Abstract
RNase P catalyzes 5′-maturation of tRNAs in all three domains of life. This primary function is accomplished by either a ribozyme-centered ribonucleoprotein (RNP) or a protein-only variant (with one to three polypeptides). The large, multicomponent archaeal and eukaryotic RNase P RNPs appear disproportionate to the simplicity of their role in tRNA 5′-maturation, prompting the question of why the seemingly gratuitously complex RNP forms of RNase P were not replaced with simpler protein counterparts. Here, motivated by growing evidence, we consider the hypothesis that the large RNase P RNP was retained as a direct consequence of multiple roles played by its components in processes that are not related to the canonical RNase P function.
Keywords: RNase P, RNase MRP, ribonucleoprotein, evolution
RNase P is found in all domains of life, and its endoribonucleolytic activity to cleave the 5′ leader of precursor tRNAs is manifested in either a ribozyme-powered ribonucleoprotein (RNP) or a protein form (Evans et al. 2006; Altman 2007; Esakova and Krasilnikov 2010; Jarrous and Gopalan 2010; Lai et al. 2010; Rossmanith 2012; Lechner et al. 2015). The RNP form relies on a universally conserved catalytic RNA. The structural and functional similarity of this RNA in all three domains of life points to its ancient origins tracing back to the putative last universal common ancestor (Li and Altman 2004; Evans et al. 2006; Gopalan 2007).
In bacteria, the catalytic RNA (typically, ∼120 kDa) is aided by a small protein cofactor (∼14 kDa) (Altman 2007). However, in striking contrast to the simplicity of its conserved primary function, eukaryotic RNase P is a large RNP complex containing at least nine protein components (Fig. 1; Esakova and Krasilnikov 2010; Jarrous and Gopalan 2010). The molecular masses of purified nuclear forms are ∼450 kDa. Intermediate in complexity is archaeal RNase P (∼200 kDa) composed of one RNA and up to five different protein subunits, whose presence in the genomes of at least eight of 11 archaeal phyla (Samanta et al. 2016) reflects their origins early in archaeal evolutionary history. All archaeal RNase P proteins have homologs in eukaryotic RNase P (Fig. 1). This level of subunit complexity of the archaeal and eukaryotic RNase P RNP variants appears disproportionate for hydrolysis of a single phosphodiester bond in a precursor tRNA, given that four of the five eukaryotic supergroups also possess typical, moderately sized (∼60 kDa) protein-only RNase P (PRORP) having a similar enzymatic activity (Gobert et al. 2010; Lechner et al. 2015). Moreover, genetic complementation studies in Escherichia coli and Saccharomyces cerevisiae indicate that some of the PRORPs can substitute for the RNase P RNP activity without detrimental effects on growth under laboratory conditions (Weber et al. 2014; Gößringer et al. 2017). In one genetic background and under high-salt conditions, a strain of S. cerevisiae with a protein-only form (a single polypeptide from Arabidopsis) as the functional RNase P was able to outcompete in growth a wild-type counterpart that uses the 10-subunit RNP (Weber et al. 2014).
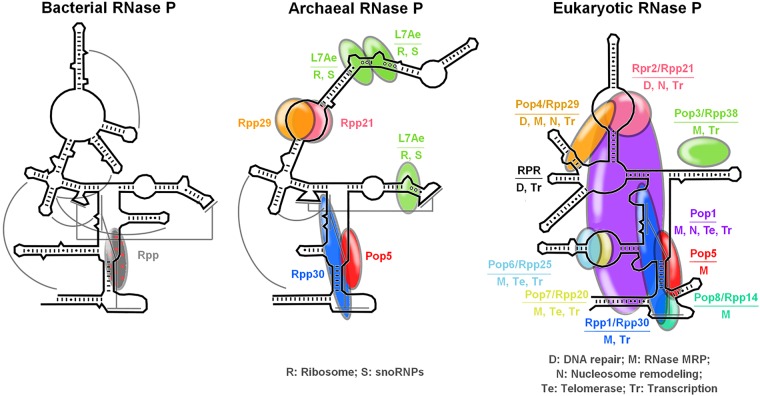
FIGURE 1.
