Abstract
In the yeast Saccharomyces cerevisiae, the exposure to mating pheromone activates a prototypic mitogen-activated protein kinase (MAPK) cascade and triggers a dose-dependent differentiation response. Whereas a high pheromone dose induces growth arrest and formation of a shmoo-like morphology in yeast cells, lower pheromone doses elicit elongated cell growth. Previous population-level analysis has revealed that the MAPK Fus3 plays an important role in mediating this differentiation switch. To further investigate how Fus3 controls the fate decision process at the single-cell level, we developed a specific translocation-based reporter for monitoring Fus3 activity in individual live cells. Using this reporter, we observed strikingly different dynamic patterns of Fus3 activation in single cells differentiated into distinct fates. Cells committed to growth arrest and shmoo formation exhibited sustained Fus3 activation. In contrast, most cells undergoing elongated growth showed either a delayed gradual increase or pulsatile dynamics of Fus3 activity. Furthermore, we found that chemically perturbing Fus3 dynamics with a specific inhibitor could effectively redirect the mating differentiation, confirming the causative role of Fus3 dynamics in driving cell fate decisions. MAPKs mediate proliferation and differentiation signals in mammals and are therapeutic targets in many cancers. Our results highlight the importance of MAPK dynamics in regulating single-cell responses and open up the possibility that MAPK signaling dynamics could be a pharmacological target in therapeutic interventions.
Keywords: cell fate decision, single-cell analysis, yeast mating response, mitogen-activated protein kinase (MAPK), differentiation, pheromone, cell signaling, imaging
Introduction
The mitogen-activated protein kinase (MAPK) cascade consists of three sequentially activated kinases, MAPKK kinase, MAPK kinase, and MAPK, and is a conserved eukaryotic signaling module that plays crucial roles in mediating responses to a variety of extracellular cues. An increasing number of studies have revealed that the temporal dynamics of MAPK activity could encode environmental information and mediate cell fate decisions (1–4). In an oft-cited example, transient activation of the MAPK ERK leads to cell proliferation, whereas sustained ERK activation results in cell differentiation (5). In the yeast Saccharomyces cerevisiae, pheromone stimulation activates a prototypical MAPK cascade composed of Ste11, Ste7, and ultimately the MAPKs Fus3 and Kss1, and it initiates the processes of mating differentiation. High concentrations of pheromone induce cell cycle arrest and the formation of a pear-shaped shmoo morphology, but lower pheromone doses elicit elongated chemotropic growth, allowing non-motile yeast cells to grow toward distant mating partners (6, 7). A previous population-level study revealed that different doses of pheromone lead to different dynamics of MAPK activation, which might be critical to cell fate determination (6). Furthermore, Fus3, but not its homolog Kss1, plays a crucial role in mediating the dose-dependent differentiation switch between elongated growth and shmoo formation (6, 7). However, because yeast cells show a high cell-to-cell variation in their responses to pheromone (8–11), it remains largely elusive how Fus3 is activated dynamically in individual cells committed to elongated growth or shmoo formation and whether Fus3 dynamics contribute to cell fate determination at the single-cell level.
Previous single-cell analyses of the mating response pathway used gene expression reporters with fluorescent proteins under the control of a mating response promoter (8–10). These reporters require multiple slow processes, including transcription, translation, and maturation of fluorescent proteins, for their expression and detection, and therefore they are not ideal for indicating the fast MAPK activation. To evaluate the regulatory role of MAPK dynamics, a direct measurement of MAPK activity in live single cells would be needed. Recently, a fluorescence resonance energy transfer (FRET) reporter has been successfully applied to monitor the mating MAPK activity and has provided new insights into the dynamics and variability of MAPK activation during the yeast pheromone response (12). However, FRET reporters in general have a low signal-to-noise ratio and require two fluorescent proteins, limiting the number of signaling events that can be monitored simultaneously in a given cell. More importantly, the FRET reporter measures the activities of both Fus3 and Kss1. It is not a specific reporter for Fus3, the single MAPK that mediates the switch between elongated growth and shmoo formation during mating differentiation (6, 7).
Another strategy for designing kinase reporters is based on the observation that phosphorylation of a substrate protein can often lead to a rapid change in its nucleocytoplasmic localization, which can then be used as a reporter for the activity of the upstream kinase (13). Using this strategy, we developed a kinase translocation reporter (KTR)2 specific for Fus3 activity in single cells. This reporter features a rapid response time, full reversibility, a high signal-to-noise ratio and, importantly, a high specificity to Fus3 activity. Using this reporter, we tracked Fus3 activity over time in single cells upon pheromone stimulation and found different dynamic patterns correlated with two distinct cell fates, elongated growth or shmoo formation. Based on these single-cell observations, we chemically perturbed the dynamics of Fus3 activation and redirected mating differentiation, demonstrating the causative role of Fus3 dynamics in cell fate determination.
