Abstract
The branch point recognition region of spliceosomal snRNA U2 is heavily modified post-transcriptionally in most eukaryotic species. We focused on this region to learn how nearby positions may interfere with each other when targeted for modification. Using an in vivo yeast Saccharomyces cerevisiae cell system, we tested the modification activity of several guide RNAs from human, mouse, the frog Xenopus tropicalis, the fruit fly Drosophila melanogaster, and the worm Caenorhabditis elegans. We experimentally verified predictions for vertebrate U2 modification guide RNAs SCARNA4 and SCARNA15, and identified a C. elegans ortholog of SCARNA15. We observed crosstalk between sites in the heavily modified regions, such that modification at one site may inhibit modification at nearby sites. This is true for the branch point recognition region of U2 snRNA, the 5′ loop of U5 snRNA, and certain regions of rRNAs, when tested either in yeast or in HeLa cells. The position preceding a uridine targeted for isomerization by a box H/ACA guide RNA is the most sensitive for noncanonical base-pairing and modification (either pseudouridylation or 2′-O-methylation). Based on these findings, we propose that modification must occur stepwise starting with the most vulnerable positions and ending with the most inhibiting modifications. We discuss possible strategies that cells use to reach complete modification in heavily modified regions.
Keywords: guide RNA, 2′-O-methylation, pseudouridylation, U2 snRNA, U5 snRNA
INTRODUCTION
The presence of post-transcriptionally modified nucleotides in spliceosomal U snRNAs has been known since the 1970s and the overall modification pattern of major and minor spliceosomal snRNAs has been extensively characterized in different species (Reddy and Busch 1988; Gu et al. 1996; Massenet et al. 1999; Patton and Padgett 2005; Deryusheva and Gall 2009; Karijolich and Yu 2010; Deryusheva et al. 2012). Many modified positions are evolutionary conserved and functionally significant (Yu et al. 1998; Dönmez et al. 2004; Zhao and Yu 2004, 2007; Yang et al. 2005; Wu et al. 2016). The most abundant modifications in snRNAs are pseudouridylation and 2′-O-methylation. The positioning of these modifications typically involves guide RNAs, the so-called box H/ACA and box C/D snoRNAs, respectively. At the same time, a guide RNA-independent mechanism is also possible at certain positions (Massenet et al. 1999; Zhao et al. 2002; Ma et al. 2003; Behm-Ansmant et al. 2006; Wu et al. 2011; Deryusheva and Gall 2017). Although the corresponding guide RNAs and/or guide RNA-independent modification enzymes have been assigned to most modified positions (Karijolich and Yu 2010; Deryusheva and Gall 2017), little is known about how the modification pattern is established as a whole. Do modifications occur at random, or is there a series of sequential events that occur in a hierarchical order? A sequential order of post-transcriptional modification has been demonstrated for certain positions in yeast 25S rRNA (Lapeyre and Purushothaman 2004), archaeal tRNATrp (Singh et al. 2004, 2008), and eukaryotic tRNATyr (Nishikura and De Robertis 1981). As for spliceosomal snRNAs, modifications mediated by stand-alone proteins are slower than guide RNA-mediated modifications (Zhao et al. 2002) and modification of the branch point recognition region of U2 snRNA occurs at a higher rate than modification of the 5′ terminal region (Zhao and Yu 2004). However, no systematic analysis has been done to test the coordinated stepwise modification of snRNAs.
Although U2 snRNA pseudouridylation appears to be an autonomous process—pseudouridine formation at one site does not require prior modification at another site or sites (Patton et al. 1994; Deryusheva and Gall 2013)—in our previous studies we observed that pseudouridylation of U2 snRNA at certain positions can inhibit formation of pseudouridine at another nearby position (Deryusheva and Gall 2013, 2017). This made us think that within heavily modified regions the positioning of modifications must occur in a stepwise fashion. To test this hypothesis, we focus here on the branch point recognition region of U2 snRNA. In higher eukaryotes and the fission yeast Schizosaccharomyces pombe this region is heavily modified (Fig. 1). Conversely, in the budding yeast Saccharomyces cerevisiae U2 snRNA is modified normally at three positions only; for each position, the modification mechanism is well characterized (Massenet et al. 1999; Ma et al. 2003, 2005). Using an in vivo yeast cell system, we tested several guide RNAs from various species for their ability to modify U2 and U5 snRNA. Based on these data, we propose the existence of crosstalk between different modified positions. We discuss possible mechanisms that cells utilize to reach the required post-transcriptional modification patterns.
FIGURE 1.
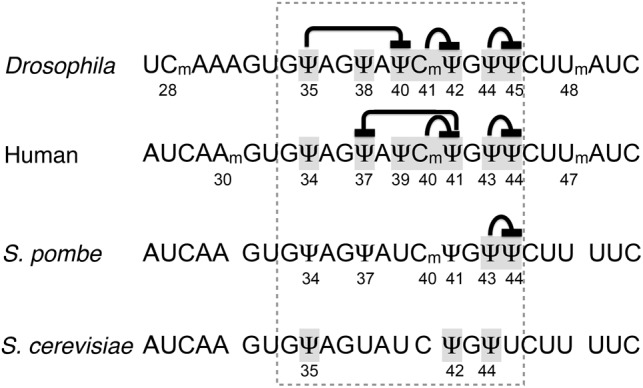
Post-transcriptional modification at the branch point recognition region of U2 snRNA from different species. The recognition region is highlighted with a dashed line. Positions examined in this and previous studies are shadowed. Proposed interactions between the tested positions are indicated.
RESULTS
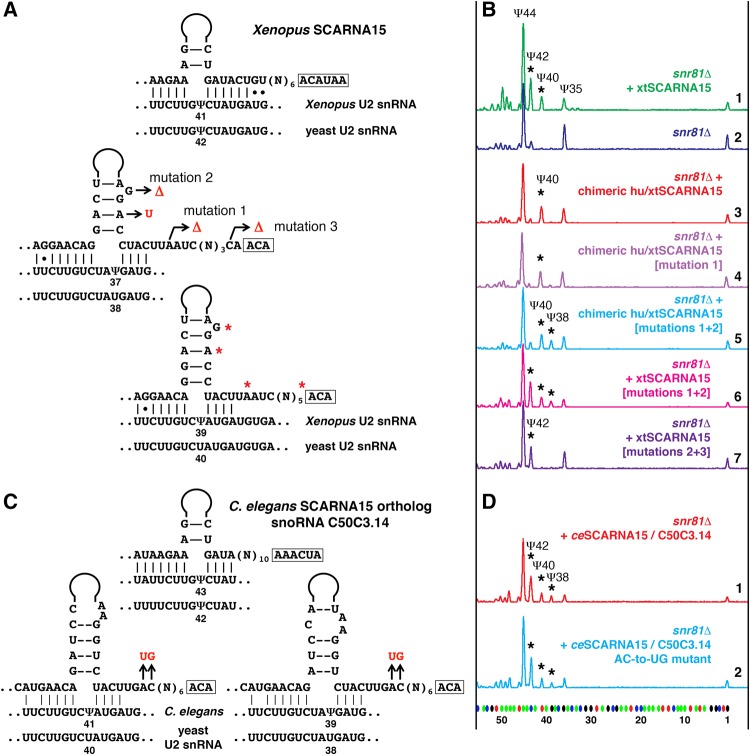
Guide activity of SCARNA15/ACA45 RNA from different species
In our previous study of Drosophila guide RNAs (Deryusheva and Gall 2013), we found a triple guide, scaRNA:ΨU2-38.40.42, that in vitro and in a yeast cell system mediated pseudouridylation of U2 snRNA at position 40 much less efficiently than expected, especially when position 35 was modified. In human and other vertebrate species, box H/ACA SCARNA15 (also called ACA45) originally was annotated as a guide RNA for position 37 (equivalent to position 38 in Drosophila) of U2 snRNA (Kiss et al. 2004; Lestrade and Weber 2006); a more recent prediction for the same scaRNA assigned position 39 (position 40 in Drosophila) instead of position 37 as a potential target (Jorjani et al. 2016). As soon as we reanalyzed the alignment of human SCARNA15 and U2 snRNA, we recognized a pseudouridylation pocket similar to one in Drosophila scaRNA:ΨU2-38.40.42. In this pocket, two alternative configurations target two positions of U2 snRNA for pseudouridylation; the positioning of Ψ37 requires a noncanonical configuration of the pseudouridylation pocket with 3 nucleotides (nt) left unpaired at the target position (Fig. 2A). We expressed human SCARNA15 in S. cerevisiae wild-type and mutant pus7Δ and snr81Δ strains. Yeast U2 snRNA became pseudouridylated at positions 40 and 38 (equivalent to positions 39 and 37 in human U2 snRNA), although, surprisingly, Ψ38 was evident only in the snr81Δ strain, which had no pseudouridine at position 42 (Fig. 2B, traces 2, 3, and 4). In contrast to Drosophila scaRNA:ΨU2-38.40.42, neither presence nor absence of Ψ35 had any effect on modification of positions 38 and 40 directed by human SCARNA15 in yeast cells. These data are summarized in Figure 1.
