Abstract
Transcription factor 19 (TCF19) has been reported as a type 1 diabetes–associated locus involved in maintenance of pancreatic β cells through a fine-tuned regulation of cell proliferation and apoptosis. TCF19 also exhibits genomic association with type 2 diabetes, although the precise molecular mechanism remains unknown. It harbors both a plant homeodomain and a forkhead-associated domain implicated in epigenetic recognition and gene regulation, a phenomenon that has remained unexplored. Here, we show that TCF19 selectively interacts with histone 3 lysine 4 trimethylation through its plant homeodomain finger. Knocking down TCF19 under high-glucose conditions affected many metabolic processes, including gluconeogenesis. We found that TCF19 overexpression represses de novo glucose production in HepG2 cells. The transcriptional repression of key genes, induced by TCF19, coincided with NuRD (nucleosome-remodeling-deacetylase) complex recruitment to the promoters of these genes. TCF19 interacted with CHD4 (chromodomain helicase DNA-binding protein 4), which is a part of the NuRD complex, in a glucose concentration–independent manner. In summary, our results show that TCF19 interacts with an active transcription mark and recruits a co-repressor complex to regulate gluconeogenic gene expression in HepG2 cells. Our study offers critical insights into the molecular mechanisms of transcriptional regulation of gluconeogenesis and into the roles of chromatin readers in metabolic homeostasis.
Keywords: gene transcription, gluconeogenesis, glucose metabolism, histone modification, nucleosome remodeling deacetylase (NuRD), PHD finger, transcription factor, transcription repressor, histone H3 lysine 4 trimethylation, histone deacetylation
Introduction
The liver is the critical organ that maintains metabolic homeostasis in the body. Particularly in glucose homeostasis, the liver maintains a fine balance between plasma and hepatic glucose levels by regulating the uptake, storage, and production of glucose (1). Under well-fed conditions, excess glucose (i.e. the amount of glucose left after uptake by skeletal muscle, red blood cells, and brain tissue) is stored in the form of glycogen. Prolonged fasting conditions induce de novo glucose synthesis from the liver by reactions that essentially reverse the glycolytic machinery. Three key enzymes (glucose-6-phosphatase (G6PC),3 fructose-1,6-bisphosphatase (FBP1), and pyruvate carboxyl kinase 1 (PCK1)) are responsible for reversing glycolysis. The expression of these enzymes is controlled by an array of transcription regulators, which respond to hormones and the signaling molecules insulin, glucagon, epinephrine, and cAMP (2). Important transcriptional regulators have been implicated in this regulation, including CREB-binding protein (CBP)/p300, CREB-regulated transcription co-activator 2 (CRTC2), peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α), and protein arginine methyltransferases (3). In addition to these, histone modification enzymes, such as histone deacetylases (HDAC1 and HDAC2) and sirtuins, are important regulators that act as transcriptional switches of genes in response to various metabolic and hormonal cues (4, 5).
Modification of chromatin states to either facilitate or inhibit transcriptional machinery is an efficient and reversible means of adapting to a metabolic environment. The elevated glucose levels in cells alter the epigenetic landscape by affecting histone modifications (methylation and acetylation) as well as DNA methylation and contribute to activation of several factors and signaling pathways (6, 7). For example, the promoter methylation status of PGC-1α was found to be different in diabetic patients (8). Interestingly, the DNA methylation status of genes involved in insulin and calcium signaling is differentially modulated in patients with a familial history of type 2 diabetes (T2D) (9, 10). Further, alteration in H3K4Me2/3 status in the adipocyte cells has been reported in T2D patients (11). In the current study, we focus on a previously unexplored role of transcription factor 19 (TCF19) as an important regulator of the key gluconeogenic genes. TCF19 was discovered as a serum-stimulated trans-activating factor with maximum expression at the G1/S boundary of the cell cycle (12). In recent years, the protein has been implicated in various genome-wide association studies, indicating a possible role in various physiological disorders, specifically type 2 and type 1 diabetes (13–15). We report here that the PHD finger has a unique preference for the lysine 4 trimethylation of histone H3, an epigenetic signature canonically recognized by plant homeodomains (16–18). Microarray analysis on TCF19-depleted cells showed a global effect on metabolic pathways, and interestingly, the gluconeogenic genes were significantly up-regulated. Physical interaction of TCF19 with CHD4 and MTA1 and their co-recruitment onto promoters of gluconeogenic genes in high-glucose conditions suggest that the observed repression is possibly mediated in concert with NuRD complex, of which CHD4 and MTA1 are an integral part (19, 20). More in-depth analysis revealed how TCF19 exerts a repressive effect on the gluconeogenic genes by integrating the hormonal and metabolic cues via its PHD finger interactions with chromatin. Thus, TCF19 could be an important target in modulating the glucose homeostasis in cells.
Results
PHD finger of TCF19 specifically interacts with histone H3K4Me3
Transcription factor 19 is a putative trans-activating factor, found ubiquitously in all eukaryotes. The protein harbors two conserved domains: PHD and Forkhead-associated (FHA) domain (Fig. 1A). PHDs in general have the capacity to interact with H3K4Me3, H3K4Me0 (18), H3K14Ac (21), or H4K16Ac (22), as dictated by the sequence and structure of the interacting module. We performed in vitro pulldown assays of purified GST-tagged PHD finger of TCF19 (supplemental Fig. S1A) with core histones, purified from chicken erythrocytes, and we found strong association with histone H3 and H4 (Fig. 1B). Histone H3 and H4 could also be immunoprecipitated with FLAG-conjugated full-length TCF19, but not with the PHD finger–deleted construct. More specifically, an interaction of FLAG-conjugated TCF19 full-length protein, but not PHD-deleted protein, with H3K4Me3 could be detected (Fig. 1C). The expression levels for full-length and PHD-deleted FLAG-conjugated constructs are represented in supplemental Fig. S2A). The extent of overexpression as quantified was between 2- and 3-fold of the endogenous TCF19 (supplemental Fig. S2B).
Figure 1.
PHD finger of TCF19 binds to histone H3K4Me3. A, schematic representation of TCF19 protein with domain arrangement. FHA, Forkhead-associated domain. B, purified GST-PHD protein incubated with core histones isolated from chicken erythrocyte, immunoblotted with the anti-H3 and anti-H4 antibody. C, preferential interaction of TCF19-PHD (FLAG) with H3K4Me3. HEK293 cells were transiently transfected with full-length TCF19 (FLAG) and TCF19-ΔPHD (FLAG), followed by M2-agarose pulldown, and probed with anti-FLAG, anti-H3K4Me3, anti-H3, and anti-H4 antibody. Deletion of PHD domain abrogates interaction of TCF19 with H3K4Me3 as well as H3 and H4. CONT, empty vector–transfected; TRANS, FLAG-tagged full-length TCF19/ΔPHD–transfected. D, interaction of biotinylated peptides with TCF19 PHD-GST. Pulldown was performed with streptavidin beads, followed by immunoblotting by the anti-GST antibody. Purified GST-PHD domain showed preferential interaction with H3K4Me3 peptide. E, binding isotherms obtained from steady-state fluorescence spectroscopy of TCF19-PHD and indicated peptides. Below, interaction of the PHD finger with increasing multiples of the equilibrium dissociation constant (0.5, 1, 2, 3, 4, and 5 times Kd) showed incremental binding to H3K4Me3. F, GST-tagged PHD finger of TCF19 shows a preference for first the H3 lysine 4 trimethylation, as opposed to other H3 trimethylations. G, multiple-sequence alignment shows conserved features of TCF19-PHD and other H3K4Me3-binding PHD fingers. Zinc-coordinating sites are highlighted in cyan; other residues for lysine trimethyl recognition are highlighted in magenta. ZN1, zinc 1; ZN2, zinc 2. H, molecular modeling of TCF19 PHD domain followed by docking of H3K4Me3 N-terminal peptide (amino acids 1–8). Trp-316 and Trp-307 are proximal to the H3K4Me3 peptide in the docked structure. I, interaction of the indicated biotinylated peptides with wild-type TCF19 PHD-GST and W316A/W307A mutants of PHD-GST. Mutation in tryptophan at position 316 compromised the binding ability of the PHD domain. The Coomassie Blue gel image represents the amount of purified wild-type and mutant protein PHD finger (for all Western blots). Amido Black–stained spots below represent an equal amount of peptide loading. J, TCF19 interacts preferentially with H3K4Me3 compared with other H3 trimethylations. Lysate from HepG2 cells grown under high-glucose (40 mm glucose) conditions was immunoprecipitated with anti-TCF19 antibody and probed with TCF19, H3K4Me3, H3K9Me3, H3K27Me3, and H3K36Me3 antibody. Inp, input.
