Abstract
The question of whether RNA is more stable or unstable compared to DNA or other nucleic acids has long been a subject of extensive scrutiny and public attention. Recently, thermodynamically stable and degradation-resistant RNA motifs have been utilized in RNA nanotechnology to build desired architectures and integrate multiple functional groups. Here we report the effects of phosphorothioate deoxyribonucleotides (PS-DNA), deoxyribonucleotides (DNA), ribonucleotides (RNA), 2′-F nucleotides (2′-F), and locked nucleic acids (LNA) on the thermal and in vivo stability of the three-way junction (3WJ) of bacteriophage phi29 motor packaging RNA. It was found that the thermal stability gradually increased following the order of PS-DNA/PS-DNA < DNA/DNA < DNA/RNA < RNA/RNA < RNA/2′-F RNA < 2’-F RNA/2′-F RNA < 2′-F RNA/LNA < LNA/LNA. This proposition is supported by studies on strand displacement and the melting of homogeneous and heterogeneous 3WJs. By simply mixing different chemically modified oligonucleotides, the thermal stability of phi29 pRNA 3WJ can be tuned to cover a wide range of melting temperatures from 21.2°C to over 95°C. The 3WJLNA was resistant to boiling temperature denaturation, urea denaturation, and 50% serum degradation. Intravenous injection of fluorescent LNA/2′-F hybrid 3WJs into mice revealed its exceptional in vivo stability and presence in urine. It is thus concluded that incorporation of LNA nucleotides, alone or in combination with 2′-F, into RNA nanoparticles derived from phi29 pRNA 3WJ can extend the half-life of the RNA nanoparticles in vivo and improve their pharmacokinetics profile.
Keywords: three-way junction, thermal stability, in vivo stability, RNA nanotechnology, chemical modification, RNA 4D structure
INTRODUCTION
The “central dogma” states that DNA makes RNA then makes protein, and protein plays a major role in life function (Crick 1958). However, the completion of the first human genome sequencing argued that only 1.5% of the human genome coded for protein. A significant percentage of the human genome was believed to be a junk DNA scaffold. But more and more evidence from the subsequent studies shows that part of the so-called junk DNA actually code for small (Moss 2001; Calin and Croce 2006; Ghildiyal and Zamore 2009; Tiemann and Rossi 2009; Zhang 2009; Liu and Paroo 2010; Wiedenheft et al. 2012), or long (Mercer et al. 2009; Morlando et al. 2014) noncoding RNAs that regulate cellular activities. It has been speculated that in addition to the two milestones of chemical and protein drugs in the history of drug development, a third milestone is predicted to be RNA as drugs or drugs targeting to RNA (Shu et al. 2014). The versatility in RNA structures, low energy in RNA folding, and amenability in structure control provide feasibility in RNA drug development. However, the question of whether RNA is more stable or unstable compared to DNA or other nucleic acids has long been under extensive scrutiny and public attention.
The field of RNA nanotechnology has emerged rapidly (Guo 2010; Jasinski et al. 2017) since the proof-of-concept in 1998, when it was found that RNA dimer, trimer, tetramer, and hexamer can be produced via bottom-up assembly of fabricated RNA fragments derived from the pRNA subunit of phi29 DNA packaging motor (Guo et al. 1998). Currently, the concept of RNA nanotechnology has been defined (Guo and Haque 2013; Jasinski et al. 2017): RNA nanotechnology is the bottom-up self-assembly of nanometer scale RNA architectures with its major frame composed of RNA. In RNA nanoparticles, the scaffolds, ligands, therapeutics, regulators can all be composed of RNA. RNA nanotechnology is a unique field that is distinct from the classical studies on RNA structure and function, which focus on intra-RNA interactions and 2D/3D structures. RNA nanotechnology focuses on inter-RNA interactions and quaternary (4D) structures (Guo et al. 1998; Jaeger and Leontis 2000; Jaeger et al. 2001; Chworos et al. 2004; Shu et al. 2004, 2011; Afonin et al. 2008, 2014; Grabow and Jaeger 2014; Li et al. 2015; Sharma et al. 2015).
Two major engineering steps have been used to construct RNA nanoparticles. The first step is the utilization of the spontaneous self-folding property of RNA into defined structures by base/base interaction. Based on the calculation of the Gibbs free energy ΔG, a variety of RNA sequences can be predicted via computational algorithms and tested empirically based on the prediction of the secondary or tertiary structure (Gai et al. 2004; Yingling and Shapiro 2007; Afonin et al. 2010; Geary et al. 2014). The second step is the use of the naturally occurring RNA motifs as building blocks to assemble RNA nanoparticles based on the predicted quaternary architectures (Lundbaek and Andersen 1994; Westhof et al. 1996; Shu et al. 2004, 2011; Severcan et al. 2010; Dibrov et al. 2011; Grabow et al. 2011; Khisamutdinov et al. 2014, 2016; Li et al. 2016).