FIGURE 2.
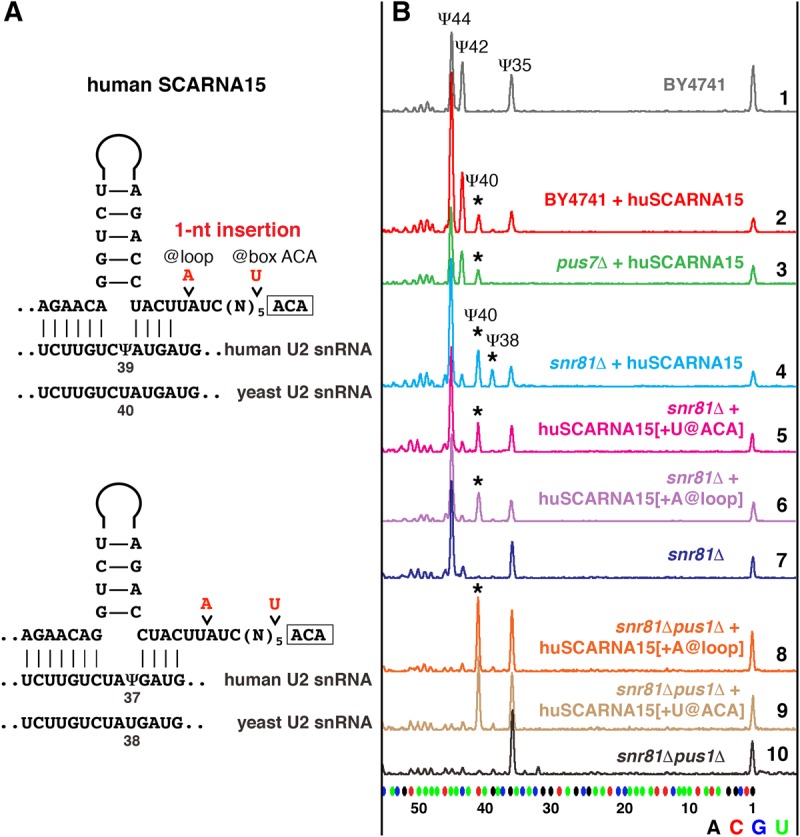
Pseudouridylation of yeast U2 snRNA induced by human SCARNA15. (A) Predicted base-pairing of huSCARNA15 with human and yeast U2 snRNA. Point mutations introduced into huSCARNA15 are shown in red. (B) Mapping of pseudouridines by fluorescent primer extension reactions. Top (gray) trace: Wild-type S. cerevisiae (BY4741) U2 snRNA is normally modified at positions 44, 42, and 35. Traces 2 (red) and 3 (green): When huSCARNA15 was expressed in the wild-type (red) or mutant pus7Δ strains (green), position 40 became pseudouridylated, as indicated by the star. Trace 4 (light blue): In the snr81Δ strain, huSCARNA15 induces pseudouridylation of positions 40 and 38 (stars). Traces 5 (magenta) and 6 (violet): 1-nt insertions between the box ACA and the upper stem (+U@ACA, or +A@loop) eliminate modification at position 38. Trace 7 (blue): Control snr81Δ strain without exogenous guide RNA. Traces 8–10: When expressed in the snr81Δpus1Δ mutant strain, the mutated RNAs produce only Ψ40 (star).
In most vertebrate species, except eutherians, SCARNA15 contains a pseudouridylation pocket for position 41 of U2 snRNA (Fig. 3A), which makes this RNA a triple guide similar to Drosophila scaRNA:ΨU2-38.40.42. We chose Xenopus SCARNA15 to test in the yeast cell system. In the X. tropicalis genome two genes encode SCARNA15: One is expressed ubiquitously, whereas the other is detected only in oocytes. We tested yeast expression constructs for both copies of X. tropicalis SCARNA15. Because these constructs behaved similarly, we focus here mostly on the ubiquitously expressed copy of SCARNA15. We expected to see U2 snRNA pseudouridylation at positions 40 and 38 in addition to the rescue of Ψ42 in the snr81Δ yeast mutant strain. Intriguingly, we could detect only Ψ42 and Ψ40, when Xenopus SCARNA15 was introduced into yeast cells (Fig. 3B, top two traces). The first and most plausible explanation was that Ψ42 somehow inhibited the formation of Ψ38, as we observed when human SCARNA15 was expressed in snR81-positive yeast strains (Fig. 2B, traces 2 and 3). To eliminate the U2-Ψ42 guide activity from Xenopus SCARNA15, we replaced the 5′ terminal stem, which is associated with this modification, with the corresponding human segment, which does not show guide activity for positioning Ψ42 in yeast U2. However, the chimeric SCARNA15 did not induce the formation of Ψ38 (Fig. 3B, trace 3). Two other structural differences between human and Xenopus SCARNA15 were (i) the distance from the ACA box to the base of the upper stem in the configuration for positioning Ψ38: the canonical 15 nt in human (Fig. 2A) versus 16 nt in Xenopus (Fig. 3A) and (ii) the stability of the upper stem: strong base-pairing in human (Fig. 2A; Supplemental Fig. S1) versus noncanonical A–A pairing and 1-nt bulge in Xenopus (Fig. 3A; Supplemental Fig. S1). In fact, an extra nucleotide inserted in human SCARNA15 between the 3′-terminal antisense element and box ACA made the pocket nonfunctional for positioning Ψ38 in yeast U2 snRNA (Fig. 2B, traces 5 and 6). The positions of inserted nucleotides are depicted in Figure 2A. We also expressed the mutated human SCARNA15 in the snr81Δpus1Δ strain. In this strain, no preceding stop signals disguise the detection of the test pseudouridines; still Ψ40 but not Ψ38 was detected (Fig. 2B, three bottom traces). Intriguingly, a 1-nt deletion in Xenopus SCARNA15 that mimics the human SCARNA15 loop structure (Fig. 3A, mutation 1; Supplemental Fig. S1) failed to create a functional pseudouridylation pocket for positioning Ψ38 (Fig. 3B, trace 4). Two additional mutations that generate strong canonical base-pairing in the upper stem (Fig. 3A, mutation 2; Supplemental Fig. S1) were required for the guide activity at position 38 of U2 snRNA in yeast cells (Fig. 3B, trace 5); these mutations alone were not sufficient to gain the activity. Remarkably, when the distance from the ACA box to the base of the upper stem and the upper stem structure were fixed in Xenopus SCARNA15, this RNA became fully functional at all three predicted positions in yeast U2 snRNA (Ψ42, Ψ40, and Ψ38) with no interference between Ψ42 and Ψ38 (Fig. 3B, trace 6).
FIGURE 3.
Pseudouridylation of yeast U2 snRNA induced by Xenopus SCARNA15 and by the C. elegans ortholog, snoRNA C50C3.14. (A,C) Predicted base-pairing between SCARNA15 and Xenopus U2 snRNA (A) or C. elegans U2 snRNA (C). The equivalent yeast U2 snRNA sequences are shown below. Mutated nucleotides are highlighted in red. (B,D) Fluorescent primer extension reactions. (B) Traces 1 (green) and 2 (blue): Wild-type Xenopus SCARNA15 modifies yeast U2 snRNA at positions 42 and 40 when expressed in the snr81Δ mutant yeast strain. Trace 3 (red): Chimeric SCARNA15 (human 5′ terminal hairpin and Xenopus 3′ terminal hairpin) modifies only position 40 of yeast U2 snRNA. Trace 4 (violet): Mutation 1 alone did not change pseudouridylation of position 38. Traces 5 (light blue) and 6 (magenta): A combination of mutations 1 and 2 is required to make the Xenopus 3′ terminal pseudouridylation pocket functional at position 38 in U2 snRNA. Trace 7 (dark violet): Mutation 3 makes the 3′-terminal domain of Xenopus SCARNA15 nonfunctional on yeast U2 snRNA; only the 5′ terminal pocket functions to position Ψ42. Stars indicate induced modifications. (D) Traces 1 (red) and 2 (blue): Both wild-type (red) and mutant C. elegans SCARNA15 (blue) are fully functional at all three predicted positions in yeast U2 snRNA: Ψ42, Ψ40, and Ψ38 (stars).