We performed peptide pulldown assays with N-terminal histone peptides and likewise found a preferential interaction of TCF19 PHD (GST) with histone H3K4Me3 (amino acids 1–25) (Fig. 1D). Selective interaction of histone H3 lysine 4 methylation was quantified by employing fluorescence spectroscopy with PHD finger (N-terminal GST tag cleaved) of TCF19 (protein profile shown in supplemental Fig. S1B). The progressive quenching of fluorescence of tryptophan residues present in TCF19 PHD, upon the addition of increasing concentrations of histone peptides, arises from the association between them. The dissociation constant determined corroborates the results from the pulldown assay. A significant interaction was scored with histone H3K4Me3(1–25) (Kd ∼6.7 μm); in contrast, a weaker interaction could be detected with the remaining histones (Fig. 1E and Table 1). In addition to evaluation of the dissociation constant for the TCF19-H3K4Me3 complex, this observation further indicates either a change in conformation of TCF19 or alteration in the electronic environment of the tryptophan residue(s) upon its association with H3K4Me3 peptide. The specificity of binding of the PHD finger with the lysine trimethylation was also reflected in peptide pulldown based on the dissociation constant of the interaction (Kd). The pulldown and fluorescence titration showed consistent results (Fig. 1E, bottom). The interaction was found to be specific for the H3K4Me3 modification only, and other methylations showed marginal binding, as shown biochemically (Fig. 1F).
Table 1.
Apparent dissociation constant was obtained from steady-state fluorescence spectroscopy
At 25 °C, the Kd values were averaged over three separate titration experiments, with error calculated as S.D. between runs.
| Peptides | Kd |
|---|---|
| μm | |
| H3K4Me3 | 6.7 ± 1.2 |
| H3K4Me2 | 31.2 ± 3.6 |
| H3K4Me1 | NDa |
| H3(1–21) | ND |
| H4(1–21) | 33.6 ± 1.7 |
a ND, not determined.
Sequence alignment shows sequence similarity of TCF19 with other H3K4Me3-binding proteins (Fig. 1G). Along with the zinc-coordinating sites, tryptophan of TCF19 at the 316 position appears to be a highly conserved residue. To understand TCF19-PHD and histone H3K4Me3 interaction and to determine the key residues mediating it, the homology model of TCF19-PHD finger was generated using JARID1B-PHD (Jumonji/ARID domain-containing protein 1A) as a template. The negative and low value of free energy of binding indicates a strong favorable interaction between TCF19-PHD finger and H3K4Me3 peptide in most favorable conformations. The docking revealed that the tryptophan residues of the TCF19 PHD domain (positions 307 and 316) are in close proximity to the lysine 4 trimethyl moiety, hence indicating their importance in recognition of the modification (Fig. 1H).
Individual contribution of the two Trp residues (Trp-316 and Trp-307) of TCF19-PHD in H3K4Me3 recognition was further investigated through site-directed mutagenesis. Whereas W316A shows no interaction with either unmodified histone H3 or H3K4Me3, W307A behaved like wild-type protein (Fig. 1I). These results clearly indicate that the Trp-316 residue in the PHD finger of TCF19 plays a critical role in histone H3K4Me3 recognition.
Endogenous H3K4Me3 was also co-immunoprecipitated with TCF19 from HepG2 cells, only under high-glucose conditions (Fig. 1J). Under low glucose conditions, TCF19 showed a weaker interaction with endogenous H3K4Me3 (supplemental Fig. S3). Similar interaction of TCF19 with other trimethylated histone H3 modifications (H3K9Me3, H3K27Me3, and H3K36Me3) could not be detected in either normal- or high-glucose conditions. The results confirm the ability of TCF19 to show selective interaction with H3K4Me3 in a cellular context.
TCF19 controls gluconeogenic gene expression and influences critical cellular pathways in high-glucose conditions
We have observed a selective interaction of TCF19 with H3K4Me3 in high-glucose conditions. This led us to explore the expression levels of TCF19 and its epigenetic interactor in a differential glucose milieu. To check for the expression level and stability of TCF19 in high-glucose conditions, HepG2 cells were maintained in 40 mm glucose for 48 h and then shifted back to low glucose level (5.5 mm) and thereafter maintained in that condition for 3 consecutive days. NF-κB was used as a positive control for high-glucose stress, and upon high-glucose induction, its protein level increased. TCF19 protein levels were markedly increased in a high-glucose environment too but subsequently came down to basal level when cells were shifted to low-glucose conditions. No further decrease in protein level was observed in consecutive days of low glucose incubation, unlike NFκB, whose expression showed a dramatic reduction and then subsequently increased to attain a basal level upon transfer to a low-glucose medium (Fig. 2A). Altered metabolic state of the cells also affects the histone post-translational modifications, thus directly influencing transcription regulation and signal transduction (23). We observed a significant increase in H3K4Me3 levels when cells were shifted from low-glucose (5.5 mm) to high-glucose conditions (40 mm). Notably, the other histone modifications did not show significant alteration (Fig. 2B). This result corroborates our observation that TCF19, which is also endogenously overexpressed in high-glucose conditions, stoichiometrically interacts with increased levels of H3K4Me3 under the same conditions.
Figure 2.
Regulation of metabolic processes by TCF19. A, HepG2 cells were cultured in LG conditions of 5 mm glucose, followed by HG exposure at 40 mm for 48 h, and subsequently reverting to the LG state for 2, 3, and 4 days. Cell lysates were probed with anti-TCF19, anti-NF-κB (as a positive control for high-glucose conditions), and β-actin (loading control). Below, quantification of protein levels are expressed as -fold change over loading control. Nor. Int., normalized intensity. B, alteration in histone H3 trimethylation status in HepG2 cells maintained at LG (5.5 mm glucose) and HG (40 mm) conditions. Numerical values below each band represent normalized values of protein level over β-tubulin. C, heat maps of expression values for differentially expressed genes on TCF19 knockdown under high-glucose conditions (p ≤ 0.05, -fold change ≥ 1.5). Down-regulated genes are marked in green, and up-regulated genes are marked in red. D, effect of knockdown of TCF19 in HepG2, Huh7, and HepaRG cells on core gluconeogenic genes. TCF19 siRNA was transfected in the above-mentioned cells under low-glucose conditions (a) and high-glucose conditions (b). Non-targeting siRNA was used as a negative control, and 18S rRNA was used for normalization. Experiments were repeated three times, and error bars represent S.D. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01). E, table showing a list of enriched GO terms, common between BinGO and DAVID analysis tools. Genes from TCF19 knockdown (high-glucose conditions) with p ≤ 0.05 and -fold change ≥ 1.5 were used for analysis. F, selected GO categories showing the highest enrichment of differentially expressed genes from TCF19 knockdown in high-glucose conditions, analyzed by the PANTHER classification tool.
To understand the role of TCF19 in modulation of global gene expression in normoglycemia (LG)/hyperglycemia (HG), we knocked down TCF19 levels in HepG2 cells by TCF19 siRNA in the presence of low or high glucose and performed microarray analysis. The level of depletion of TCF19 mRNA and protein (in normal and high-glucose conditions) is shown in supplemental Figs. S4 and S5, respectively. The cluster diagram for TCF19 knockdown in high-glucose conditions is shown in Fig. 2C. A large number of differentially expressed genes (DEGs) were obtained from the microarray data (23 in low-glucose conditions and 3035 in high-glucose conditions) (p ≤ 0.05, -fold change ≥ 1.5). In an effort to identify various biological processes associated with the DEGs obtained, we performed gene ontology enrichment analysis using a gene ontology (GO) enrichment tool, BinGO, which enumerates the statistically enriched GO processes from the list of DEGs and forms a cytoscape network based on it. Important biological process networks like the cell cycle network (supplemental Fig. S6A), metabolic network (supplemental Fig. S6B), transcription control network (supplemental Fig. S6C), and insulin signaling network (supplemental Fig. S6D) were found to be differentially expressed upon silencing TCF19. Within the metabolic network, the process of gluconeogenesis was found to be highly dysregulated upon TCF19 depletion. Gluconeogenesis is activated in a low-glucose environment to maintain the energy requirement of cells. However, with increased glucose flux in cells, gluconeogenesis is stopped, as cells can get a direct ATP supply by breaking down glucose. Abnormal hepatic gluconeogenesis in diabetic patients contributes to high concentration of circulating blood glucose (24). We have monitored the expression of three key gluconeogenic enzymes, G6PC, FBP1, and PCK1, upon depletion of TCF19 in a low- or high-glucose environment in HepG2, Huh7, and HepaRG cells. Absence of TCF19 has a comparatively more significant effect on gluconeogenic genes in high-glucose conditions (Fig. 2D, a and b). Concordant results after TCF19 knockdown in two different hepatoma cell lines (HepG2 and Huh7) and a primary hepatocyte-like cell line (HepaRG) indicate that repression of gluconeogenic gene expression by TCF19 is a hepatic phenomenon, rather than a cell-specific event. Using other GO-based tools like DAVID and PANTHER, we were able to tabulate the most significant (in terms of p value) and common GO processes enriched on depletion of TCF19 in high-glucose conditions (Fig. 2, E and F). Overall, the above results indicate that TCF19 helps to shut down expression of gluconeogenic genes in HepG2 cell lines under high-glucose conditions.