Diversity of the ribonucleoprotein variants of RNase P, and inventory of the multifunctional protein subunits of archaeal/eukaryal RNase P. Overlaid on the secondary structure of the respective RNA subunit are the different RNase P protein subunits. The approximate placement of the protein subunits is based on data reported elsewhere (Tsai et al. 2006; Pulukkunat and Gopalan 2008; Xu et al. 2009; Chen et al. 2010; Reiter et al. 2010; Hipp et al. 2012; Khanova et al. 2012; Fagerlund et al. 2015; Lai et al. 2017). For subunits that have been demonstrated to have roles other than RNase P catalysis, these are indicated using one- or two-letter codes (see key for explanation of the codes). Protein sizes are not exact, and the schematic is not drawn to scale.
The apparent functional equivalence of PRORPs and RNase P RNPs raises the intriguing question of why evolution has favored retention of complex catalytic RNPs that could have been substituted with simpler protein enzymes. Here, we consider how chance and necessity in evolution may have led to the presence and concurrent use of both RNP- and protein-based forms of RNase P.
Constructive neutral evolution (Gray et al. 2010; Stoltzfus 2012; Weber et al. 2014) merits consideration as a possible driving force for the runaway complexity of the protein makeup of archaeal and eukaryotic RNase P RNPs. In this scenario, fortuitous binding of proteins unrelated to RNase P may have provided additional means to stabilize the catalytic RNA's tertiary structure and thus helped suppress subsequent mutations that would have been otherwise deleterious. Such an interdependency would have lessened the evolutionary pressure to maintain auxiliary, scaffolding RNA elements, leading to the eventual loss of these parts of the RNA, but at the cost of an absolute reliance on structural/functional roles of the newly acquired protein component(s). A comparison of the RNase P RNP variant from the three domains of life lends support to this notion. The tertiary structure of the RNA component of bacterial RNase P RNP is stabilized by an intricate network of tertiary RNA–RNA interactions, and the structural role of the sole protein component appears limited (Reiter et al. 2010). The increase in the complexity of the protein complement of the RNase P RNPs from bacteria to archaea to eukarya is paralleled by a loss of the RNA elements that play scaffolding roles in bacterial RNase P, presumably because such structural roles were delegated to proteins in archaeal/eukaryotic RNase P (Pulukkunat and Gopalan 2008; Fagerlund et al. 2015).
While the constructive neutral evolution-driven acquisition of additional protein components (“complexity by chance”) is plausible, such changes may also be important to ensure the proper localization of the RNP, to facilitate crosstalk with other cellular machineries and/or to prevent the activation of the innate immune response that is triggered by long stretches of “naked” dsRNA (Hull and Bevilacqua 2016) (“complexity by necessity”). Several recent observations support the claim that the retention and persistence of large eukaryotic RNase P RNPs is likely a direct consequence of essential roles played by RNase P components in processes unrelated directly to its primary task as an endoribonuclease responsible for cleavage of the 5′-leader of precursor tRNAs.
First, recent studies show that RNase P or its subassemblies are connected to chromatin structure and function. In human cells, variants of nuclear RNase P RNPs bind to rRNA and small noncoding RNA genes transcribed by RNA polymerase (Pol) I and Pol III, respectively (Reiner et al. 2006, 2008; Serruya et al. 2015). This RNase P RNP is incorporated into Pol III initiation complexes that transcribe 5S rRNA genes (Serruya et al. 2015). Three protein subunits of human RNase P (Rpp21, Rpp29, and Pop1) repress histone H3.3 nucleosome deposition and mediate transcriptional silencing (Newhart et al. 2016). In Drosophila, nuclear RNase P is implicated in expression of tRNA and piRNA gene clusters (Molla-Herman et al. 2015), presumably through indirect effects related to transcription-replication conflicts, which arise when the crosstalk between RNase P and Pol III is weakened by mutations. A variant of human RNase P, containing the Rpp21, Rpp29, and RPPH1 RNA, but not Rpp14, Rpp25, and Rpp38, promotes homology-directed DNA repair of double-strand breaks induced by irradiation (Abu-Zhayia et al. 2017). Moreover, human RNase P activity in tRNA processing increases in response to DNA replication stress induced by depleting the WRN helicase (Orlovetskie et al. 2017), a RecQ DNA helicase with multiple tasks in DNA fork progression during replication, proofreading, base excision repair, and transcription-bubble progression (Croteau et al. 2014). The exact molecular mechanisms underlying these novel functions of human RNase P (either in whole or as subassemblies) in replication stress and DNA repair await further exploration.
