Abstract
To silence target mRNAs, small RNAs and Argonaute (Ago) proteins need to be assembled into RNA-induced silencing complexes (RISCs). Although the assembly of Drosophila melanogaster RISC was recently reconstituted by Ago2, the Dicer-2/R2D2 heterodimer, and five chaperone proteins, the absence of a reconstitution system for mammalian RISC assembly has posed analytical challenges. Here we describe reconstitution of human RISC assembly using Ago2 and five recombinant chaperone proteins: Hsp90β, Hsc70, Hop, Dnaja2, and p23. Our data show that ATP hydrolysis by both Hsp90β and Hsc70 is required for RISC assembly of small RNA duplexes but not for that of single-stranded RNAs. The reconstitution system lays the groundwork for further studies of small RNA-mediated gene silencing in mammals.
Keywords: Argonaute, RISC, chaperone, siRNA, microRNA
INTRODUCTION
Small interfering RNAs (siRNAs) and microRNA (miRNAs) are ∼21–22 nucleotides (nt) small RNAs, which silence gene expression from their complementary target mRNAs through cleavage, translation repression, deadenylation, and/or decapping (Ghildiyal and Zamore 2009; Ha and Kim 2014; Iwakawa and Tomari 2015). Small RNAs act via the effector RNA–protein complex called RNA-induced silencing complex (RISC), of which a member of Argonaute (Ago) family proteins lies at the core. Although many auxiliary proteins have been identified to be associated with Ago proteins (Meister et al. 2005; Höck et al. 2007; Landthaler et al. 2008), a small RNA and an Ago protein can constitute the minimal RISC.
siRNAs and miRNAs are born as small RNA duplexes, which are processed from long double-stranded RNAs and precursor miRNA (pre-miRNA) hairpins, respectively, by Dicer and its partner double-stranded RNA-binding protein (Ghildiyal and Zamore 2009; Ha and Kim 2014). RISC is assembled through at least two-steps: loading of a small RNA duplex into Ago protein (duplex loading) followed by the removal of one of the two strands called the passenger strand from Ago (passenger strand ejection). In the duplex loading step, the small RNA duplex is incorporated into Ago protein with the aid of ATP (Nykänen et al. 2001; Kawamata et al. 2009; Yoda et al. 2010) and the Hsp70/Hsp90 chaperone machinery (Iki et al. 2010; Iwasaki et al. 2010; Johnston et al. 2010; Miyoshi et al. 2010) to form “pre-RISC.” Then, pre-RISC ejects the passenger strand via cleavage of the strand by Ago's catalytic activity (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Leuschner et al. 2006; Kim et al. 2007) and/or with the help of mismatches in the duplex (Kawamata et al. 2009, 2011; Yoda et al. 2010), forming mature RISC containing the single-stranded guide strand.
We have recently succeeded in reconstituting the assembly of Drosophila melanogaster Ago2-RISC using Ago2, Dicer-2 (one of two fly Dicers), R2D2 (the partner protein of Dicer-2), and five chaperone machinery proteins: Hsp83 (Hsp90 homolog), Hsc70-4, Hop, Droj2 (Hsp40 homolog), and p23. This system allowed us to precisely track the RISC assembly pathway at the single-molecule level (Iwasaki et al. 2015). In contrast, mammalian RISC assembly has so far been recapitulated only in crude cell lysate (Leuschner et al. 2006; Yoda et al. 2010), blurring the requirement of individual factors in RISC assembly. Here, we developed a method that faithfully reconstitutes human RISC assembly, using immunopurified Ago2 and five components of the Hsc70/Hsp90 chaperone machinery: Hsp90β, Hsc70, Hop, Dnaja2, and p23. Our system lays the groundwork for investigation of chaperone-mediated RISC assembly and its regulation in mammals.
RESULTS AND DISCUSSION
As a starting point to reconstitute human RISC assembly in vitro, we expressed Flag-tagged human Ago2, the catalytically active Ago protein among the four human paralogs, in HEK293T cells and immunopurified it on magnetic beads (Fig. 1A). We then incubated immunopurified Flag-Ago2 with let-7 siRNA duplex in the presence or absence of naive HEK293T cell lysate at 37°C to allow RISC assembly, washed the beads extensively, and further incubated with a radiolabeled complementary target RNA to monitor its cleavage. No target cleavage was observed with immunopurified Flag-Ago2 and let-7 siRNA duplex alone, but supplementing them with naive HEK293T cell lysate during RISC assembly restored efficient target cleavage (Supplemental Fig. 1). Thus, the target cleavage assay can be used as a sensitive method to evaluate the reconstitution of human RISC assembly in vitro.