PHD finger is essential for TCF19 to modulate expression of gluconeogenic genes
Gluconeogenic genes are subjected to a tight hormonal regulation by cAMP and insulin. To understand the role of TCF19 in regulating expression of the gluconeogenic genes in the presence of known activators (cAMP/dexamethasone) and repressor (insulin), full-length TCF19 FLAG construct was transfected in HepG2 cells. Transfected cells were incubated in the presence or absence of the above-mentioned gluconeogenesis activators or repressors (Fig. 3A). Relative mRNA level of the three key gluconeogenic genes showed a significant decrease in full-length TCF19–overexpressed conditions, even in the presence of cAMP/dexamethasone. The presence of insulin in TCF19-overexpressed conditions decreases the mRNA levels for all three genes even further. Under TCF19 ΔPHD-overexpressed conditions, however, the inhibitory effect of TCF19 was not significant, and interestingly, the addition of insulin in this case also did not produce significant transcription repression of gluconeogenic genes. Treatment of HepG2 cells with gluconeogenic repressor under TCF19-depleted conditions also failed to repress gluconeogenic genes (Fig. 3B). We also replicated our results in Huh7 cells, another well-studied glucocorticoid-responsive cell line for metabolic studies. Similar results were also obtained in Huh7 cell lines (Fig. 3, C and D). A similar experimental setup was used in HepG2 and Huh7 cells, where forskolin was used instead of cAMP as a gluconeogenic activator. The results show that the mode of transcription regulation by TCF19 of gluconeogenic genes is independent of the type of activator used (supplemental Fig. S7, A–D). To negate any abnormal metabolic phenotype pertaining to the malignant state of both the HepG2 and Huh7 cell lines, we used differentiated HepaRG cells as a substitute model for primary human hepatocytes. HepaRG cells were cultured under conditions to induce its differentiation to primary hepatocytes for 21 days (25–27) (Fig. 3E, a and b). Increased mRNA levels of hepatocyte differentiation markers apolipoprotein B (APOB), albumin (ALB), and hepatocyte nuclear factor 4α (HNF4α) (28) after 21 days indicated substantial primary cell-like characteristics (Fig. 3E, c). Similar to HepG2 and Huh7 cells, gluconeogenic gene transcription was significantly decreased on full-length TCF19 overexpression in differentiated HepaRG cells, and the addition of insulin further contributed to the inhibitory effect (Fig. 3F). The results thus indicate that gluconeogenic gene suppression by TCF19 is a consistent hepatocyte phenomenon. Morphological changes in HepaRG cells upon differentiation at different time points are represented in supplemental Fig. S8 (A–D).
Figure 3.
TCF19 inhibits transcription of key gluconeogenic genes. A, transcriptional activity of three key gluconeogenic genes in HepG2 cells (G6PC, FBP1, and PCK1) was quantified in the presence of known activators (dexamethasone/cMAP) and repressor (insulin), either singly or in combination, with overexpression of either full-length TCF19 (TCF19 FL) or TCF19 ΔPHD FLAG-tagged constructs. After 16 h of transfection, cells were treated with the above-mentioned activator and repressor for 6 h in glucose production buffer. -Fold change was calculated over mRNA levels of HepG2 cells under low-glucose medium, and 18S rRNA was used for normalization. Experiments were repeated three times, and error bars represent S.D. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01). B, transcription levels of the three gluconeogenic genes were quantified by real-time PCR with mRNA isolated from HepG2 treated with TCF19 siRNA, under the influence of gluconeogenic activator (Dexamethasone/cMAP) or repressor (Insulin) either singly or in combination. -Fold change calculated over mRNA levels of HepG2 cells treated with scrambled siRNA and 18S rRNA was used for normalization. Experiments were repeated three times, and error bars represent S.D. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01). C, similar experiments as in A were repeated in Huh7 cells under gluconeogenic activator or repressor influence, transfected with either FLAG-tagged full-length TCF19 or TCF19ΔPHD construct. D, transcriptional activity of the gluconeogenic genes were quantified in Huh7 cells under the influence of gluconeogenic activator or repressor after TCF19 siRNA transfection. E, HepaRG cells were chosen as a non-cancerous model to study the effect of TCF19 on gluconeogenesis. HepaRG cells were cultured under specialized conditions to induce differentiation to primary-like cells. A difference in cell morphology was observed between cells after 24 h of plating (a) and after 21 days of culture to induce differentiation (b). Images were acquired in an Invitrogen EVOS FL cell imaging system, under 20× magnification. c, mRNA levels of hepatocyte differentiation markers were checked for cells at day 21 versus cells at day 1. APOB, apolipoprotein B; ALB, albumin; HNF4α, hepatocyte nuclear factor 4α. 18S rRNA was used for normalization. Experiments were repeated three times, and error bars represent S.D. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01). F, expression of the key gluconeogenic genes were quantified in differentiated HepaRG cells, transfected with FLAG-tagged full-length TCF19 protein under the influence of gluconeogenic activators dexamethasone and cAMP (Dex/cAMP) either singly or in combination with repressor insulin. G, firefly luciferase reporter construct containing an upstream G6PC promoter region was used to assess promoter binding affinity of full-length TCF19 or ΔPHD constructs under the influence of gluconeogenic activators or repressors. H, G6PC reporter construct containing critical mutations at the insulin response element in the promoter region was used to assess the specificity of TCF19 for insulin-mediated recruitment at the G6PC promoter. Full-length TCF19 or TCF19 ΔPHD was transiently transfected into HepG2 cells, and a luciferase assay was performed after the indicated treatments. The activities are shown as mean -fold enhancement compared with the empty vector after normalization with Renilla luciferase activity. Each transfection was performed in triplicate, and the experiments were repeated three times. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01).
Based on these findings, we made an effort to determine whether TCF19 binds to the promoter-proximal region of G6PC. Luciferase assay was conducted on lysate from HepG2 cells containing a test vector (G6PC proximal promoter cloned upstream of firefly luciferase gene) and Renilla luciferase construct as control. To assess the role of PHD finger in recruitment of TCF19 on the G6PC promoter region, full-length TCF19 and TCF19ΔPHD were overexpressed independently. Even under the influence of cAMP/dexamethasone, the full-length protein displayed remarkable ability to repress the G6PC promoter activity. TCF19-mediated repression was enhanced in the presence of insulin. In the case of the PHD finger truncated protein, the repressive ability, although present, was compromised, and the addition of insulin had no synergistic effect on the repression (Fig. 3G). The G6PC promoter region contains cis-acting sequences known as insulin response elements (IREs), which confer the transcription regulatory of insulin (29). To ascertain whether TCF19 has any affinity for the IRE, we further performed a luciferase assay with the G6PC promoter region containing key G to C nucleotide mutations in the IRE at sites −187 (IRS1) and −173 (IRS2) and an A to C nucleotide mutation at −165 (IRS3) position from the transcription start site, which render it insulin-unresponsive (30). The results clearly indicate that the mutated promoter is unresponsive to overexpressed levels of TCF19, in either the presence or absence of insulin (Fig. 3H). This observation suggests that TCF19 specifically binds to the insulin response sequences under the influence of insulin to exert its inhibitory effect on gluconeogenic genes.
