Abstract
Sulfotransferase 4A1 (SULT4A1) belongs to the cytosolic sulfotransferase (SULT) superfamily of enzymes that catalyze sulfonation reactions with a variety of endogenous and exogenous substrates. Of the SULTs, SULT4A1 was shown to have the highest sequence homology between vertebrate species, yet no known function or enzymatic activity has been identified for this orphan SULT. To better understand SULT4A1 function in mammalian brain, two mutant SULT4A1 mouse strains were generated utilizing clustered regulatory interspaced short palindromic repeats (CRISPR)–content-addressable storage (Cas) 9 technology. The first strain possessed a 28-base pair (bp) deletion (Δ28) in exon 1 that resulted in a frameshift mutation with premature termination. The second strain possessed a 12-bp in-frame deletion (Δ12) immediately preceding an active site histidine111 common to the SULT family. Homozygous pups of both strains present with severe and progressive neurologic symptoms, including tremor, absence seizures, abnormal gait, ataxia, decreased weight gain compared with littermates, and death around postnatal days 21–25. SULT4A1 immunostaining was decreased in brains of heterozygous pups and not detectable in homozygous pups of both SULT4A1 mutants. SULT4A1 localization in subcellular fractions of mouse brain showed SULT4A1 associated with mitochondrial, cytosolic, and microsomal fractions, a novel localization pattern for SULTs. Finally, primary cortical neurons derived from embryonic (E15) CD-1 mice expressed high levels of SULT4A1 throughout the cell except in nuclei. Taken together, SULT4A1 appears to be an essential neuronal protein required for normal brain function, at least in mammals. Mouse models will be valuable in future studies to investigate the regulation and functions of SULT4A1 in the mammalian brain.
Introduction
In 2000, Falany et al. identified and cloned sulfotransferase 4A1 (SULT4A1) from the brains of humans and rats. Sequence analysis revealed that SULT4A1 possessed key structural domains characteristic of SULTs, such as the “KXXXFTVXXXE” dimerization domain, an active site histidine (His111), and a 3′-phosphoadenosine-5′-phosphosulfate (PAPS) binding site, yet SULT4A1 lacked detectable in vitro enzymatic activity (Falany et al., 2000). Immunohistochemical studies showed SULT4A1 to be predominately expressed in neurons of the human cerebral cortex, cerebellum, thalamus, pituitary, medial temporal lobe, choroid plexus, and lentiform nucleus (Liyou et al., 2003). In addition, SULT4A1 is highly conserved among vertebrate species, more so than all other SULTs. Human SULT4A1 shares >97.5% sequence identity with chimpanzee, rabbit, rat, and murine SULT4A1s. Moreover, 118 DNA samples of ethnically diverse people were sequenced for single-nucleotide polymorphisms in 10 different SULT genes, and SULT4A1 was the least genetically diverse with only five polymorphisms and none located within the coding region of the gene, suggesting an evolutionary conserved function in vertebrate brain (Allali-Hassani et al., 2007).
Although a validated function or substrate has not been identified, SULT4A1 has been implicated in several neurologic disorders, such as schizophrenia and Phelan-McDermid syndrome (PMS). The association of SULT4A1 with schizophrenia was first reported in transmission disequilibrium studies. A microsatellite marker (D22s1749E) in the 5′-UTR of SULT4A1 was linked to schizophrenia development, and later studies showed several intronic polymorphisms were associated with a worse psychopathology and poorer performance in cognition tests (Brennan and Condra, 2005; Meltzer et al., 2008). PMS is caused by deletions to the distal long arm of chromosome 22 (22q13.3 deletion syndrome) and is characterized by global developmental delay, intellectual disabilities, and autistic-like behaviors (Phelan, 2008). Approximately 30% of patients with PMS have a deletion encompassing SULT4A1 (Sarasua et al., 2014). Patients <4 years of age with a SULT4A1 deletion were shown to have developmental quotients lower than patients having two intact SULT4A1 alleles (Zwanenburg et al., 2016). Recently, nine patients were documented with interstitial 22q13.3 deletions, and SULT4A1 was among the genes within the minimal critical region of all patients examined in this study. Interestingly, these patients presented with symptoms similar to PMS, including developmental and speech delays (Disciglio et al., 2014). Currently, it is unclear the extent to which SULT4A1 contributes to the pathology observed in these nine patients, although SULT4A1 was singled out as a potential candidate gene for this disorder (Disciglio et al., 2014).