Results
Synthetic reporter for Fus3 activity based on translocation
Pheromone stimulation activates two parallel MAPKs, Fus3 and Kss1 (14), and induces a differentiation switch depending on pheromone concentrations. Whereas a high concentration of pheromone leads to cell cycle arrest and shmoo formation, lower doses trigger elongated chemotropic growth (6, 7). Previous studies showed that Fus3, but not Kss1, is required for mediating the switch between these two distinct cell fates (6, 7). To investigate how Fus3 regulates the fate determination in live single cells, we set out to develop a specific reporter for Fus3 activity (Fig. 1A). To this end, we first considered natural Fus3 substrates that undergo phosphorylation-regulated translocation and found a candidate protein, Fus2, which is a key cell fusion regulator that translocates from the nucleus to the cytoplasm in response to pheromone stimulation (15). Researchers have identified a minimal region of Fus2 (Fus2(54–99)) that contains both the nuclear localization sequence (NLS) and nuclear export sequence (NES) and is necessary and sufficient for regulated nucleocytoplasmic translocation (16). The MAPK Fus3 phosphorylates Ser-84 to drive its nuclear export, whereas a cyclin-dependent kinase (CDK) phosphorylates Ser-67 to drive its nuclear import. When Ser-67 is mutated to Ala (Fus2(54–99)-S67A), nucleocytoplasmic translocation is solely dependent on Fus3 activity (Fig. 1B) (16). We expressed this fragment of 46 amino acids with mCherry at the C terminus under the control of a constitutive promoter PADH1 and observed a weak localization change from the nucleus to the cytoplasm in response to pheromone stimulation (Fig. S1).
Figure 1.
Development of a KTR for Fus3 activity. A, top panel, diagram of the MAPK signaling pathway that mediates mating differentiation in yeast. Bottom panel, representative images of cells undergoing elongated growth and shmoo formation. B, schematic of the KTR for Fus3 developed in this study. Green regions, NLS; blue regions, NES. C, representative time-lapse images showing the reporter translocation in response to pheromone stimulation. D, average time traces of reporter translocation response to a 30-min pulse of 1 μm pheromone treatment. The reporter response was quantified as the cytoplasmic over nuclear fluorescence intensities (C/N ratio) and was normalized by the basal level. Means and S.E. are shown for WT (blue curve), kss1Δ (red curve), and fus3Δ (green curve). E, average time trace of reporter response in the FUS3as, KSS1 strain (blue curve). Cells were exposed to a constant 1 μm pheromone treatment, and 10 μm 1-NM-PP1 was added 30 min after pheromone stimulation. The response from cells without pheromone exposure was shown as a control (gray curve).
To increase the dynamic range of the localization change, we further tested a series of reporter variants (Fig. S1). We incorporated different truncated forms of Fus2(54–99)-S67A to the N or C terminus of the original fragment to enhance the strength of regulated localization signals. One of the variants with an additional regulated NLS at the N terminus of Fus2(54–99)-S67A significantly increases the dynamic range of localization change (Fig. 1B). In response to a high dose of pheromone treatment (1 μm), this variant was exported from the nucleus to the cytoplasm rapidly and robustly (Fig. 1, C, bottom panels, and D, blue curve). Upon the removal of pheromone, it returned to the nucleus quickly (Fig. 1D, blue curve). We chose to use this construct as the kinase translocation reporter for Fus3 activity in our single-cell analysis.
To examine the specificity of this reporter for Fus3 activity, we monitored the reporter responses in strains with either the MAPK Fus3 or Kss1 deleted. In kss1Δ cells, the reporter quickly translocated from the nucleus to the cytoplasm with the time trace closely resembling that of the WT cells (Fig. 1D, red curve versus blue curve). In contrast, in fus3Δ cells, although Kss1 is highly activated in response to pheromone stimulation (14, 17), the reporter remained enriched in the nucleus and did not undergo translocation (Fig. 1D, green curve). These results demonstrated that, unlike previously reported MAPK reporters in yeast (12, 18), our reporter specifically measures the activity of Fus3, but not Kss1, and hence is particularly useful for studying the Fus3-mediated differentiation switch. We also examined the effect of Fus3 inhibition on the reporter response using an analog-sensitive mutant FUS3as (FUS3-Q93G) (19). We observed that, similar to the response in WT, the reporter translocated from the nucleus to the cytoplasm in response to pheromone stimulation. When the inhibitor for Fus3 activity was added, the reporter rapidly returned from the cytoplasm to the nucleus-enriched basal level (Fig. 1E). These results further confirmed the reversibility of the reporter and its specificity to Fus3 activity. Although Kss1 is activated to a significantly higher extent when Fus3 is inhibited (17), the reporter only responded to the drop in Fus3 activity but not to the increase in Kss1 activity.