From these experiments one might conclude that a strong upper stem and an exact 15-nt distance from box ACA to the target position are necessary and sufficient elements for fully functional pseudouridylation pockets, especially for those with a noncanonical “3-nt-unpaired” configuration. However, the situation is not quite so simple. When we deleted 1 nt adjacent to the box ACA in Xenopus SCARNA15 (Fig. 3A, mutation 3; Supplemental Fig. S1), the 3′ terminal hairpin with the “exact-15-nt-distance” and optimized upper stem no longer formed a functional pseudouridylation pocket for any position in U2 snRNA. This mutated guide RNA was expressed at an unusually low level in yeast cells and supported pseudouridylation of U2 snRNA only at position 42, the target of the 5′-terminal pseudouridylation pocket (Fig. 3B, trace 7). The most intriguing observation came when we tested the homologous guide RNA from the worm Caenorhabditis elegans. We identified C. elegans snoRNA C50C3.14 as an ortholog of SCARNA15. The actual 5′ terminus of this RNA, as verified by 5′ RACE, is 5-nt longer than the annotated sequence. The extended 5′ terminus forms a pseudouridylation pocket for position 43 in C. elegans U2 snRNA (equivalent to position 41 in vertebrates or position 42 in yeast). The 3′ terminal pocket in two alternative configurations base-pairs with U2 to positions Ψ41 and Ψ39 (equivalent to Ψ39 and Ψ37 in vertebrates). The C. elegans RNA has an imperfect upper stem and a 16-nt distance from box ACA to the target position (Fig. 3C). However, when expressed in yeast cells, this RNA was fully functional at all three predicted positions in yeast U2 snRNA: 42, 40, and 38 (Fig. 3D, top trace). Then we compared software-predicted secondary structures of all tested wild-type and mutated variants of the guide RNAs for positioning Ψ38 and Ψ40 in yeast U2 snRNA (Supplemental Fig. S1). Based on this analysis, we hypothesized that the formation of a prominent bulge between the lower and upper stems might be critical for efficient base-pairing of guide RNA with its substrate and the positioning of pseudouridine. However, mutations in C. elegans C50C3.14 RNA that generated a stiff hairpin without obvious loop formation (Fig. 3C; Supplemental Fig. S1) did not affect guide RNA modification activity on yeast U2 snRNA (Fig. 3D, bottom trace).
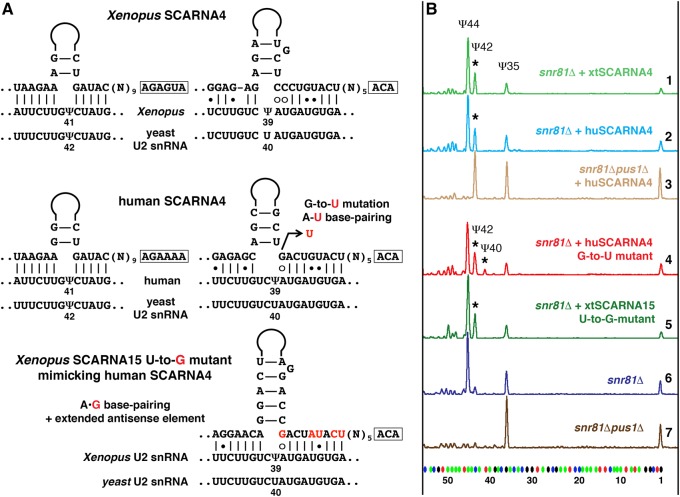
Human and Xenopus SCARNA4/ACA26 guide RNA activity in the yeast cell system
Human SCARNA4 (also known as ACA26) was also annotated as a guide for U2 snRNA pseudouridylation at positions 39 and 41 (Kiss et al. 2004). However, both human and Xenopus SCARNA4, when tested in the yeast cell system, modified only position 42 in yeast U2 snRNA (equivalent to position 41 in vertebrates) (Fig. 4B, top three traces). In fact, we consider the configuration of the U2-Ψ39 pseudouridylation pocket too weak. It contains several noncanonical features: three unpaired nucleotides at the target position, four U–G base pairs, and an A–G mismatch adjacent to the target U at the 3′ side of the loop (Fig. 4A). A single G-to-U mutation in human SCARNA4, which allows A–U base-pairing to substitute for A–G, permitted this guide RNA to modify position 40 in yeast U2 snRNA (Fig. 4B, trace 4). Contrariwise, a single U-to-G mutation in Xenopus SCARNA15, which converted the canonical A–U pairing into an A–G mismatch (Fig. 4A), made the resultant pseudouridylation pocket nonfunctional (Fig. 4B, trace 5). This was true even if the remaining base-pairing at the 3′ side of the mutated pocket was additionally extended (Fig. 4A). These data allow us to postulate that efficient base-pairing at the position adjacent to the targeted uridine is one of the most essential elements of functional pseudouridylation pockets. Accordingly, we could not identify position 39 of U2 snRNA as a target for human SCARNA4. Table 1 summarizes previously known and newly verified mechanisms of post-transcriptional modification within the branch point recognition region of U2 snRNA in different species.
FIGURE 4.
Pseudouridylation of yeast U2 snRNA induced by Xenopus and human SCARNA4. (A) Predicted base-pairing of SCARNA4 with vertebrate and yeast U2 snRNA. A point mutation in human SCARNA4 that converts the A–G mismatch into a canonical A–U base pair is shown in red. A mutated Xenopus SCARNA15 that mimics base-pairing of human SCARNA4 with U2 snRNA is also shown; mutated positions are highlighted in red. (B) Fluorescent primer extension reactions. Traces 1 (green), 2 (blue), and 3 (pale brown): Xenopus and human SCARNA4 rescue yeast U2 snRNA pseudouridylation at position 42 (star) in the snr81Δ and snr81Δpus1Δ mutant strains. Trace 4 (red): Human SCARNA4 shows no activity on position 40 unless G in the 3′-terminal pocket is mutated to U. Trace 5 (dark green): A–G base-pairing within the 3′ terminal pseudouridylation pocket of the mutated Xenopus SCARNA15 makes the 3′-terminal domain nonfunctional on yeast U2 snRNA. Note absence of Ψ40. Traces 6 (dark blue) and 7 (brown): Controls without exogenous RNAs.
TABLE 1.