FIGURE 1.
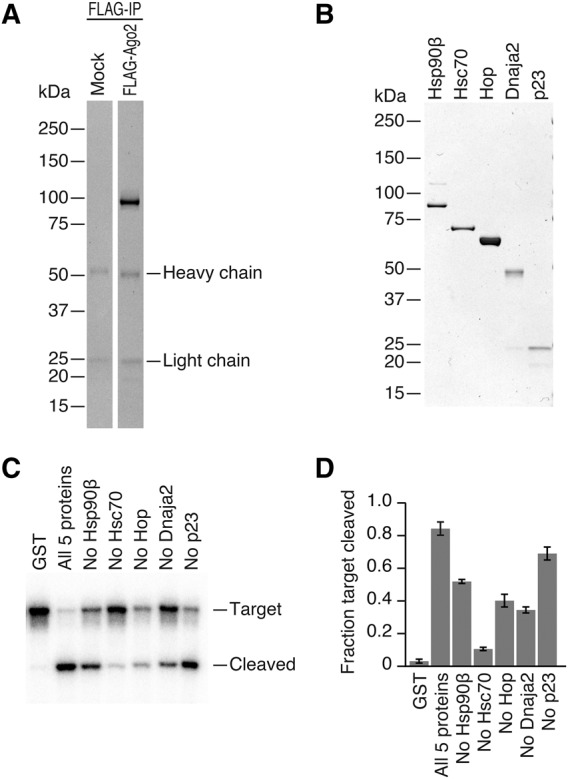
In vitro reconstitution of human Ago2-RISC assembly by recombinant proteins. (A,B) Coomassie staining of immunopurified Ago2 and recombinant Hsp70/Hsp90 chaperone machinery proteins. (C) Target RNA cleavage assay by reconstituted human Ago2-RISC. The highest cleavage activity was observed when all five chaperone proteins were present. (D) Quantification of target cleavage in Figure 1C. The fraction target cleaved is the amount of cleaved target divided by the sum of full-length target and cleaved target. The mean ± SD from three independent experiments is shown.
It has been reported that various chaperone inhibitors block human RISC assembly in cells as well as in vitro (Iwasaki et al. 2010; Johnston et al. 2010; Pare et al. 2013). Encouraged by our previous reconstitution of Drosophila Ago2-RISC assembly with recombinant Hsp70/Hsp90 chaperone proteins (Iwasaki et al. 2015), we hypothesized that human RISC assembly could also be reconstituted using the same chaperone orthologs. Given that the human genome encodes many homologs of chaperone and co-chaperone proteins, we surveyed the published proteome of Ago interacting proteins (Meister et al. 2005; Höck et al. 2007; Landthaler et al. 2008) for candidate chaperone proteins and selected Hsp90β (constitutively expressed paralog of Hsp90), Hsc70, Hop, Dnaja2 (Hsp40 family protein), and p23 for further analysis.
Indeed, recombinant proteins of the five chaperone factors could reconstitute human Ago2-RISC assembly; incubation of all five chaperone proteins (Fig. 1B) with immunopurified human Ago2 and let-7 siRNA duplex enabled efficient target RNA cleavage (Fig. 1C,D), whereas the negative control GST produced only marginal cleavage products. This background cleavage was most likely caused by contaminated chaperones in immunopurified Ago2 despite our best efforts to eliminate those, since it was blocked by chaperone inhibitors (data not shown). Omitting each component of the five chaperone proteins from the reaction reduced cleavage efficiency (Fig. 1C,D). Thus, the five chaperone components are required and sufficient to program catalytically active human Ago2-RISC.