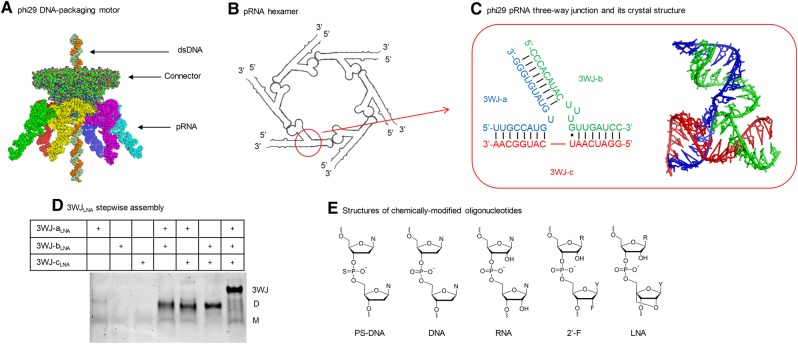
Many types of RNA three-way junctions and four-way junctions have been found in nature (Duckett et al. 1995; Felden et al. 1996; Rettberg et al. 1999; Diamond et al. 2001; Hohng et al. 2004; Leontis et al. 2006; Lescoute and Westhof 2006; de la Peña et al. 2009; Ouellet et al. 2010) and can be used for the fabrication of RNA nanoparticles. In particular, the phi29 bacteriophage DNA motor packaging RNA (pRNA) three-way junction (Fig. 1A–C) was found to efficiently self-assemble from three RNA fragments and exhibit an exceptional thermodynamic stability. RNA and DNA folding is dependent on energy landscape as described by ΔG = ΔH − TΔS. While enthalpy change (ΔH) is more relevant to heat, and entropy change (ΔS) relates to the structuring, the two factors are significant in considering RNA and DNA complex production and assembly. Duplex hybridization is believed to be driven by a lower enthalpy value (Gyi et al. 1996; Rauzan et al. 2013); however, our previous study showed pRNA 3WJ assembly was governed more by entropy change than by enthalpy change (Binzel et al. 2014, 2016a). That is, the structuring factor plays a key role in 3WJ assembly. It has been reported that the three fragments of the pRNA 3WJ coassembled with extraordinary speed and affinity via a two-step mechanism. The first step between 3WJ-b and 3WJ-c is highly dynamic since these two fragments only contain eight complementary base pairs. Though enthalpy change plays an important role at this step, we believe entropy change is key to the idea of “irreversibility.” More specifically, formation of the 3WJ-b + 3WJ-c dimer generates a special structure with a fixed angle and two protruding sequences (Zhang et al. 2013; Binzel et al. 2016a). Such a structure is an entropically favorable receptor for 3WJ-a, which contains 17-nt complementary to the formed dimer complex. Thus, the second step occurs at a very high association rate and locks the unstable dimer into a highly stable 3WJ (Binzel et al. 2016a). Consequently, the pRNA 3WJ is more stable than any of the dimer species, as shown in the much more rapid association rate and smallest dissociation rate constant, creating a more irreversible association of three strands. These findings support the claim that pRNA 3WJ assembly was governed more by entropy (Binzel et al. 2014, 2016a). Furthermore, this robust RNA 3WJ stays intact at ultralow concentration, is resistant to a high molar urea denaturation (Shu et al. 2011), and also remains stable under high mechanical rupture forces (Xu et al. 2017). In addition, substituting uridine and cytidine with their 2′-fluoro-modified counterparts further increases nanoparticle stability and prevents digestion by RNases (Layzer et al. 2004). 2′-F modified phi29 pRNA 3WJ thus greatly enhances the thermodynamic and enzymatic stability of RNA nanoparticles and has paved the way for medical applications of RNA nanotechnology (Cui et al. 2015; Shu et al. 2015; Binzel et al. 2016b; Zhang et al. 2017).
FIGURE 1.
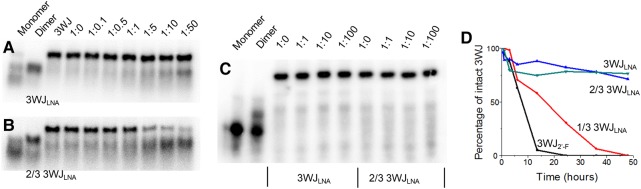
Structures of phi29 DNA-packaging motor, pRNA hexamer, pRNA 3WJ, stepwise assembly of homogeneous 3WJLNA, and structures of five different chemically modified oligonucleotides. (A) Structure of phi29 DNA-packaging motor. (B) Packaging RNA (pRNA) hexamer. (C) Phi29 pRNA 3WJ and its crystal structure. (D) Stepwise assembly of 3WJLNA. (Short single-stranded oligonucleotides and some chemically modified oligonucleotides respond to EtBr staining differently. Thus, LNA single strands are stained weakly and are not very clear to see in this example.) M indicates the migration of monomer. D indicates the migration of dimer. 3WJ indicates the migration of three-way junction. (E) Structures of chemically modified oligonucleotides (2′-F denotes 2′-fluoro-U/C-modified oligonucleotides and LNA denotes LNA-U/C-modified oligonucleotides).
Recent clinical trials of chemically modified oligonucleotides (Burnett and Rossi 2012; Lundin et al. 2015; Khvorova and Watts 2017) and the FDA approval of a phosphorothioate-modified oligonucleotide (Stein and Castanotto 2017) clearly indicate the importance of chemical modifications in medical applications of RNA nanotechnology. Extensive studies have been performed on the effects of chemical modifications on the thermodynamic stability (De Mesmaeker et al. 1995; Vester and Wengel 2004; Karkare and Bhatnagar 2006) and chemical stability (Kawasaki et al. 1993; Heidenreich et al. 1994; Allerson et al. 2005) of duplex and triplex (Betts et al. 1995; Kaur et al. 2006; Hansen et al. 2009; Piao et al. 2013). For example, in a DNA duplex, each 2′-methoxy- (2′-OMe-) or 2′-fluoro- (2′-F-) modified nucleoside has been shown to increase the Tm by 1°C–2°C (Kawasaki et al. 1993). Additionally, modifications with phosphorothioate-DNA (PS-DNA), 2′-F, 2′-OMe, and locked nucleic acid (LNA) have been shown to increase the rather short half-life seen in unmodified siRNAs (Shaw et al. 1991; Kawasaki et al. 1993; Elmen et al. 2005). While few studies have been published on more complex RNA structures (e.g., RNA 3WJs), a more complete understanding of how chemically modified oligonucleotides affect the thermal and in vivo stability of the pRNA three-way junction is essential for future clinical applications of RNA nanotechnology.
To this end, we have previously measured thermodynamic parameters for phi29 pRNA 3WJ composed of DNA, RNA, and 2′-F RNA. However, in vitro and in vivo stability against enzymatic degradation was not discussed (Binzel et al. 2014). We herein extend our previous efforts in 3WJ thermodynamics to two well-known nuclease-resistant oligonucleotides, PS-DNA and LNA. We assess and compare the melting temperatures of homogeneous and heterogeneous 3WJs using temperature-gradient gel electrophoresis (TGGE), as well as modification-specific oligonucleotide affinity in strand displacement between single-stranded oligonucleotides and assembled 3WJs (Fig. 1D; Supplemental Fig. S1). In addition, we investigate the effects of chemical modifications (Fig. 1E) on the in vivo stability of highly thermostable phi29 pRNA 3WJ.
RESULTS AND DISCUSSION
Melting of homogeneous 3WJs
The melting of homogeneous 3WJs was assessed on TGGE, where a linearly increasing temperature gradient was applied perpendicular to the electrical current. Thus, each separate lane of the 3WJ sample was subjected to heating at a gradually increased temperature across the gel. Each lane on the gel was then integrated using ImageJ, thereby producing a melting curve for each 3WJ sample. The melting temperature (Tm) was determined when 50% of the assembled 3WJs were dissociated (Binzel et al. 2014). Dissociation would result in a relatively sharp transition of the band, while the gradual migration rate increase from left to right was most notably due to the swelling of gel pores at elevated temperatures.