Branch point recognition region of U2 snRNA: modified positions and corresponding modification mechanisms
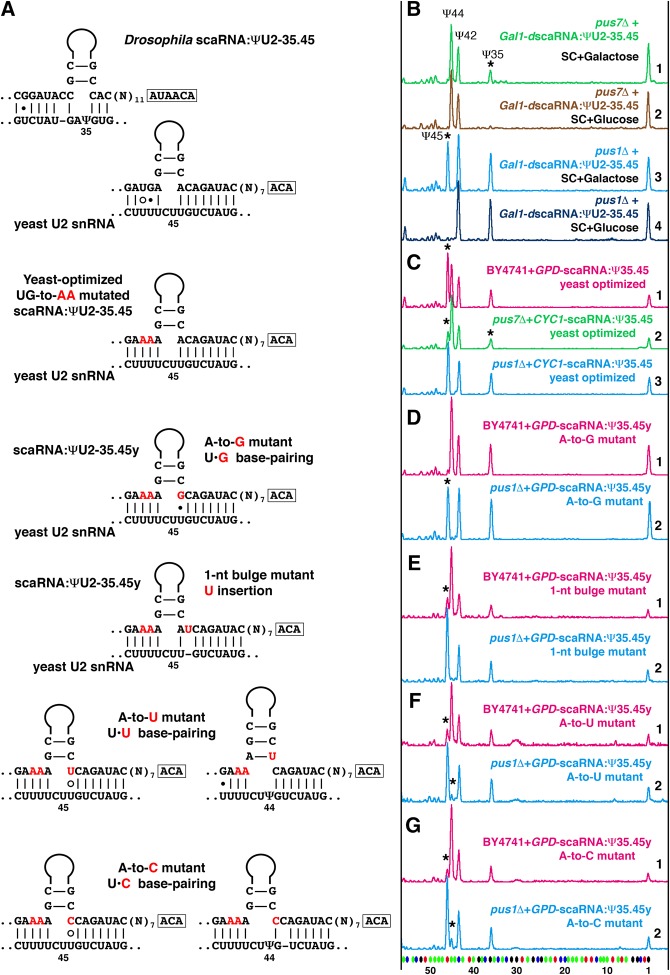
U2-Ψ43 and Ψ44 interference
In our previous study, we observed that in an S. cerevisiae yeast cell system Ψ44 in U2 snRNA interfered with the positioning of Ψ45 mediated by the Drosophila guide RNA, scaRNA:ΨU2-35.45 (Deryusheva and Gall 2013). Positions 44 and 45 in Drosophila and S. cerevisiae U2 snRNA correspond to positions 43 and 44 in vertebrates and S. pombe (Fig. 1). Furthermore, a preexisting Ψ44 completely inhibited pseudouridylation at position 45 when Drosophila scaRNA:ΨU2-35.45 was expressed in yeast cells stepwise from an inducible Gal promoter (Fig. 5B). One could argue that this interference resulted from a suboptimal antisense element in the Drosophila pugU2-35/45 RNA (Fig. 5A). We made a new construct that expresses Drosophila scaRNA:ΨU2-35.45 optimized for positioning Ψ45 in yeast U2 snRNA (Fig. 5A). We could now reproduce in S. cerevisiae the Ψ44Ψ45 modification pattern found in higher eukaryotes, but only when the guide RNA was overexpressed (Fig. 5C, top trace). The same optimized guide RNA did not efficiently mediate pseudouridylation at position 45 in Pus1p-positive strains, when it was expressed stepwise from an inducible Gal promoter or when expressed constitutively, but at lower levels, from a plasmid with a weak CYC1 promoter (Fig. 5C, traces 2 and 3). That is, a prior Ψ44 formation and A-Ψ base-pairing between the guide and substrate RNAs hinders modification. We saw the same phenomenon (Fig. 6A) in similar experiments using mouse SCARNA8, an established guide RNA for positions 44, 43, and 34 in vertebrate U2 snRNA (Deryusheva and Gall 2017).
FIGURE 5.
Pseudouridylation of U2 snRNA at position 44 interferes with pseudouridylation at position 45 in the yeast cell system. (A) Base-pairing of wild-type and mutated variants of Drosophila scaRNA:ΨU2-35.45 with yeast U2 snRNA. Mutated nucleotides are shown in red. (B–G) Fluorescent primer extension reactions. (B) Traces 1 (green) and 2 (light brown): Drosophila scaRNA:ΨU2-35.45 cannot induce pseudouridylation at position 45 when expressed stepwise from an inducible Gal promoter in the pus7Δ yeast strain. Rescue of pseudouridylation at position 35 (star) serves as an internal positive control. Traces 3 (blue) and 4 (dark blue): In the same experimental setup, Drosophila scaRNA:ΨU2-35.45 is active at position 45 (star) in the pus1Δ strain, which has no Ψ44 in U2 snRNA. (C) Trace 1 (magenta): scaRNA:ΨU2-35.45 optimized for positioning Ψ45 in yeast U2 snRNA efficiently modifies position 45 in wild-type yeast strain only when overexpressed. Trace 2 (green): The same yeast-optimized RNA modifies position 45 inefficiently in Pus1p-positive strains, such as pus7Δ. Trace 3 (blue): This RNA is fully functional at position 45 in the pus1Δ strain. (D–G) Traces 1 (magenta): Point mutations in the 3′ side of the yeast-optimized U2-Ψ45 pseudouridylation pocket affect the corresponding modification activity, especially in the presence of U2-Ψ44 in a “wild-type” yeast strain. Traces 2 (blue): The U2-Ψ45 modification activity is evident in the absence of U2-Ψ44 in the pus1Δ yeast strain. Stars indicate guide RNA-induced pseudouridines.
FIGURE 6.
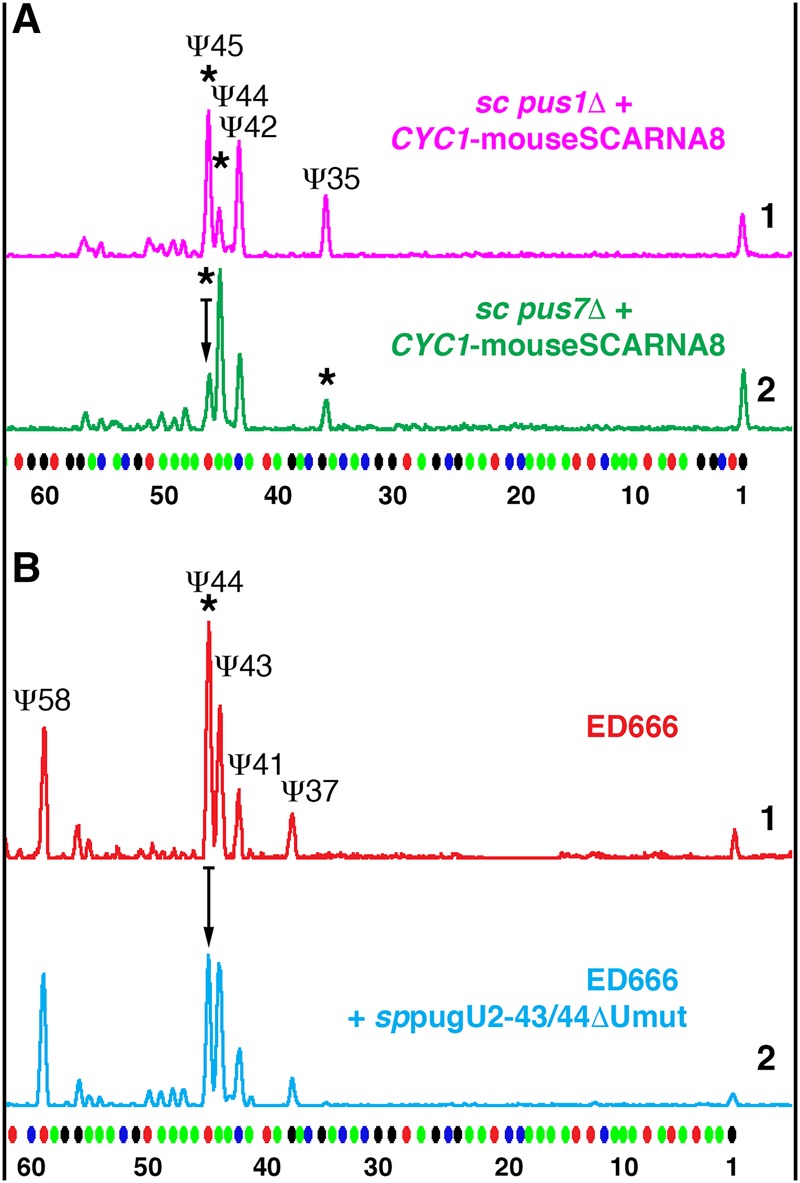
Prior pseudouridylation of U2 snRNA at position 43 inhibits positioning of Ψ44 by mouse SCARNA8 and S. pombe pugU2-43/44, SPNCRNA.1709. Fluorescent primer extension reactions. (A) Mouse SCARNA8 was expressed in the S. cerevisiae strains pus1Δ (top magenta trace) and pus7Δ (bottom green trace) from a plasmid with a weak CYC1 promoter that allowed prior modification of position 44 in yeast U2 snRNA by endogenous Pus1p. In the pus7Δ strain, modification of position 45 was reduced relative to that in the pus1Δ strain (arrow shows reduction in peak height). Stars indicate SCARNA8-induced pseudouridylation. Modification of position 35 in the pus7Δ strain served as an internal positive control. (B) Top trace (red): Pseudouridylation pattern of U2 snRNA in wild-type S. pombe (strain ED666). Bottom trace (blue): Overexpression of pugU2-43/44ΔUmut RNA caused reduction of pseudouridylation at position 44 (arrow shows reduction in peak height).