Second, there is an increased appreciation of the sharing of protein subunits of RNase P with other RNPs. Three protein subunits of yeast nuclear RNase P, Pop1, Pop6, and Pop7 (homologs of human Pop1, Rpp25, and Rpp20, respectively), were recently shown to be essential components of the telomerase RNP (Lemieux et al. 2016). This subunit overlap appears to be related to a common structural platform in these three RNPs. In this regard, archaeal RNase P also presents parallels: Ribosomal protein L7Ae, which helps remodel the H/ACA and C/D box guide snoRNAs for better binding to either target substrates or other protein subunits (Omer et al. 2002; Youssef et al. 2007), is a bona fide subunit of RNase P (Cho et al. 2010).
Eukaryotic nuclear RNase P has long been recognized to share most of its protein components with another essential RNP enzyme, nucleolar RNase MRP, which is widespread in eukaryotes (Chang and Clayton 1987; Chamberlain et al. 1998; Salinas et al. 2005; Esakova and Krasilnikov 2010). RNase MRP has distinct RNA processing tasks in the cell, including rRNA maturation (Esakova and Krasilnikov 2010), and its substrate specificity does not overlap with that of RNase P (Esakova et al. 2011). Sharing of components between RNase P RNP and RNase MRP does not appear to extend to the RNPs found in organelles, perhaps accounting at least partly for differences in evolution of organellar RNase P. This scenario holds at least in yeast where the protein inventories of mitochondrial and nucleolar RNase P as well as RNase MRP RNPs are distinct (Martin and Lang 1997; Lu et al. 2010). Such divergence would disincentivize evolutionary retention of the exceedingly complex RNase P/MRP RNPs in organelles due to various reasons including import-related complications (Howard et al. 2013). In cases when an organelle did retain a large RNP with a unique protein makeup, one might expect that the recruited protein(s) have some additional role(s) unrelated to the canonical RNP activity. While further studies are required to test this conjecture, especially in organisms other than yeast, this claim is supported by dual-targeted Rpm2p, a subunit of yeast mitochondrial RNase P RNP that doubles up as a transcriptional activator in the nucleus (Stribinskis et al. 2005).
By having and sharing critical protein–protein and RNA–protein interactions, the RNase P RNP is linked to other RNPs, such as the evolutionarily related RNase MRP and telomerase as well as other molecular machineries implicated in RNA metabolism like the exosome (Koonin et al. 2001; Jiang and Altman 2002) and snoRNPs (Salgado-Garrido et al. 1999; Fernandez et al. 2004). Whether the linkages between these divergent RNPs relate to shared intracellular regulatory circuits or even physical networks remain to be tested. If indeed a similar organizing center is used to build a common architecture in functionally divergent archaeal/eukaryal RNPs, such intricate interdependencies would not only have fixed seemingly fortuitous RNA–protein interactions but also rendered difficult (if not impossible) wholesale and exclusive remodeling of any one of these interconnected RNPs (including RNase P). Consistent with an earlier suggestion (Weber et al. 2014), it is clear that an evolutionary substitution of nuclear RNase P RNA with a PRORP in yeast will not offset the requirement of multiple RNase P protein components for non-RNase P functions, thus nullifying the potential benefits of a PRORP substitution.
The above observations that emphasize the basis for retention of the larger RNP isoforms of RNase P need to be squared with the successful genetic replacement of the RNP with a protein-only form in E. coli and yeast (Weber et al. 2014; Gößringer et al. 2017). This neutral swap indicates the elasticity to rewire a central housekeeping enzyme, but its biological impact remains to be determined since genetic or environmental factors could engender different organismal robustness outcomes within the framework of this “neutral” mutation. This neutral swap also motivates questions related to biogenesis cost and functional repertoires. For instance, the bacterial RNase P RNP is no more difficult for the cell to produce than a typical protein enzyme. Indeed, aside from the small (only ∼14 kDa) protein component, bacterial RNase P is just a ∼400-nt long noncoding RNA, which is similar in size to an mRNA encoding a ∼14 kDa protein but does not require translation. Thus, from the perspective of biogenesis cost, the RNP form of bacterial RNase P may have a modest advantage over a PRORP. In archaea and eukarya, chance could have favored either form, but it is likely that (at least in specific subcellular locales) functional/regulatory necessities tilted the balance toward the larger RNP form that is ensconced within a larger framework of macromolecular interactomes.