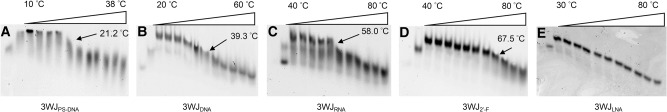
With the exception of 3WJLNA, all other homogeneous 3WJs showed a relatively sharp increase in migration rate, suggesting the dissociation of one or two strands (Fig. 2). The Tms generated from TGGE at the concentration of 10 µM were calculated to be 21.2°C, 39.3°C, 58.0°C, and 67.5°C for homogeneous 3WJPS-DNA, 3WJDNA, 3WJRNA and 3WJ2′-F, respectively (Table 1), consistent with our previous observations (Binzel et al. 2014). The 3WJLNA did not show a sharp transition in migration rate on the gel, indicating it did not melt at 80°C heating, as this is the temperature limitation of the TGGE. Clearly, the thermal stability of phi29 pRNA 3WJ, expressed as Tm, was increased by chemical modifications in the order of PS-DNA < DNA < RNA < 2′-F RNA < LNA.
FIGURE 2.
Melting of homogeneous 3WJs on TGGE. (A) Melting of 3WJPS-DNA from 10°C to 38°C. (B) Melting of 3WJDNA from 20°C to 60°C. (C) Melting of 3WJRNA from 40°C to 80°C. (D) Melting of 3WJ2′-F from 40°C to 80°C. (E) Melting of 3WJLNA from 30°C to 80°C. Black arrow and number indicate Tm.
TABLE 1.
Tm summary
Melting of heterogeneous 3WJs
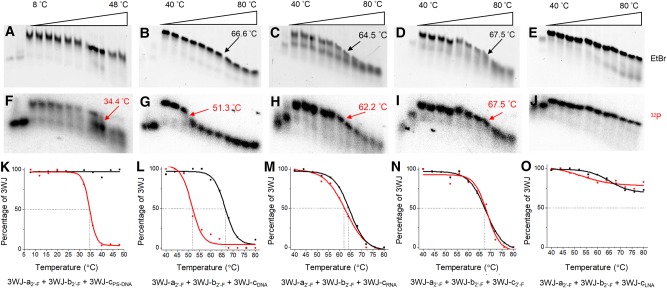
To further confirm the contribution of different chemically modified oligonucleotides to the thermal stability of the phi29 pRNA 3WJ, TGGE assays were performed on heterogeneous 3WJs with only one variant strand. The 3WJ-c was labeled with 32P for tracking the dissociation of the 3WJ. The resulting gels were stained by EtBr first to evaluate the melting of overall hybrid 3WJ structure before phosphorimaging. Assembly of the hybrid 3WJs was confirmed through a stepwise assembly gel with monomer and dimer controls. Phosphorimaging clearly showed the dissociation of 3WJ-cPS-DNA, 3WJ-cDNA, 3WJ-cRNA, and 3WJ-c2′-F from each hybrid 3WJ at 34.4°C, 51.3°C, 62.2°C, and 67.5°C, respectively (Table 1). The 3WJ-cLNA strand still did not yield a transition below 80°C (Fig. 3F–J). This further confirms the order of the stabilizing effect (PS-DNA < DNA < RNA < 2′-F RNA < LNA) of the five oligonucleotides within phi29 pRNA 3WJ.
FIGURE 3.
Melting of hybrid 3WJs (3WJ-c is 32P-labeled). The first two lanes of each sample are monomer and dimer as controls. Red curve is melting curve from phosphorimaging; black curve is melting curve from EtBr channel. Red arrow and number indicate Tm of c strand in hybrid 3WJs. Black arrow and number indicate Tm of 3WJ-a2′-F + 3WJ-b2′-F. (A,F,K) Melting of PS-DNA hybrid 3WJ. (B,G,L) Melting of DNA hybrid 3WJ. (C,H,M) Melting of RNA hybrid 3WJ. (D,I,N) Melting of 2′-F homogeneous 3WJ. (E,J,O) Melting of LNA hybrid 3WJ. Bottom graphs are overlap of melting curves of 3WJ-c (phosphorimaging, F–J) and the hybrid 3WJ (EtBr staining, A–E) for each hybrid 3WJ.
Surprisingly, EtBr imaging displayed only one clear melting transition with a Tm around 64.5°C–67.5°C for all hybrid 3WJs except for the LNA hybrid 3WJ (Fig. 3A–E), even including PS-DNA and DNA hybrid 3WJs in which the 3WJ-c had a much lower Tm. The difference in these results could be due to the similar mobility of the 3WJ-a2′-F + 3WJ-b2′-F dimer and the hybrid 3WJ2′-F/PS-DNA or 3WJ2′-F/DNA on 12% native PAGE (Supplemental Fig. S2). In this case, the only Tm derived from migration was from the melting of 3WJ-a2′-F + 3WJ-b2′-F dimer (Supplemental Fig. S3). However, phosphorimaging provided a confident marker to assay the dissociation of the labeled strand. This led to the elucidation of a distinctive two-step dissociation for PS-DNA and DNA hybrid 3WJs through the combination of the EtBr staining and phosphorimaging of the gels. On the other hand, 3WJ-cRNA dissociated from RNA hybrid 3WJ was seen only slightly before the melting of 3WJ-a2′-F + 3WJ-b2′-F dimer, indicating RNA and 2′-F RNA had a similar contribution to the thermal stability of 3WJ. Such a small difference in Tm would easily be omitted by other assays, demonstrating the advantage of TGGE in monitoring the exact behavior of a specific component in a multiple-stranded system and distinguishing a small difference of melting temperatures. The melting curve of individual 3WJ-c2′-F was nearly identical to the overall melting of 3WJ2′-F, suggestive of a cooperative melting of homogeneous 3WJ due to the close affinities of three 2′-F RNA strands. LNA hybrid 3WJ exhibited 20% to 30% dissociation based on the melting of individual 3WJ-cLNA or the overall 3WJ on the gel, suggestive of a Tm higher than 80°C.
Strand displacement assay by chemically modified replacement strands
The strand displacement of preassembled pRNA 3WJ by individual chemically modified replacement strands was assessed to examine the variance in strand affinity over the competitor 3WJ strand. To investigate the strand displacement between different single-stranded oligonucleotide and assembled 3WJs, 32P-labeled 3WJ-cDNA, 3WJ-cRNA, 3WJ-c2′-F, and 3WJ-cLNA were incubated with assembled nonlabeled homogeneous 3WJDNA, 3WJRNA, 3WJ2′-F, and 3WJLNA at 37°C for 3 h, respectively. PS-DNA was not included in this study because the Tms of 3WJPS-DNA and 3WJ-cPS-DNA in its hybrid 3WJ were lower than 37°C, limiting its potential for in vivo applications.