TCF19 inhibits endogenous glucose production in HepG2, Huh7, and differentiated HepaRG cell lines
Following earlier observations of TCF19 inhibiting the transcription levels of key gluconeogenic genes, we went ahead and performed a glucose production assay to investigate the effect of TCF19 on endogenous glucose production by hepatic cell lines. Under full-length TCF19 overexpression conditions, gluconeogenesis was compromised in HepG2 cells, even in the presence of activators like cAMP/dexamethasone. The addition of insulin led to a further decrease in glucose production under these conditions. Conversely, truncation of the PHD domain did not alter the glucose production to a similar extent, and the presence of insulin showed no synergistic effect (Fig. 4A). Silencing of TCF19 led to an increase in glucose output by HepG2 cells, even in the presence of insulin (Fig. 4B). Similar observations were obtained in the Huh7 cell line, under identical treatment conditions (Fig. 4, C and D). A glucose production assay was performed in differentiated HepaRG cell line as well, under full-length TCF19–overexpressed conditions. Overexpression of TCF19 compromised glucose production in HepaRG cells as well, and the synergism between insulin and TCF19 was also visible here (Fig. 4E). The glucose production assay was also repeated in HepG2 and Huh7 cells, using forskolin instead of cAMP as an gluconeogenesis activator. A similar inhibitory role of TCF19 was observed in both cell lines (supplemental Fig. S9, A–D). Because our observations indicate that TCF19 responds to hormonal activators and repressors of gluconeogenesis, we further proceeded to investigate the occupancy of TCF19 at gluconeogenic gene promoters under the influence of either cAMP/dexamethasone (activator) or insulin (repressor). The results in Fig. 4F clearly show that TCF19 is significantly enriched in the gluconeogenic gene promoters under the influence of insulin. Thus, the results highlight the unique role of TCF19 in mediating repression of gluconeogenic genes under the influence of high glucose as well as insulin.
Figure 4.
TCF19 inhibits endogenous glucose production in cancerous (HepG2 and Huh7) and primary-like (differentiated HepaRG) hepatocytes. A, a glucose production assay was done to measure endogenous glucose production in HepG2 cells under overexpression of either TCF19-FLAG or TCF19 (ΔPHD)-FLAG by transient transfection. Cells were maintained in glucose production buffer and treated with 5 μm dexamethasone + 10 mm cAMP with or without 100 nm insulin for 8 h. B, glucose production was also measured in HepG2 cells under TCF19-depleted conditions (TCF19 siRNA transfected versus scrambled siRNA). Cells were maintained in glucose production buffer and treated with 5 μm dexamethasone + 10 mm cAMP with or without 100 nm insulin for 8 h. Similar experiments were replicated in Huh7 cells under FLAG TCF19 overexpression (full-length or ΔPHD) (C) or TCF19 depletion (TCF19 siRNA) (D). E, a glucose production assay was also performed in differentiated HepaRG cells following the same protocol under FLAG full-length TCF19 overexpression. F, TCF19 occupancy was investigated at the gluconeogenic gene promoters in HepG2 cells treated with gluconeogenic activator (5 μm dexamethasone + 10 mm cAMP) or in combination with repressor (100 nm insulin). NAV1.2 gene promoter was used as a control non-gluconeogenic gene promoter. Values are shown as S.D. of biological triplicate in each case. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01).
TCF19 binds to promoters of gluconeogenic genes in high-glucose conditions, along with NuRD complex
Previous reports have shown that repression of gluconeogenic gene transcription is mediated by NuRD (nucleosome-remodeling-deacetylase) complex recruitment (31), but questions regarding NuRD recruitment and the precise mechanism of resulting epigenetic changes were unanswered. To determine whether TCF19 associates with NuRD components, we studied chromatin occupancy of TCF19 and NuRD members: MTA1, CHD4, and HDAC1. ChIP was performed on HepG2 cell lysate maintained in either low- or high-glucose conditions. Interestingly, we observed a significant increase in the occupancy of TCF19 upon high-glucose induction in the gluconeogenic gene promoters. The NAV1.2 gene promoter proximal region was used as a non-glucose-responsive negative control. Upon high-glucose induction, there was no significant recruitment of TCF19 in the NAV1.2 promoter site. A similar increased recruitment of CHD4, MTA1, and HDAC1 was also observed (Fig. 5, A–D). To ascertain that TCF19 is essential for recruitment of CHD4 to gluconeogenic promoters, CHD4 occupancy in the above promoters was evaluated after TCF19 knockdown in HepG2 cells in high-glucose conditions. Knockdown of TCF19 resulted in reduced occupancy of CHD4 on the three gluconeogenic gene promoters (Fig. 5E). Co-immunoprecipitation assays to detect the TCF19-CHD4 complex were performed, in HepG2 cells, in both low- and high-glucose conditions (Fig. 5F). Immunoprecipitating TCF19, CHD4 was detected, and vice versa. Changes in glucose level apparently had no effect on the binding.
Figure 5.
TCF19 associates with NuRD complex and regulates gluconeogenic genes. ChIP assays were done in HepG2 cells maintained under low-glucose conditions (5 mm glucose) or high-glucose conditions (40 mm glucose). As a template for recruitment, ∼500 bp of the region upstream of the transcription start site of each of the three key gluconeogenic genes was selected, and previously reported primers were used. pp, proximal promoter region. The 3′-UTRs of individual genes were used as a negative control. NAV1.2 promoter is used as a glucose–non-responsive control for TCF19. Recruitment of TCF19 was found to be enhanced in high-glucose conditions (A). High-glucose conditions also resulted in increased chromatin occupancy of CHD4 (B), MTA1 (C), and HDAC1 (D), which are components of the NuRD complex. E, binding of CHD4 to promoter region was reduced on siRNA-mediated knockdown of TCF19 in HepG2 cells under high-glucose conditions. Values are shown as S.D. of biological triplicates in each case. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01). F, TCF19 and CHD4 interact with each other. Endogenous IP was done in HepG2 cells in both LG and HG conditions. IP with anti-CHD4 was probed for TCF19, and IP with anti-TCF19 was probed for CHD4. IgG used as negative control in both cases.
TCF19 knockdown under high-glucose conditions leads to activation of the gluconeogenic gene transcription via activating histone modifications
Under high-glucose conditions, enrichment of nucleosome was observed in the gluconeogenic promoters, which is indicative of a repressed chromatin state (Fig. 6, A and B). The H3K4Me3 mark was surprisingly unaltered following the glycemic shift (Fig. 6C). Consequently, under high-glucose conditions in the same promoter region, a significant reduction in occupancy was noticed in the case of H4K5Ac and H4K8Ac modifications, which are mostly found in the euchromatin region. The reduction in acetylation is probably due to NuRD recruitment to the promoter region (Fig. 6, D and E). We also tested the gluconeogenic gene promoters for the H3K9Ac mark, which is a well-documented mark for activation of gluconeogenic genes (32). Under high-glucose conditions, occupancy of H3K9Ac was also reduced, indicating a repressive state at the promoter (Fig. 6F). The results are concordant with previous reports that show how the acetylation marks are sensitive to metabolic environment, whereas methylation marks remain stable (33).
Figure 6.
A and B, hyperglycemia induced changes in histone modifications at the gluconeogenic gene promoters. A and B, increased chromatin occupancy of H3 and H4 unmodified histone, indicating chromatin compaction on increasing glucose concentration. C, no significant change was observed for the H3K4Me3 mark in the target promoter region upon high-glucose shift. D–F, depletion of activation marks at the promoter site; H4K8Ac, H4K5Ac, and H3K9Ac also indicate overall chromatin compaction. pp, proximal promoter region. The 3′-UTRs of individual genes were used as a negative control. Values are shown as S.D. of biological triplicates in each case. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01).
