Abstract
The bacterial tubulin FtsZ polymerizes to form a discontinuous ring that drives bacterial cell division by directing local cell wall synthesis. FtsZ comprises a polymerizing GTPase domain, an intrinsically disordered C-terminal linker (CTL), and a C-terminal conserved peptide (CTC). FtsZ protofilaments align circumferentially in the cell, with the CTC mediating attachment to membrane-associated division proteins. The assembly of FtsZ protofilaments into dynamic clusters is critical for cell division, but the interactions between protofilaments and regulatory mechanisms that mediate cluster assembly and dynamics are unknown. Here, we describe a role for the CTL of Caulobacter crescentus FtsZ as an intrinsic regulator of lateral interactions between protofilaments in vitro. FtsZ lacking its CTL (ΔCTL) shows a dramatically increased propensity to form long multifilament bundles compared with wild type (WT). ΔCTL also displays a reduced GTP hydrolysis rate compared with WT, but this altered activity does not account for bundle formation, as reducing protofilament turnover in WT is not sufficient to induce bundling. Surprisingly, binding of the membrane-anchoring protein FzlC disrupts ΔCTL bundling in a CTC-dependent manner. Moreover, the CTL affects the ability of the FtsZ curving protein FzlA to promote formation of helical bundles. We conclude that the CTL of FtsZ influences polymer structure and dynamics both through intrinsic effects on lateral interactions and turnover and by influencing extrinsic regulation of FtsZ by binding partners. Our characterization of CTL function provides a biochemical handle for understanding the relationship between FtsZ-ring structure and function in bacterial cytokinesis.
Keywords: bacteria, cytokinesis, cytoskeleton, electron microscopy (EM), intrinsically disordered protein, Caulobacter crescentus, FtsZ, bundling
Introduction
Canonical cytoskeletal proteins in animal cells polymerize to form structural elements that provide shape and mechanical integrity to the cell. In bacteria, however, the cell wall is the primary structural element, maintaining cell shape and preventing lysis. The role of bacterial cytoskeletal proteins that impact cell shape is in the spatial and temporal regulation of cell wall synthesis (1, 2). Cytoskeletal polymer assembly, structure, and dynamics collectively regulate local shape changes by constraining and/or directing cell wall remodeling enzymes (2). During cell division, the cytoskeletal protein FtsZ polymerizes to form the cytokinetic ring or “Z-ring” at the incipient division site and recruits over two dozen proteins, including cell wall enzymes (3–5). The Z-ring comprises discontinuous clusters of circumferentially aligned FtsZ protofilaments that are highly dynamic (6–9). How protofilaments are arranged within the resolution-limited clusters and how they interact with each other is largely unknown.
Recent studies in Escherichia coli and Bacillus subtilis demonstrated that FtsZ protofilament clusters in the Z-ring undergo treadmilling motion (8, 9). FtsZ treadmilling, in turn, drives the circumferential movement of cell wall enzymes, thereby directing local cell wall synthesis and remodeling toward constriction. The treadmilling of FtsZ clusters is presumably driven by the polymerization and depolymerization of FtsZ protofilaments, because GTP hydrolysis mutants that have reduced turnover in vitro lead to slower movement of clusters in vivo (8, 9). In addition to turnover of polymers, other aspects of the Z-ring such as conformational changes within FtsZ, lateral interactions between protofilaments, and interaction with FtsZ-binding proteins likely regulate cluster movement and thereby local cell wall remodeling (4, 10, 11). For example, FtsZ-binding proteins such as ZapA in E. coli and FzlA in Caulobacter crescentus that cause cross-linking or bundling of FtsZ protofilaments in vitro are important for efficient cytokinesis through unclear mechanisms (12–14).
FtsZ has a tubulin-like GTPase domain that polymerizes on binding GTP (Fig. 1A) (15–17). GTP hydrolysis is stimulated by polymerization and destabilizes the monomer–monomer interface, leading to depolymerization (17, 18). Under physiologically relevant conditions and in the presence of GTP, FtsZ predominantly forms straight or gently curved single protofilaments and/or double protofilament bundles in vitro by transmission electron microscopy (19, 20). In cells, FtsZ protofilaments are recruited to the membrane by membrane-anchoring proteins that bind FtsZ through the conserved peptide at its extreme C terminus (CTC2 or C-terminal conserved peptide) (3, 21–24). Between the GTPase domain and the CTC is an unstructured region called the C-terminal linker or CTL that varies widely in length (2–330 amino acids) and sequence across species (21). FtsZ from most bacteria with a peptidoglycan cell wall that have a CTC have a minimum CTL of 9 amino acids (21). Studies in E. coli and B. subtilis suggest length-, flexibility-, and/or disorder-dependent roles for the intrinsically disordered CTL in determining FtsZ assembly and function in vivo (25–27).
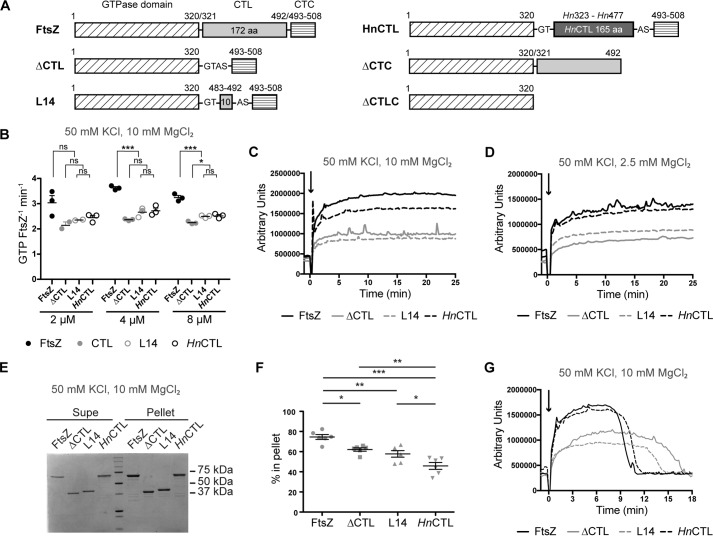
Figure 1.
CTL affects FtsZ polymer dynamics. A, schematic showing different regions of FtsZ truncations and CTL variants. The numbers shown above the schematic correspond to the amino acid numbers in wild type (CcFtsZ). Hn323–Hn477 represent amino acid numbers in H. neptunium FtsZ. (G, T, A, and S represent amino acids Gly, Thr, Ala, and Ser introduced due to restriction sites added for cloning). B, GTP hydrolysis rates of CTL variants at 2, 4, and 8 μm concentrations with 2 mm GTP, 50 mm KCl, and 10 mm MgCl2. Line and error bars represent average and standard deviation, respectively. *, p < 0. 05; **, p < 0.01; ***, p < 0.001; ns, p > 0.05 for Tukey's multiple comparison test (one-way analysis of variance). C, right angle light scatter at 350 nm over time for 4 μm FtsZ CTL variants with 2 mm GTP (added at time = 0 min) and 10 mm MgCl2 (mean of three replicates). D, right angle light scatter at 350 nm over time for 4 μm FtsZ CTL variants with 2 mm GTP (added at time = 0 min), 50 mm KCl, and 2.5 mm MgCl2 (mean of at least three replicates). E, representative Coomassie-stained SDS-polyacrylamide gel of a pelleting assay showing relative amounts of FtsZ CTL variants in supernatant (Supe) or pellet for 4 μm FtsZ CTL variant with 2 mm GTP, 50 mm KCl, and 10 mm MgCl2, 15 min after addition of GTP. F, quantification of the percentage of FtsZ CTL variants in pellet corresponding to experiment in E (n = 6). *, p < 0. 05; **, p < 0.01; ***, p < 0.001 for non-parametric t tests. G, right angle light scatter at 350 nm over time for 4 μm FtsZ CTL with 0.5 mm GTP (added at time = 0 min), 50 mm KCl, and 10 mm MgCl2.