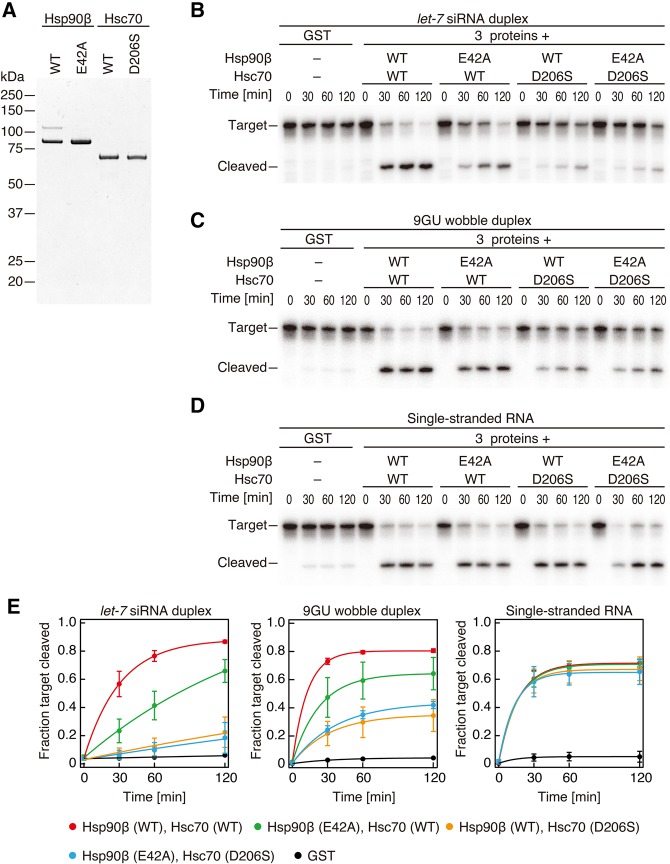
Duplex loading is an ATP-dependent reaction (Nykänen et al. 2001; Kawamata et al. 2009; Yoda et al. 2010). Hsp90β and Hsc70 are only ATPases in the reaction, although they utilize ATP in completely opposite manners; Hsc70 binds to the client protein in its ADP-bound form, whereas Hsp90β does so in its ATP-bound form (Saibil 2013). Thus, simple ATP depletion or using nonhydrolyzable ATP analog would create a complicated situation for the evaluation of ATP-dependency of duplex loading. To avoid such complexity, we mutated crucial residues in Hsp90β and Hsc70 for their ATP hydrolyses (E42A for Hsp90β and D206S for Hsc70) (Fig. 2A; Flaherty et al. 1990; Prodromou et al. 2000) and asked whether their ATPase activities are necessary for RISC formation. Substitution of wild-type Hsp90β or Hsc70 protein into its ATPase mutant significantly reduced the target cleavage activity (Fig. 2B,E). We concluded that our reconstitution system reflects the ATP-mediated RISC assembly reaction as previously reported in cell lysate (Nykänen et al. 2001; Yoda et al. 2010).
FIGURE 2.
ATPase activities of Hsp90β and Hsc70 are necessary for RISC assembly initiated with small RNA duplex but not with single-stranded RNA. (A) Coomassie staining of recombinant ATPase mutants of Hsp90β and Hsc70. (B–D) Target RNA cleavage assay by reconstituted human Ago2-RISC using ATPase mutants of Hsp90β and Hsc70 with let-7 siRNA duplex (B), 9GU wobble duplex (C), and single-stranded RNA (D). (E) Quantification of target cleavage in B–D. The mean ± SD from three independent experiments is shown.
In Drosophila miRNAs and siRNAs are sorted into Ago1 and Ago2, respectively, primarily according to the presence and the position of internal mismatches and G•U wobbles (Tomari et al. 2007; Kawamata et al. 2009; Okamura et al. 2009). Mammals, however, do not have such a strict small RNA sorting system, and all Ago1–4 in mammals can incorporate both perfectly complementary siRNAs and partially complementary miRNAs (Su et al. 2009; Yoda et al. 2010). To monitor RISC assembly of miRNA-type small RNAs, we prepared a small RNA duplex with a single G•U wobble at position 9 in the central region of let-7 siRNA duplex (9GU). The predicted thermodynamic stability of this 9GU wobble duplex is identical to the perfectly complementary siRNA duplex (−30.89 kcal/mol by RNAcofold [Gruber et al. 2008]). Interestingly, however, the requirement of the ATPase activities of Hsp90β or Hsc70 was slightly but significantly lower for the 9GU wobble duplex than for the complementary siRNA duplex (Fig. 2C,E). Given that G•U wobble pairs are known to confer structural flexibility (Varani and McClain 2000), the degree of the chaperone-mediated conformational change required for duplex loading might be partially alleviated by the G•U wobble, a possibility that warrants future investigation of the molecular dynamics during RISC assembly.