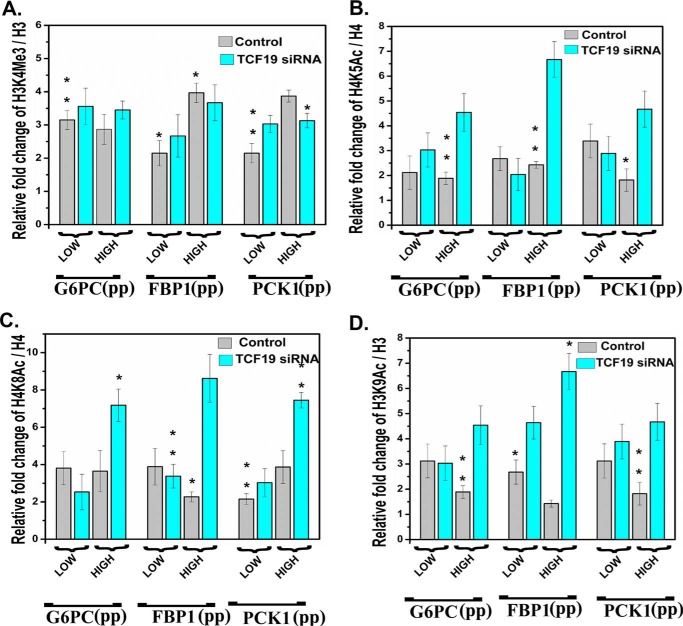
Taking a cue from the above results, we proceeded to further validate the connection between recruitment of TCF19 at the gluconeogenic promoter sites and the change in histone post-translational modifications at that locus. Accordingly, we checked the occupancy of H3K4Me3, H4K5Ac, H4K8Ac, and H3K9Ac at the promoter locus, under TCF19 depletion in both low-glucose and high-glucose conditions. TCF19 depletion had no significant effect on H3K4Me3 under either normal- or high-glucose conditions (Fig. 7A). But under high-glucose conditions only, depletion of TCF19 led to an increased occupancy of activating histone marks H4K5Ac (Fig. 7B), H4K8Ac (Fig. 7C), and H3K9Ac (Fig. 7D). These observations clearly indicate that TCF19 is instrumental in maintaining the repression of gluconeogenic genes under high-glucose conditions specifically.
Figure 7.
TCF19 depletion in HepG2 cells leads to increase in histone acetylation at gluconeogenic promoters. siRNA-mediated depletion of TCF19 in HepG2 cells maintained in either low-glucose or high-glucose conditions causes no significant alteration of the H3K4Me3 mark at the promoter region of the key gluconeogenic genes (A), whereas activation marks like H4K5Ac (B), H4K8Ac (C), and H3K9Ac (D) were significantly increased upon TCF19 knockdown in high-glucose conditions, indicating derepression of the gluconeogenic genes. pp, proximal promoter region. Unpaired Student's t test was used to determine p value (*, p < 0.05; **, p < 0.01).
Discussion
Blood glucose homeostasis is established by a fine balance of transcriptional regulation by specific hormones and circulating nutrients. Especially, expression of glycolytic, gluconeogenic, and lipolytic enzymes is regulated by the availability of circulating carbohydrates as well as important pancreatic hormones, insulin and glucagon (5, 6). Hormones (insulin and glucagon) and metabolites (e.g. glucose) exert their regulatory effect via a number of transcription factors (34, 35). Important metabolic genes like FAS, l-pyruvate kinase, PCK, G6PC, FBP1, and ACC (acetyl-CoA carboxylase) are controlled either by independent action of insulin and glucose or by the presence of both (2, 36). One of the major contributors of high blood glucose in long term type 2 diabetes is dysregulated elevation of gluconeogenesis in the liver (24, 37), and key enzymes regulating glucose flux in this reaction are PCK1, FBP1, and G6PC. Besides several signal-dependent transcription factors that have been implicated in regulation of gluconeogenesis in fasting and feeding states, epigenetic changes also have a major role in controlling assembly of transcription machinery. Dysregulation of these changes has been shown to contribute to diabetes pathology. Epigenetic marks like H3K4Me3, H4K5Ac, H4K8Ac, and H3K9Ac have been shown to effect the transcription of gluconeogenic genes directly and control physiological glucose flux (32, 38). Therefore, interpretation of these marks through appropriate reader proteins is essential in maintaining glucose homeostasis.
The current study demonstrates a novel function of PHD finger–containing protein TCF19 in transcriptional regulation of gluconeogenic genes. PHD finger is a versatile epigenome reader implicated in transcription regulation (18). One of the key modifications recognized by PHD finger is lysine 4 trimethylation at histone H3, which is a transcription activation mark. Reports in literature suggest that PHD finger can also mediate transcription repression through H3K4Me3 recognition (39). ING2 (inhibitor of growth) tumor suppressor protein, in the case of humans, has been shown to interact with the H3K4Me3 modification at the promoters of proliferation genes and recruit mSin3a-HDAC1 complex, to mediate active gene repression in response to DNA damage (16). In the case of yeast, the Set3 histone deacetylase complex (Set3C) has also been reported to be recruited to H3K4Me2-rich promoter regions of meiotic genes and repress their transcription (30). Although PHD fingers have been implicated in cell proliferation, recombination, DNA damage response, etc., which are mostly associated with cancer (40), no previous reports of PHD finger–mediated metabolic disorders have been elucidated. Here we show that TCF19 interacts with an activating histone modification, thereby recruiting NuRD corepressor complex at gluconeogenic gene promoters, leading to their repression upon high-glucose conditions. However, the mechanism of selective recognition of TCF19 to the gluconeogenic gene promoters requires further investigation.
TCF19 has been implicated as an important candidate for T2D in recent genome-wide association studies (41). Our microarray results showed differential regulation of genes involved in various metabolic pathways (like gluconeogenesis, insulin signaling, nucleotide metabolism, etc.). Here we have explored the role of TCF19 in de novo glucose production from the perspective of a histone-interacting protein. TCF19 protein level in HepG2 cells shows a strong and positive correlation with glucose concentration of culture medium, which is indicative of it being a glucose-sensing transcription regulator.
Simultaneous increase in occupancy of TCF19 and H3/H4 indicates chromatin compaction. Subsequent H4 deacetylation due to NuRD recruitment via TCF19 also emphasizes the repressive role of TCF19. Interestingly, this is the first report that elucidates NuRD recruitment to gluconeogenic genes through a PHD finger association with an activating histone post-translational modification. Knockdown of TCF19 in HepG2, Huh7, and HepaRG cell lines resulted in increased expression of key gluconeogenic genes. Glucose production in the above-mentioned cell lines was reduced by a synergistic repressive effect of TCF19 overexpression and insulin treatment. The PHD domain truncated protein displayed a lack of synergism with insulin; hence, no significant change in glucose output was observed. We speculate that the PHD finger possibly acts as a tether for the protein to identify and bind to a preferred chromatin state. The rest of the protein acts as a scaffold to stabilize the interaction and possibly help in recruitment of the accessory repressive factors to chromatin. Our results, therefore, describe a previously unexplored importance of PHD fingers in physiological glucose production. These results also hint toward a unique role of TCF19 in integrating regulatory signals from both circulating metabolites and hormonal signals, to control gluconeogenesis. Significantly, our gene expression analysis clearly indicates that TCF19 has an important contribution in several other key biological processes, including cell cycle, transcription regulation, nucleotide metabolism, carbohydrate metabolism, protein localization, etc. In conformity, a recent study has acknowledged the role of TCF19 in diverting glycolytic intermediates to other biosynthetic pathways producing protein, nucleotides, lipids, and carbohydrates (42).
Taken together, our study demonstrates for the first time the remarkable level of versatility and functional heterogeneity of TCF19, as dictated by the PHD finger. To date, only a few studies have demonstrated the link between PHD finger-H3K4Me3 identification and transcription repression (16, 43). Our study answers several mechanistic questions regarding transcription regulation of gluconeogenesis and elucidates the importance of chromatin readers in metabolic homeostasis and disease conditions. Further exploration of functional links between histone modifications, glucose homeostasis, and diabetes is bound to shed light on the importance of chromatin readers as pharmacological targets for diabetes medication and provide future therapeutic options.
Experimental procedures
Protein purification and site-directed mutagenesis
Competent bacterial cells, after transformation, were grown until OD reached 0.6, and 1 m isopropyl 1-thio-β-d-galactopyranoside was used to induce the culture, followed by overnight incubation in a shaker incubator at 20 °C and 180 rpm. The lysis buffer used contained 20 mm Tris-HCl (pH 8), 150 mm NaCl, 0.05% Nonidet P-40, 1 mm DTT, 2 mm PMSF, 1× protease inhibitor mixture (EDTA-free). After sonication, the lysate was spun down twice at 13,000 rpm at 4 °C for 30 min. Glutathione beads were added to the supernatant and incubated for 2 h at 4 °C. The beads were separated by centrifugation and washed with a buffer containing 20 mm Tris-HCl (pH 8), 500 mm NaCl, 1 mm DTT. The bead-bound proteins were eluted with elution buffer (20 mm Tris-HCl (pH 8), 150 mm NaCl, 100 mm reduced glutathione, 2 mm DTT). For a further concentration of eluted protein, Amicon Filter tubes (Millipore) was used, and a Superdex75 column (GE Healthcare) was used for further purification. The point mutation was generated using the QuikChange 2 site-directed mutagenesis kit from Agilent (catalog no. 200523) as per standard protocol (44).
