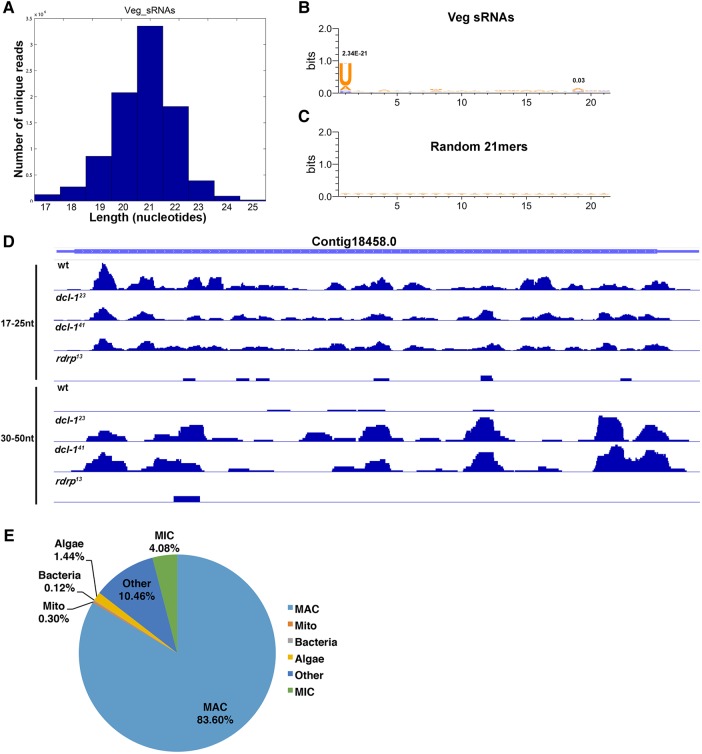
FIGURE 2.
Oxytricha MAC sRNAs do not lead to post-transcriptional gene silencing. (A) The length distribution profile for sRNAs shows a peak at 21 nt. Sequence logos for (B) 21-nt sRNAs versus (C) random 21-nt words generated from the macronuclear sequence show a strong preference for U at the first position (P-value = 2.3 × 10−21) and a modest increase in A at the 19th position (P-value = 0.03); P-values noted above the 1st and 19th bases. (D) 17- to 25-nt and 30- to 50-nt sRNA mapping to representative MAC Contig18458.0 for wt, dcl-1 and rdrp cells show reads mapping to the entire chromosome (excluding telomeres), including the CDS (thickest bars) and nongenic, subtelomeric regions (thin, flanking lines). (E) sRNA mapping statistics reveal that the majority of sRNAs map to the MAC genome (MIC, germline-limited; Mito, mitochondria). Of the fraction of sRNAs that map to germline-limited regions, 0.5% map to IESs and 1.6% to transposons or other MIC-limited repeats. Sequences in the “Other” category do not map to any Oxytricha genome.