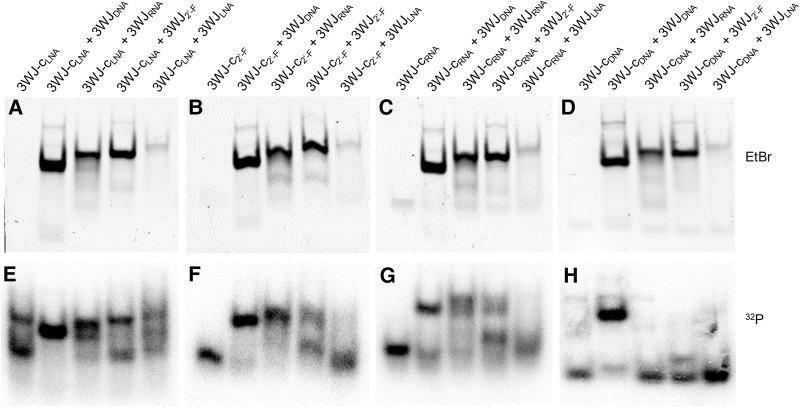
As expected, 3WJ-cLNA was able to completely displace 3WJ-cDNA and 3WJ-cRNA from their homogeneous 3WJs, with partial displacement of the 3WJ-c2′-F on the 3WJ2′-F. The 3WJ-c2′-F also displayed strong thermodynamic stability and was able to fully displace 3WJ-cDNA and 3WJ-cRNA from their homogeneous 3WJs, and to partially displace 3WJ-c2′-F within the 3WJ2′-F. In contrast, no 3WJ-cLNA from the 3WJLNA was displaced by 3WJ-c2′-F, demonstrating the higher stabilization of the 3WJ composed of LNA and affinity of LNA over 2′-F RNA to the remaining two strands on the 3WJ. Continuing the same stability trends seen in the TGGE studies, 3WJ-cRNA demonstrated replacement in the 3WJDNA, partially for the 3WJRNA, and weakly for the 3WJ2′-F; that is, a decreasing displacement yield from 3WJDNA to 3WJ2′-F. Again, the 3WJLNA was not vulnerable to such strand displacement and replacement. Finally, each of the 3WJs was resistant to 3WJ-cDNA displacement with the exception of the 3WJDNA (Fig. 4). The strand displacement results clearly show that LNA has the strongest stabilizing effect among the four different oligonucleotides, while DNA has the weakest. The displacement profiles for 2′-F RNA and RNA are similar, but RNA replaces a smaller percentage of the competitor strand compared to 2′-F RNA for 3WJDNA and 3WJRNA. This may indicate that the difference of the stabilizing effect for these two oligonucleotides is not as dramatic between RNA/DNA or LNA/2′-F RNA, with RNA slightly less than 2′-F RNA. These data are consistent with the melting of homogeneous 3WJs, as the Tm difference between 3WJ2′-F and 3WJRNA is smaller than that of 3WJRNA to 3WJDNA and 3WJLNA to 3WJ2′-F. This is also evidenced by the difference in Tms of different 3WJ-c strands in hybrid 3WJs, as the Tm difference between 3WJ-c2′-F and 3WJ-cRNA is the smallest among all c strands in hybrid 3WJs (Table 1). Overall, the strand displacement results are consistent with the melting data, and further confirm that the stabilizing trend of these oligonucleotides to the pRNA 3WJ was DNA < RNA < 2′-F RNA < LNA.
FIGURE 4.
Strand displacement between radiolabeled single-stranded 3WJ-c and assembled homogeneous 3WJs. (A,E) LNA displaces 3WJs. (B,F) 2′-F RNA displaces 3WJs. (C,G) RNA displaces 3WJs. (D,H) DNA displaces 3WJs. (A–D) EtBr channel; (E–H) phosphorimaging.
In vitro characterization of LNA/2′-F hybrid 3WJs
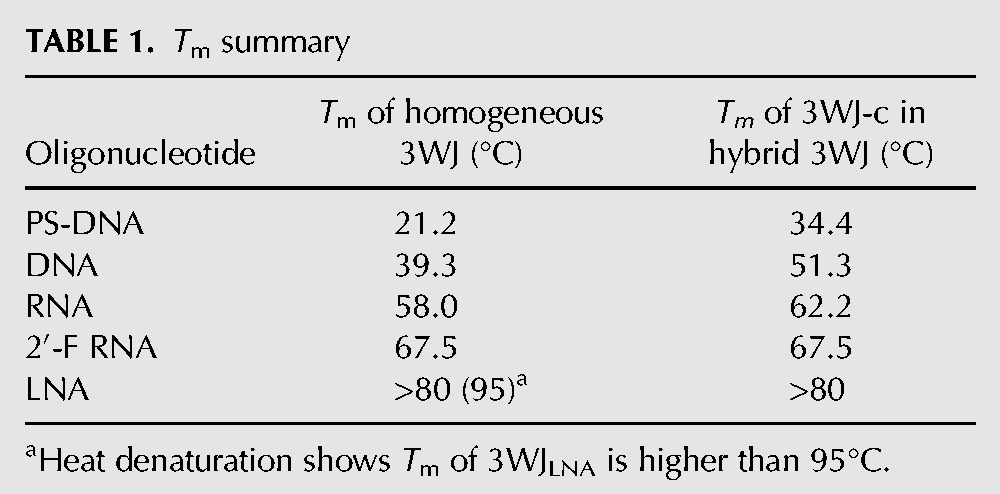
The exceptional stabilizing effect of LNA led to characterization of the 3WJLNA further through more in depth thermodynamic analysis to gain a better understanding of the melting temperature of LNA modified 3WJs. As Tm is concentration dependent, we attempted to measure Tm for the 3WJLNA at lowered concentrations; however, its Tm still remained above 95°C even at the lowest concentration detectable. This rendered the accurate Tm measurement difficult. Thus, we studied heat denaturation of 32P-labeled LNA/2′-F hybrid 3WJs at a low concentration of 16 nM in the presence of 50-fold excess nonradiolabeled competitors to prevent renaturation. Even at this lowered concentration, homogeneous 3WJLNA was found to be resistant to heat denaturation at 95°C for at least 5 min, as only ∼16% of 32P-labeled 3WJ-cLNA was displaced by the excess nonradiolabeled 3WJ-cLNA. In order to examine a melting temperature, a series of 3WJLNA/2′-F hybrids were created, and it was found that decreasing the LNA content resulted in a lowered Tm. Consequently, a 2/3 3WJLNA (3WJ-a2′-F + 3WJ-bLNA + 3WJ-cLNA) appeared vulnerable to heat denaturation with 50-fold excess competitors (Fig. 5A,B), but still did not display a melting profile below 80°C by TGGE analysis. Further reduction of the ratio of LNA in 3WJ to 1/3 (3WJ-a2′-F + 3WJ-b2′-F + 3WJ-cLNA) led to a sharp drop of Tm to 74°C, while the 3WJ2′-F at 16 nM melted at 57°C. Despite distinctive melting temperatures, all four LNA/2′-F hybrid 3WJs showed similar apparent equilibrium dissociation constants in the low nanomolar range (Table 2). The high melting temperatures of homogeneous and heterogeneous LNA 3WJs led us to select them for resistance against denaturing reagents. Like the heat denaturation experiment, these two 3WJs (3WJLNA and 2/3 3WJLNA) were found resistant to 8 M urea at 37°C in an assay in the presence of 100-fold excess competitors (Fig. 5C). Taken together, the high Tm, low Kd and resistance to 8 M urea suggest that the high thermodynamic stability of LNA modified 3WJs.