Single-cell Fus3 dynamics during mating differentiation
Previous population-level MAPK analysis suggested that the dose-dependent dynamics of Fus3 is important for switch-like mating differentiation responses (6). Using our new reporter, we could evaluate the role of Fus3 dynamics in individual cells. To this end, we monitored the single-cell reporter responses upon two different doses of pheromone, which induced two distinct cell fates, elongated growth and shmoo formation, respectively.
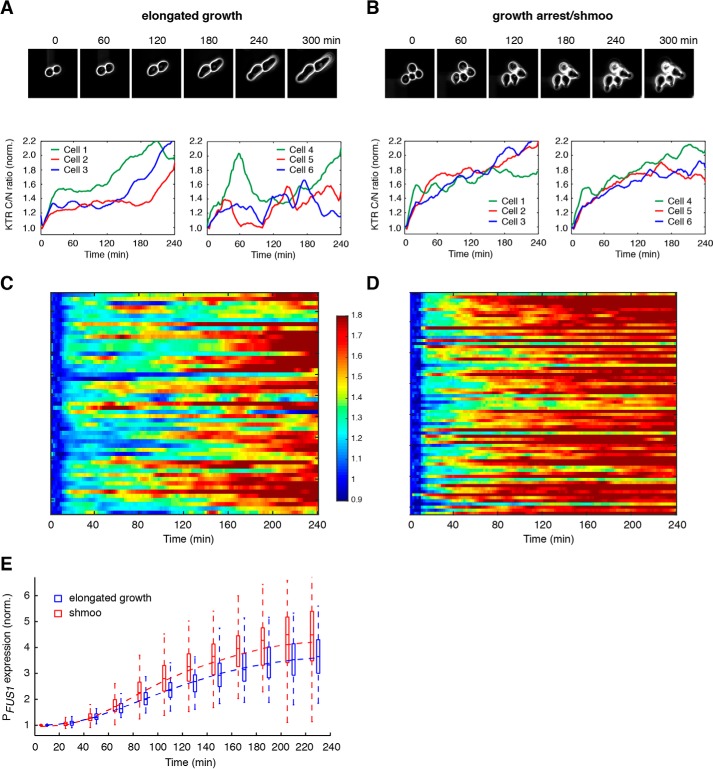
In response to 0.25 μm pheromone treatment, about 60% of cells initiated elongated growth, whereas the rest of the cells remained rounded morphology. We focused on the cells undergoing elongated growth (Fig. 2A, top panel) and observed two different dynamic patterns of Fus3 activation. A large portion (∼70%) of elongated growing cells showed a rapid rise in Fus3 activity within 10 min, followed by a delayed elevation in the kinase activity after 2–3 h (see representative single-cell time traces in Fig. 2A, bottom left panel). Intriguingly, the other ∼30% of elongated growing cells exhibited pulsatile Fus3 activation, with two or three strong pulses during the 4-h experiments (see representative single-cell time traces in Fig. 2A, bottom right panel). We note that these Fus3 pulses were largely irregular and were not correlated with cell division or cell cycle progression, indicating that the pulsatile dynamics were not an artifact from nuclear division or reorganization. Fus3 dynamics for cells remaining round-shaped are shown in Fig. S2. By contrast, in response to 1 μm pheromone treatment, all of the cells underwent cell cycle arrest and formed a shmoo-like morphology (Fig. 2B, top panel). Most of these cells showed an initial rapid rise in Fus3 activity, followed by a sustained Fus3 activation with a gradual increase in amplitude (see representative single-cell time traces in Fig. 2B, bottom panels).
Figure 2.