In our previous study characterizing CTL function in C. crescentus, we observed that C. crescentus FtsZ lacking its 172-amino acid-long CTL (ΔCTL) forms highly bundled protofilaments in vitro (Fig. 1A) (28). This mutant of ftsZ when expressed in vivo leads to filamentation due to cytokinesis failure but also causes dominant lethal local cell wall defects leading to cell envelope bulges and rapid lysis. We were surprised to find that a variant of FtsZ with a 14-amino acid CTL (L14, Fig. 1A) caused filamentation but no bulging or lysis. Moreover, the L14 variant formed less bundled structures in vitro (28). We hypothesized that the aberrant assembly properties of ΔCTL in vitro underlie the defects in the structure and function of the Z-ring formed by ΔCTL in vivo (28).
In this study, we sought to fully characterize the contributions of the CTL to the polymerization properties of FtsZ in vitro. To this end, we compared polymers formed by wild-type FtsZ with ΔCTL and L14. We also included in our in vitro characterization a chimeric FtsZ variant with the Hyphomonas neptunium CTL in place of the C. crescentus CTL (HnCTL) (Fig. 1A). HnCTL is inefficient but functional for cytokinesis; while the HnCTL gene can replace ftsZ at its genomic locus in C. crescentus, the resulting cells have slower doubling time and heterogeneous cell length (28). Our in vitro characterization of the CTL variants of FtsZ and their interactions with FtsZ-binding factors has revealed that the CTL is important for protofilament turnover and lateral interaction. ΔCTL tends to form long extended bundles that require GTP binding but not hydrolysis for their formation. The membrane-anchoring protein FzlC can disrupt bundle formation in a CTC-dependent manner, but the CTC itself does not contribute to bundling. Moreover, the FtsZ curving protein FzlA can no longer robustly form helical bundles with ΔCTL. Overall, our study provides a biochemical framework to connect the assembly properties of these CTL variants in vitro with their in vivo Z-ring structures, dynamics, and effects on the regulation of cell wall modeling.
Results
FtsZ-CTL contributes to polymer dynamics
To better understand the contributions of the CTL to FtsZ polymerization, we performed a series of biochemical assays using purified CTL variants, FtsZ (WT), ΔCTL, L14, and HnCTL, in vitro. Because polymerization and depolymerization are coupled to GTP binding and hydrolysis, we first determined the GTP hydrolysis rate for each variant using a malachite green assay to detect inorganic phosphate liberated over time. We observed a statistically significant reduction in the rate of GTP hydrolysis for each CTL variant as compared with WT (Fig. 1B), with ΔCTL exhibiting the slowest rate (Fig. 1B and supplemental Fig. 1) (28). At 10 mm MgCl2, WT FtsZ had a GTP hydrolysis rate of 3.25 ± 0.17 GTP FtsZ−1 min−1, whereas HnCTL, L14, and ΔCTL had significantly slower rates of 2.51 ± 0.10, 2.49 ± 0.09, and 2.24 ± 0.06 GTP FtsZ−1 min−1, respectively (determined from measurements at 2, 4, and 8 μm protein concentrations and 2 mm GTP concentration, supplemental Fig. 1).
To monitor polymerization over time for the FtsZ CTL variants, we used right angle light scattering. Light scattering has been used as a readout of FtsZ polymerization, as polymers scatter more light than monomers and the amount of light scatter increases with the size of polymers (19, 20). It is important to keep in mind, however, that light scattering is also influenced by monomer size and structure and by formation of higher order filament assemblies such as bundles. Using this method for reactions containing 10 mm MgCl2 and 2 mm GTP, we found that all variants readily polymerized upon addition of GTP with no detectable lag phase at our time resolution, and we did not observe any obvious differences in their initial rates of increase in scatter (Fig. 1C). However, whereas WT and HnCTL resulted in similar light scatter values at steady state, steady-state light scatter values for ΔCTL and L14 were significantly lower (Fig. 1C). This trend was also true at the lower MgCl2 concentration of 2.5 mm (Fig. 1D).
Curiously, when we determined the extent of polymerization of the FtsZ CTL variants using high-speed pelleting in the presence of 10 mm MgCl2, we were surprised to observe a distinct trend for steady-state polymer mass. Although each CTL variant showed a reduced percentage in the pellet (representing polymer fraction) compared with WT, ΔCTL and L14 were enriched in the pellet compared with HnCTL (Fig. 1, E and F). We reason that the longer CTLs of WT and HnCTL may significantly contribute to the light-scattering properties of their resulting polymers and that formation of higher order structures by one or more of the CTL variants may also influence light scattering or propensity to pellet, leading to different results in these two assays.
Finally, we used limiting concentrations of GTP to monitor the decrease in light scatter as filaments depolymerize following consumption of GTP. We found that ΔCTL, with the slowest GTP hydrolysis rates at this condition (Fig. 1B), took longer to return to baseline compared with WT and the other CTL variants, suggesting a correlation between the effect of the CTL on GTP hydrolysis rates and polymer dynamics, as we would predict (Fig. 1G).
CTL influences lateral interaction between protofilaments
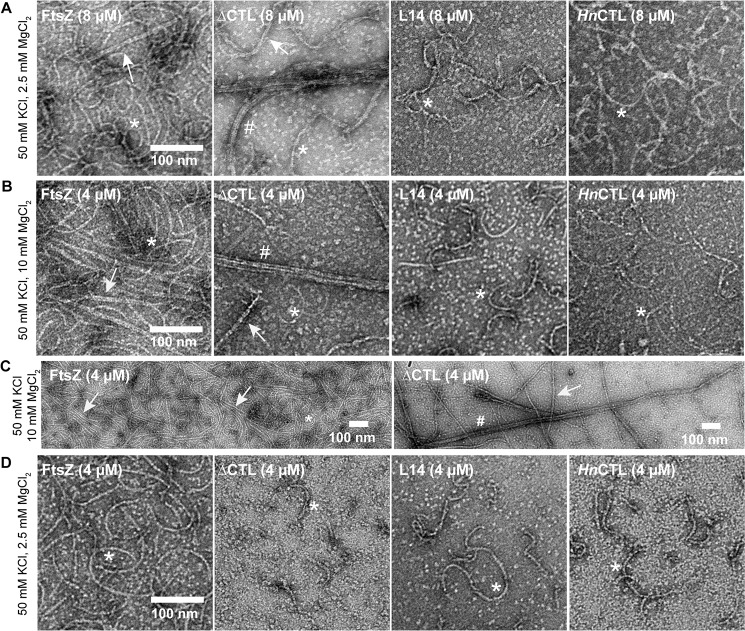
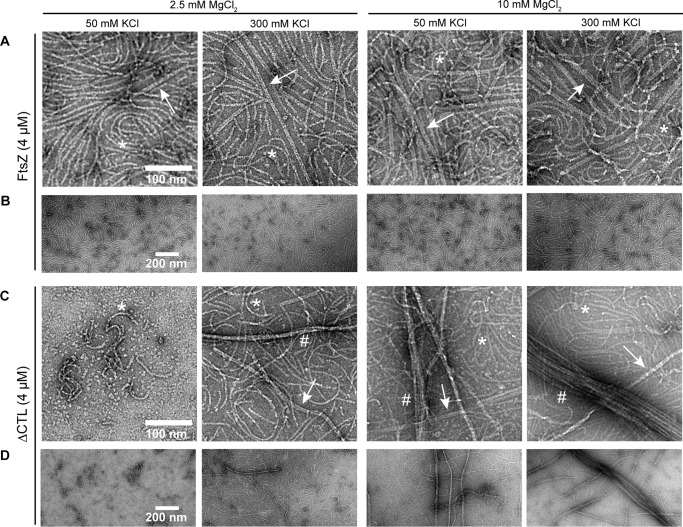
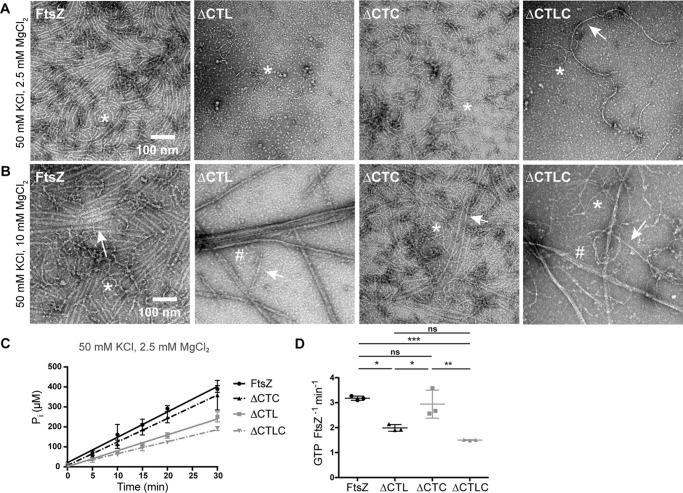
We next assessed the filament structures formed by the four CTL variants using negative stain transmission electron microscopy (TEM). We observed polymers with similar protofilament widths for the four variants but with distinct appearance and abundance depending on the nature of the CTL. Similar to previous reports for C. crescentus FtsZ (12, 20, 29, 30), at 8 μm protein concentration with GTP and 2.5 mm MgCl2 WT FtsZ formed gently curved single filaments and occasional straight double-filament bundles (Fig. 2A). HnCTL and L14 predominantly formed sparse, short single filaments under the same conditions. Strikingly, as reported earlier (28), ΔCTL predominantly forms long multifilament bundles at 8 μm protein concentration and 2.5 mm MgCl2. We did not observe these long bundled structures for the other variants (Fig. 2A).