FIGURE 5.
Thermal stability and stability against 50% serum of selected 3WJs. (A) Resistance to heat denaturation for homogeneous 3WJLNA. (B) Resistance to heat denaturation for hybrid 2/3 3WJLNA (For A and B, lane 1: monomer, 32P-labeled 3WJ-cLNA; lane 2: dimer, 32P-labeled 3WJ-cLNA + 3WJ-aLNA; lane 3: 32P-labeled 3WJ without heat denaturation; lanes 4–10: heat-denaturated 3WJ samples in the presence of increasing nonradiolabeled competitor up to 50-fold). (C) Resistance to 8 M urea denaturation for homogeneous LNA 3WJ and 2/3 LNA hybrid 3WJ (all samples were incubated with 8 M urea, lane 1: monomer, 32P-labeled 3WJ-cLNA; lane 2: dimer, 32P-labeled 3WJ-cLNA + 3WJ-aLNA; lanes 3–6: 3WJLNA in the presence of increasing nonradiolabeled competitor up to 100-fold; lanes 7–10: 2/3 3WJLNA in the presence of increasing nonradiolabeled competitor up to 100-fold). (D) Stability of 3WJs in 50% serum (green: homogeneous 3WJLNA, 3WJ-aLNA + 3WJ-bLNA + 3WJ-cLNA; blue: hybrid 2/3 3WJLNA, 3WJ-a2′-F + 3WJ-bLNA + 3WJ-cLNA; red: hybrid 1/3 3WJLNA, 3WJ-a2′-F + 3WJ-b2′-F + 3WJ-cLNA; black: homogeneous 3WJ2′-F, 3WJ-a2′-F + 3WJ-b2′-F + 3WJ-c2′-F).
TABLE 2.
Summary of LNA/2′-F 3WJs
To further evaluate the potential application of LNA/2′-F hybrid 3WJs for in vivo applications, we investigated their stabilities in 50% of fetal bovine serum, mimicking the in vivo serum concentrations. Serum stability of the 3WJs was assayed by electrophoretic analysis of RNA nanoparticles incubated in the serum over varying time points. Increasing the ratio of LNA content in hybrid 3WJs resulted in dramatic improved stability of 3WJs. Incorporation of one LNA strand extended the half-life of homogeneous 3WJ2′-F from 7.7 to 16.6 h. The 2/3 3WJLNA and homogeneous 3WJLNA exhibited similar half-lives of above 48 h assayed in these studies (Fig. 5D). Overall, these findings led us to the conclusion that LNA 3WJs were found superiorly thermodynamically stable and resistant to serum degradation, overcoming the two major hurdles commonly seen in other nanotechnology platforms, thus providing promise in use for in vivo drug carrier applications.
In vivo stability of LNA/2′-F 3WJs and urine assay upon IV injection
As a drug delivery platform, RNA nanoparticles usually integrate multiple functional modules, including cancer cell-targeting motif, signal-reporting group and therapeutic reagent. High thermodynamic stability and strong resistance to degradation are thus desired to keep RNA nanoparticles intact and remain as an all-in-one carrier of necessary modules. To assess the in vivo stability of LNA/2′-F 3WJs, intravenous injections of fluorescent 2/3 3WJLNA (3WJ-aLNA + 3WJ-bLNA + 3WJ-c2′-F labeled with Alexa647) and homogeneous 3WJ2′-F (3WJ-a2′-F + 3WJ-b2′-F + 3WJ-c2′-F labeled with Alexa647) were performed in BALB/c mice. Whole-body imaging (Supplemental Fig. S4) displayed rapid biodistribution of the nanoparticles throughout the mice with a rapid clearance of ∼40% of nanoparticles within the first hour by urinary excretion, similar to the study of antisense oligonucleotides (Zhang et al. 1995). In order to examine the stability of the 3WJs in vivo, urine samples were collected from the mice during this period of rapid clearance. Analysis of collected urine by native PAGE showed the majority of homogeneous 3WJ2′-F was degraded within the first 1 h. This result is not surprising, as similar chemically modified oligonucleotides were reported to be mostly degraded to products with lower molecular weights, via urine assays, though the oligonucleotides absorbed into body tissues were shown to remain stable at 6 h (Agrawal et al. 1995). Most strikingly, no significant degradation of 2/3 LNA hybrid 3WJ was observed within the first hour compared to the injected samples, and ∼20% of 2/3 LNA hybrid 3WJ was still found to be intact after urine excretion at 3 h (Fig. 6), suggestive of the super resistance of LNA 3WJ to in vivo degradation. In vitro incubation of both 3WJ samples in mouse urine further confirmed these results as no degradation was examined. While it cannot be concluded if the 2′-F RNA nanoparticles were degraded during blood circulation or within the urine excretion, it can be concluded that a high percentage of the LNA modified 3WJ samples were able to remain stable throughout the entirety of the in vivo studies. Thus, it is very exciting to find out that 3WJLNA is significantly more resistant to in vivo degradation and it can potentially improve drug delivery efficiency as it survives longer in vivo and may have a larger chance to deliver intact nanoparticles to the targets.
FIGURE 6.