Cells with different fates exhibit distinct dynamics of Fus3 activation. A and B, time traces of Fus3 activation in individual cells with different fates. Top panels, time-lapse phase images for representative cells undergoing elongated growth (A) or shmoo formation (B). Bottom panels, time traces of Fus3 activation in representative single cells undergoing elongated growth (A) or shmoo formation (B). C and D, single-cell color map trajectories of Fus3 activation for all of the cells that underwent elongated growth in response to 0.25 μm pheromone treatment (C) or all of the cells committed to growth arrest and shmoo formation in response to 1 μm pheromone treatment (D). Each row represents the time trace of a single cell. Color represents the normalized C/N ratio, as indicated in the color bar. E, boxplot showing the time-dependent distributions of gene expression responses in cells undergoing elongated growth (blue) and shmoo formation (red). Data are from the same cells in C and D. In the plot, the bottom and top of the box are first (the 25th percentile of the data, q1) and third quartiles (the 75th percentile of the data, q3); the band inside the box is the median; the dashed curve is the mean; the whiskers cover the range between q1 − 1.5 × (q3-q1) and q3 + 1.5 × (q3-q1).
In summary, our single-cell analysis revealed that cells committed to distinct fates, elongated growth or shmoo formation, exhibited a similar initial rapid rise in Fus3 activity but strikingly different long-term dynamics (Fig. 2, compare C and D). These long-term dynamic patterns of Fus3 activation might serve a determining role in mating differentiation.
Furthermore, to measure the downstream gene expression simultaneously with Fus3 activity, we had incorporated a reporter expressing a destabilized GFP under the FUS1 promoter (20) in the same cells with the Fus3 reporter. We found that the gene expression response exhibited a very high cell-to-cell variability, and as a result, cells with distinct fates, while showing different Fus3 dynamics, displayed largely overlapping gene expression outputs (Fig. 2E). These results suggested that, due to its inherent stochasticity, the downstream gene expression machinery might not be effective for faithfully decoding Fus3 dynamics. Additional mechanisms are needed to convert the divergent Fus3 dynamics to switch-like cell fate decisions (see under “Discussion” for more information).
Redirecting mating differentiation by chemically perturbing Fus3 dynamics
Our single-cell observations revealed that different Fus3 dynamics are correlated with distinct cell fates. To further test the causative role of Fus3 dynamics on cell fate determination, we artificially perturbed Fus3 dynamics and examined the resultant differentiation responses. To control the activity of Fus3 in cells, we employed an engineered strain carrying an analog-sensitive mutation in FUS3 (FUS3-Q93G) that renders the selective inhibition by a cell-permeable inhibitor 1-NM-PP1 (19, 21, 22). KSS1 was deleted in the strain to eliminate its potential influence on cell morphology (6, 7, 14, 17).
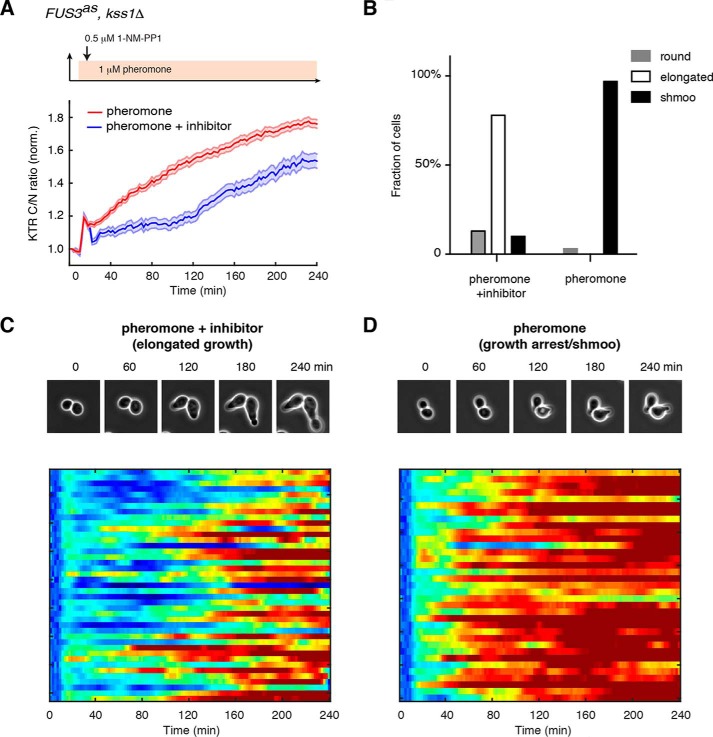
As shown above in Fig. 2, yeast cells with distinct fates exhibited a similar initial rise in Fus3 activity but different subsequent long-term dynamics; cells undergoing elongated growth showed a significantly delayed increase in Fus3 activity, compared with the cells committed to growth arrest and shmoo formation. We reasoned that the long-term increase in Fus3 activity might be driven by some positive feedback regulation (11, 12); thereby, a partial inhibition of Fus3 activity after its initial rise might be sufficient to delay the subsequent gradual increase in the kinase activity. Based on this hypothesis, we designed our perturbation experiment in which cells were first exposed to a high dose of pheromone treatment (1 μm), and a constant low level (0.5 μm) of inhibitor treatment was applied 10 min after the pheromone addition (after the initial rise in Fus3 activity; see Fig. 3A, top panel).