Owing to the high homology of SULT4A1 between species and the apparent absence of in vitro enzymatic activity, animal models represent an excellent system to investigate the functional role of SULT4A1 in vivo. Our laboratory recently reported that transient knockdown of zebrafish SULT4A1 using morpholino oligomers in 72-hour post fertilization (hpf) embryos resulted in an upregulation of phototransduction genes (Crittenden et al., 2014). These genes were related to cone function and were the first identified cellular process associated with SULT4A1. Subsequent studies using transcription activator-like effector nucleases (TALEN)-induced SULT4A1 knockout (KO) zebrafish models, homozygous SULT4A1-KO adult zebrafish were shown to have an activity suppression phenotype during daylight hours compared with wild-type (WT) zebrafish (Crittenden et al., 2015). The homozygous SULT4A1-KO zebrafish appeared normal, were able to readily reproduce and possessed normal lifespans (Crittenden et al., 2015). Like humans, zebrafish are diurnal organisms and are more active during daylight hours. Although these observations represent the first biologic pathways associated with SULT4A1, zebrafish brains lack several key brain regions characteristic of mammals, such as a cerebral cortex and an expanded telencephalon (Parker et al., 2013). Currently, there is no mammalian in vivo model developed to investigate the function of SULT4A1. Therefore, clustered regulatory interspaced short palindromic repeats (CRISPR)– content-addressable storage (Cas) 9 was used to generate two distinct SULT4A1 mutant mouse models. Homozygous SULT4A1-deficient mice exhibited a progressive and severe neurologic deterioration resulting in death prior to adulthood, and immunoblotting demonstrated a lack of detectable SULT4A1 expression in these mice. Interestingly, subcellular fractionation of WT brain homogenates showed SULT4A1 localized to mitochondrial, cytosolic, and microsomal fractions, but not to nuclei. Finally, SULT4A1 protein expression and localization was validated in primary cortical neuron cultures, an important in vitro model system for future molecular and biochemical studies. The behavioral phenotype and lethality in postnatal mice that lack functional SULT4A1 suggests that this orphan enzyme is necessary for normal neurologic development during postnatal development.
Materials and Methods
Generation of SULT4A1 Mouse Models.
SULT4A1 mutant mice were custom generated by the University of Alabama at Birmingham (UAB) Transgenic Facility using CRISPR-Cas9 gene-editing technology in a C57BL/6J mouse background (details in Supplemental Methods). Exon 1 (Δ28) was targeted to generate a SULT4A1 mutation, whereas Exon 3 (Δ12) was targeted to disrupt the structure around the active site His111. Female founders were used to generate breeding colonies for each model to generate SULT4A1 WT, heterozygous, and homozygous mutants for this study. Genotyping primers were as follows: Δ28 forward (5′-TCGGCCTGTAAAC-3′) and reverse (5′-GAGAGCAAGTACT-3′) and Δ12 forward (5′-CAGATGGCTTACG-3′) and reverse (5′-TGCTCAACTGTGA-3′). Breeding colonies were maintained in air-filtered cages under barrier conditions and kept on a 12 hour light/dark cycle with ad libitum access to food and water. Breeding colonies were maintained in accordance with the UAB Institutional Animal Care and Use Committee (IACUC) guidelines and all procedures were approved by the UAB Committee on Animal Research.
Subcellular Fractionation of Brain Tissue.