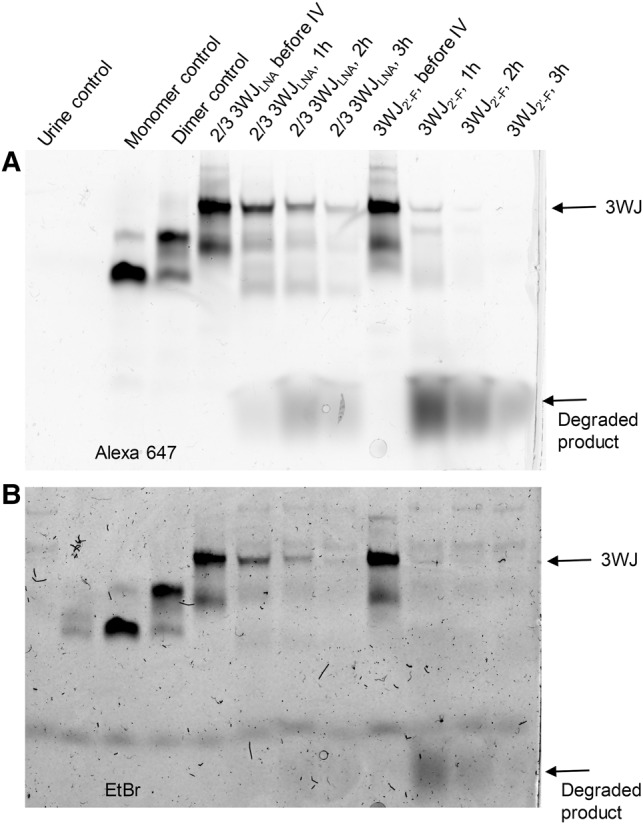
In vivo stability of hybrid 2/3 3WJLNA and homogeneous 3WJ2′-F. (A) Gel image under Alexa 647 channel. (B) Gel image under EtBr channel. Each lane is loaded with the same volume (1 µL) of samples to give a comparison of the fluorescence intensity of each sample.
Conclusion
Studies on strand displacement, heat dissociation, chemical denaturation, serum assay, in vivo trial, as well as melting of homogeneous and hybrid complex lead to the conclusion that the order of the stabilizing effect of five different chemically modified oligonucleotides for phi29 pRNA 3WJ is PS-DNA < DNA < RNA < 2′-F RNA < LNA. The thermal stability of phi29 pRNA 3WJ can be easily tuned by incorporating varying oligonucleotides, resulting in a wide range of Tms from 21.2°C to over 95°C. Incorporation of LNA into 3WJ would greatly protect it from rapid in vivo degradation, solving the instability problem of RNA nanoparticles. The exceptional stabilizing effect of LNA to phi29 pRNA 3WJ may be helpful to overcome pharmaceutical formulation difficulty, increase drug delivery efficacy, and render RNA nanotechnology clinically practical.
MATERIALS AND METHODS
DNA oligonucleotides and phosphorothioate-DNA strands were purchased from Integrated DNA Technologies. RNA strands, 2′-F U/C-substituted RNA strands (2′-F), and LNA-U/C-substituted RNA strands (LNA) were synthesized on AZCO DNA synthesizer using standard phosphoramidite chemistry with doubled coupling and oxidation time for LNA phosphoroamidites. RNA and 2′-F phosphoroamidites were purchased from Bioautomation, and LNA phosphoroamidites were purchased from Exiqon. Other chemicals for oligonucleotide synthesis were obtained from Novabiochem and used without further purification. All gels were scanned on Typhoon FLA 7000 (GE). The temperature-gradient gel electrophoresis (TGGE) system was from Biometra. Oligonucleotide concentration was calculated from the absorbance at 260 nm on Nanodrop 2000 (Fisher Scientific) and its extinction coefficient at 260 nm. Fetal bovine serum was obtained from Sigma-Aldrich. [γ-32P]ATP was obtained from PerkinElmer. Mice were ordered from Taconic Farms, Inc. All other chemicals were from Fisher Scientific.
Assembly of 3WJs
Assembly of 3WJs was performed by mixing equimolar molarity of corresponding strands in TMS buffer (50 mM TRIS pH = 8.0, 100 mM NaCl, 10 mM MgCl2 unless otherwise specified) and annealed at 85°C for 5 min followed by slow cooling to 4°C over ∼40 min. The 3WJ formations were confirmed on a 15% native PAGE in TBM running buffer (89 mM Tris, 200 mM boric acid, and 5 mM MgCl2) run at 120 V at 4°C for 90 min. Gels were stained with ethidium bromide (EtBr).
Temperature-gradient gel electrophoresis (TGGE)
All assembled 3WJ samples were run on 12% native PAGE in TMS buffer for 1 h at 100 V. The 3WJ-c was 5′ end radiolabeled using [γ-32P]ATP (PerkinElmer) in the hybrid 3WJs. To compare the dissociation of 3WJ, 3WJ-c (monomer) and 3WJ-a + 3WJ-c (dimer) were used as control samples. The concentration of each sample was 10 µM in the experiments. Gels were stained with EtBr and scanned prior to being dried and imaged by phosphorimaging. The concentration of each sample was 16 nM in the characterization of LNA/2′-F hybrid 3WJs and gels were only imaged by phosphorimaging. All gel images were analyzed by ImageJ. All samples were repeated at least three times.
Strand displacement
Preassembled homogeneous 3WJs of different oligonucleotides were mixed with 32P-labeled 3WJ-cLNA, 3WJ-c2′-F, 3WJ-cRNA, and 3WJ-cDNA, respectively. The final concentration of 3WJs and 32P-labeled 3WJ-c were 3 µM in 9 µL. All samples were incubated at 37°C for 3 h followed by 12% Native PAGE separation at 110 V for 100 min at 4°C. The gels were stained by EtBr and visualized on a Typhoon scanner prior to being dried and imaged by phosphorimaging.
Kd measurement
The apparent equilibrium dissociation constant (Kd) for assembled 3WJs was measured by titrating 3WJ-a + 3WJ-b into 32P-labeled 3WJ-c. The concentrations of 3WJ-a and 3WJ-b were identical, ranging from 0 to 256 nM, and the 3WJ-c had a constant concentration of 1 nM. Each sample was heated at 95°C for 5 min and cooled to 4°C slowly over 50 min in TMS buffer. The formed 3WJs were resolved on 12% native PAGE at 4°C under the voltage of 110 V for 90 min. Fractions of monomer, dimer, and 3WJ on each lane were quantified using ImageJ and fitted into a Hill1 equation in Origin 8 following our previous publications (Binzel et al. 2014). All samples were repeated at least three times.