Figure 2.
CTL affects lateral interaction between FtsZ protofilaments. Electron micrographs of polymers formed by FtsZ CTL variants spotted on grids 15 min after addition of GTP and stained with uranyl formate. A, 8 μm FtsZ CTL variant with 2 mm GTP, 50 mm KCl, and 2.5 mm MgCl2. B, 4 μm FtsZ CTL variant with 2 mm GTP, 50 mm KCl, and 10 mm MgCl2. C, low magnification micrographs of FtsZ or ΔCTL polymers with 2 mm GTP, 50 mm KCl, and 10 mm MgCl2. D, 4 μm FtsZ CTL variant with 2 mm GTP, 50 mm KCl, and 2.5 mm MgCl2. Scale bars, 100 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles.
Increasing the concentration of MgCl2 has been reported to promote lateral interactions between FtsZ protofilaments. We therefore explored the effect of high MgCl2 concentration on polymer superstructure for the four FtsZ variants. WT FtsZ frequently formed two- or three-filament bundles (7–15 nm wide) at 10 mm MgCl2 (Fig. 2, B and C). In contrast, under the same conditions ΔCTL bundles often consisted of more than four filaments (∼20–200 nm in width) and appeared as very long, straight structures frequently extending longer than 1 μm (Fig. 2, B and C). At 4 μm protein and 2.5 mm MgCl2 concentration, we did not observe bundles for ΔCTL (Fig. 2D). Compared with WT protofilaments, L14 and HnCTL protofilaments were sparse on the grids and were predominantly gently-curved single filaments at either MgCl2 concentration (Fig. 2, B and D).
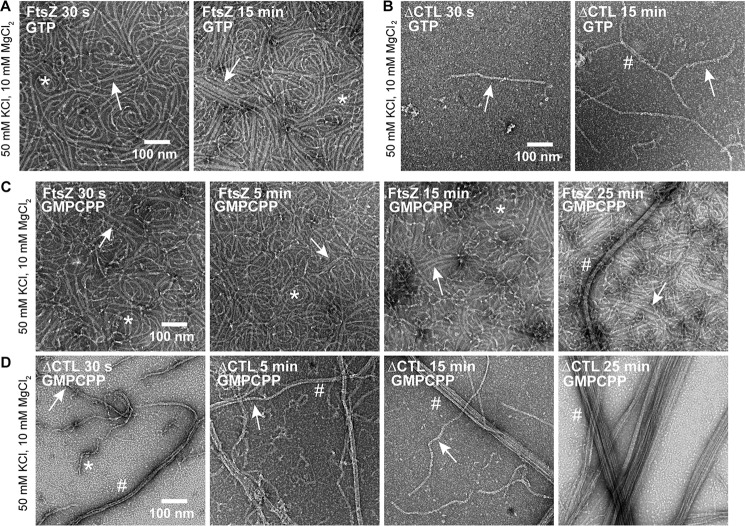
We further investigated whether polymer stability and/or slow GTP hydrolysis contributed to the increased bundling observed for ΔCTL in vitro. To test this, we used GMPCPP, a slowly hydrolyzed analog of GTP, to form stable protofilaments at high and low concentrations of MgCl2. By TEM, at 2.5 mm MgCl2 concentration, the polymer structures were not significantly different from GMPCPP compared with GTP (Fig. 3A). At 10 mm MgCl2, we readily observed large multifilament bundles of ΔCTL protofilaments with GMPCPP similar to those observed with GTP (Fig. 3B). Although we observed more protofilaments for all the FtsZ CTL variants, and more interaction between protofilaments for WT, we failed to find the long, wide multifilament bundles characteristic of ΔCTL for the other variants. We conclude that the ability to form long multifilament bundles is specific to ΔCTL.
Figure 3.
Effects of the CTL on protofilament lateral interaction are independent of GTP hydrolysis. Electron micrographs of polymers formed by FtsZ CTL variants spotted on grids 15 min after addition of GMPCPP and stained with uranyl formate. A, 4 μm FtsZ CTL variant with 0.2 mm GMPCPP, 50 mm KCl, and 2.5 mm MgCl2. B, 4 μm FtsZ CTL variant with 0.2 mm GMPCPP, 50 mm KCl, and 2.5 mm MgCl2. C, low magnification micrographs of FtsZ or ΔCTL polymers with 0.2 mm GMPCPP, 50 mm KCl, and 10 mm MgCl2. Scale bars, 100 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles.
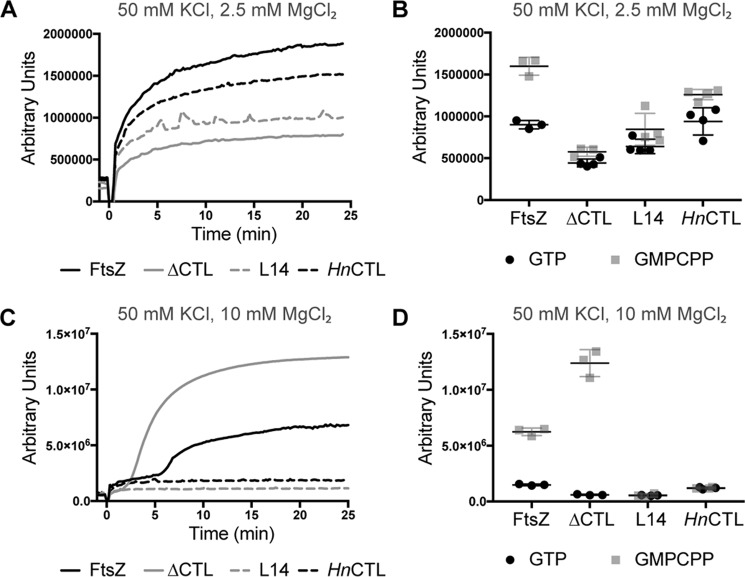
The influence of CTL on lateral interaction was also evident from the differences in light scatter profiles of the CTL variants with GMPCPP (Fig. 4). At 2.5 mm MgCl2, we did not observe large differences in the kinetics or extent of light scatter over time with GTP compared with GMPCPP for the CTL variants (Fig. 4, A and B). Only WT showed significantly higher scatter with GMPCPP compared with GTP (Fig. 4B). However, at 10 mm MgCl2 with GMPCPP, we observed a two-step increase in light scatter for WT and ΔCTL (Fig. 4C). For WT we observed an initial rapid increase immediately following the addition of nucleotide (similar to light scatter with GTP) and then a second slower, much larger increase about 5 min later, likely corresponding to polymerization into protofilaments and formation of bundles, respectively. For ΔCTL, the initial rapid increase was almost immediately followed by the second increase (after about 2 min). Moreover, the secondary increase in light scatter was much faster and larger for ΔCTL compared with WT (Fig. 4, C and D). HnCTL and L14 only undergo a rapid one-step increase in light scatter after addition of GMPCPP, not obviously different from GTP (Fig. 4, C and D) likely due to the absence of higher order assemblies of protofilaments under these conditions.