To test further how base-pairing at the 3′-side of a pseudouridylation pocket regulates the efficiency of guide RNA function, we generated four mutant variants of the yeast-optimized guide RNA with noncanonical U–G, U–U, U–C base-pairing and a 1-nt bulge at the 3′-side of the pocket (Fig. 5A). These mutated guide RNAs were fully functional only in the absence of U2-Ψ44 in the pus1Δ strain, even if they were overexpressed (Fig. 5D–G, compare traces 1 and 2). It is worth noting that in the pus1Δ mutant strain, two of four mutated guide RNAs were partially active at position 44 in addition to position 45 (Fig. 5F,G, traces 2). Alternative base-pairing within the corresponding pseudouridylation pocket is shown (Fig. 5A), as this is most likely associated with the positioning of Ψ44. Importantly, Northern blot analysis showed that all mutated variants of the guide RNA were expressed at the same level in the wild-type and mutant strains. Thus, we observed that imperfect base-pairing at the position preceding the target uridine hindered guide RNA modification activity, especially when the preceding position was also pseudouridylated.
The interference between positions 44 and 45 of U2 snRNA during their pseudouridylation was also observed when we tested a recently identified S. pombe guide RNA pugU2-43/44 (now annotated as SPNCRNA.1709 in the S. pombe genome assembly) in an S. cerevisiae system (positions 43 and 44 in S. pombe U2 snRNA correspond to positions 44 and 45 in S. cerevisiae). In this case, modification activity on position 45 was lost when the yeast-specific double-guide RNA was mutated to make the configuration for positioning Ψ44 more favorable (Deryusheva and Gall 2017). However, in all these experiments, the evidence of interference between the two adjacent pseudouridylated positions in U2 snRNA was limited to observations made using S. cerevisiae cells, where U2 snRNA is normally modified at position 44 but not at position 45. To assess this phenomenon in a species where U2 snRNA is endogenously modified at the two corresponding positions, we expressed a mutated sppugU2-43/44 RNA (sppugU2-43/44ΔUmut) in the wild-type S. pombe strain; this construct is active only on position 44 in S. cerevisiae cells (Deryusheva and Gall 2017). Overexpression of wild-type sppugU2-43/44 RNA served as a control. Although the effect of the mutant guide RNA expression in S. pombe was not very dramatic, the modification of U2 snRNA at position 44 was reduced relative to modification of nearby positions (Fig. 6B). This result suggests that in S. pombe, prior pseudouridylation of position 43 by overexpression of the mutant guide RNA inhibited positioning of Ψ44 by the endogenous guide RNA.
Cm40Ψ41 interference
In fission yeast and higher eukaryotes, there is a 2′-O-methylated cytosine adjacent to a pseudouridine in the branch point recognition region of U2 snRNA—dinucleotide Cm40Ψ41 (Cm41Ψ42 in Drosophila) depicted in Figure 1. In S. cerevisiae, 2′-O-methylated residues have not been detected in any spliceosomal snRNAs and in this species, the corresponding dinucleotide in U2 snRNA is unmethylated C41Ψ42. To test whether induction of 2′-O-methylation of C41 could affect the positioning of Ψ42 mediated by endogenous snR81 guide RNA, we overexpressed Drosophila scaRNA:MeU2-C41 in the wild-type yeast strain. We previously confirmed that this RNA is a genuine guide RNA for U2 snRNA modification (Deryusheva and Gall 2009); no orthologs have been identified so far in any other species. In the wild-type yeast strain transformed with a plasmid for overexpression of Drosophila scaRNA:MeU2-C41, the induced 2′-O-methylation of C41 led to a dramatic reduction of the pseudouridylation level at position 42 (Fig. 7A). A similar effect was observed when human SCARNA4 substituted for snR81 RNA in the snr81Δ yeast strain. Furthermore, in HeLa cells, overexpression of an artificial guide RNA for 2′-O-methylation of U2 snRNA at position 40 resulted in a slight but highly reproducible reduction of U2 snRNA pseudouridylation at position 41 (Fig. 7B).
FIGURE 7.
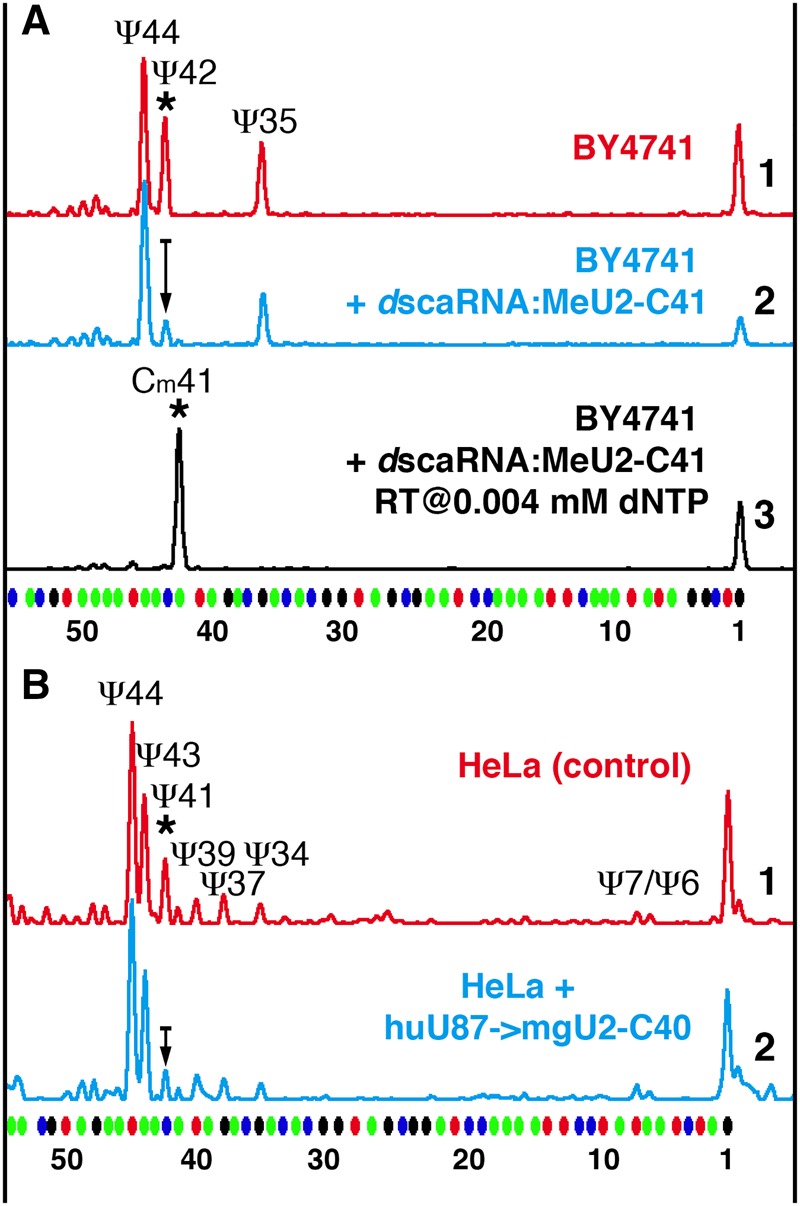
Prior 2′-O-methylation of a position in the branch point recognition region inhibits pseudouridylation of the following position. Fluorescent primer extension reactions. (A) Trace 3 (black): Expression of Drosophila scaRNA:MeU2-C41 in the wild-type S. cerevisiae strain BY4741 results in 2′-O-methylation of yeast U2 snRNA at position 41. Traces 1 (red) and 2 (blue): Methylation at position 41 results in dramatic reduction of pseudouridylation at position 42 (trace 2) relative to control (trace 1). (B) Traces 1 (red) and 2 (blue): In HeLa cells, overexpression of the methylation guide RNA for position C40 of U2 snRNA (trace 2) leads to a lower level of U2 snRNA pseudouridylation at position 41 than in control cells transfected with an empty vector (trace 1). The arrow in trace 2 shows reduction in peak height.
Positioning of Cm45Ψ46 in U5 snRNA
A modified dinucleotide (CmΨ) has been found in the 5′ loop of U5 snRNA in many eukaryotes (Szkukalek et al. 1995), except for S. cerevisiae (Massenet et al. 1999). The corresponding modification guide RNA is a compound box C/D and box H/ACA RNA U85 (currently annotated as SCARNA10). It was one of the first guide RNAs discovered and its modification activity at the predicted target positions was experimentally verified (Jády and Kiss 2001). As expected, human and Xenopus U85 RNAs could modify a CU dinucleotide in a fragment of human U5 snRNA, when these guide RNAs and their substrate RNA U87–U5[37–57] were expressed in yeast cells (Fig. 8). However, of the two annotated copies of Xenopus U85 RNAs we tested, only one supported modification of both positions, generating Cm45Ψ46 (Fig. 8B–D, trace 2; 8E, trace 1); the other induced only 2′-O-methylation at position 45 (Fig. 8B–D, trace 4; 8E, trace 3). Importantly, pseudouridylation of position 46 became evident when an antisense element for 2′-O-methylation of U5–C45 was mutated in the latter copy of Xenopus U85 RNA (Fig. 8B–D, trace 3; 8E, trace 2). Thus, in the case of modification of the 5′ loop in U5 snRNA, we see again that prior 2′-O-methylation of position 45 might inhibit pseudouridylation of adjacent position 46. Surprisingly, the two copies of Xenopus U85 RNAs have similar sequences; the domains containing U5-Ψ46 pseudouridylation pockets are 80.9% identical. The binding affinity with the substrate for Ψ46 positioning is even stronger in the least functional copy of Xenopus U85 (Fig. 8A). What might make this pocket weaker are alterations in secondary structure: slightly weaker base-pairing in the upper stem compared to human U85 and impaired H box-containing hinge compared to human and the other copy of Xenopus U85 (Supplemental Fig. S2).