Brain tissue from male mice were immediately harvested after euthanasia from 25-day-old Δ28 and Δ12 SULT4A1 WT, heterozygous, and homozygous male mice and washed in ice-cold PBS. Tissue was snap-frozen in liquid N2 and stored at −80°C. Frozen brain tissue was placed in chilled subcellular fractionation buffer (20 mM HEPES pH 7.4, 10 mM KCL, 2 mM MgCl2, 1 mM of EDTA, and 1 mM EGTA) with 1 mM dithiothreitol and cOmplete protease inhibitor, mini (Roche Diagnostics, Indianapolis, IN) added immediately prior to use. A Potter-Elvehjem homogenizer was used to generate crude cell homogenates. The subcellular fractionation procedure was performed as follows: Brain homogenates were subjected to centrifugation at 720g to isolate the nuclear pellet. The supernate was centrifuged at 6200g to separate the mitochondrial pellet. The nuclear and mitochondrial pellets were washed twice in subcellular fractionation buffer. The cytosolic fraction was isolated after centrifugation at 100,000g for 1 hour, at the same time the resulting microsomal fraction was washed and recentrifuged. Protein concentrations were determined by Bradford assay and standard curves were generated using 1 mg/ml γ-globulin. Subcellular fractions (50 μg) were separated by SDS-PAGE (12% acrylamide) gel electrophoresis and transferred to nitrocellulose. SULT4A1 protein was detected using rabbit anti-SULT4A1 polyclonal IgG (1:1000; Proteintech, Chicago, IL) and β-actin (1:1000; Cell Signaling, Danvers, MA) was used as a loading control, followed by a goat anti-rabbit horseradish peroxidase-conjugated secondary Ab (1:55,000; Southern Biotech, Birmingham, AL), and developed with SuperSignal West Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA). Purity of fractions were determined by reprobing with the following antibodies: 1) nuclear (Histone H3 1:2500 rabbit polyclonal; Cell Signaling), 2) mitochondria (Tom20 1:1000 rabbit polyclonal; Proteintech), 3) cytosol (GAPDH 1:1000 rabbit monoclonal; Cell Signaling), and 4) microsomes (Na+/K+ ATPase 1:1000 rabbit polyclonal; Cell Signaling).
Generation of Primary Neuronal Cultures.
Primary cortical neuronal cultures were prepared from gestational day-15 CD-1 mice. Briefly, the cortical regions of the embryonic brains were aseptically dissociated, freed of meninges and dissociated in minimum essential medium containing 10% fetal bovine serum. Cortical neurons were cultured in Neurobasal medium (Thermo Fisher Scientific) supplemented with B27, sodium pyruvate, and 2% horse serum. Cells were plated to a density of 5 × 105 in six-well plates (Nalge Nunc International, Rochester, NY) previously coated with 0.1 mg/ml sterile poly-l-ornithine (Sigma-Aldrich, St. Louis, MO). The cultures were maintained at 37°C in 5% CO2 and in a humidified incubator. On day-in-vitro (DIV) 3, the cultures were treated with 5-fluoro-2-deoxyuridine (40 μM) to inhibit glial growth and proliferation. Experiments were performed at DIV 12. Under these conditions, mature neurons represent greater than 95% of the cells in the culture.
Immunocytochemistry.
For immunofluorescence studies, primary cortical neurons were grown on poly- l-ornithine-coated cover slips for 12 days. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) then fixed in 4% (w/v) paraformaldehyde for 20 minutes at room temperature. Next, cells were washed three times with PBS and permeabilized with 0.2% (v/v) Triton X-100 in 10% donkey serum for 1 hour. The permeabilized cells were then incubated with primary Ab (SULT4A1 1:500; Proteintech) overnight at 4°C. After incubation with primary Ab, cells were washed with 1× PBS and incubated with the secondary antibody conjugated with Alexa Fluor 555 (Thermo Fisher Scientific/Invitrogen) [1:1500 in 1% (w/v) donkey serum in PBS] in dark for 1 hour at room temperature. Nuclei were visualized using Hoechst (Thermo Fisher Scientific) diluted in PBS (1:20,000). Finally, cells were washed with PBS and mounted onto a glass slide using Immuno-Mount (Thermo Fisher Scientific). Cells were visualized and images were captured using Nikon confocal microscope with a 100 1.35 NA objective (Wetzler, Hassen, Germany).