Figure 3.
Chemical perturbation of Fus3 dynamics redirects cell fate decision. A, average time traces of Fus3 activity in response to 1 μm pheromone treatment (red curve) or 1 μm pheromone treatment + 0.5 μm inhibitor treatment (blue curve). Top panel, schematic showing the perturbation pattern: 0.5 μm 1-NM-PP1 was added after 10 min of 1 μm pheromone treatment. Means and S.E. are shown for both the control and perturbation conditions. B, bar graphs showing fractions of cells exhibiting round, elongated, or shmoo morphology upon 1 μm pheromone treatment or 1 μm pheromone treatment + 0.5 μm inhibitor treatment. Data are from the same cells in A. C and D, lower panels, single-cell color map trajectories of Fus3 activity for all of the cells that underwent elongated growth in response to 1 μm pheromone treatment + 0.5 μm inhibitor treatment (C) or all of the cells committed to growth arrest and shmoo formation in response to 1 μm pheromone treatment (D). The color scheme is identical to that in Fig. 2, C and D. Top panels, time-lapse phase images for representative cells under the control or the perturbation condition.
As shown in Fig. 3, the inhibitor treatment after the initial rise of Fus3 activity indeed caused a dramatically delayed increase of kinase activity (Fig. 3, A and C), largely resembling the dynamic response observed in elongated growing cells under a lower pheromone dose. Importantly, we observed that a large fraction of yeast cells (∼78%) with the chemically perturbed Fus3 dynamics underwent elongated growth, even under a high dose of pheromone treatment (Fig. 3B). In contrast, the same pheromone stimulation without the perturbation induced sustained Fus3 activation (Fig. 3, A and D), resulting in growth arrest and shmoo formation in the vast majority of treated cells (Fig. 3B). Cells treated with the inhibitor alone in the absence of pheromone remained in vegetative growth throughout the experiment.
These results demonstrated that a rationally-designed perturbation of Fus3 dynamics could effectively redirect cell fate decision and therefore provided direct evidence about the specific and causative role of Fus3 dynamics in driving cell fate determination during mating differentiation.
Discussion
We are particularly interested in the cell fate determination between elongated growth and shmoo formation, triggered by alterations in pheromone concentration. It has been found that, whereas both Kss1 and Fus3 promote the transition from vegetative to elongated growth, only Fus3 is required to mediate the differentiation switch between elongated growth and shmoo formation (6, 7). To investigate this Fus3-driven cell fate decision in single cells, we would need to specifically measure its activity during mating differentiation.
Previous studies of the yeast mating MAPK pathway have been primarily focused on the average response in a cell population. Recently, a growing number of studies showed that genetically identical cells could respond very differently to the same pheromone stimuli (8–11, 23); however, traditional population-based assays are insufficient to further unravel the regulatory mechanisms and physiological consequences of these cell-to-cell variations. To study signaling response at the single-cell level, researchers have successfully developed FRET-based or translocation-based reporters to monitor MAPK activity in individual yeast cells (12, 18). However, these reporters measure the activities of both Fus3 and Kss1. Neither of them is specific to Fus3 (12, 18). In fact, Conlon et al. (12) have used the FRET reporter to monitor MAPK signaling during the mating response. But because their reporter measured the combined activity of Fus3 and Kss1, they only correlated the reporter dynamics with the phenotypic transition from vegetative growth and elongated growth, which are mediated by both Fus3 and Kss1 and occurred in a very low pheromone dose range. In our study, we focused on the differentiation switch between elongated growth and shmoo formation that occurs at higher pheromone doses and is driven by Fus3 only. Therefore, we developed a new reporter specific for Fus3 activity and combined this reporter with microfluidics and time-lapse microscopy to track Fus3 activity in differentiating yeast cells. Using this approach, we found that cells undergoing elongated growth and shmoo formation showed similar initial Fus3 activation but strikingly divergent long-term dynamics, suggesting that different Fus3 dynamics might determine cell fate during mating differentiation.