Figure 4.
Polymerization kinetics of CTL variants with GMPCPP. A, right angle light scatter at 350 nm over time for 4 μm FtsZ CTL variants with 0.2 mm GMPCPP (added at time = 0 min), 50 mm KCl, and 2.5 mm MgCl2 (mean of three replicates). B, extent of light scatter (quantified as the difference between light scatter at steady state (time = 25 min) and light scatter prior to addition of nucleotide (time = −1 min)) for FtsZ CTL variants with GTP (from Fig. 1D) or GMPCPP (Fig. 4A). C, right angle light scatter at 350 nm over time for 4 μm FtsZ CTL variants with 0.2 mm GMPCPP (added at time = 0 min), 50 mm KCl, and 10 mm MgCl2 (mean of three replicates). D, extent of light scatter (quantified as the difference between light scatter at steady state (time = 25 min) and light scatter prior to addition of nucleotide (time = −1 min)) for FtsZ CTL variants with GTP (from Fig. 1C) or GMPCPP (Fig. 4C).
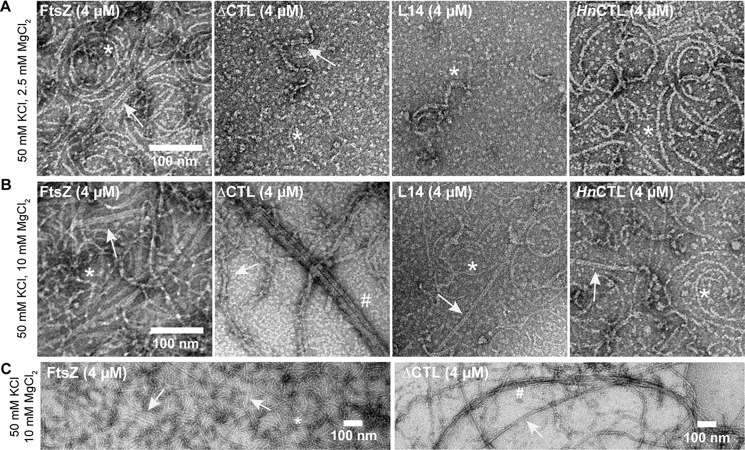
We confirmed that the second increase in light scatter for WT and ΔCTL with GMPCPP at 10 mm MgCl2 was indeed due to increased lateral interaction by observing polymer structure over time (Fig. 5). WT incubated with GTP and 10 mm MgCl2 forms predominantly short single filaments after 30 s and both single filaments and double- or triple-filament bundles after 15 min (Fig. 5A). In contrast, ΔCTL incubated with GTP and 10 mm MgCl2 forms few straight bundles that become longer and thicker with time (Fig. 5B). The difference in polymer structure for WT and ΔCTL over time was more apparent with GMPCPP and 10 mm MgCl2 (Fig. 5, C and D). Although WT largely formed single gently curved protofilaments after incubation with GMPCPP for 30 s, we observed more double- and triple-filament bundles after 5 min or a longer incubation (Fig. 5C). We also observed a few long four- or five-filament bundles for WT after incubation with GMPCPP for 25 min. In contrast, we almost exclusively observe long four- or five-filament bundles for ΔCTL after incubation with GMPCPP for 30 s (Fig. 5D). ΔCTL multifilament bundles were more prominent, longer and thicker after a longer incubation with GMPCPP. Taking our observations together, we infer that the CTL is important for reducing lateral interaction between FtsZ protofilaments and that a minimal CTL of 14 amino acids is sufficient to perform this role.
Figure 5.
ΔCTL becomes more bundled with time. Electron micrographs of polymers formed by FtsZ or ΔCTL spotted on grids at the indicated amount of time after addition of nucleotide and stained with uranyl formate. A, 4 μm FtsZ with 2 mm GTP, 50 mm KCl, and 10 mm MgCl2. B, 4 μm ΔCTL with 2 mm GTP, 50 mm KCl, and 10 mm MgCl2. C, 4 μm FtsZ with 0.2 mm GMPCPP, 50 mm KCl, and 10 mm MgCl2. D, 4 μm ΔCTL with 0.2 mm GMPCPP, 50 mm KCl, and 10 mm MgCl2. Scale bars, 100 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles.
Curiously, HnCTL does not form prominent double- or triple-filament bundles under any condition tested despite having WT-like light scatter with GTP. The decreased ability of HnCTL to form higher order protofilament assemblies is further suggested by the lower percentage of HnCTL in the polymer fraction as observed by high-speed pelleting (Fig. 1, E and F). A technical explanation for the sparse, unbundled filaments observed for HnCTL by TEM might be that HnCTL does not stick to the grids for TEM as well as WT FtsZ. However, with GMPCPP it is clear from light scattering that WT FtsZ forms higher order structures, although HnCTL does not. Thus, the sequence of the CTL seems to play a role in maintaining optimal interfilament interactions in a way that is altered for HnCTL.
High KCl and MgCl2 concentrations favor bundle formation
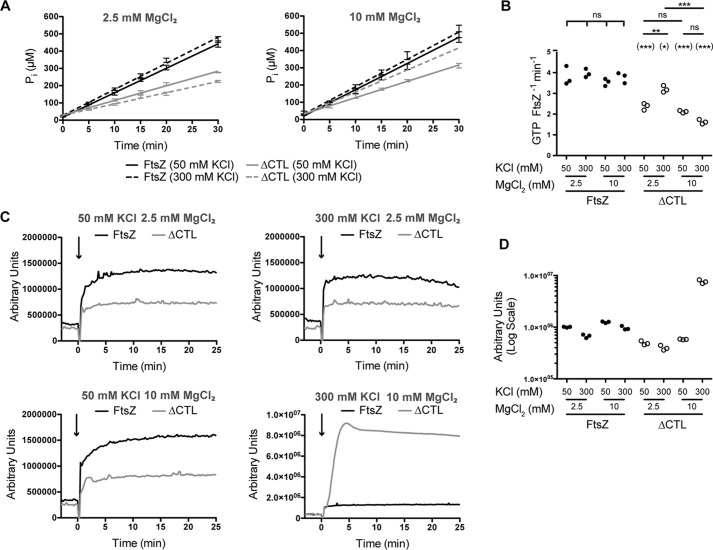
Because we observe large bundles of ΔCTL at a higher concentration of MgCl2 (10 mm), we decided to further investigate the effect of salt on lateral interactions between protofilaments at higher KCl concentrations. We performed GTP hydrolysis rate measurements, light scattering, and EM imaging of polymers formed by WT FtsZ or ΔCTL at low (50 mm) or high (300 mm) KCl concentrations and low (2.5 mm) or high (10 mm) MgCl2 concentrations. We did not observe significant differences in the GTP hydrolysis rates of WT at the different KCl and MgCl2 concentrations (Fig. 6, A and B). However, unlike WT, we observed significant differences in GTP hydrolysis rates of ΔCTL depending on KCl and MgCl2 concentrations. At 50 mm KCl concentration, ΔCTL displayed significantly reduced GTP hydrolysis rates compared with WT at both 2.5 and 10 mm MgCl2 concentrations (Fig. 6, A and B). At 2.5 mm MgCl2 concentration, we were surprised to see a significant increase in GTP hydrolysis rates of ΔCTL at 300 mm KCl concentration compared with 50 mm KCl concentration (Fig. 6, A and B). In fact, ΔCTL displayed the most similar (but still statistically significantly different) GTP hydrolysis rates compared with WT at 300 mm KCl and 2.5 mm MgCl2 concentrations (Fig. 6B). In contrast, at 10 mm MgCl2 concentration, there was a significant decrease in GTP hydrolysis rate of ΔCTL at 300 mm KCl concentration compared with 50 mm KCl concentration (Fig. 6, A and B), suggesting that the reduction in GTP hydrolysis rate of ΔCTL compared with WT is further enhanced at 300 mm KCl and 10 mm MgCl2. Thus, we conclude that KCl concentration affects GTP hydrolysis rates of FtsZ and ΔCTL differently, and in a MgCl2 concentration-dependent manner.