The genetic complementation experiments that established functional equivalence of the two forms of RNase P suggest that the ability to catalyze a reaction alone or the size/complexity of one variant is insufficient cause for its retention in an organelle/organism. These outcomes were likely not swayed solely by chance, but driven by the necessity of each cellular piece finding its place in a more intricate network entailing fundamental cellular processes; in the case of RNase P, for example, the linkage is between processing and transcription, replication, and DNA repair. It is instructive to consider nuclear and mitochondrial RNase P in S. cerevisiae and humans, two species that have evolved along separate paths for nearly a billion years. While both utilize a shared RNP variant in the nucleus, S. cerevisiae appears to use a distinct mitochondrial RNase P RNP (with protein components not related to those of the nuclear counterpart) and humans have a protein-only organellar variant. Albeit challenging, untangling the complex evolution of RNase P and its components and related RNPs, as well as elucidating their multifaceted roles, will continue to offer exciting prospects.
ACKNOWLEDGMENTS
We thank Phil Bevilacqua (Penn State) for valuable comments and suggestions. We are grateful to Sara Lung (OSU) for assistance with preparation of the figure, and Lien Lai (OSU) for help and input with this illustration. A.K., N.J., and V.G. gratefully acknowledge funding for their research from the National Institutes of Health (GM085149 [A.K.], GM120582 [V.G.], and NS096600 [V.G.]), the US-Israel Binational Science Foundation (#2015/157 [N.J., V.G.]), and the Israel Science Foundation (#1205/2017 [N.J.]).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063107.117.
REFERENCES
- Abu-Zhayia ER, Khoury-Haddad H, Guttmann-Raviv N, Serruya R, Jarrous N, Ayoub N. 2017. A role of human RNase P subunits, Rpp29 and Rpp21, in homology directed-repair of double-strand breaks. Sci Rep 7: 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S. 2007. A view of RNase P. Mol Biosyst 3: 604–607. [DOI] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. 1987. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science 235: 1178–1184. [DOI] [PubMed] [Google Scholar]
- Chen WY, Pulukkunat DK, Cho IM, Tsai HY, Gopalan V. 2010. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res 38: 8316–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. 2010. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci 107: 14573–14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Popuri V, Opresko PL, Bohr VA. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem 83: 519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS. 2010. Of proteins and RNA: the RNase P/MRP family. RNA 16: 1725–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Perederina A, Quan C, Berezin I, Krasilnikov AS. 2011. Substrate recognition by ribonucleoprotein ribonuclease MRP. RNA 17: 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR. 2006. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci 31: 333–341. [DOI] [PubMed] [Google Scholar]
- Fagerlund RD, Perederina A, Berezin I, Krasilnikov AS. 2015. Footprinting analysis of interactions between the largest eukaryotic RNase P/MRP protein Pop1 and RNase P/MRP RNA components. RNA 21: 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CF, Pannone BK, Chen X, Fuchs G, Wolin SL. 2004. An Lsm2-Lsm7 complex in Saccharomyces cerevisiae associates with the small nucleolar RNA snR5. Mol Biol Cell 15: 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P. 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744. [DOI] [PubMed] [Google Scholar]
- Gößringer M, Lechner M, Brillante N, Weber C, Rossmanith W, Hartmann R. 2017. Protein-only RNase P function in Escherichia coli: viability, processing defects and differences between PRORP isoenzymes. Nucleic Acids Res 45: 7441–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan V. 2007. Uniformity amid diversity in RNase P. Proc Natl Acad Sci 104: 2031–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. 2010. Cell biology. Irremediable complexity? Science 330: 920–921. [DOI] [PubMed] [Google Scholar]