Results and Discussion
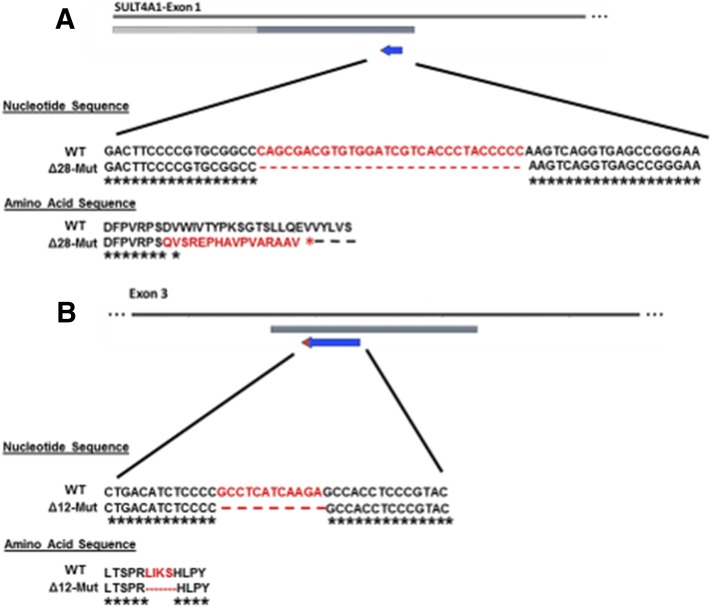
SULT4A1 remains an orphan SULT with an unknown function. In zebrafish eyes, SULT4A1 mRNA was detected by real-time quantitative polymerase chain reaction in 72 hpf embryos, and morpholino-mediated SULT4A1 knockdown resulted in upregulation of phototransduction genes related to cone function (Crittenden et al., 2014). TALEN-induced SULT4A1-KO adult zebrafish were shown to be less active during daylight hours, yet appeared normal, reproduced, and thrived (Crittenden et al., 2015). To determine the function of SULT4A1 in a mammalian system, CRISPR-Cas9 was used to mutate/delete SULT4A1 in C57BL/6J mice. Analysis of founder mice yielded two female mice with SULT4A1 mutations of interest and were selected for breeding. The first founder possessed a 28-bp deletion (Δ28) in exon 1 of the SULT4A1 gene causing a frameshift mutation beginning at amino acid (AA) 46 and premature stop codon 16 AA downstream. (Fig. 1A). The second founder female carried a 12-bp (Δ12) in-frame deletion on exon 3 of the SULT4A1 gene (Fig. 1B). These four AAs (L-I-K-S) form a β-sheet immediately adjacent to the active site His111 and structurally align the His within the substrate binding pocket. Although SULT4A1 lacks sulfonation activity in vitro, a possibility remains that the active site His111 is integral to its function in vivo. Furthermore, dynamic modeling suggested the 4 AA in-frame deletion would displace the active site His111 in such a manner as to be nonfunctional.
Fig. 1.
Schematic representation of SULT4A1 mutations generated by CRISPR-Cas9. (A) Diagram of the Δ28 mutant SULT4A1 mouse line. CRISPR-Cas9 technology induced a 28-bp deletion within exon 1 that resulted in a frameshift mutation and premature stop codon at AA 62. (B) Diagram of the Δ12 mutant SULT4A1 mouse line. Gene editing induced a 12-bp in-frame deletion of the 4 AA immediately preceding the active site His. For both lines, SULT4A1 female founders were used to establish breeding colonies.
Heterozygous mice of both strains appeared to develop normally to adulthood, reproduced without noticeable deficits, and lacked any readily observable phenotype. Interestingly, select offspring from heterozygote breeding pairs of both the Δ28 and Δ12 mice appeared normal at birth; however 8–10 days postnatal, several of the pups developed a slight tremor. As these pups aged, tremor increased in intensity and other symptoms appeared, including absence seizures, abnormal gait, ataxia, and decreased weight gain compared with WT and heterozygous littermates. Symptoms progressed in intensity until the mice were unable to move or feed and were euthanized according to IACUC guidelines around postnatal days 21–25. Videos of day-17 Δ28 (Supplemental Video 1) and Δ12 (Supplemental Video 2) homozygous pups exhibiting some of the neurologic phenotypes are included in the supplemental data. DNA sequencing confirmed the pups exhibiting the phenotype were Δ28- and Δ12-homozygous mutants and possessed the same genomic deletions as the founder females (Supplemental Fig. 1). The neurologic phenotype is consistent in all homozygous pups for both strains. Since homozygotes do not reach adulthood, these pups must be generated by mating heterozygous SULT4A1 mutant male and females.
Homozygous SULT4A1 mutant mice presenting with severe neurologic deficits was an unexpected novel finding. On the basis of the behavioral phenotype of SULT4A1-KO zebrafish and the lack of effect on breeding, morphology, or lifespan, the SULT4A1 mutant mice were not predicted to exhibit a severe neurologic phenotype. Extensive behavioral testing was anticipated to identify a behavioral phenotype in mice. The lethality of the pups during postnatal development suggests SULT4A1 has a critical role in mammalian postnatal brain development. Primary symptoms manifested by both Δ28- and Δ12-homozygous mice were tremor, rigidity, and seizure. In general, seizure disorders are thought to manifest because of an imbalance between neuronal excitation and inhibition, that leads to hyperexcitability and manifestation of a seizure (Scharfman, 2007). Although not the singular cause, functional SULT4A1 would seem to be a protein critical to the homeostatic regulation of balanced neuronal signaling. Further studies are needed to fully characterize this behavioral phenotype in mice with absent or nonfunctional SULT4A1. Owing to the severe phenotype, heterozygote mice of both strains will be carefully analyzed to determine whether loss of one functional SULT4A1 allele results in haploinsufficiency.