Figure 6.
Effect of salt concentrations on FtsZ or ΔCTL polymerization dynamics. A, inorganic phosphate (Pi) concentration in solution over time in reactions containing 4 μm FtsZ or ΔCTL with 2 mm GTP at 2.5 mm MgCl2 or 10 mm MgCl2, and 50 mm KCl or 300 mm KCl concentrations. Error bars represent standard deviation. Straight lines indicate linear fits of averages. B, comparison of GTP hydrolysis rates (determined as the slope of linear fit for matched replicates of inorganic phosphate concentration over time) for FtsZ or ΔCTL corresponding to experiments in A. *, p < 0. 05; **, p < 0.01; ***, p < 0.001; ns, p > 0.05 for Tukey's multiple comparison test (one-way analysis of variance). C, right angle light scatter at 350 nm over time for 4 μm FtsZ or ΔCTL with 2 mm GTP (added at time = 0 min) at 2.5 mm MgCl2 or 10 mm MgCl2 and 50 mm KCl or 300 mm KCl concentrations. D, extent of light scatter (quantified as the difference between light scatter at steady state (time = 25 min) and light scatter prior to addition of nucleotide (time = −1 min) for FtsZ or CTL corresponding to experiments in C.
Although the differences in the effects of KCl concentration on GTP hydrolysis rates of ΔCTL at low and high MgCl2 concentrations appear counter-intuitive, our observations from light scattering and EM imaging helped provide a possible explanation. At 50 mm KCl concentration, ΔCTL displayed significantly reduced light scatter compared with WT following addition of GTP, irrespective of MgCl2 concentration (Fig. 6C). This reduction in light scatter was also observable at 300 mm KCl and 2.5 mm MgCl2 concentrations (Fig. 6C). However, at 300 mm KCl and 10 mm MgCl2, we observed a large increase in light scatter for ΔCTL compared with WT following addition of GTP (Fig. 6, C and D). This increase appeared comparable with the secondary increase observed for ΔCTL following addition of GMPCPP at 50 mm KCl and 10 mm MgCl2 concentrations (Fig. 4, C and D).
Next, we observed by EM the polymers formed by WT or ΔCTL at the different KCl and MgCl2 concentrations after incubation with GTP for 15 min. Although FtsZ forms predominantly gently-curved single filaments at 50 mm KCl and 2.5 mm MgCl2 concentrations, we observed double- or triple-filament bundles of FtsZ more predominantly with higher KCl and/or MgCl2 concentrations (Fig. 7, A and B, and supplemental Fig. 2A). This effect of KCl and MgCl2 concentrations on lateral interaction between protofilaments was even more dramatic with ΔCTL. Although ΔCTL filaments were predominantly short, curved filaments at 50 mm KCl and 2.5 mm MgCl2 concentrations, we observed long, straight filament bundles along with single gently-curved filaments at 300 mm KCl and 2.5 mm MgCl2 concentrations (Fig. 7, C and D, and supplemental Fig. 2B). Overall, at 2.5 mm MgCl2 concentration, there appeared to be more filaments on the grid at 300 mm KCl concentration compared with 50 mm KCl concentration for ΔCTL (Fig. 7, C and D). Moreover, these filaments appeared longer at the higher KCl concentration (Fig. 7, C and D, and supplemental Fig. 2B). Taking this observation together with the results from GTP hydrolysis rate measurements, we conclude that at 2.5 mm MgCl2 the higher KCl concentration potentiates ΔCTL polymerization.
Figure 7.
ΔCTL bundle formation is enhanced at higher KCl concentrations. Electron micrographs of polymers formed by FtsZ (A and B) or ΔCTL (C and D) on grids 15 min after addition of 2 mm GTP and stained with uranyl formate at 2.5 mm MgCl2 or 10 mm MgCl2, and 50 mm KCl or 300 mm KCl concentrations as indicated. A and C, scale bars, 100 nm. B and D, scale bars, 200 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles.
Interestingly, despite the increased GTP hydrolysis rate of ΔCTL at 300 mm KCl and 2.5 mm MgCl2, we still observed long, straight multifilament bundles that are distinct from the double- or triple-filament bundles observed for either ΔCTL (on the same grid) or WT. Moreover, these bundles are comparable in thickness to the structures observed for ΔCTL at 50 mm KCl and 10 mm MgCl2. Most strikingly, we observed large multifilament bundles that are much longer and thicker than those observed in the presence of GTP under all other conditions tested for ΔCTL at 300 mm KCl and 10 mm MgCl2 concentrations (Fig. 7, C and D, andsupplemental Fig. 2B), providing an explanation for the large increase in light scatter observed for ΔCTL at this condition. We also observed a large number of single protofilaments, similar to those we typically observe for WT, coating the grid in the background under the high KCl, high MgCl2 condition for ΔCTL. This likely reflects increased polymerization of ΔCTL at higher KCl concentration, but it may also reflect an increased propensity of polymers to interact with the EM grid.
In light of these observations, we propose that the decreased GTP hydrolysis rate observed for ΔCTL at this condition (300 mm KCl and 10 mm MgCl2 concentrations) is likely due to decreased turnover of monomers in the abundant, large protofilament bundles observed by EM. Alternatively, increasing KCl concentration could have a stabilizing effect on protofilament bundles by negatively affecting GTP hydrolysis rate at high MgCl2 concentrations. The latter possibility is less likely because KCl appears to have a positive effect on ΔCTL's GTP hydrolysis rate at low MgCl2 concentration, and it has no observable effect on WT FtsZ's GTP hydrolysis rate irrespective of MgCl2 concentration.
High MgCl2 concentration does not increase polymerization of ΔCTL
We observed bundles of ΔCTL with GTP or GMPCPP more readily at 10 mm MgCl2 compared with 2.5 mm MgCl2. This effect of MgCl2 on increasing lateral interaction has been documented before for full-length E. coli FtsZ, with more two- or three-filament bundles at higher MgCl2 concentrations (19). We see the same effect for C. crescentus FtsZ (WT) (compare TEM shown in Figs. 2, B and D, or 3, A and B). However, we see fewer protofilaments for ΔCTL by EM and lower signal by light scattering at 50 mm KCl and 2.5 mm MgCl2 as compared with 10 mm MgCl2. We reasoned that the effects of increased divalent cation concentration might be the result of increased polymer formation (i.e. increase in total protofilament mass), increased bundling, or both. To differentiate between these possibilities, we used the polymerization-specific fluorescence reporter, a tryptophan mutant of FtsZ (FtsZL72W), developed by the Erickson lab (29). This mutant has a tryptophan residue engineered on the polymerizing interface of FtsZ, the environment of which changes on polymerization resulting in a polymerization-specific increase in fluorescence signal. Unlike light scattering, the increase in tryptophan fluorescence on polymerization is minimally affected by bundling and is a more reliable readout of number of the longitudinal interactions.