Since our reconstitution system is essentially free of contamination of nonspecific nucleases, we can directly compare RISC assembly by small RNA duplexes and single-stranded RNAs, which are too fragile to be examined in cells or lysate-based in vitro systems. In contrast to duplex RNA, neither of the ATPase activities of Hsp90β or Hsc70 was required for loading of a single-stranded RNA with the same sequence as the guide strand of let-7 siRNA duplex (Fig. 2D,E). This agrees well with the idea that the flexibility of single-stranded RNAs allows them to “sneak” into Ago proteins without the help of the chaperone-mediated dynamic conformational change (Rivas et al. 2005; Yoda et al. 2010; Iwasaki et al. 2015).
Assembly of Drosophila Ago2-RISC requires not only the five chaperone proteins; Dicer-2 and R2D2 are prerequisites for fly Ago2-RISC assembly even when initiated by pre-diced siRNA duplexes (Liu et al. 2003, 2006; Lee et al. 2004; Pham et al. 2004; Tomari et al. 2004; Pham and Sontheimer 2005; Nishida et al. 2013). In contrast, we and others have previously reported that Dicer and its partner protein TRBP/PACT depletions in mammals do not hinder siRNA-initiated RISC assembly of human Ago proteins (Martinez et al. 2002; Kanellopoulou et al. 2005; Murchison et al. 2005; Betancur and Tomari 2012; Kim et al. 2014; Suzuki et al. 2015). To rule out any contamination of endogenous Dicer or TRBP from HEK293T cells, we expressed and purified Flag-Ago2 from Dicer1−/− mouse embryonic fibroblast cells (Yang et al. 2010). Still, the target cleavage activity was well supported by the addition of five chaperone factors (Fig. 3C). Moreover, an addition of recombinant Dicer/TRBP (Fig. 3A), which was fully active for dicing of pre-let-7 into mature let-7 (Fig. 3B), did not affect the efficiency of RISC assembly judged by target cleavage (Fig. 3C,D).
FIGURE 3.
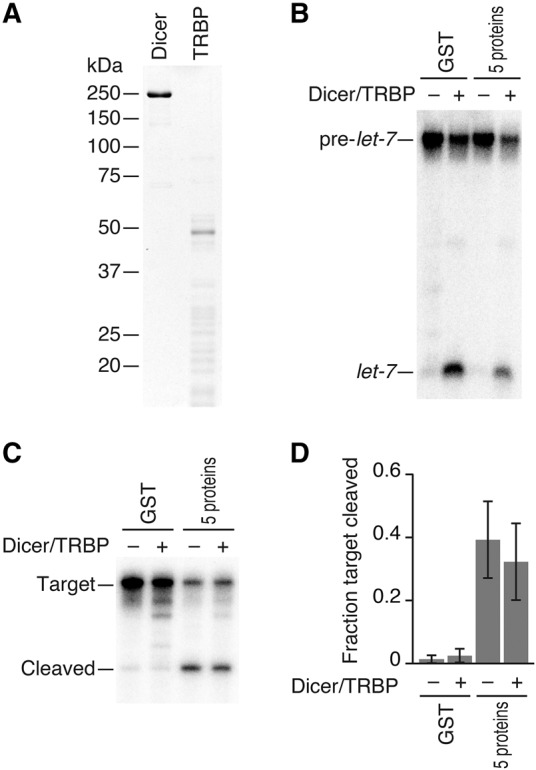
Dicer/TRBP is dispensable for RISC assembly in the reconstituted system. (A) Coomassie staining of recombinant Dicer and TRBP proteins. (B) Dicing assay by recombinant Dicer/TRBP. (C) Target RNA cleavage assay by human Ago2-RISC programmed in the presence or absence of Dicer/TRBP. Ago2 was immunopurified from Dicer1−/− MEF cells. Supplementation of Dicer/TRBP did not enhance RISC assembly in the reconstituted system. (D) Quantification of target cleavage in C. The mean ± SD from three independent experiments is shown.
Along with our previous report, we established the reconstitution system for both Dicer-dependent (fly Ago2) (Iwasaki et al. 2015) and Dicer-independent (human Ago2) RISC assembly. Strikingly, regardless of the Dicer-dependency, the requirement of the five chaperone proteins (Hsp90, Hsc70, Hop, Hsp40, and p23) and the ATPase activities of Hsp90 and Hsc70 in both systems indicates that the mechanism of duplex loading is highly conserved; a conformational change of Ago protein by the chaperone machinery allows incorporation of bulky small RNA duplexes into Ago proteins, just like the same chaperone machinery assists steroid receptors to accommodate their ligands (Smith and Toft 2008). Although it needs to be answered in the future how and why Dicer-dependent and Dicer-independent RISC assembly pathways are different, our reconstitution systems will provide a versatile framework for further studies of small RNA-mediated gene silencing.