Resistance to heat denaturation
LNA/2′-F hybrid 3WJs were assembled in TMS buffer and the concentration of 3WJ is 16 nM. The 3WJ-cLNA was 5′ end labeled using [γ-32P]ATP in the formed 3WJ. LNA 3WJ was mixed with nonradiolabeled 3WJ-cLNA over a range of concentrations from 0 to 800 nM (0–50 equivalents). The mixed samples were heated at 95°C for 5 min and cooled to 4°C quickly followed by resolving on 12% native PAGE in TBM buffer. Control samples were 3WJ-cLNA (monomer), 3WJ-cLNA + 3WJ-aLNA (dimer), and 3WJ-cLNA + 3WJ-aLNA + 3WJ-bLNA (trimer, 3WJ) without heat denaturation. The 3WJ-cLNA was radiolabeled in the control samples. Gels were exposed to phosphorimaging screen overnight and visualized on a Typhoon scanner. All samples were repeated at least three times.
Resistance to 8 M urea denaturation
LNA/2′-F hybrid 3WJs were assembled in TMS buffer and the 3WJ-cLNA was 5′ end labeled using [γ-32P]ATP in the formed 3WJ. The 3WJ samples were mixed with nonradiolabeled 3WJ-cLNA over a range of concentrations from 0 to 1600 nM (0 to 100 equivalents). All 3WJ samples were mixed with 12 M urea to give a final concentration of 16 nM and the urea concentration was 8 M. The mixed samples were incubated at 37°C for 30 min prior to 8 M urea denaturing gel analysis. Gels were exposed to phosphorimaging screen overnight and visualized on a Typhoon scanner. All samples were repeated at least three times.
Stability against 50% serum
LNA/2′-F hybrid 3WJs were assembled in TMS buffer and mixed with FBS. The final concentrations of 3WJ samples were 1 µM and FBS were 50%. Samples were incubated at 37°C for a maximum of 48 h. Samples were taken out of a thermocycler and stored at −80°C until analysis on 15% native PAGE. Gels were run at 4°C under the voltage of 110 V for 90 min and stained with EtBr.
In vivo stability
3WJs were assembled in PBS buffer and the 3WJ-c2′-F was labeled with Alexa647. One hundred microliters of 20 µM assembled 3WJs were injected into BALB/c mice and the urine was completely collected. The urine samples were resolved on 15% native PAGE. Gels were run at 100 V for 100 min at 4°C. The gels were stained by EtBr and scanned under both the Alexa647 channel and EtBr channel.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The research in P.G.’s laboratory was supported by National Institutes of Health grants R01EB019036, U01CA207946, and UH3TR000875. We thank Hongran Yin, Zhefeng Li, Mario Vieweger, and Congcong Xu for their helpful discussion and assistance in experiments. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G. is the consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd., as well as the cofounder of Shenzhen P&Z Bio-medical Co. Ltd. and its subsidiary US P&Z Biological Technology LLC.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063057.117.
REFERENCES
- Afonin KA, Cieply DJ, Leontis NB. 2008. Specific RNA self-assembly with minimal paranemic motifs. J Am Chem Soc 130: 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. 2010. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, et al. 2014. Multifunctional RNA nanoparticles. Nano Lett 14: 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Zhang X, Lu Z, Zhao H, Tamburin JM, Yan J, Cai H, Diasio RB, Habus I, Jiang Z, et al. 1995. Absorption, tissue distribution and in vivo stability in rats of a hybrid antisense oligonucleotide following oral administration. Biochem Pharmacol 50: 571–576. [DOI] [PubMed] [Google Scholar]
- Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. 2005. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J Med Chem 48: 901–904. [DOI] [PubMed] [Google Scholar]
- Betts L, Josey JA, Veal JM, Jordan SR. 1995. A nucleic acid triple helix formed by a peptide nucleic acid-DNA complex. Science 270: 1838–1841. [DOI] [PubMed] [Google Scholar]
- Binzel DW, Khisamutdinov EF, Guo P. 2014. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry 53: 2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzel DW, Khisamutdinov E, Vieweger M, Ortega J, Li J, Guo P. 2016a. Mechanism of three-component collision to produce ultrastable pRNA three-way junction of Phi29 DNA-packaging motor by kinetic assessment. RNA 22: 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzel D, Shu Y, Li H, Sun M, Zhang Q, Shu D, Guo B, Guo P. 2016b. Specific delivery of miRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol Ther 24: 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ. 2012. RNA-based therapeutics: current progress and future prospects. Chem Biol 19: 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. 2006. MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. 2004. Building programmable jigsaw puzzles with RNA. Science 306: 2068–2072. [DOI] [PubMed] [Google Scholar]
- Crick FH. 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163. [PubMed] [Google Scholar]
- Cui D, Zhang C, Liu B, Shu Y, Du T, Shu D, Wang K, Dai F, Liu Y, Li C, et al. 2015. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Sci Rep 5: 10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña M, Dufour D, Gallego J. 2009. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA 15: 1949–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mesmaeker A, Altmann KH, Waldner A, Wendeborn S. 1995. Backbone modifications in oligonucleotides and peptide nucleic acid systems. Curr Opin Struct Biol 5: 343–355. [DOI] [PubMed] [Google Scholar]
- Diamond JM, Turner DH, Mathews DH. 2001. Thermodynamics of three-way multibranch loops in RNA. Biochemistry 40: 6971–6981. [DOI] [PubMed] [Google Scholar]
- Dibrov SM, McLean J, Parsons J, Hermann T. 2011. Self-assembling RNA square. Proc Natl Acad Sci 108: 6405–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett DR, Murchie AI, Lilley DM. 1995. The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell 83: 1027–1036. [DOI] [PubMed] [Google Scholar]
- Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, et al. 2005. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 33: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B, Florentz C, Giegé R, Westhof E. 1996. A central pseudoknotted three-way junction imposes tRNA-like mimicry and the orientation of three 5′ upstream pseudoknots in the 3′ terminus of tobacco mosaic virus RNA. RNA 2: 201–212. [PMC free article] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119: 47–60. [DOI] [PubMed] [Google Scholar]
- Geary C, Rothemund PW, Andersen ES. 2014. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 345: 799–804. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow WW, Jaeger L. 2014. RNA self-assembly and RNA nanotechnology. Acc Chem Res 47: 1871–1880. [DOI] [PubMed] [Google Scholar]
- Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. 2011. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett 11: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. 2010. The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Haque F. 2013. RNA nanotechnology and therapeutics. CRC Press, Boca Raton, FL. [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, Garver K. 1998. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2: 149–155. [DOI] [PubMed] [Google Scholar]
- Gyi JI, Conn GL, Lane AN, Brown T. 1996. Comparison of the thermodynamic stabilities and solution conformations of DNA • RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry 35: 12538–12548. [DOI] [PubMed] [Google Scholar]