FIGURE 8.
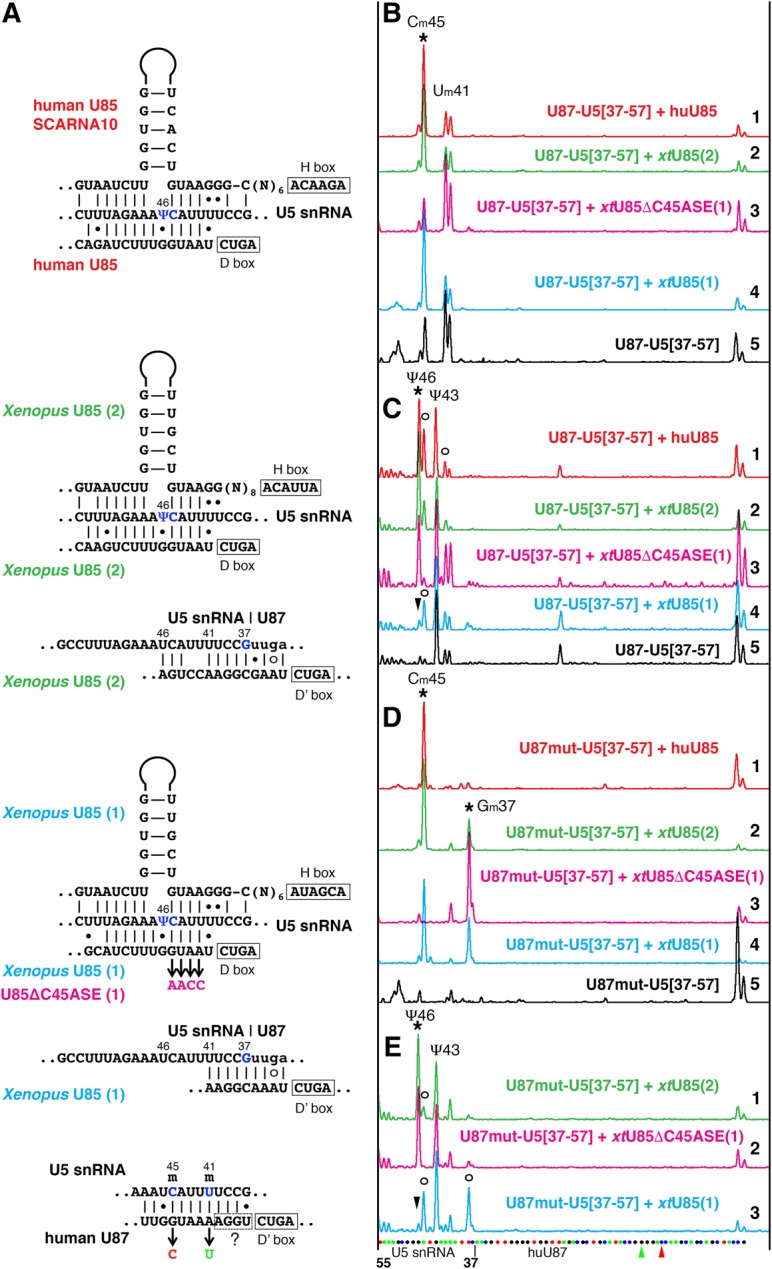
U85 scaRNA modification activity on U5 snRNA. (A) Predicted interactions of human U85, Xenopus U85, and human U87 scaRNAs with U5 snRNA. Targeted positions are shown in blue. Mutations introduced in antisense elements of Xenopus U85 and human U87 RNAs are color-coded to match colors used in B–E. (B–E) Fluorescent primer extension reactions. (B,D) 2′-O-methylation and (C,E) pseudouridylation of the 5′ loop of vertebrate U5 snRNA (nucleotides 37–57) inserted into U87 RNA and expressed in wild-type yeast cells. Stars indicate peaks that correspond to modifications induced by human U85 (traces 1, red in B–D) and two Xenopus wild-type U85 (traces 2, green and 4, blue in B–D; traces 1, 3 in E), and Xenopus U85 with mutated Cm45 antisense element, U85ΔC45ASE (traces 3, magenta in B–D; trace 2 in E). Arrowheads point to missing Ψ46. Trace 5, black in B, C, and D: Control samples of U87–U5 substrate RNAs expressed alone in yeast cells. In the tested artificial U5 snRNA substrate, U87–U5[37–57], position 43 is pseudouridylated by an unknown endogenous yeast snoRNA (C,E) and positions 41 and 45 are 2′-O-methylated by U87 RNA itself (B). (D) When the antisense element for positioning Um41 and Cm45 was removed from U87, U87mut-U5[37–57], an additional position became 2′-O-methylated by Xenopus U85; human U85 could not induce this modification. Open circles in C and E indicate stops at 2′-O-methylated positions. In the interest of space, only three Xenopus U85 traces are shown in E; the pattern induced by human U85 (not shown) is very similar to that in trace 1 (green) of Xenopus U85(2); the control U87mut-U5[37–57] alone (not shown) looks as expected, with one peak at Ψ43 and no extra peak at Um41. The sequence of U87–U5 substrate RNA and mutated positions are shown at the bottom of E.
It is worth describing here another observation made in these experiments. The U87 RNA (SCARNA5) that we use to make stable artificial substrates for expression in yeast cells is itself a guide RNA for 2′-O-methylation of U5 snRNA at position 41; in the yeast cell system it also inefficiently modifies U5–C45 (Fig. 8B, traces 3 and 5). Intriguingly, when these modification activities were removed from U87–U5[37–57] substrate RNA by point mutations (Fig. 8A), Xenopus U85 RNA became a methylation guide for an additional position in a U5 snRNA fragment (Fig. 8D); potential base-pairing is shown in Figure 8A. In this case, 2′-O-methylation at a nearby position might impede 2′-O-methylation mediated by a guide RNA with a weak antisense element. Notably, the absence of Um41 did not improve pseudouridylation activity of the intact inactive copy of Xenopus U85 (Fig. 8E, trace 3), but likely facilitated pseudouridylation by its mutated active variant (Fig. 8E, trace 2).
DISCUSSION
Since the first discovery of modification guide RNAs for spliceosomal U snRNAs (Tycowski et al. 1998), most known modifications in snRNAs have been matched with the corresponding guide RNAs (https://www-snorna.biotoul.fr/index.php, Lestrade and Weber 2006; Karijolich and Yu 2010). In higher eukaryotes, the guide RNA assignment is generally based on a computational search for sequence complementarity between guide and substrate RNAs, but only a few of these predictions have been experimentally verified (Jády and Kiss 2001; Zhao et al. 2002; Deryusheva and Gall 2009, 2013, 2017). Even in these cases, the guide RNA assays are usually limited to certain predicted target positions, excluding noncanonical interactions from the analysis. For example, two positions are targeted by alternative configurations of the 3′-terminal pseudouridylation pocket in vertebrate SCARNA8, but only one was originally tested (Zhao et al. 2002; Deryusheva and Gall 2017). At the same time, software-based predictions often cannot be verified experimentally (Deryusheva and Gall 2009; Xiao et al. 2009). Thus, experimental verification of each guide RNA assignment is necessary.