MATERIALS AND METHODS
General methods
Lysis buffer [30 mM HEPES-KOH pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, and 1 mM dithiothreitol (DTT)], 40× reaction mix (containing ATP, ATP regeneration system, and RNase inhibitor), small RNA duplexes, formamide dye, and cap-labeled target RNAs were prepared as previously described (Nykänen et al. 2001; Haley et al. 2003; Yoda et al. 2010; Kawamata and Tomari 2011). Small RNAs (5′-UGAGGUAGUAGGUUGUAUAGU-3′ [guide strand for let-7 siRNA duplex and 9GU wobble duplex], 5′-UAUACAACCUACUACCUCUCU-3′ [passenger strand for let-7 siRNA duplex] and 5′-UAUACAACCUUCUACCUCUCU-3′ [passenger strand for 9GU wobble duplex]) were 5′ mono-phosphorylated by T4 polynucleotide kinase (Takara). The template DNA for the target RNA was amplified by PCR from pGL3-Basic (Promega) using a forward primer (5′-GCGTAATACGACTCACTATAGTCACATCTCATCTACCTCC-3′) and a reverse primer (5′-CCCATTTAGGTGACACTATAGATTTACATCGCGTGGATCTACTGGTCTGCCTAAAGAAGGTTGAGGTAGTAGGTTGTATAGTGAAGAGAGGAGTTCATG-3′).
Plasmid constructions
pColdI-His-SUMOstar-Hsp90β, Hsp90β (E42A), Hsc70, Hop, Dnaja2, and pCold-His-SUMO-Hsc70 (D206S)
DNA fragments coding SUMOstar-tagged human Hsp90β, Hsc70, Hop, or Dnaja2 were amplified from pE-SUMOstar or pE-SUMOpro (LifeSensors) and cDNAs obtained from RIKEN BioResource Center and cloned into pColdI (Takara) at the NdeI site by In-Fusion HD cloning kit (Clontech). E42A in Hsp90β and D206S in Hsc70 were introduced by site-directed mutagenesis.
pCAGEN-SBP-p23, Dicer, and TRBP
DNA fragment coding streptavidin-binding peptide (SBP)-tagged human p23 was amplified from pASW (Iwasaki et al. 2010) and cDNA obtained from the RIKEN BioResource Center, and cloned into pCAGEN at the EcoRI site by In-Fusion HD cloning kit (Clontech). To make pCAGEN-SBP-Dicer and TRBP, cDNA clones coding human Dicer and TRBP (kind gifts from R. Fukunaga and P.D. Zamore) were used.
Expression and purification of recombinant proteins
Recombinant Hsp90β, Hsp90β (E42A), Hsc70, Hsc70 (D206S), Hop, and Dnaja2 were expressed as N-terminal His- and SUMOstar-tagged or SUMO-tagged proteins in Escherichia coli BL21 strain. The E. coli cells transformed with each plasmid were cultivated in 1-L culture with 100 µg/mL ampicillin to an OD600 of 0.4 at 37°C, induced by 1 mM isopropyl-b-D-thiogalactoside (IPTG), and then grown at 15°C for 24 h. The cell pellets were collected by centrifugation, resuspended in His A buffer [30 mM HEPES-KOH pH 7.4, 200 mM KOAc, 2 mM Mg(OAc)2, 5% glycerol, 20 mM imidazole, and 0.2 mM TCEP] containing 1× EDTA-free protease inhibitor cocktail (Roche), 1 mM PMSF, 20 µg/mL Pefabloc, 5 µg/mL Leupeptin, 5 µg/mL Aprotinin, and 2 µg/mL Pepstatin, sonicated, and then clarified by centrifugation at 10,000g for 20 min at 4°C. The supernatant was loaded onto HisTrap FF crude 1 mL (GE Healthcare) and eluted with linear gradient from His A buffer to His B buffer (His A buffer containing 400 mM imidazole). The peak fractions were collected, buffer-exchanged into His A buffer by PD-10 (GE Healthcare), digested with 150 U of SUMOstar proteinase 1 or SUMO proteinase 1 (Lifesensors) for overnight at 