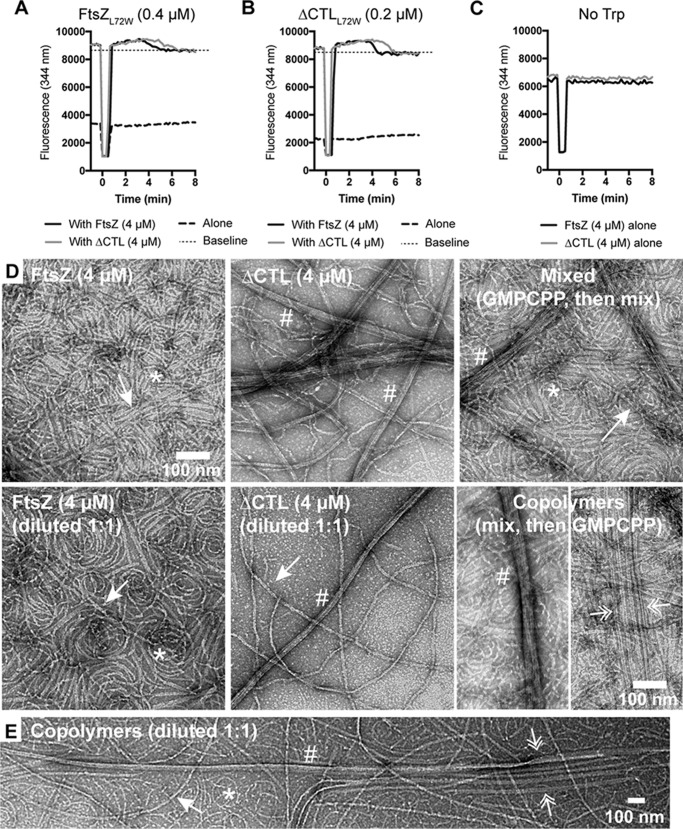
Using this assay, we first added 0.1 mm GMPCPP to reactions containing 4 μm FtsZ (with 10% FtsZL72W) or ΔCTL (with 10% ΔCTLL72W) and 50 mm KCl and then initiated formation of polymers with 2.5 mm MgCl2 (Fig. 8A). We observed a polymerization-dependent increase in tryptophan fluorescence for both WT and ΔCTL, as expected. Then we added MgCl2 up to 10 mm and continued to measure the fluorescence intensity. Because the excitation and emission wavelengths for right angle light scatter and tryptophan fluorescence are similar (350–350 nm compared with 295–344 nm respectively), we also included FtsZ alone or ΔCTL alone (with no tryptophan mutation) as controls for a possible increase in intensity readout as a result of scatter. For WT, with or without the tryptophan mutant, we failed to observe a change in intensity when MgCl2 concentration was increased, suggesting that increased MgCl2 does not change the polymer mass at steady state under these conditions. For ΔCTL, we did observe an increase in signal when we increased the MgCl2 concentration. However, this signal increase was observed for the ΔCTL alone control, as well, indicating that it was the result of bundling-induced light scatter. Indeed, when we subtract the fluorescence signal observed for the ΔCTL alone control from that of ΔCTL with the tryptophan mutant, we observe no additional increase in signal on increasing MgCl2 concentration (Fig. 8B). These observations argue against the possibility that the increased bundling seen by EM and increased signal observed by light scattering at 50 mm KCl and high MgCl2 concentration are due to increased protofilament mass.
Figure 8.
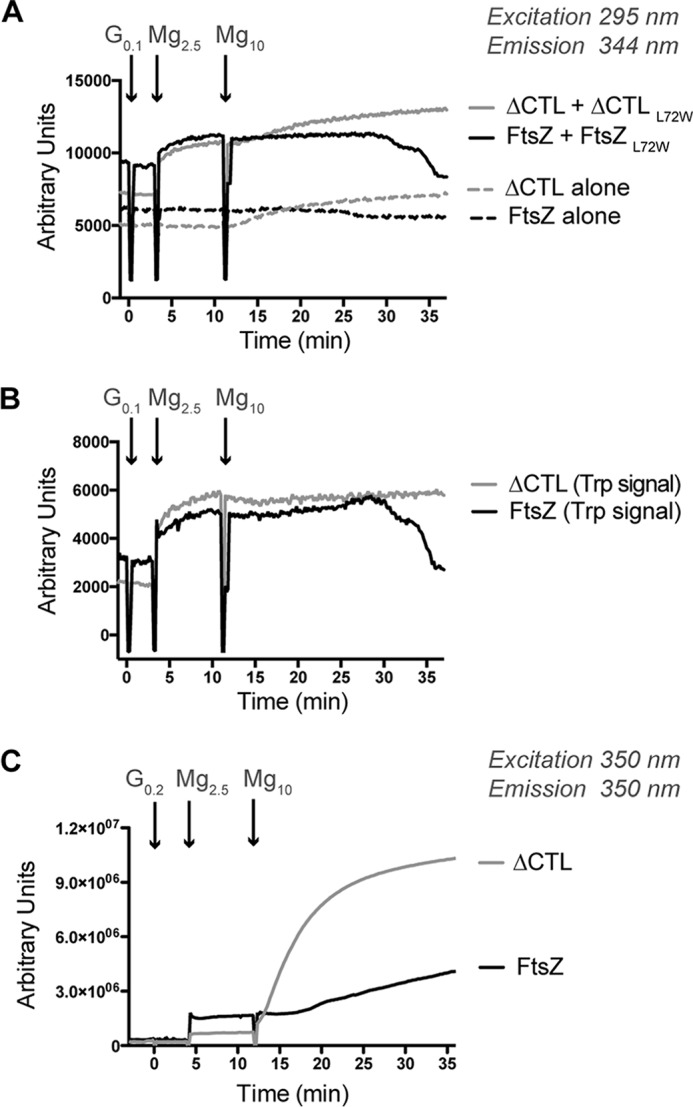
Effect of high MgCl2 concentration on polymerization of FtsZ or ΔCTL. A, fluorescence signal emitted over time (emission at 344 nm after excitation at 295 nm) for FtsZ or ΔCTL with 0.1 mm GMPCPP added at time = 0 min in HEK50 buffer (50 mm KCl). MgCl2 was added to the reaction to a concentration of 2.5 mm at time = 3 min and subsequently to a concentration of 10 mm at time = 11 min (mean of three replicates). Total concentration of protein in each reaction was 4 μm. L72W mutants when used were at 10% total protein concentration (3.6 μm FtsZ + 0.4 μm FtsZL72W, 3.6 μm ΔCTL + 0.4 μm ΔCTLL72W, 4 μm FtsZ alone, or 4 ΔCTL alone). B, signal specific to tryptophan fluorescence (calculated by subtracting mean of signal for FtsZ or ΔCTL alone from mean of corresponding signal for FtsZ + FtsZL72W or ΔCTL + ΔCTLL72W, respectively) for experiment in Fig. 6A. C, right angle light scatter (emission at 350 nm after excitation at 350 nm) over time for 4 μm FtsZ or ΔCTL with 0.2 mm GMPCPP added at time = 0 min in HEK50 buffer (50 mm KCl). MgCl2 was added to reaction to a concentration of 2.5 mm at time = 4 min and subsequently to a concentration of 10 mm at time = 12 min (mean of at least three replicates).
We also observed a drop in tryptophan fluorescence signal back to baseline after some time for WT, potentially due to the eventual depletion of GMPCPP through slow hydrolysis (Fig. 8, A and B). In the same time frame, we did not observe a similar decrease for ΔCTL likely because it has a slow nucleotide hydrolysis rate compared with WT.
To confirm that the increase in signal observed for ΔCTL with GMPCPP following the increase in MgCl2 concentration is due to increased light scatter, we measured the amount of light scattered (at 350 nm excitation and 350 nm emission wavelengths) by 4 μm FtsZ or 4 μm ΔCTL and 50 mm KCl after addition of 0.2 mm GMPCPP and subsequent additions of MgCl2 to 2.5 mm concentration and finally to 10 mm concentration. As predicted from our tryptophan fluorescence assay, we observed a large increase in light scatter for ΔCTL immediately following the increase in MgCl2 concentration to 10 mm (Fig. 8C). In comparison, we only observed a modest increase in light scatter for WT (Fig. 8C) similar to our prior observation with 0.2 mm GMPCPP at 10 mm MgCl2 concentration (Fig. 4, C and D). Taken together, our results indicate that at 50 mm KCl, increasing MgCl2 concentration promotes higher order assembly of ΔCTL protofilaments without significantly increasing longitudinal interactions.