Peptide pulldown assay
Assays were performed following standard protocols (45). Briefly, GST-tagged protein was incubated with biotinylated histone peptides in IP buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.05% Nonidet P-40, 1 mm DTT) at 4 °C overnight. The following day, streptavidin bead interaction was performed for 2 h at 4 °C followed by washes with IP buffer, and the bead-bound complex was analyzed by Western blotting.
Commercial peptides procured from Anaspec and details are provided in supplemental Table S1. Antibodies used in the subsequent Western blotting are listed in supplemental Table S2.
Protein–protein interaction studies
Standard protocols were followed (46). Briefly, proteins were incubated in equal molar ratio in binding buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.05% Nonidet P-40, 1 mm DTT, and the reaction mixture was subjected to pulldown by preblocked glutathione-Sepharose beads. Washing was done in binding buffer, and the protein complex was analyzed by Western blotting.
Fluorescence spectroscopy and analysis of the binding data
The fluorescence measurements were performed in a Perkin-Elmer LS 55 luminescence spectrometer at 25 °C. Increasing concentrations of the indicated peptides were incubated with 0.5 μm TCF19 PHD (GST tag–cleaved) dissolved in buffer containing 20 mm Tris-HCl, pH 8, 150 mm NaCl, 10 μm ZnCl2, 0.01% Tween 20, 1 mm DTT. The protein contains two tryptophan residues, and the peptides do not have any tryptophan residue. Therefore, the association between them could be checked and quantitatively monitored by fluorescence spectroscopy. Excitation of the tryptophan residues in the protein (Trp-316 and Trp-307) was carried out at 295 nm. The emission spectrum of free protein in the presence of the peptide(s) was monitored in the range 310–400 nm. Emission intensity at 345 nm was monitored as a function of increasing peptide concentration until no change in fluorescence intensity was observed. The final data were averaged over three scans, and for each spectrum, the corresponding buffer baseline was subtracted. Binding analysis of the fluorimetric titrations was carried out using non-linear curve fitting analysis, as reported in earlier literature (47, 48).
Molecular modeling and docking studies
A homology model of TCF19-PHD domain was generated using the C-terminal PHD finger of Jumonji/ARID domain-containing protein 1A (Protein Data Bank entry 2KGI) as a template, as it has 50% sequence similarity to TCF19-PHD. Modeling was done using SWISS-MODEL (49). The structural quality of the homology model was assessed using PROCHECK via the Ramachandran plot. Geometry was investigated by WHATIF (50). Structure was visualized using PyMOL. Docking with H3K4Me3 peptide was done with the FlexPepDock server (51).
Western blotting
Whole-cell extracts were prepared with Laemmli buffer (4% SDS, 20% glycerol, and 120 mm Tris-HCl, pH 6.8) and sonicated followed by boiling at 100 °C. The samples were analyzed by a standard Western blot. Primary antibody used has been listed in supplemental Table S2.
Cell culture and treatment
HepG2, HEK293, and Huh7 cells were maintained in Dulbecco's modified Eagle's medium (Gibco). All cell lines were supplemented with 10% fetal bovine serum (Gibco) and penicillin/streptomycin (10 μl/ml of medium, Gibco) at 37 °C in 5% (v/v) CO2.
For high-glucose induction, HepG2 and Huh7 cells were seeded in DMEM overnight, and for the following 2 days, fresh medium was supplemented with 40 mm glucose. To create gluconeogenic conditions in the indicated experiments, cells were maintained in DMEM supplemented with 10% FBS overnight, and at 90% confluence, medium was replaced with glucose production buffer (serum, glucose, phenol red–free DMEM (Invitrogen), supplemented with 20 mm sodium lactate, and 2 mm sodium pyruvate). Treatment was done for the indicated time periods with dexamethasone (5 μm), dibutyryl cAMP (10 mm), and insulin (100 nm). In the case of forskolin treatment, 10 μm forskolin (dissolved in DMSO) treatment was used for 16 h, and results were compared with DMSO-treated control. All cell culture additives were procured from Sigma. Because dexamethasone was dissolved in DMSO, results from dexamethasone-treated cells were normalized with DMSO-treated control cells. HepaRG cells were procured from Gibco and cultured in William's E medium, containing HepaRG thaw and plate general purpose medium supplement (Thermo Fisher Scientific) and Glutamax (Gibco). Medium was also supplemented with penicillin/streptomycin (10 μl/ml of medium, Gibco). After plating, HepaRG cells were maintained as reported previously (26, 52). Differentiated HepaRG cells were used in further experiments. High-glucose induction of HepaRG cell lines was done in William's E medium, in a similar manner as HepG2 and Huh7.
ChIP
ChIP assays were performed as per standard protocol (54). Briefly, 1% formaldehyde was used for cross-linking, and 0.125 m glycine was used to stop the reaction. Cells were lysed using cell lysis buffer (5 mm PIPES, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40 supplemented with protease inhibitor mixture (Roche Applied Science)), followed by nuclear lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS supplemented with protease inhibitor). After ultrasound shearing, preclearing was done with normal sheep serum. Antibodies used for immunoprecipitation are listed in supplemental Table S2. Preblocked DYNA beads were added for binding to pulled chromatin complex. Beads were washed with buffers in the following order: radioimmune precipitation assay buffer, high-salt buffer, LiCl buffer, and Tris-EDTA. After RNase A and proteinase K treatment, the beads were kept for decross-linking at 65 °C. DNA was extracted using the phenol-chloroform method and dissolved in deionized water. It was stored at 4 °C until further analysis by quantitative RT-PCR. Previously reported primers were used for the G6PC, PCK1 (55), and FBP1 (31) promoter regions. All experiments were performed three times, and error was calculated as ±S.D. between biological triplicates.
Co-IP
Previously established protocols were followed for co-immunoprecipitation (56). Briefly, cross-linked cells were lysed using a buffer containing 50 mm HEPES (pH 7.5), 150 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 0.5% sodium deoxycholate, 5% glycerol, 1 mm DTT, 2 mm PMSF, 1 mm trichostatin A, 5 mm sodium butyrate, and complete protease inhibitor mixture. This was followed by incubation on ice for 1 h and centrifugation at 13,000 rpm for 10 min at 4 °C. The precleared lysate was incubated with primary antibodies overnight, followed by washing with the same buffer and analysis by Western blotting. The antibodies used in the Western blots are listed in supplemental Table S2.
Gene silencing
Cells were transfected with TCF19 siRNA (sc-63113, Santa Cruz Biotechnology, Inc.) or negative control siRNA using interferin transfection reagent (Polyplus) according to the manufacturer's protocol for 24 h. RNA was extracted using the RNA-Xpress reagent (Himedia) following the manufacturer's protocol. Reverse transcription was performed with Revertaid Fast strand cDNA synthesis kit (Thermo Scientific). Previously reported real-time PCR primers were used for G6PC, PCK1 (55), and FBP1 (31). Rests of the primers were designed using the NCBI-Primer BLAST tool (supplemental Table S3).
Glucose production assay
Cellular glucose production in HepG2, Huh7, and HepaRG cells was estimated under three different conditions: overexpression of either TCF19 full-length or TCF19 ΔPHD (PHD domain–truncated) FLAG construct and TCF19-depleted conditions by specific siRNA. A previously established protocol was followed to estimate glucose production (57). Cells were washed three times with PBS to remove glucose, incubated for 16 h in 2 ml of glucose production medium (glucose- and phenol red–free DMEM containing gluconeogenic substrates, 20 mm sodium lactate, and 2 mm sodium pyruvate) and in the presence of 100 nm insulin (100 nm), dexamethasone (5 μm), dibutyryl cAMP (10 mm). A quantity of 300 μl of medium was sampled for measurement of glucose concentration using a glucose assay kit (GAGO20, Sigma). Glucose concentration was normalized with cellular protein concentration.
