;35:2960-74. 10.1200/JCO.2017.72.4401 [DOI] [PubMed] [Google Scholar]
- 3.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. 10.1200/JCO.2007.13.9030 [DOI] [PubMed] [Google Scholar]
- 4.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Offin M, Chabon JJ, Razavi P, et al. Capturing Genomic Evolution of Lung Cancers through Liquid Biopsy for Circulating Tumor DNA. J Oncol 2017;2017:4517834. 10.1155/2017/4517834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dongen JJ, Seriu T, Panzer-Grumayer ER, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998;352:1731-8. 10.1016/S0140-6736(98)04058-6 [DOI] [PubMed] [Google Scholar]
- 8.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. 10.1126/scitranslmed.aab0021 [DOI] [PubMed] [Google Scholar]
- 10.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis DR, Check DP, Caporaso NE, et al. US lung cancer trends by histologic type. Cancer 2014;120:2883-92. 10.1002/cncr.28749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. 10.1126/scitranslmed.aan2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razavi P, Li BT, Abida W, et al. Performance of a high-intensity 508-gene circulating-tumor DNA (ctDNA) assay in patients with metastatic breast, lung, and prostate cancer. J Clin Oncol 2017;35:LBA11516 10.1200/JCO.2017.35.18_suppl.LBA11516 [DOI] [Google Scholar]
- 15.Lee SM, Falzon M, Blackhall F, et al. Randomized Prospective Biomarker Trial of ERCC1 for Comparing Platinum and Nonplatinum Therapy in Advanced Non-Small-Cell Lung Cancer: ERCC1 Trial (ET). J Clin Oncol 2017;35:402-11. 10.1200/JCO.2016.68.1841 [DOI] [PubMed] [Google Scholar]