FtsZ and ΔCTL can copolymerize in vitro
Because ΔCTL has a dominant effect on cell shape and division in cells, we hypothesized that ΔCTL and WT FtsZ co-polymerize in vivo and that ΔCTL exerts its propensity for increased lateral interaction in the resulting copolymers. Because the polymerizing GTPase domain is intact in ΔCTL, we expected that WT and ΔCTL could still copolymerize. To test this formally, first, we estimated the minimum concentrations required for polymerization (i.e. critical concentration) of the tryptophan mutants FtsZL72W and ΔCTLL72W by determining the concentration below for which no increase in tryptophan fluorescence was observed on addition of GTP. We observed no significant difference in tryptophan fluorescence with or without GTP for protein concentrations less than 0.6 μm for FtsZL72W and 0.4 μm for ΔCTLL72W at 2.5 mm MgCl2 and 50 mm KCl concentrations (data not shown). We used these concentrations as estimated critical concentrations in the following experiments. As expected, using only FtsZL72W below its estimated critical concentration for polymerization, we observed no increase in fluorescence on addition of GTP at 2.5 mm MgCl2 concentration (Fig. 9A). When we added WT FtsZ to this reaction mixture, pushing the total protein concentration above the critical concentration for FtsZ polymerization, we observed a significant GTP-dependent increase in fluorescence (Fig. 9A). We inferred from this observation that WT FtsZ can copolymerize with FtsZL72W and pushes the overall monomer concentration above the critical concentration, thereby resulting in polymerization and an increase in tryptophan fluorescence. Importantly, we also observed this fluorescence increase when we added ΔCTL to the reaction mixture instead of FtsZ, indicating that FtsZL72W could copolymerize with ΔCTL as well as WT. We observed the same effect when we used ΔCTLL72W instead of FtsZL72W further confirming that FtsZ and ΔCTL can copolymerize (Fig. 9B). Because we used limiting concentrations of GTP (50 μm), we could also observe the decrease in fluorescence intensity due to depolymerization following depletion of GTP. The amount of time taken until depolymerization was exactly as estimated from the individual GTP hydrolysis rates, dominated by the predominant species of monomers (FtsZ or ΔCTL), further adding confidence to our results. We did not observe any significant increase in fluorescence signal at 344 nm on addition of GTP for 4 μm FtsZ or ΔCTL at 2.5 mm MgCl2 (Fig. 9C), suggesting that the signal observed in the presence of tryptophan mutants are indicative of polymerization and not background scatter from polymerization of the proteins lacking the tryptophan.
Figure 9.
FtsZ and ΔCTL can copolymerize to form very long bundles. A–C, tryptophan fluorescence increase (emission at 344 nm after excitation at 295 nm) for FtsZ CTL variants with 50 μm GTP (added at time = 0 min), 2.5 mm MgCl2, and 50 mm KCl. Straight black dotted line above 8000 indicating baseline after depolymerization has been included to better visualize the increase in fluorescence after addition of GTP because we observed a small but significant drop in intensity following the addition of GTP due to dilution of the proteins in solution. We define the baseline as the intensity following depletion of GTP. A, fluorescence signal from 0.4 μm FtsZL72W in reaction with 4 μm FtsZ or ΔCTL or by itself (n = 3). B, fluorescence signal from 0.2 μm ΔCTLL72W in reaction with 4 μm FtsZ or ΔCTL or by itself (n = 3). C, fluorescence signal from 4 μm FtsZ or ΔCTL (n = 1). D, electron micrographs showing polymers formed by FtsZ or ΔCTL individually, mixed together (after polymerization individually), or FtsZ/ΔCTL copolymers with 0.2 mm GMPCPP, 10 mm MgCl2, and 50 mm KCl spotted on EM grids 15 min after addition of GMPCPP and stained with uranyl formate. Mixed represents micrographs of 4 μm FtsZ and 4 μm ΔCTL polymerized individually and then mixed prior to spotting on grids. FtsZ (diluted to 1:1) or ΔCTL (diluted 1:1) represent polymers obtained by diluting GMPCPP-stabilized polymers of 4 μm FtsZ or ΔCTL, respectively, with polymerization buffer (1:1 dilution) prior to spotting on grids. Copolymer represents micrographs of polymers formed by FtsZ (2 μm) and ΔCTL (2 μm) both in the same reaction mixture (total FtsZ variant concentration, 4 μm) with GMPCPP. E, low magnification micrographs of copolymers formed by 2 μm FtsZ and 2 μm ΔCTL (initial total FtsZ variant concentration, 4 μm), diluted 1:1 with polymerization buffer (final total FtsZ variant concentration, 2 μm) following 15 min of incubation with 0.2 mm GMPCPP, 10 mm MgCl2, and 50 mm KCl prior to spotting on grids. Dilution of GMPCPP-stabilized polymers reduces crowding on EM grids and helps in better visualization of polymer structures. Scale bar, 100 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles; pair of double arrowheads, groups of two- or three-filament bundles that run parallel.
On confirming that FtsZ and ΔCTL can copolymerize, we compared the polymer structures formed by the copolymers at different ratios of FtsZ to ΔCTL using negative stain TEM (supplemental Fig. 3). We compared the structures observed for mixtures of ΔCTL and WT FtsZ with those formed at the same concentrations of WT FtsZ alone at 50 mm KCl and 10 mm MgCl2 concentrations with GTP. We observed more polymer density on the EM grids with increasing FtsZ/ΔCTL ratios (supplemental Fig. 3A). We also observed more bundles for FtsZ/ΔCTL combinations compared with WT alone (compare supplemental Fig. 3, A and 1st panel of B). We saw prominent long ΔCTL-like bundles at the lowest FtsZ/ΔCTL ratio (1:9). At higher ratios (1:3 or 1:1), the polymers were predominantly double- or triple-filament bundles similar to WT. However, the very tight packing of WT FtsZ on the grids made it difficult to distinguish possible differences in the length and abundance of bundles formed by FtsZ alone versus copolymers. The slowed polymer turnover of ΔCTL could potentially lead to a higher ratio of FtsZ/ΔCTL in the polymer at steady state compared with initial conditions, further complicating the interpretation of these observations by TEM.
To more clearly visualize whether a 1:1 copolymer of FtsZ/ΔCTL exhibits different filament architecture compared with individual polymers of FtsZ or ΔCTL, we compared GMPCPP-stabilized protofilaments formed with a mixture of 2 μm FtsZ and 2 μm ΔCTL to those formed by 2 μm FtsZ or 2 μm ΔCTL alone (Fig. 9D). The use of GMPCPP-stabilized filaments allowed us to also visualize a 1:1 mixture of separately pre-polymerized protofilaments of FtsZ and ΔCTL to better identify structures unique to copolymers. We observed polymer structures characteristic of both FtsZ (predominantly gently curved single filaments or two- or three-filament bundles) and ΔCTL (long multifilament bundles) on the grids with a mixture of pre-polymerized protofilaments (Fig. 9D, mixed (2 μm each)). FtsZ/ΔCTL copolymers had a range of polymer structures from individual curved protofilaments to long double-filament bundles to large multifilament bundles (Fig. 9, D, copolymers (4 μm), and E). Uniquely, the FtsZ/ΔCTL copolymers also formed very long, evenly spaced double protofilament bundles that were not seen for mixed or individual polymers of FtsZ and/or ΔCTL (Fig. 9, D and E, double arrow). These long double-filament bundles were conspicuous, as they often were observed in evenly spaced parallel arrays on the grid.
Overall, our results confirm that FtsZ and ΔCTL can copolymerize and that FtsZ/ΔCTL copolymers can have distinct structural properties, most notably propensity to bundle and length of polymers, depending on the ratio of FtsZ/ΔCTL. Although it is not clear whether ΔCTL's propensity to form multifilament bundles is relevant in vivo, these observations are in agreement with the increased time taken to elicit the ΔCTL bulging and lysis phenotype in the presence of WT FtsZ compared with in its absence (28).
It is also interesting to note that we still see long thick ΔCTL bundles on EM grids on dilution after polymerization with GMPCPP (compare ΔCTL (4 μm) to ΔCTL (diluted 1:1) in Fig. 9D), whereas WT FtsZ double- or triple-filament bundles were less obvious on dilution after polymerization with GMPCPP (compare FtsZ (4 μm) to FtsZ (diluted 1:1) in Fig. 9D). This further suggests that the increased ΔCTL interaction is not an artifact of crowding on the EM grids but an ability to form stable lateral interactions that is lacking in WT FtsZ.