In this study, we used an in vivo yeast cell system to analyze six different guide RNAs from human, mouse, Xenopus, Drosophila, and C. elegans. Our data and previous studies have demonstrated that extended stretches of complementarity with substrate RNA do not necessarily predict guide RNA functionality. In fact, minor alterations in RNA secondary structure may turn a “perfect” guide RNA into a nonfunctional one. Thus, the stability of the upper and lower stems and the correct distance (15 ± 1 nt) between the target position and the box ACA (or H) play important roles in the formation of a functional pseudouridylation pocket (Ni et al. 1997; Bortolin et al. 1999; Xiao et al. 2009; Deryusheva and Gall 2017, this study). However, these structural requirements are neither absolutely essential nor sufficient to create a functional pseudouridylation pocket, as seen in the case of wild-type and mutant SCARNA15 tested in this study (Fig. 3). We also learned from our analysis that noncanonical base-pairing and post-transcriptional modifications placed within the region of interaction between guide and substrate RNAs may significantly impair modification activity. For instance, we found base-pairing at the position preceding the uridine targeted for isomerization especially sensitive to mismatches and modifications. It is not clear how modified substrate RNA mechanistically affects box H/ACA pseudouridylation guide RNP function. While being capable of pairing with any of the four major bases, pseudouridine may increase the backbone rigidity and stabilize base stacking (Spenkuch et al. 2014). Ribose methylation is also known to cause conformational changes and increased stability of RNA (Helm 2006). Does it change interaction with a guide RNA and/or cause incorrect positioning of the target uridine at the active site? To our knowledge no structural studies on RNA-guided pseudouridine synthases have been done using premodified substrate or guide RNAs.
Thus, prior modification at certain positions may inhibit modification of nearby nucleotides. Nevertheless, in some guide RNAs, “suboptimal” configurations are well preserved during evolution. Furthermore, most noncanonical base pairs and bulges within guide-substrate base-paired regions coincide with positions of known post-transcriptionally modified nucleotides in rRNAs and snRNAs. We postulate that these “suboptimal” guide RNAs must have been “optimized” for desired post-transcriptional modification in heavily modified regions. We argue that these minor alterations in primary and secondary structure of guide RNAs may help generate heterogeneous populations of modified RNA molecules. A region within the unstructured loop in the central domain of human 18S rRNA is an example of this interplay. Based on the binding affinity of guide RNAs SNORA24, SNORA28, and SNORD98 for positioning Ψ863, Ψ866, and Gm867 in human 18S rRNA (Fig. 9A), we predict that prior modification of position 866 and/or position 867 would inhibit pseudouridylation of position 863; at the same time Ψ863, if positioned first, would interfere with positioning of Gm867, but not Ψ866 (Fig. 9B, top line). Thus, one would expect to find two differentially modified rRNA molecules: those having Ψ863 and Ψ866, or Ψ866 and Gm867, but none having all three positions modified in the same molecule (Fig. 9B, bottom two lines). Precisely this pattern was demonstrated in early biochemical studies of mammalian 18S rRNA modification (Maden and Wakeman 1988). The existence of differentially modified rRNA molecules has been well established (Buchhaupt et al. 2014; Taoka et al. 2016). The same is presumably true for all post-transcriptionally modified RNA species, including spliceosomal snRNAs. However, the functional significance of this heterogeneity is still an open question.
FIGURE 9.
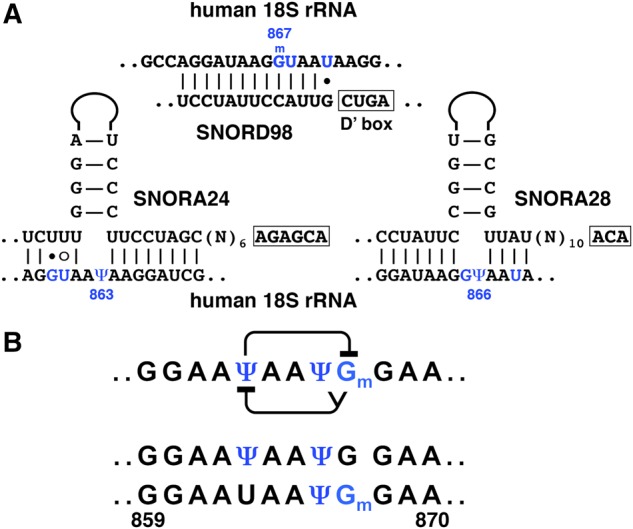
Interactions within a heavily modified region of human 18S rRNA. (A) Base-pairing between human 18S rRNA and guide RNAs SNORA24, SNORA28, and SNORD98 assigned for modification of positions 863, 866, and 867, respectively. (B) Predicted interference between Ψ863, Ψ866, and Gm867 (top row) and differentially modified fragments (second and third rows) experimentally detected by classical biochemical techniques (Maden and Wakeman 1988). Modified positions are colored in blue.
At the same time, modification levels often reach >95%, even in regions where modification of certain positions inhibits subsequent modification of nearby positions (Taoka et al. 2016). How do cells regulate this uniform modification process? Very probably these modifications occur in a strictly coordinated manner, starting from the positions most compromised by their neighbors. One possibility is that cells express multiple guide RNAs at different levels. The guide RNAs for modification of U2-Cm41Ψ42 in Drosophila provide one of many examples. Another way to achieve coordinated modification is to combine two machineries with different kinetics: fast and efficient guide RNA-mediated modification and slower modification by stand-alone enzymes; the trinucleotide Um2921Gm2922Ψ2923 in the A-loop of yeast 25S rRNA (Lapeyre and Purushothaman 2004) and Ψ43Ψ44 in U2 snRNA (Deryusheva and Gall 2017) are typical examples. Still a third possibility is to combine multiple modification activities in one guide RNA and to vary the efficiency of each guide domain according to the required order for stepwise positioning of these modifications. SCARNA15 and SCARNA10/U85 tested in this study represent such multitarget guide RNAs. Combinations of these mechanisms are also exploited. For example, Pus1p and a double guide RNA are both used to modify U2-Ψ43Ψ44 in vertebrates and S. pombe (Deryusheva and Gall 2017). Another example involves the double guide RNA U85 to modify U5-Cm45Ψ46 and an independent guide RNA, U89/SCARNA12, to modify the more vulnerable position U5-Ψ46.
Because the modification machinery is highly conserved for evolutionary conserved sites, the two must have co-evolved in each species. Comparison of the modifications in human (Deryusheva et al. 2012) and Drosophila U2 snRNA (Deryusheva and Gall 2013) provide an example. Coincident with the difference in 2′-O-methylation upstream of the branch point recognition region, predominant and inhibited positioning of pseudouridines within the branch point recognition region differs between human and Drosophila (Fig. 1).
Perspectives
Our data strongly suggest that interconnections between structure and function of modification guide RNAs are more complex than generally anticipated. The substrate RNA and its overall modification pattern also contribute to guide RNA functionality. RNA modification occurs in a highly coordinated manner, especially in heavily modified regions. Diversified activities of modification guide RNAs serve as key regulators in generating the pool of properly modified RNA molecules in the cell; either uniformly or differentially modified pools can be considered as the properly modified RNAs. The presence or absence of certain modifications may trigger (i) a degradation of correspondingly modified RNA or (ii) extra stability of innate RNA/RNP, as well as (iii) an assembly of RNP with new protein counterparts, which is essential for (iv) a new function/configuration. All these scenarios are equally plausible and exploitable. Use of innovative techniques that allow RNA modification mapping at single molecule level will give us new insights into RNA modification diversity and function.
MATERIALS AND METHODS
Guide RNA sequence and structure analysis
We retrieved guide-substrate base-pairing annotations available for human and other vertebrate species (https://www-snorna.biotoul.fr, http://www.bioinf.uni-leipzig.de/publications/supplements/15-065). We also reanalyzed guide RNA secondary structures using the Mfold and Vienna RNAfold web servers for RNA folding prediction (http://mfold.rna.albany.edu, http://rna.tbi.univie.ac.at) followed by visual analysis for some adjustments. Standard RACE-PCR was used to verify 5′ and 3′ ends of C. elegans snoRNA C50C3.14.