4°C, and loaded on to HisTrap FF 1 mL. Flow though fraction was diluted tenfold with 30 mM HEPES-KOH pH 7.4, 2 mM Mg(OAc)2, 5% glycerol, and 1 mM DTT, loaded onto MonoQ 5/50 GL 1 mL (GE Healthcare), and eluted with linear gradient from MonoQ A buffer [30 mM HEPES-KOH pH 7.4, 20 mM KCl, 2 mM Mg(OAc)2, 5% glycerol, and 1 mM DTT] to MonoQ B buffer (MonoQ A buffer containing 1 M KCl). The peak fractions were collected, buffer-exchanged to 30 mM HEPES-KOH pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 10% glycerol, and 1 mM DTT with NAP-5 (GE Healthcare). For Hsp90β and Hsp90β(E42A), glycerol and the reducing reagent were omitted from entire steps. p23, Dicer, and TRBP were expressed as N-terminal SBP-tagged proteins in HEK293T cells. A day before transfection, 1 × 106 cells/mL of HEK293T cells were placed on 10-cm dishes. After 19 h, the cells were transfected with 21.7 µL of Lipofectamine 3000 (Thermo Fisher Scientific), 14 µg of each plasmid, 28 µL of P3000 reagent (Thermo Fisher Scientific). At 38 h post-transfection, cell lysate was prepared as described previously (Yoda et al. 2010). Typically, 400 µL of the lysate was incubated with 100 µL of Streptavidin Sepharose beads (GE Healthcare) equilibrated with lysis buffer at 4°C for 2 h. Beads were washed five times with lysis buffer containing 0.5 M NaCl and 0.5 M CaCl2 for p23 or lysis buffer containing 0.5 M NaCl for Dicer and TRBP and then rinsed twice with lysis buffer. The proteins were eluted with 80 µL of lysis buffer containing 10% glycerol and 2.5 mM biotin. Column chromatography was performed using AKTA purifier (GE Healthcare). All purified proteins were flash-frozen in liquid nitrogen and stored at −80°C.
Immunopurification of Flag-Ago2
For Figures 1 and 2, N-terminal 1× Flag-tagged human Ago2 was transiently expressed in HEK293T cells by transfection of pIRESneo-Flag-HA-Ago2 using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions. The cell lysate was prepared as previously reported (Yoda et al. 2010). Typically for 15 reactions, 30 µL Dynabeads Protein G (Invitrogen) were equilibrated with lysis buffer, incubated with 1 µL of 1 mg/mL anti-Flag M2 antibody (F1804, Sigma) at 4°C for 30 min, washed three times with lysis buffer, and incubated with 60 µL of lysate from HEK293T cells expressing Flag-Ago2 in incubation buffer (lysis buffer containing 1 M NaCl, 5% glycerol, 0.5% Triton X-100, and 2.5 mM EDTA) for 1 h at 4°C. Beads were briefly washed three times with wash buffer (lysis buffer containing 2 M NaCl, 10% glycerol, 1% Triton X-100, and 5 mM EDTA), incubated with wash buffer for 15 min at 4°C, rinsed four times with lysis buffer, and then split into tubes for further RISC assembly and target cleavage assay. For Figure 3, N-terminal 1× Flag-tagged human Ago2 was expressed in Dicer1−/− MEF cells by transfection of pIRESneo-Flag-HA-Ago2 using Lipofectamine 3000 according to the manufacturer's instructions. Lysate preparation and Flag-Ago2 immunopurification were performed similarly as above, except that 16 µL Dynabeads Protein G (Invitrogen) and 40 µL of lysate in incubation buffer were used for four reactions.