Reporter construct and luciferase assay
The 600-bp upstream promoter region of human glucose-6-phosphatase catalytic subunit was amplified by PCR using human genomic DNA as template and then cloned into a pGL3 basic vector (Promega) at the KpnI/HindIII site. The primer sequences used to clone the promoter region are listed in supplemental Table S3, where the restriction enzyme sites are underlined. All constructs were sequenced to confirm their identity. The mutation in the promoter region of G6PC was generated using the above construct as a template. The primer for generating the mutation is mentioned in supplemental Table S3.
A previously established protocol was used to perform transfection and the luciferase assay (47). At the end of 16 h, cells were washed with PBS three times and incubated with glucose production buffer and the indicated supplements for 6 h. Each transfection was performed in triplicate, and the experiments were repeated three times. For the luciferase assay with promoter mutated sequence, pGL3 vector containing wild-type promoter sequence was mutated using mutation primer as mentioned in supplemental Table S3.
Microarray analysis and validation
RNA isolation and microarray analysis
Total cellular RNA was extracted using TRIzol reagent (Invitrogen) as per the manufacturer's instructions, and RNA quantity and quality were determined using a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Microarray analysis
Affymetrix raw data were robust multiarray average–normalized, and baseline transformation was done to the median of all samples using GeneSpring GX version 12.5 software (Agilent Technologies Inc., Santa Clara, CA).
Statistical analysis and differentially expressed genes
Differentially expressed probe sets or genes upon TCF19 knockdown treatment in comparison with control cells were identified by applying a volcano plot using a -fold change threshold of absolute -fold change ≥1.1 and a statistically significant t test p value threshold adjusted for a false discovery rate of <0.001. Statistically significant, enriched genes with a p value adjusted for a false discovery rate <0.05 derived using the hypergeometric distribution test corresponding to differentially expressed genes were determined using a t test. Unsupervised hierarchical clustering of the differentially expressed genes upon TCF19 knockdown treatment in comparison with control was done using Pearson's centered algorithm with average linkage rule to determine the cluster of genes whose expression levels are significantly repeated across the replicates.
Gene regulatory network modeling
Statistically significant dysregulated biological categories (gene ontology and pathways) along with the genes were used as input for distinguishing the regulatory connections. The output file was imported to Cytoscape version 2.8 to visualize the connections along with modeling the network. Genes were subjected to color codes in accordance with their -fold change, and the major biological processes were highlighted to discern the gene regulation. BiNGO, a Java-based plugin for Cytoscape that determines GO categories statistically overrepresented in a set of genes, was used to generate a biological network (59). DAVID (60, 61) and PANTHER (53, 58) classification tools were also used to ascertain the functional classification of the differentially expressed genes.
The microarray data have been deposited to the GEO database and are available under accession ID GSE107471.
Author contributions
C. D. conceived the study, designed experiments, analyzed data, and wrote the manuscript. S. Sen designed the experiments, performed all experiments, analyzed data, and wrote the manuscript. D. D. and S. R. designed biophysical and biochemical experiments. S. Sanyal performed ChIP experiments. D. K. S. performed mutations and helped with a luciferase assay. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge Prof. Jessica K. Tyler (Weill Cornell Medical College) and Prof. Michelle C. Barton (MD Anderson Cancer Center) for critical comments on the manuscript. We also thank Anirban Roy of Bose Institute (Kolkata) and Dr. Moitri Basu, Debashis Mukherjee, and Payel Mondal (SINP, Kolkata) for help. We also acknowledge Dr. Suvendra Nath Bhattacharyya's laboratory at CSIR-IICB for help in performing the luciferase assay. We also thank Payal Chakraborty and Madavan Vasudevan (Genome Informatics Research and Development Unit, Bionivid Technology Pvt. Ltd.) for help in microarray data analysis.
This work was supported by a research grant entitled Biomolecular Assembly, Recognition and Dynamics (BARD) (Grant 12-R&D-SIN-5.04-0103) from the Department of Atomic Energy (DAE), Government of India (to C. D.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1–S3 and Figs. S1–S9.
The microarray data have been deposited to the GEO database and are available under accession ID GSE107471.
- G6P/G6PC
- glucose-6-phosphatase
- TCF19
- transcription factor 19
- PHD
- plant homeodomain
- FBP1
- fructose-1,6-bisphosphatase
- PCK1
- pyruvate carboxyl kinase 1
- NuRD
- nucleosome remodeling deacetylase
- CHD4
- chromodomain helicase DNA-binding protein 4
- CREB
- cAMP-response element-binding protein
- CRTC2
- CREB-regulated transcription co-activator 2
- PGC-1α
- peroxisome proliferator-activated receptor γ co-activator 1α
- HDAC
- histone deacetylase
- NAV1.2
- sodium channel, voltage-gated, type II
- IRE
- insulin response element
- T2D
- type 2 diabetes
- LG
- low glucose
- HG
- high glucose
- GO
- gene ontology
- DEG
- differentially expressed gene
- IP
- immunoprecipitation
- H3 and H4
- histone H3 and H4, respectively
- Me2 and Me3
- di- and trimethylation, respectively
- Ac
- acetylation.
References
- 1. Nordlie R. C., Foster J. D., and Lange A. J. (1999) Regulation of glucose production by the liver. Annu. Rev. Nutr. 19, 379–406 [DOI] [PubMed] [Google Scholar]
- 2. Foufelle F., and Ferré P. (2002) New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. Biochem. J. 366, 377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He L., Sabet A., Djedjos S., Miller R., Sun X., Hussain M. A., Radovick S., and Wondisford F. E. (2009) Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell 137, 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gray S. G., and De Meyts P. (2005) Role of histone and transcription factor acetylation in diabetes pathogenesis. Diabetes Metab. Res. Rev. 21, 416–433 [DOI] [PubMed] [Google Scholar]
- 5. Chalkiadaki A., and Guarente L. (2011) Metabolic signals regulate SIRT1 expression. EMBO Rep. 12, 985–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donohoe D. R., and Bultman S. J. (2012) Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell. Physiol. 227, 3169–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vaulont S., Vasseur-Cognet M., and Kahn A. (2000) Glucose regulation of gene transcription. J. Biol. Chem. 275, 31555–31558 [DOI] [PubMed] [Google Scholar]
- 8. Barrès R., Osler M. E., Yan J., Rune A., Fritz T., Caidahl K., Krook A., and Zierath J. R. (2009) Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 10, 189–198 [DOI] [PubMed] [Google Scholar]
- 9. Volkmar M., Dedeurwaerder S., Cunha D. A., Ndlovu M. N., Defrance M., Deplus R., Calonne E., Volkmar U., Igoillo-Esteve M., Naamane N., Del Guerra S., Masini M., Bugliani M., Marchetti P., Cnop M., Eizirik D. L., and Fuks F. (2012) DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 31, 1405–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirchner H., Osler M. E., Krook A., and Zierath J. R. (2013) Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends Cell Biol. 23, 203–209 [DOI] [PubMed] [Google Scholar]
- 11. Jufvas A., Sjödin S., Lundqvist K., Amin R., Vener A. V., and Strålfors P. (2013) Global differences in specific histone H3 methylation are associated with overweight and type 2 diabetes. Clin. Epigenetics 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ku D. H., Chang C. D., Koniecki J., Cannizzaro L. A., Boghosian-Sell L., Alder H., and Baserga R. (1991) A new growth-regulated complementary DNA with the sequence of a putative trans-activating factor. Cell Growth Differ. 2, 179–186 [PubMed] [Google Scholar]