A previous study discovered oscillatory activation of Fus3 in synchronized cell populations (24). Interestingly, we also found that a fraction of elongated growing cells displayed pulsatile Fus3 dynamics, the pattern of which is similar to that observed at the population level. These single-cell dynamic patterns are, however, not correlated with cell cycle progression or the formation of multiple mating projections. Further analysis is needed to elucidate the mechanisms that underlie these pulsatile dynamics in single cells. As the population-level oscillations require negative pathway regulators Sst2 and Msg5, it would be interesting to examine their individual and combined contributions in single cells. Intriguingly, Sst2 also suppresses cell-cell variability in the pheromone response (23). A careful analysis of Sst2 might lead to a further understanding about the influence of Fus3 oscillations on the heterogeneity of mating response.
We have previously combined chemical genetics with microfluidics to dynamically perturb PKA signaling and examine how PKA dynamics control gene expression responses (25–27). In this study, we employed a similar strategy to perturb Fus3 dynamics and test the causative role of these dynamics in driving cell fate determination. Whereas yeast cells exposed to a high dose of pheromone were supposed to undergo growth arrest and shmoo formation, we reshaped Fus3 dynamics using a well-controlled inhibitor treatment to mimic the dynamics observed in elongated growing cells at a lower pheromone dose. As a result, the majority of cells initiated elongated growth, instead of shmoo formation, demonstrating that perturbing Fus3 dynamics could effectively redirect mating differentiation. We would like to emphasize that the differentiation switch that we investigated here is different from another well-studied developmental switch between the nutrient-dependent filamentous invasive growth and pheromone-induced mating response (14, 17, 28–30). In that case, Kss1 is needed for filamentous invasive growth, whereas Fus3 simultaneously suppresses invasive growth and promotes mating. In contrast, the differentiation switch in this study requires only Fus3, which promotes elongated chemotropic growth or shmoo formation, depending on the pheromone concentration. Because Kss1-mediated filamentous growth and Fus3-mediated elongated growth share some similar morphological features, we have deleted KSS1 in the perturbation experiment to avoid its influence on cell morphology.
A remaining question is how downstream molecular processes decode different Fus3 dynamics to induce appropriate cell fates. One possibility is that different kinase dynamics could lead to distinct gene expression programs that determine cell fates. As our single-cell analysis of a reporter gene (Fig. 2E) suggests that the inherently stochastic gene expression machinery might not be sufficient to faithfully mediate cell fate decision, a more comprehensive genome-wide analysis would be needed to characterize the gene expression programs associated with distinct cell fates and further evaluate their roles in mediating differentiation. Another possibility is that Fus3 dynamics can be decoded by some specific substrates. For example, the CDK inhibitor Far1 might be a potential candidate. Far1 constitutes a bistable switch in pheromone-driven cell cycle control (31, 32) and functions in regulating polarized cell growth via the interaction with Cdc24 (33). It would be interesting to track Fus3 and Far1 in the same cells and test the role of Far1 in decoding Fus3 dynamics. Finally, the coordination of the pheromone-response pathway with other cellular regulatory pathways might also contribute to the differentiation switch. We have recently identified a pheromone-driven repression of the ribosomal biogenesis pathway (34). Interestingly, this cross-regulation is dose-dependent, only occurring at high pheromone concentrations that induce growth arrest and shmoo formation. Future work is needed to examine the role of Fus3 in mediating this cross-inhibition and how it contributes to cell fate determination during mating differentiation.
Experimental procedures
Reporter construction
The reporter constructs were made by cloning DNA fragments carrying the ADH1 promoter, the reporter variant ORFs, and mCherry into the integrating plasmid pRS305. PCR products carrying the ADH1 promoter (600 bp), ORFs for reporter variants, and mCherry were amplified separately and assembled into a single DNA fragment using fusion PCR. The assembled product was then inserted between XhoI and SacI in pRS305. The ORF for Fus2(54–99)-S67A was amplified from the plasmid pMR6161, PGAL1-FUS2(54–99)-S67A-3xGFP (16). The ORFs of other reporter variants were constructed by assembling different truncated forms of Fus2(54–99)-S67A to the N or C terminus of the original fragment with a flexible linker (Gly-Gly-Ser-Gly-Gly) in between (Fig. S1A). The resultant plasmids were linearized using AflII and integrated into the leu2 locus in yeast.