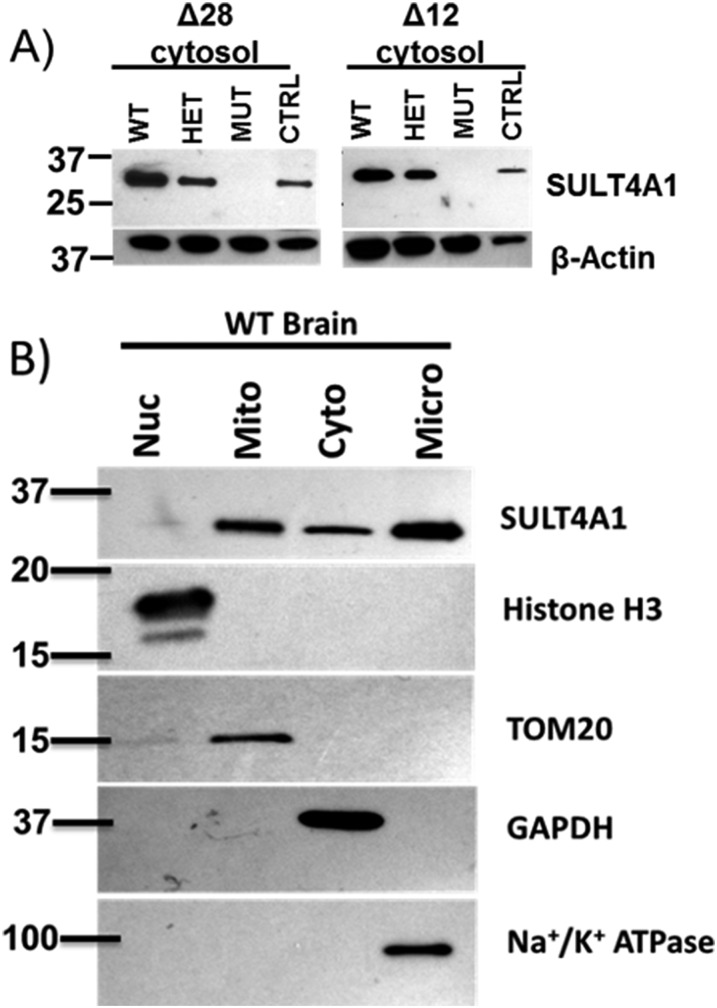
To confirm if homozygous mutant pups lacked SULT4A1, cytosol was prepared from male brains of each genotype and immunoblotted to determine SULT4A1 protein levels in both the strains. As shown in Fig. 2A, for both strains, heterozygotes showed a modest decrease in SULT4A1 protein levels compared with WT littermates. For Δ28- and Δ12-homozygous mutant pups, SULT4A1 protein was not detectable by immunoblotting. The lack of SULT4A1 staining in the Δ12-homozygous samples was surprising on the basis of dynamic modeling studies (data not shown). Multiple antibodies were used to probe Δ12-homozygous mutant brain samples, and the results showed a consistent lack of immunostaining, suggesting rapid SULT4A1 protein degradation.
Fig. 2.
Characterization of SULT4A1 protein expression in Δ28 and Δ12 mutant mouse brains and subcellular localization of WT SULT4A1. WT, heterozygous, and homozygous Δ28 and Δ12 25-day-old mice were euthanized and whole brains were harvested. (A) WT, heterozygous (HET), and homozygous (MUT) Δ28 and Δ12 mice were probed for SULT4A1 protein expression. Human brain cytosol was used as a positive control (CTRL) for SULT4A1. For both strains, WT and heterozygotes showed positive staining for SULT4A1; however, Δ28 and Δ12 homozygotes were negative for SULT4A1. (B) Subcellular fractionation was used to generate nuclear (Nuc), mitochondrial (Mito), cytosolic (Cyto) and microsomal (Micro) fractions from WT adult mouse brain homogenates. Aliquots of each fraction (50 μg) were subjected to SDS-PAGE and immunoblotted for SULT4A1. To determine the purity of subcellular fractions, marker proteins specific to each fraction were probed. Histone H3 was used to probe nuclear fraction purity, mitochondrial import receptor subunit TOM20 homolog (TOM20) was used as a marker protein for the mitochondrial fraction, GAPDH was used as a marker for the cytosolic fraction, and Na+/K+ ATPase was used to determine the microsomal fraction purity. SULT4A1 protein was detected in the mitochondrial, cytosolic, and microsomal fractions, and marker proteins demonstrated pure subcellular fractions.