- Hipp K, Galani K, Batisse C, Prinz S, Böttcher B. 2012. Modular architecture of eukaryotic RNase P and RNase MRP revealed by electron microscopy. Nucleic Acids Res 40: 3275–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MJ, Liu X, Lim WH, Klemm BP, Fierke CA, Koutmos M, Engelke DR. 2013. RNase P enzymes: divergent scaffolds for a conserved biological reaction. RNA Biol 10: 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Bevilacqua PC. 2016. Discriminating self and non-self by RNA: roles for RNA structure, misfolding, and modification in regulating the innate immune sensor PKR. Acc Chem Res 49: 1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Gopalan V. 2010. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res 38: 7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Altman S. 2002. A protein subunit of human RNase P, Rpp14, and its interacting partner, OIP2, have 3′-->5′ exoribonuclease activity. Proc Natl Acad Sci 99: 5295–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanova E, Esakova O, Perederina A, Berezin I, Krasilnikov AS. 2012. Structural organizations of yeast RNase P and RNase MRP holoenzymes as revealed by UV-crosslinking studies of RNA-protein interactions. RNA 18: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res 11: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Vioque A, Kirsebom LA, Gopalan V. 2010. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett 584: 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Tanimoto A, Lai SM, Chen WY, Marathe IA, Westhof E, Wysocki VH, Gopalan V. 2017. A novel double kink-turn module in euryarchaeal RNase P RNAs. Nucleic Acids Res 45: 7432–7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Rossmanith W, Hartmann RK, Thölken C, Gutmann B, Giegé P, Gobert A. 2015. Distribution of ribonucleoprotein and protein-only RNase P in eukarya. Mol Biol Evol 32: 3186–3193. [DOI] [PubMed] [Google Scholar]
- Lemieux B, Laterreur N, Perederina A, Noël JF, Dubois ML, Krasilnikov AS, Wellinger RJ. 2016. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell 165: 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Altman S. 2004. In search of RNase P RNA from microbial genomes. RNA 10: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wierzbicki S, Krasilnikov AS, Schmitt ME. 2010. Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components. RNA 16: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NC, Lang BF. 1997. Mitochondrial RNase P: the RNA family grows. Nucleic Acids Symp Ser: 42–44. [PubMed] [Google Scholar]
- Molla-Herman A, Vallés AM, Ganem-Elbaz C, Antoniewski C, Huynh JR. 2015. tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription. EMBO J 34: 3009–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A, Powers SL, Shastrula PK, Sierra I, Joo LM, Hayden JE, Cohen AR, Janicki SM. 2016. RNase P protein subunit Rpp29 represses histone H3.3 nucleosome deposition. Mol Biol Cell 27: 1154–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci 99: 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlovetskie N, Serruya R, Abboud-Jarrous G, Jarrous N. 2017. Targeted inhibition of WRN helicase, replication stress and cancer. Biochim Biophys Acta 1867: 42–48. [DOI] [PubMed] [Google Scholar]
- Pulukkunat DK, Gopalan V. 2008. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res 36: 4172–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. 2006. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev 20: 1621–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N. 2008. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS One 3: e4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmanith W. 2012. Of P and Z: mitochondrial tRNA processing enzymes. Biochim Biophys Acta 1819: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J 18: 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas K, Wierzbicki S, Zhou L, Schmitt ME. 2005. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J Biol Chem 280: 11352–11360. [DOI] [PubMed] [Google Scholar]
- Samanta MP, Lai SM, Daniels CJ, Gopalan V. 2016. Sequence analysis and comparative study of the protein subunits of archaeal RNase P. Biomolecules 6: E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruya R, Orlovetskie N, Reiner R, Dehtiar-Zilber Y, Wesolowski D, Altman S, Jarrous N. 2015. Human RNase P ribonucleoprotein is required for formation of initiation complexes of RNA polymerase III. Nucleic Acids Res 43: 5442–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. 2012. Constructive neutral evolution: exploring evolutionary theory's curious disconnect. Biol Direct 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V, Heyman HC, Ellis SR, Steffen MC, Martin NC. 2005. Rpm2p, a component of yeast mitochondrial RNase P, acts as a transcriptional activator in the nucleus. Mol Cell Biol 25: 6546–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. 2006. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc Natl Acad Sci 103: 16147–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Hartig A, Hartmann RK, Rossmanith W. 2014. Playing RNase P evolution: swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms. PLoS Genet 10: e1004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. 2009. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J Mol Biol 393: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef OA, Terns RM, Terns MP. 2007. Dynamic interactions within sub-complexes of the H/ACA pseudouridylation guide RNP. Nucleic Acids Res. 35: 6196–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]