Strain construction
Standard methods for the growth, maintenance, and transformation of yeast and bacteria and for manipulation of DNA were used throughout. The yeast strains used in this study are derived from the W303 background (ADE+ MATa trp1 leu2 ura3 his3 can1 GAL+ psi+). In all the strains used in this study, BAR1 was deleted using a pop-in/pop-out method, and NHP6a was C-terminally tagged with a yeast codon optimized iRFP from a pKT vector (used as a nuclear marker for image analysis). To make the kss1Δ mutant, a CgURA3 fragment was amplified using PCR and transformed to replace the KSS1 ORF by homologous recombination. Similarly, the FUS3 ORF was replaced with CgURA3 to make the fus3Δ mutant. To make the analog-sensitive FUS3 strain (FUS3as), a Q93G mutation was introduced into the FUS3 ORF using in vivo site-directed mutagenesis (35). The PFUS1-UBI-YΔkGFP reporter was integrated into yeast as described previously (20). A list of strains is provided in Table S1.
Microfluidics and time-lapse microscopy
The experimental setup for microfluidics devices was performed as described previously (25–27, 36). Time-lapse microscopy experiments were performed using a Nikon Ti-E inverted fluorescence microscope with Perfect Focus, coupled with an EMCCD camera (Andor iXon X3 DU897). The light source is a Spectra X LED system. Images were taken using a CFI Plan Apochromat Lambda DM ×60 oil immersion objective (NA 1.40 WD 0.13MM). In all experiments, the microscope was programmed to acquire images for each fluorescence channel every 2 min. The pheromone stock used in this study was ordered from Genscript (catalog no. RP01002). We note that pheromone stocks from different sources differ in their effective concentrations.
Image analysis and quantification
Fluorescence images were processed with a custom MATLAB code as described previously (25–27, 36). The cytoplasm and the nucleus of single cells were identified by thresholding the iRFP nuclear marker and the phase image. For each individual cell, the mean fluorescence intensities for the cytoplasm and the nucleus were then quantified and smoothed separately. The ratio of the cytoplasmic over nuclear intensity (KTR C/N ratio) was calculated and normalized by the basal level at the time 0.
Author contributions
Y. L. formal analysis; Y. L., J. R., and Z. A. investigation; Y. L. methodology; Y. L. and N. H. writing-original draft; Y. L., J. R., Z. A., and N. H. writing-review and editing; N. H. conceptualization; N. H. supervision; N. H. funding acquisition; N. H. project administration.
Supplementary Material
Acknowledgment
We thank Dr. Mark D. Rose (Princeton University) for generously providing the pMR6161 plasmid.
This work was supported by National Institutes of Health Grant R01 GM111458. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Table S1.
- KTR
- kinase translocation reporter
- NLS
- nuclear localization sequence
- NES
- nuclear export sequence
- CDK
- cyclin-dependent kinase
- C/N
- cytoplasmic over nuclear fluorescence.
References
- 1. Behar M., and Hoffmann A. (2010) Understanding the temporal codes of intra-cellular signals. Curr. Opin. Genet. Dev. 20, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Purvis J. E., and Lahav G. (2013) Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Santos S. D., Verveer P. J., and Bastiaens P. I. (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 9, 324–330 [DOI] [PubMed] [Google Scholar]
- 4. Albeck J. G., Mills G. B., and Brugge J. S. (2013) Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marshall C. J. (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185 [DOI] [PubMed] [Google Scholar]
- 6. Hao N., Nayak S., Behar M., Shanks R. H., Nagiec M. J., Errede B., Hasty J., Elston T. C., and Dohlman H. G. (2008) Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol. Cell 30, 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Erdman S., and Snyder M. (2001) A filamentous growth response mediated by the yeast mating pathway. Genetics 159, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poritz M. A., Malmstrom S., Kim M. K., Rossmeissl P. J., and Kamb A. (2001) Graded mode of transcriptional induction in yeast pheromone signalling revealed by single-cell analysis. Yeast 18, 1331–1338 [DOI] [PubMed] [Google Scholar]
- 9. Wang X., Hao N., Dohlman H. G., and Elston T. C. (2006) Bistability, stochasticity, and oscillations in the mitogen-activated protein kinase cascade. Biophys. J. 90, 1961–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colman-Lerner A., Gordon A., Serra E., Chin T., Resnekov O., Endy D., Pesce C. G., and Brent R. (2005) Regulated cell-to-cell variation in a cell-fate decision system. Nature 437, 699–706 [DOI] [PubMed] [Google Scholar]
- 11. Paliwal S., Iglesias P. A., Campbell K., Hilioti Z., Groisman A., and Levchenko A. (2007) MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 446, 46–51 [DOI] [PubMed] [Google Scholar]