To date, no sulfonation activity has been reported for SULT4A1 in the literature. In addition, a thorough investigation of the cellular location of SULT4A1 in brain has not been reported. Therefore, WT male mouse brain homogenates were subjected to subcellular fractionation via differential centrifugation to investigate the localization of SULT4A1. In addition to a cytosolic localization, SULT4A1 was detected in both mitochondrial and microsomal fractions (Fig. 2B). Nuclear, mitochondrial, and microsomal pellets were washed twice and resuspended to ensure purity of the fraction. The mechanism of SULT4A1 localization to these fractions remains unknown; however, several possibilities exist to explain this occurrence. First, it is possible that SULT4A1 has a binding partner that localizes to the mitochondrial or microsomal fraction. A second possibility involves an unknown post-translational modification, altering the localization of SULT4A1. This is not unprecedented as the peptidylprolyl cis/trans isomerase, NIMA-interacting 1 protein (PIN1) was shown to bind to SULT4A1 by coimmunoprecipitation (Co-IP) studies and binding negatively regulated SULT4A1 protein stability (Mitchell and Minchin, 2009). SULT4A1 was shown to be phosphorylated by extracellular signal-regulated kinase 1 on a Thr residue, which facilitated PIN1 binding and increased SULT4A1 degradation (Mitchell et al., 2011). Importantly, these experiments were carried out in vitro and the in vivo significance of this interaction in neurons is still unknown. Future Co-IP studies will be conducted to determine binding partners in each fraction using mouse brain and/or fetal cortical neurons.
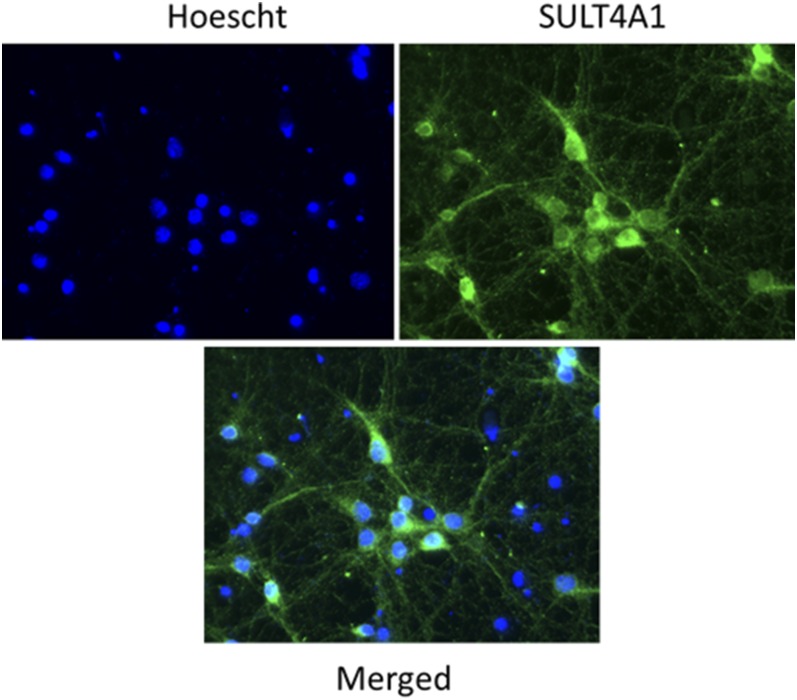
Active SULT4A1 protein requires proper splicing, since reports indicate SULT4A1 mRNA is detectable in various non-neuronal tissues but is not correctly spliced and is nonfunctional (Falany et al., 2000; Sidharthan et al., 2014). Low levels of SULT4A1 have been reported to be endogenously expressed by immunoblot in established cell lines, including LN229 glioblastoma and SH-SY5Y neuroblastoma cells (Sun et al., 2012; Sidharthan et al., 2014). The question remains if these immortalized cancer cell lines recapitulate the neuronal environment in which SULT4A1 exerts its normal function, as this protein is found to be highly localized to neurons. Therefore, the expression and localization of SULT4A1 in primary mouse cortical neuron culture was investigated, as these primary cultures may provide a more suitable in vitro model to study SULT4A1. Primary cortical neurons were harvested on gestational day-15 CD-1 mice and grown on coverslips for 12 days to become neuron-enriched. Figure 3 shows that SULT4A1 (green) is highly expressed in DIV-12 primary cortical neurons, indicating SULT4A1 is present early in embryonic neuron development. In addition, SULT4A1 immunoreactivity is absent in the nuclei (blue) but is detected in neurite projections, as well as in the cell body of the neuron. Although the SULT4A1 mutant mice were generated in a C57BL/6J and these primary cultures were derived from CD-1 mice, we expected that this result would be readily reproducible in our strain, even though SULT4A1 expression was also detected in mitochondrial, microsomal, and cytosolic fractions, but not nuclei, generated from the primary cultured neurons by immunoblotting similar to the pattern observed with mouse brains (data not shown).