- 13. Krautkramer K. A., Linnemann A. K., Fontaine D. A., Whillock A. L., Harris T. W., Schleis G. J., Truchan N. A., Marty-Santos L., Lavine J. A., Cleaver O., Kimple M. E., and Davis D. B. (2013) Tcf19 is a novel islet factor necessary for proliferation and survival in the INS-1 β-cell line. Am. J. Physiol. Endocrinol. Metab. 305, E600–E610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nair A. K., Muller Y. L., McLean N. A., Abdussamad M., Piaggi P., Kobes S., Weil E. J., Curtis J. M., Nelson R. G., Knowler W. C., Hanson R. L., and Baier L. J. (2014) Variants associated with type 2 diabetes identified by the transethnic meta-analysis study: assessment in American Indians and evidence for a new signal in LPP. Diabetologia 57, 2334–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheung Y. H., Watkinson J., and Anastassiou D. (2011) Conditional meta-analysis stratifying on detailed HLA genotypes identifies a novel type 1 diabetes locus around TCF19 in the MHC. Hum. Genet. 129, 161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., and Patel D. J. (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanchez R., and Zhou M.-M. (2011) The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai A. Y., and Wade P. A. (2011) Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alqarni S. S. M., Murthy A., Zhang W., Przewloka M. R., Silva A. P. G., Watson A. A., Lejon S., Pei X. Y., Smits A. H., Kloet S. L., Wang H., Shepherd N. E., Stokes P. H., Blobel G. A., Vermeulen M., et al. (2014) Insight into the architecture of the NuRD complex: structure of the RbAp48-MTA1 subcomplex. J. Biol. Chem. 289, 21844–21855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dreveny I., Deeves S. E., Fulton J., Yue B., Messmer M., Bhattacharya A., Collins H. M., and Heery D. M. (2014) The double PHD finger domain of MOZ/MYST3 induces α-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Res. 42, 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Y., and Grummt I. (2005) The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 15, 1434–1438 [DOI] [PubMed] [Google Scholar]
- 23. Katada S., Imhof A., and Sassone-Corsi P. (2012) Connecting threads: epigenetics and metabolism. Cell 148, 24–28 [DOI] [PubMed] [Google Scholar]
- 24. Boden G., Chen X., and Stein T. P. (2001) Gluconeogenesis in moderately and severely hyperglycemic patients with type 2 diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 280, E23–E30 [DOI] [PubMed] [Google Scholar]
- 25. Marion M.-J., Hantz O., and Durantel D. (2010) The HepaRG cell line: biological properties and relevance as a tool for cell biology, drug metabolism, and virology studies. Methods Mol. Biol. 640, 261–272 [DOI] [PubMed] [Google Scholar]
- 26. Samanez C. H., Caron S., Briand O., Dehondt H., Duplan I., Kuipers F., Hennuyer N., Clavey V., and Staels B. (2012) The human hepatocyte cell lines IHH and HepaRG: models to study glucose, lipid and lipoprotein metabolism. Arch. Physiol. Biochem. 118, 102–111 [DOI] [PubMed] [Google Scholar]
- 27. Kanebratt K. P., and Andersson T. B. (2008) Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab. Dispos. 36, 1444–1452 [DOI] [PubMed] [Google Scholar]
- 28. Saha S. K., Parachoniak C. A., Ghanta K. S., Fitamant J., Ross K. N., Najem M. S., Gurumurthy S., Akbay E. A., Sia D., Cornella H., Miltiadous O., Walesky C., Deshpande V., Zhu A. X., Hezel A. F., et al. (2014) Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature 513, 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo S., Rena G., Cichy S., He X., Cohen P., and Unterman T. (1999) Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 274, 17184–17192 [DOI] [PubMed] [Google Scholar]
- 30. Vander Kooi B. T., Streeper R. S., Svitek C. A., Oeser J. K., Powell D. R., and O'Brien R. M. (2003) The three insulin response sequences in the glucose-6-phosphatase catalytic subunit gene promoter are functionally distinct. J. Biol. Chem. 278, 11782–11793 [DOI] [PubMed] [Google Scholar]
- 31. Pan D., Mao C., and Wang Y.-X. (2013) Suppression of gluconeogenic gene expression by LSD1-mediated histone demethylation. PLoS One 8, e66294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ravnskjaer K., Hogan M. F., Lackey D., Tora L., Dent S. Y. R., Olefsky J., and Montminy M. (2013) Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. J. Clin. Invest. 123, 4318–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mews P., Zee B. M., Liu S., Donahue G., Garcia B. A., and Berger S. L. (2014) Histone methylation has dynamics distinct from those of histone acetylation in cell cycle reentry from quiescence. Mol. Cell. Biol. 34, 3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lerner R. G., Depatie C., Rutter G. A., Screaton R. A., and Balthasar N. (2009) A role for the CREB co-activator CRTC2 in the hypothalamic mechanisms linking glucose sensing with gene regulation. EMBO Rep. 10, 1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., and Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1a and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 36. Vidal-Puig A., and O'Rahilly S. (2001) Metabolism: controlling the glucose factory. Nature 413, 125–126 [DOI] [PubMed] [Google Scholar]
- 37. Magnusson I., Rothman D. L., Katz L. D., Shulman R. G., and Shulman G. I. (1992) Increased rate of gluconeogenesis in type II diabetes mellitus: a 13C nuclear magnetic resonance study. J. Clin. Invest. 90, 1323–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winkler R., Benz V., Clemenz M., Bloch M., Foryst-Ludwig A., Wardat S., Witte N., Trappiel M., Namsolleck P., Mai K., Spranger J., Matthias G., Roloff T., Truee O., Kappert K., et al. (2012) Histone deacetylase 6 (HDAC6) is an essential modifier of glucocorticoid-induced hepatic gluconeogenesis. Diabetes 61, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howe F. S., Fischl H., Murray S. C., and Mellor J. (2017) Is H3K4me3 instructive for transcription activation? Bioessays 39, 1–12 [DOI] [PubMed] [Google Scholar]
- 40. Baker L. A., Allis C. D., and Wang G. G. (2008) PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat. Res. 647, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harder M. N., Appel E. V. R., Grarup N., Gjesing A. P., Ahluwalia T. S., Jørgensen T., Christensen C., Brandslund I., Linneberg A., Sørensen T. I. A., Pedersen O., and Hansen T. (2015) The type 2 diabetes risk allele of TMEM154-rs6813195 associates with decreased beta cell function in a study of 6,486 Danes. PLoS One 10, e0120890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trapé A. P., Katayama M. L. H., Roela R. A., Brentani H., Ravacci G. R., de Araujo Lima L., and Brentani M. M. (2012) Gene expression profile in response to doxorubicin-rapamycin combined treatment of HER-2-overexpressing human mammary epithelial cell lines. Mol. Cancer Ther. 11, 464–474 [DOI] [PubMed] [Google Scholar]
- 43. Kuzmichev A., Zhang Y., Erdjument-Bromage H., Tempst P., and Reinberg D. (2002) Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 22, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu H., and Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wysocka J. (2006) Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods 40, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B.-J., Li C., Chen R., Li W., Wang Y., et al. (2011) Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D. K., Roy S., and Das C. (2016) Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 291, 2664–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chakrabarti S., Roy P., and Dasgupta D. (1998) Interaction of the antitumor antibiotic chromomycin A3 with glutathione, a sulfhydryl agent, and the effect upon its DNA binding properties. Biochem. Pharmacol. 56, 1471–1479 [DOI] [PubMed] [Google Scholar]
- 49. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Davis I. W., Murray L. W., Richardson J. S., and Richardson D. C. (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. London N., Raveh B., Cohen E., Fathi G., and Schueler-Furman O. (2011) Rosetta FlexPepDock web server: high resolution modeling of peptide–protein interactions. Nucleic Acids Res. 39, W249–W253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nibourg G. A. A., Chamuleau R. A. F. M., van der Hoeven T. V., Maas M. A. W., Ruiter A. F. C., Lamers W. H., Oude Elferink R. P. J., van Gulik T. M., and Hoekstra R. (2012) Liver progenitor cell line HepaRG differentiated in a bioartificial liver effectively supplies liver support to rats with acute liver failure. PLoS One 7, e38778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thomas P. D., Kejariwal A., Guo N., Mi H., Campbell M. J., Muruganujan A., and Lazareva-Ulitsky B. (2006) Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34, W645–W650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsai W.-W., Wang Z., Yiu T. T., Akdemir K. C., Xia W., Winter S., Tsai C.-Y., Shi X., Schwarzer D., Plunkett W., Aronow B., Gozani O., Fischle W., Hung M.-C., Patel D. J., and Barton M. C. (2010) TRIM24 links a non-canonical histone signature to breast cancer. Nature 468, 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singh B. K., Sinha R. A., Zhou J., Xie S. Y., You S.-H., Gauthier K., and Yen P. M. (2013) FoxO1 deacetylation regulates thyroid hormone-induced transcription of key hepatic gluconeogenic genes. J. Biol. Chem. 288, 30365–30372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhan X., Shi X., Zhang Z., Chen Y., and Wu J. I. (2011) Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc. Natl. Acad. Sci. U.S.A. 108, 12758–12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao D., Nong S., Huang X., Lu Y., Zhao H., Lin Y., Man Y., Wang S., Yang J., and Li J. (2010) The effects of palmitate on hepatic insulin resistance are mediated by NADPH oxidase 3-derived reactive oxygen species through JNK and p38MAPK pathways. J. Biol. Chem. 285, 29965–29973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., and Narechania A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 60. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.