- 12. Conlon P., Gelin-Licht R., Ganesan A., Zhang J., and Levchenko A. (2016) Single-cell dynamics and variability of MAPK activity in a yeast differentiation pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E5896–E5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Regot S., Hughey J. J., Bajar B. T., Carrasco S., and Covert M. W. (2014) High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabbagh W. Jr, Flatauer L. J., Bardwell A. J., and Bardwell L. (2001) Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ydenberg C. A., and Rose M. D. (2009) Antagonistic regulation of Fus2p nuclear localization by pheromone signaling and the cell cycle. J. Cell Biol. 184, 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J., and Rose M. D. (2012) A mechanism for the coordination of proliferation and differentiation by spatial regulation of Fus2p in budding yeast. Genes Dev. 26, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hao N., Yildirim N., Nagiec M. J., Parnell S. C., Errede B., Dohlman H. G., and Elston T. C. (2012) Combined computational and experimental analysis reveals MAP kinase-mediated feedback phosphorylation as a mechanism for signaling specificity. Mol. Biol. Cell 23, 3899–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Durandau E., Aymoz D., and Pelet S. (2015) Dynamic single cell measurements of kinase activity by synthetic kinase activity relocation sensors. BMC Biol. 13, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu R. C., Pesce C. G., Colman-Lerner A., Lok L., Pincus D., Serra E., Holl M., Benjamin K., Gordon A., and Brent R. (2008) Negative feedback that improves information transmission in yeast signalling. Nature 456, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Houser J. R., Ford E., Chatterjea S. M., Maleri S., Elston T. C., and Errede B. (2012) An improved short-lived fluorescent protein transcriptional reporter for Saccharomyces cerevisiae. Yeast 29, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., Wood J. L., Morgan D. O., and Shokat K. M. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 [DOI] [PubMed] [Google Scholar]
- 22. Patterson J. C., Klimenko E. S., and Thorner J. (2010) Single-cell analysis reveals that insulation maintains signaling specificity between two yeast MAPK pathways with common components. Sci. Signal. 3, ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dixit G., Kelley J. B., Houser J. R., Elston T. C., and Dohlman H. G. (2014) Cellular noise suppression by the regulator of G protein signaling Sst2. Mol. Cell 55, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hilioti Z., Sabbagh W. Jr, Paliwal S., Bergmann A., Goncalves M. D., Bardwell L., and Levchenko A. (2008) Oscillatory phosphorylation of yeast Fus3 MAP kinase controls periodic gene expression and morphogenesis. Curr. Biol. 18, 1700–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hao N., and O'Shea E. K. (2011) Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 19, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hao N., Budnik B. A., Gunawardena J., and O'Shea E. K. (2013) Tunable signal processing through modular control of transcription factor translocation. Science 339, 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. AkhavanAghdam Z., Sinha J., Tabbaa O. P., and Hao N. (2016) Dynamic control of gene regulatory logic by seemingly redundant transcription factors. Elife 5, e18458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cook J. G., Bardwell L., and Thorner J. (1997) Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390, 85–88 [DOI] [PubMed] [Google Scholar]
- 29. Madhani H. D., and Fink G. R. (1997) Combinatorial control required for the specificity of yeast MAPK signaling. Science 275, 1314–1317 [DOI] [PubMed] [Google Scholar]
- 30. Roberts R. L., and Fink G. R. (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8, 2974–2985 [DOI] [PubMed] [Google Scholar]
- 31. Doncic A., and Skotheim J. M. (2013) Feedforward regulation ensures stability and rapid reversibility of a cellular state. Mol. Cell 50, 856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doncic A., Atay O., Valk E., Grande A., Bush A., Vasen G., Colman-Lerner A., Loog M., and Skotheim J. M. (2015) Compartmentalization of a bistable switch enables memory to cross a feedback-driven transition. Cell 160, 1182–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimada Y., Gulli M. P., and Peter M. (2000) Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat. Cell Biol. 2, 117–124 [DOI] [PubMed] [Google Scholar]
- 34. Shao B., Yuan H., Zhang R., Wang X., Zhang S., Ouyang Q., Hao N., and Luo C. (2017) Reconstructing the regulatory circuit of cell fate determination in yeast mating response. PLoS Comput. Biol. 13, e1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Storici F., Lewis L. K., and Resnick M. A. (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19, 773–776 [DOI] [PubMed] [Google Scholar]
- 36. Hansen A. S., Hao N., and O'Shea E. K. (2015) High-throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat. Protoc. 10, 1181–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.