Fig. 3.
SULT4A1 expression in embryo-derived mouse cortical neurons by confocal microscopy. Cortical neurons were harvested from embryonic day-15 mice and grown on cover slips for an additional 12 days prior to immunocytochemistry. Cortical neurons were probed for SULT4A1 expression using a rabbit polyclonal IgG at a 1:5000 dilution. Nuclei were labeled with Hoechst (1:20,000) with primary cortical neurons being positive (green) for SULT4A1 protein in both the soma and neurites, but not in the nuclei (blue).
The expression of SULT4A1 in primary neuron culture was first reported by Butcher et al., (2010) in a study investigating the transcriptional regulation of SULT4A1. The authors showed that both SULT4A1 mRNA and protein were detected in primary neuron cultures derived from embryonic day-16 mice, although the subcellular localization was not reported. Interestingly, SULT4A1 was expressed in the neurite projections of primary cortical neurons (Fig. 3). The impact of SULT4A1 on neurite growth and guidance remains unknown. Evidence of SULT4A1 expression in embryonic neurons is a promising model system to elucidating the mechanistic function of SULT4A1 in vitro. It is reasonable to infer that primary neuron cultures better represent the appropriate molecular environment than immortalized tumor-derived cell lines. Breeding pairs are established for the production of Δ28 and Δ12 WT, heterozygous, and homozygous primary neuron cultures. These studies in conjunction with behavioral analyses will provide valuable insights into potential differences in neuronal maturation, synaptic formation, or synaptic transmission.
In summary, this study reports the first published generation and characterization of SULT4A1 mutant mouse models. Homozygous mutant Δ28 and Δ12-SULT4A1 mice present with a severe and progressive neurologic phenotype resulting in postnatal death. The lack of SULT4A1 in the homozygous mutant pups detectable by immunostaining suggests that this is an essential protein required for normal development. In addition, although SULT4A1 was thought to be a cytosolic protein, its expression was detected in mitochondrial and microsomal fractions indicating potential novel function(s). SULT4A1 was also found to be expressed in embryonic mouse brain, further suggesting an essential role in development. Although, the function of SULT4A1 remains unknown, these mouse models will be invaluable to identifying and characterizing the role of SULT4A1 in mammalian brain.
Acknowledgments
The authors thank the UAB Transgenic and Genetically Engineered Models Core for assistance in generation of the SULT4A1 mutant mouse models. The authors would also like to thank Dr. Karina J. Yoon and Tracy Gamblin for their wonderful support with this study.
Abbreviations
- AA
amino acid
- CRISPR
clustered regulatory interspaced short palindromic repeats
- DIV
day-in-vitro
- hpf
hour postfertilization
- KO
knockout
- PBS
phosphate-buffered saline
- PMS
Phelan-McDermid syndrome
- SULT
sulfotransferase
- WT
wild-type
Authorship Contributions
Participated in research design: Garcia, Hossain, Andrabi, Falany.
Conducted experiments: Garcia, Hossain.
Performed data analysis: Garcia, Hossain, Andrabi, Falany.