- Hansen ME, Bentin T, Nielsen PE. 2009. High-affinity triplex targeting of double stranded DNA using chemically modified peptide nucleic acid oligomers. Nucleic Acids Res 37: 4498–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich O, Benseler F, Fahrenholz A, Eckstein F. 1994. High activity and stability of hammerhead ribozymes containing 2′-modified pyrimidine nucleosides and phosphorothioates. J Biol Chem 269: 2131–2138. [PubMed] [Google Scholar]
- Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DM, Ha T. 2004. Conformational flexibility of four-way junctions in RNA. J Mol Biol 336: 69–79. [DOI] [PubMed] [Google Scholar]
- Jaeger L, Leontis NB. 2000. Tecto-RNA: one-dimensional self-assembly through tertiary interactions. Angew Chem Int Ed Engl 39: 2521–2524. [DOI] [PubMed] [Google Scholar]
- Jaeger L, Westhof E, Leontis NB. 2001. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res 29: 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski D, Haque F, Binzel DW, Guo P. 2017. Advancement of the emerging field of RNA nanotechnology. ACS Nano 11: 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkare S, Bhatnagar D. 2006. Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA, and morpholino. Appl Microbiol Biotechnol 71: 575–586. [DOI] [PubMed] [Google Scholar]
- Kaur H, Arora A, Wengel J, Maiti S. 2006. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry 45: 7347–7355. [DOI] [PubMed] [Google Scholar]
- Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. 1993. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem 36: 831–841. [DOI] [PubMed] [Google Scholar]
- Khisamutdinov EF, Jasinski DL, Guo P. 2014. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 8: 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov EF, Jasinski DL, Li H, Zhang K, Chiu W, Guo P. 2016. Fabrication of RNA 3D nanoprism for loading and protection of small RNAs and model drugs. Adv Mater 28: 100079–100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Watts JK. 2017. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. 2004. In vivo activity of nuclease-resistant siRNAs. RNA 10: 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E. 2006. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol 16: 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. 2006. Topology of three-way junctions in folded RNAs. RNA 12: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang X, Guo P. 2015. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today 10: 631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang K, Pi F, Guo S, Shlyakhtenko L, Chiu W, Shu D, Guo P. 2016. Controllable self-assembly of RNA tetrahedrons with precise shape and size for cancer targeting. Adv Mater 28: 7501–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Paroo Z. 2010. Biochemical principles of small RNA pathways. Annu Rev Biochem 79: 295–319. [DOI] [PubMed] [Google Scholar]
- Lundbaek JA, Andersen OS. 1994. Lysophospholipids modulate channel function by altering the mechanical properties of lipid bilayers. J Gen Physiol 104: 645–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin KE, Gissberg O, Smith CI. 2015. Oligonucleotide therapies: the past and the present. Hum Gene Ther 26: 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159. [DOI] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Fatica A, Bozzoni I. 2014. The role of long noncoding RNAs in the epigenetic control of gene expression. ChemMedChem 9: 505–510. [DOI] [PubMed] [Google Scholar]
- Moss EG. 2001. RNA interference: it's a small RNA world. Curr Biol 11: R772–R775. [DOI] [PubMed] [Google Scholar]
- Ouellet J, Melcher S, Iqbal A, Ding Y, Lilley DM. 2010. Structure of the three-way helical junction of the hepatitis C virus IRES element. RNA 16: 1597–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao X, Xia X, Bong D. 2013. Bifacial peptide nucleic acid directs cooperative folding and assembly of binary, ternary, and quaternary DNA complexes. Biochemistry 52: 6313–6323. [DOI] [PubMed] [Google Scholar]
- Rauzan B, McMichael E, Cave R, Sevcik LR, Ostrosky K, Whitman E, Stegemann R, Sinclair AL, Serra MJ, Deckert AA. 2013. Kinetics and thermodynamics of DNA, RNA and hybrid duplex formation. Biochemistry 52: 765–772. [DOI] [PubMed] [Google Scholar]
- Rettberg CC, Prere MF, Gesteland RF, Atkins JF, Fayet O. 1999. A three-way junction and constituent stem-loops as the stimulator for programmed –1 frameshifting in bacterial insertion sequence IS911. J Mol Biol 286: 1365–1378. [DOI] [PubMed] [Google Scholar]
- Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. 2010. A polyhedron made of tRNAs. Nat Chem 2: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Haque F, Pi F, Shlyakhtenko L, Evers BM, Guo P. 2015. Controllable self-assembly of RNA dendrimers. Nanomedicine 12: 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JP, Kent K, Bird J, Fishback J, Froehler B. 1991. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res 19: 747–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Moll WD, Deng Z, Mao C, Guo P. 2004. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 4: 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. 2011. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6: 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. 2014. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev 66: 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, Xu R, Guo P. 2015. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano 9: 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, Castanotto D. 2017. FDA-approved oligonucleotide therapies in 2017. Mol Ther 25: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann K, Rossi JJ. 2009. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med 1: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Wengel J. 2004. LNA (Locked Nucleic Acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 43: 13233–13241. [DOI] [PubMed] [Google Scholar]
- Westhof E, Masquida B, Jaeger L. 1996. RNA tectonics: towards RNA design. Fold Des 1: R78–R88. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Xu Z, Sun Y, Weber JK, Cao Y, Wang W, Jasinski D, Guo P, Zhou R, Li J. 2017. Directional mechanical stability of Bacteriophage phi29 motor's 3WJ-pRNA: extraordinary robustness along portal axis. Sci Adv 3: e1601684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling YG, Shapiro BA. 2007. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett 7: 2328–2334. [DOI] [PubMed] [Google Scholar]
- Zhang C. 2009. Novel functions for small RNA molecules. Curr Opin Mol Ther 11: 641–651. [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lu Z, Zhao H, Zhang X, Diasio RB, Habus I, Jiang Z, Iyer RP, Yu D, Agrawal S. 1995. In vivo stability, disposition and metabolism of a “hybrid” oligonucleotide phosphorothioate in rats. Biochem Pharmacol 50: 545–556. [DOI] [PubMed] [Google Scholar]
- Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YY. 2013. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 19: 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Leonard M, Shu Y, Yang Y, Shu D, Guo P, Zhang X. 2017. Overcoming tamoxifen resistance of human breast cancer by targeted gene silencing using multifunctional pRNA nanoparticles. ACS Nano 11: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.