Expression in yeast cells
To express exogenous guide RNAs in yeast cells, we amplified the coding sequences from genomic DNA and cloned them into YEplac181 [LEU2 2µ] or YEplac195 [URA3 2µ] vectors containing a GPD promoter, an RNT1 cleavage site, and an snR13 terminator (Huang et al. 2011). These constructs were made for all known copies of SCARNA4/ACA26, SCARNA15/ACA45, SCARNA10/U85 from human and X. tropicalis genomes, C. elegans C50C3.14 RNA and Drosophila scaRNA:MeU2-C41. Drosophila scaRNA:U2-Ψ35.45 and mouse SCARNA8 expression constructs were previously generated (Deryusheva and Gall 2013, 2017). Fragments containing the listed guide RNA sequences with the flanking regulatory elements, RNT1 cleavage site, and snR13 terminator, were additionally cloned into p415CYC1 and p415GAL-S vectors. Other constructs were made in which Drosophila scaRNA:MeU2-C41 replaced a box C/D snoRNA (snR18) in the S. cerevisiae EFB1 gene; and sppugU2-43/44/ SPNCRNA.1709, wild-type or mutated, replaced a box H/ACA snoRNA (snR191) in the NOG2 gene. These chimeric fragments were also cloned into S. pombe expression vectors pJB114 [LEU2 ARS] or pJB115 [URA4 ARS] containing an ADH-promoter and an ACT1 terminator. To express an artificial substrate RNA that contains a sequence of the highly conserved 5′ loop of U5 snRNA (nucleotides 37–57 of human U5 snRNA), a new U87 scaRNA-based construct was generated as previously described (Deryusheva and Gall 2013, 2017). To make various mutant and chimeric guide RNA constructs we used PCR-based mutagenesis and overlap-extension PCR.
Yeast cells were transformed using standard lithium acetate methods. The following yeast strains were used in this study: S. pombe ED666 strain, S. cerevisiae “wild-type” BY4741 strain, and mutant strains snr81Δ, pus7Δ, pus1Δ, and snr81Δpus1Δ.
Transfection of HeLa cells
First, the chimeric fragment of yeast EFB1 gene expressing Drosophila scaRNA:MeU2-C41 (made for yeast expression) was subcloned into the pCS2-CMV vector. Unexpectedly, this construct did not produce a stable guide RNA in HeLa cells. A highly abundant artificial guide RNA for 2′-O-methylation of C40 in U2 snRNA was generated from the U87 scaRNA construct described elsewhere (Deryusheva and Gall 2013) by antisense element replacement and subcloning the resultant fragment into pCS2-CMV vector. HeLa cells were transfected with these constructs using ViaFect transfection reagent (Promega). After 48–96 h cultivation the transiently transfected cells were collected for RNA extraction.
RNA extraction
RNA from yeast cells was isolated by hot acid phenol extraction. TRIzol reagent was used to isolate RNA from HeLa cells, X. tropicalis oocytes and liver, and whole C. elegans worms. Direct-zol RNA MiniPrep Kit (Zymo Research) was used for RNA purification.
Northern blot
Total RNA was separated on 8% polyacrylamide–8M urea gels and transferred onto a nylon membrane (Zeta Probe, Bio-Rad). Hybridization and detection of digoxigenin-labeled probes were performed according to standard protocols (Deryusheva and Gall 2017).
RNA modification assays
Primer extension-based methods of RNA modification mapping were used to analyze pseudouridylation and 2′-O-methylation of yeast and human snRNAs (Kiss and Jády 2004; Deryusheva and Gall 2009). Fluorescently labeled oligonucleotides specific for human and yeast U2 snRNA and artificial substrate RNA were previously described (Deryusheva et al. 2012; Deryusheva and Gall 2013, 2017). To detect pseudouridines, test RNA samples were treated first with CMC, N-cyclohexyl-N′-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (Sigma-Aldrich) for 20–30 min and then with mild alkali buffer (50 mM Na2CO3, pH 10.4) for 3–4 h at 37°C. Primer extension was performed using AMV reverse transcriptase (New England Biolabs) at 0.5 mM dNTP concentration. 2′-O-methylated nucleotides were detected in primer extension reactions with a low concentration of dNTP (0.004 mM). Reaction products were separated on an ABI3730xl capillary electrophoresis instrument (Applied Biosystems); the Gene Scan-500 LIZ Size Standard was added to each sample. RNA sequencing reactions were performed as previously described (Deryusheva and Gall 2009). The data were analyzed using GeneMapper software (Applied Biosystems). Reverse transcription-based methods of modification mapping are not truly quantitative. Hence, to compare modification levels in different RNA samples, we always perform RNA treatment and enzymatic reactions on control and experimental RNAs simultaneously, perform several replicates of each experiment, and run serial dilutions of each sample.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to the following researchers: Yi-Tao Yu (University of Rochester Medical Center) for providing S. cerevisiae mutant strains and expression vectors; Peter Espenshade (Johns Hopkins University School of Medicine) for S. pombe ED666 strain and expression vectors; and Geraldine Seydoux (Johns Hopkins University School of Medicine) for supplying wild-type C. elegans worms. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01 GM33397 to J.G.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.G.G. is an American Cancer Society Professor of Developmental Genetics.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063842.117.
REFERENCES
- Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C. 2006. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA 12: 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Ganot P, Kiss T. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J 18: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchhaupt M, Sharma S, Kellner S, Oswald S, Paetzold M, Peifer C, Watzinger P, Schrader J, Helm M, Entian KD. 2014. Partial methylation at Am100 in 18S rRNA of baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS One 9: e89640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2009. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 20: 5250–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2013. Novel small Cajal body-specific RNAs identified in Drosophila: probing guide RNA function. RNA 19: 1802–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. 2017. Dual nature of pseudouridylation in U2 snRNA: Pus1p-dependent and Pus1p-independent activities in yeasts and higher eukaryotes. RNA 23: 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Choleza M, Barbarossa A, Gall JG, Bordonné R. 2012. Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells. RNA 18: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez G, Hartmuth K, Lührmann R. 2004. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10: 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Patton JR, Shimba S, Reddy R. 1996. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA 2: 909–918. [PMC free article] [PubMed] [Google Scholar]
- Helm M. 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Karijolich J, Yu YT. 2011. Post-transcriptional modification of RNAs by artificial Box H/ACA and Box C/D RNPs. Methods Mol Biol 718: 227–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Kiss T. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J 20: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR. 2016. An updated human snoRNAome. Nucleic Acids Res 44: 5068–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. 2010. Spliceosomal snRNA modifications and their function. RNA Biol 7: 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Jády BE. 2004. Functional characterization of 2′-O-methylation and pseudouridylation guide RNAs. Methods Mol Biol 265: 393–408. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jády BE, Bertrand E, Kiss T. 2004. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 24: 5797–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Purushothaman SK. 2004. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol Cell 16: 663–669. [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhao X, Yu YT. 2003. Pseudouridylation (ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. 2005. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 24: 2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden EH, Wakeman JA. 1988. Pseudouridine distribution in mammalian 18 S ribosomal RNA. A major cluster in the central region of the molecule. Biochem J 249: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. 1999. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol 19: 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. [DOI] [PubMed] [Google Scholar]
- Nishikura K, De Robertis EM. 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol 145: 405–420. [DOI] [PubMed] [Google Scholar]
- Patton JR, Padgett RW. 2005. Pseudouridine modification in Caenorhabditis elegans spliceosomal snRNAs: unique modifications are found in regions involved in snRNA–snRNA interactions. BMC Mol Biol 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JR, Jacobson MR, Pederson T. 1994. Pseudouridine formation in U2 small nuclear RNA. Proc Natl Acad Sci 91: 3324–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Busch H. 1988. Small nuclear RNAs: RNA sequences, structure, and modification. In Structure and function of major and minor small nuclear ribonucleoprotein particles (ed. Birnstiel ML), pp. 1–37. Springer, Berlin. [Google Scholar]
- Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. 2004. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem 279: 47661–47671. [DOI] [PubMed] [Google Scholar]
- Singh SK, Gurha P, Gupta R. 2008. Dynamic guide-target interactions contribute to sequential 2′-O-methylation by a unique archaeal dual guide box C/D sRNP. RNA 14: 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenkuch F, Motorin Y, Helm M. 2014. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 11: 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkukalek A, Myslinski E, Mougin A, Lührmann R, Branlant C. 1995. Phylogenetic conservation of modified nucleotides in the terminal loop 1 of the spliceosomal U5 snRNA. Biochimie 77: 16–21. [DOI] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T. 2016. The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res 44: 8951–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, You ZH, Graham PJ, Steitz JA. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 2: 629–638. [DOI] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 30: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. 2016. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J 35: 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang C, Schattner P, Yu YT. 2009. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA 15: 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, McPheeters DS, Yu YT. 2005. ψ35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem 280: 6655–6662. [DOI] [PubMed] [Google Scholar]
- Yu Y-T, Shu MD, Steitz JA. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT. 2004. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10: 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT. 2007. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res 35: 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li ZH, Terns RM, Terns MP, Yu YT. 2002. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA 8: 1515–1525. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.