Wrote or contributed to the writing of the manuscript: Garcia, Hossain, Andrabi, Falany.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM113980]; and by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS08695301].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MD, Condra J. (2005) Transmission disequilibrium suggests a role for the sulfotransferase-4A1 gene in schizophrenia. Am J Med Genet B Neuropsychiatr Genet 139B:69–72. [DOI] [PubMed] [Google Scholar]
- Butcher NJ, Mitchell DJ, Burow R, Minchin RF. (2010) Regulation of mouse brain-selective sulfotransferase sult4a1 by cAMP response element-binding protein and activating transcription factor-2. Mol Pharmacol 78:503–510. [DOI] [PubMed] [Google Scholar]
- Crittenden F, Thomas H, Ethen CM, Wu ZL, Chen D, Kraft TW, Parant JM, Falany CN. (2014) Inhibition of SULT4A1 expression induces up-regulation of phototransduction gene expression in 72-hour postfertilization zebrafish larvae. Drug Metab Dispos 42:947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden F, Thomas HR, Parant JM, Falany CN. (2015) Activity suppression behavior phenotype in SULT4A1 frameshift mutant zebrafish. Drug Metab Dispos 43:1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disciglio V, Lo Rizzo C, Mencarelli MA, Mucciolo M, Marozza A, Di Marco C, Massarelli A, Canocchi V, Baldassarri M, Ndoni E, et al. (2014) Interstitial 22q13 deletions not involving SHANK3 gene: a new contiguous gene syndrome. Am J Med Genet A 164A:1666–1676. [DOI] [PubMed] [Google Scholar]
- Falany CN, Xie X, Wang J, Ferrer J, Falany JL. (2000) Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem J 346:857–864. [PMC free article] [PubMed] [Google Scholar]
- Liyou NE, Buller KM, Tresillian MJ, Elvin CM, Scott HL, Dodd PR, Tannenberg AE, McManus ME. (2003) Localization of a brain sulfotransferase, SULT4A1, in the human and rat brain: an immunohistochemical study. J Histochem Cytochem 51:1655–1664. [DOI] [PubMed] [Google Scholar]
- Meltzer HY, Brennan MD, Woodward ND, Jayathilake K. (2008) Association of Sult4A1 SNPs with psychopathology and cognition in patients with schizophrenia or schizoaffective disorder. Schizophr Res 106:258–264. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Butcher NJ, Minchin RF. (2011) Phosphorylation/dephosphorylation of human SULT4A1: role of Erk1 and PP2A. Biochim Biophys Acta 1813:231–237. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Minchin RF. (2009) Cytosolic Aryl sulfotransferase 4A1 interacts with the peptidyl prolyl cis-trans isomerase Pin1. Mol Pharmacol 76:388–395. [DOI] [PubMed] [Google Scholar]
- Parker MO, Brock AJ, Walton RT, Brennan CH. (2013) The role of zebrafish (Danio rerio) in dissecting the genetics and neural circuits of executive function. Front Neural Circuits 7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MC. (2008) Deletion 22q13.3 syndrome. Orphanet J Rare Dis 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasua SM, Boccuto L, Sharp JL, Dwivedi A, Chen CF, Rollins JD, Rogers RC, Phelan K, DuPont BR. (2014) Clinical and genomic evaluation of 201 patients with Phelan-McDermid syndrome. Hum Genet 133:847–859. [DOI] [PubMed] [Google Scholar]
- Scharfman HE. (2007) The neurobiology of epilepsy. Curr Neurol Neurosci Rep 7:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidharthan NP, Butcher NJ, Mitchell DJ, Minchin RF. (2014) Expression of the orphan cytosolic sulfotransferase SULT4A1 and its major splice variant in human tissues and cells: dimerization, degradation and polyubiquitination. PLoS One 9:e101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Li H, Shu XH, Shi H, Chen XY, Kong QY, Wu ML, Liu J. (2012) Distinct sulfonation activities in resveratrol-sensitive and resveratrol-insensitive human glioblastoma cells. FEBS J 279:2381–2392. [DOI] [PubMed] [Google Scholar]
- Zwanenburg RJ, Ruiter SA, van den Heuvel ER, Flapper BC, Van Ravenswaaij-Arts CM. (2016) Developmental phenotype in Phelan-McDermid (22q13.3 deletion) syndrome: a systematic and prospective study in 34 children. J Neurodev Disord 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]