RISC assembly and target RNA cleavage assay
Flag-Ago2-immobilized beads were incubated with 600 nM Hsp90β, 600 nM Hsc70, 600 nM Hop, 1500 nM Dnaja2, 600 nM p23 and lysis buffer in 18 µL for 30 min at 37°C. Then, 2 µL of 50 nM let-7 siRNA duplex, 9GU wobble duplex, or single-stranded guide RNA, 5 mM ATP, 42 mM creatine monophosphate (Fluka), 0.05 U/µL creatine kinase (Calbiochem), 1.65 U/µL RNasin Plus (Promega) in lysis buffer were added to the beads and incubated for 30 min at 37°C for RISC assembly. When proteins were omitted from the reaction, equimolar GST was supplemented to the reactions. For Figure 3, 10 nM Dicer and 10 nM TRBP (final concentrations) were added to the reaction. Following the RISC assembly reaction, 1 µL of 1 nM radiolabeled target RNA was added to the reaction and incubated at 37°C for 1.5 h or the time shown in figures. Target RNAs were purified and run on 8% UREA-TBE gel, as described previously (Haley et al. 2003). Gels were dried and imaged by FLA-7000 imaging system (Fujifilm Life Sciences). Signal intensities were quantified using MultiGauge (Fujifilm) and graphs were prepared using Excel (Microsoft) and IgorPro (WaveMetrics). For Supplemental Figure 1, typically for one reaction, 1 µL of Flag-Ago2-immobilized or mock-immunoprecipitated beads were incubated with 5 µL HEK293T naive lysate or lysis buffer, 1 µl of 100 nM let-7 siRNA duplex and 3 µl of 40× reaction mix for 30 min at 37°C. After washing with lysis buffer containing 0.8 M NaCl and 1% Triton X-100 three times and rinsing with lysis buffer twice, the immunoprecipitates were used for target cleavage assay as described previously (Yoda et al. 2010).
Dicing assay
Pre-let-7 dicing assay was performed as described previously (Tsutsumi et al. 2011) in the presence or absence of the chaperone machinery.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Tomari laboratory for discussion and technical support, T. Tuschl for the pIRESneo-Flag-HA-AGO2 plasmid, M. Mito for plasmid construction, R. Fukunaga and P.D. Zamore for human Dicer and TRBP cDNAs, and RIKEN BioResource Center (BRC) for human Hsp90β, Hsc70, Hop, Dnaja2, and p23 cDNAs. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (grant number 21115002, “Functional machinery for non-coding RNAs”; grant number 26113007, “Non-coding RNA neo-taxonomy” to Y.T.; and grant number 17H05679, “Nascent chain biology” to S.I.) and Grant-in-Aid for Young Scientists (A) (grant number 17H04998) to S.I. from The Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT).
Author contributions: K.N., E.M.S., and M.W. performed the experiments. S.I. and Y.T. supervised the study and wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063891.117.
REFERENCES
- Betancur JG, Tomari Y. 2012. Dicer is dispensable for asymmetric RISC loading in mammals. RNA 18: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. 1990. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346: 623–628. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore PD. 2003. In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30: 330–336. [DOI] [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. 2015. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25: 651–665. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Sasaki HM, Sakaguchi Y, Suzuki T, Tadakuma H, Tomari Y. 2015. Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature 521: 533–536. [DOI] [PubMed] [Google Scholar]
- Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. 2010. HSP90 protein stabilizes unloaded Argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. 2011. Native gel analysis for RISC assembly. Methods Mol Biol 725: 91–105. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y. 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Yoda M, Tomari Y. 2011. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep 12: 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW. 2007. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yeo J, Lee JH, Cho J, Seo D, Kim JS, Kim VN. 2014. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep 9: 1061–1074. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. 2006. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12: 1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. 2005. Identification of novel Argonaute-associated proteins. Curr Biol 15: 2149–2155. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC. 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Miyoshi K, Ogino A, Miyoshi T, Siomi H, Siomi MC. 2013. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell 49: 680–691. [DOI] [PubMed] [Google Scholar]
- Nykänen A, Haley B, Zamore PD. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321. [DOI] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC. 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, LaPointe P, Hobman TC. 2013. Hsp90 cochaperones p23 and FKBP4 physically interact with hAgo2 and activate RNA interference-mediated silencing in mammalian cells. Mol Biol Cell 24: 2303–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Sontheimer EJ. 2005. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem 280: 39278–39283. [DOI] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. 2004. A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O'Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J 19: 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349. [DOI] [PubMed] [Google Scholar]
- Saibil H. 2013. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol 14: 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Toft DO. 2008. Minireview: the intersection of steroid receptors with molecular chaperones: observations and questions. Mol Endocrinol 22: 2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. 2009. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev 23: 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Katsura A, Yasuda T, Ueno T, Mano H, Sugimoto K, Miyazono K. 2015. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat Struct Mol Biol 22: 512–521. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. 2011. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol 18: 1153–1158. [DOI] [PubMed] [Google Scholar]
- Varani G, McClain WH. 2000. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. 2010. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci 107: 15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. 2010. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.