Increased lateral interaction between ΔCTL protofilaments is CTC-independent
We previously reported that ΔCTL requires its extreme C terminus (C-terminal conserved peptide or CTC) to cause cell wall defects (28). In vivo, ΔCTLC (FtsZ lacking both the CTL and CTC, Fig. 1A) forms elongated structures in the cytoplasm instead of Z-rings (28). In contrast, ΔCTC (FtsZ lacking only its CTC, Fig. 1A) forms broad Z-rings. To test whether the CTC contributes to the increased lateral interaction between ΔCTL protofilaments in vitro, we compared protofilaments formed by ΔCTC and ΔCTLC to ΔCTL bundles in vitro by TEM. At low MgCl2 and 4 μm protein, ΔCTC formed WT-like protofilaments, whereas ΔCTLC formed very few long protofilaments (Fig. 10A). At high MgCl2 and 4 μm protein, although the ΔCTC protofilaments looked indistinguishable from WT protofilaments, ΔCTLC formed large multifilament bundles similar to ΔCTL (Figs. 10B and 11A). This suggests that the GTPase domain is sufficient for polymerization and bundle formation, similar to observations for the GTPase domain of B. subtilis FtsZ (31). These observations also rule out the possibility that the ΔCTL bundles are the result of a difference in orientation of the CTC with respect to the polymerizing GTPase domain. We also observed similarly decreased apparent GTP hydrolysis rates for ΔCTLC and ΔCTC compared with ΔCTL and WT, respectively, suggesting that the effects of lacking the CTC on GTP hydrolysis and polymerization are independent of the effects of lacking the CTL (Fig. 10, C and D). At 2.5 mm MgCl2, 50 mm KCl, and 4 μm protein concentrations, FtsZ, ΔCTL, ΔCTC, and ΔCTLC showed GTP hydrolysis rates of 3.18 ± 0.16, 1.99 ± 0.09, 2.94 ± 0.19, and 1.50 ± 0.06 GTP FtsZ−1 min−1, respectively. We conclude that the absence of the CTL is the primary contributor to the increased lateral interaction between protofilaments in ΔCTL and that there is no observable effect of the CTC on lateral interaction in this context.
Figure 10.
ΔCTL bundles form independent of the CTC. A and B, electron micrographs of polymers formed by 4 μm FtsZ, ΔCTL, ΔCTC, or ΔCTLC with 2 mm GTP and 2.5 mm MgCl2 (A) or 10 mm MgCl2 (B) spotted on grids 15 min after addition of nucleotide and stained with uranyl formate. Scale bar, 100 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles. C, inorganic phosphate (Pi) concentration in solution over time for reactions containing 4 μm FtsZ, ΔCTL, ΔCTC, or ΔCTLC with 2 mm GTP and 2.5 mm MgCl2 (n = 3). Error bars represent standard deviation. Straight lines indicate linear fits of averages. D, comparison of GTP hydrolysis rates (determined as the slope of linear fit for matched replicates of inorganic phosphate concentration over time) corresponding to the experiment in C. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, p > 0.05 for Tukey's multiple comparison test (one-way analysis of variance).
Figure 11.
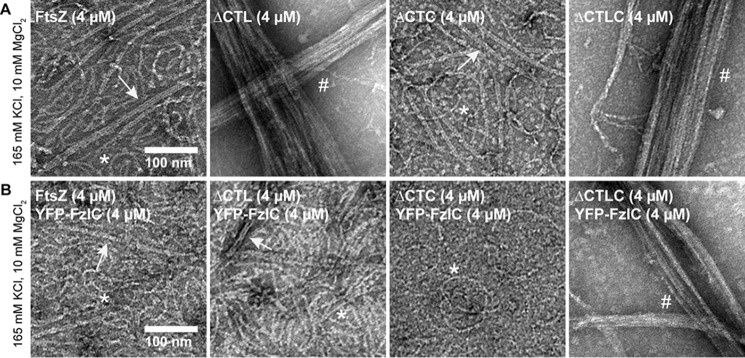
FzlC disrupts ΔCTL bundling in a CTC-dependent manner. A and B, electron micrographs of polymers formed by 4 μm FtsZ, ΔCTL, ΔCTC, or ΔCTLC with 2 mm GTP, 10 mm MgCl2, and 165 mm KCl in the absence (A) or presence (B) of 4 μm YFP-FzlC spotted on grids 15 min after addition of nucleotide. The difference in KCl concentration for this experiment is due to the high KCl concentration (300 mm) in FzlC storage buffer and low stock concentration of YFP-FzlC. Scale bars, 100 nm. *, single protofilaments; arrow, two- or three-filament bundles; #, multifilament bundles.
FzlC binding reduces lateral interaction between ΔCTL protofilaments
FtsZ-binding partners such as FzlA and ZapA have been implicated in increased lateral interaction (bundling) of protofilaments (12, 13, 32, 33). The increased bundling seen with ΔCTL appears more pronounced than the previously characterized effects of binding partners of C. crescentus FtsZ at similar polymerization conditions. Consequently, we investigated the influence of binding partners of FtsZ on the increased bundles observed for ΔCTL. Although ZapA has been implicated in increased bundling of FtsZ in E. coli, no such effect has been observed for C. crescentus ZapA (30). WT FtsZ protofilaments look indistinguishable with or without ZapA (supplemental Fig. 3C). We also failed to observe any difference in lateral interactions or structures of protofilaments formed by ΔCTL in the presence or absence of ZapA (supplemental Fig. 3C).
Next, we characterized the effects of FzlC on protofilament structure (Fig. 11). FzlC was recently identified as a membrane-anchoring protein for FtsZ in C. crescentus (12, 24). In vivo, FzlC binds specifically to FtsZ's CTC and recruits it to the membrane prior to the arrival of FtsA (12, 24). In vitro, His6-YFP-FzlC can recruit FtsZ-CFP to membranes of giant unilamellar vesicles in a CTC-dependent manner (24). At either low or high MgCl2 concentrations, we did not observe any effect of His6-YFP-FzlC (2 μm) on protofilament structure for WT FtsZ (4 μm) (Fig. 11B, FtsZ). Surprisingly, however, we found that the presence of His6-YFP-FzlC almost completely prevented bundling of ΔCTL at high MgCl2 concentrations (Fig. 11B, ΔCTL). In fact, the presence of His6-YFP-FzlC restores WT-like protofilament structures for ΔCTL. The presence of His6-YFP-FzlC did not affect the formation of multifilament bundles by ΔCTLC at high MgCl2 concentrations, suggesting that FzlC's ability to disrupt ΔCTL bundle formation requires its binding to the CTC (Fig. 11B, ΔCTLC). His6-YFP-FzlC did not have an observable effect on ΔCTC, except for an overall reduction in the number of filaments on the grid (Fig. 11B, ΔCTC). Thus, FzlC binding disrupts the increased lateral interaction in ΔCTL in a CTC-dependent manner.
ΔCTL-induced bundling is dominant to FzlA-induced bundling
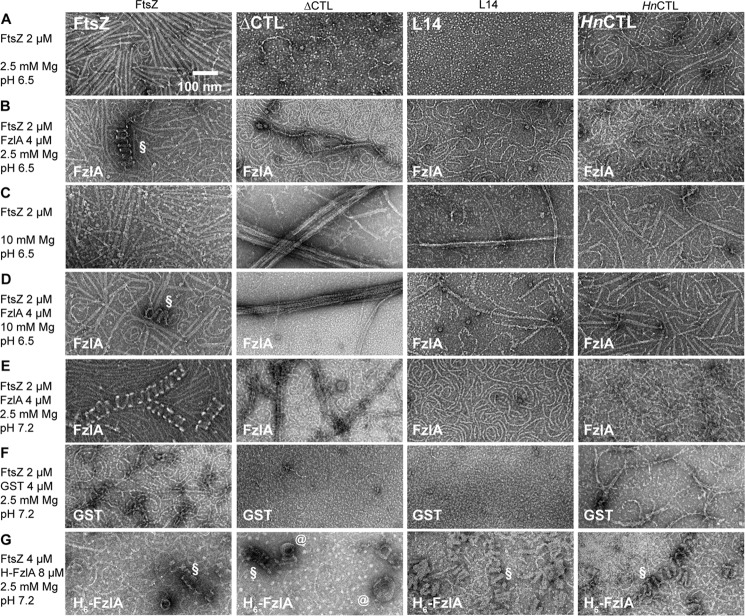
FtsZ protofilaments form helical bundles in the presence of FzlA in vitro (12). A recent characterization of FzlA from our laboratory confirmed that FzlA does not require the CTL or CTC to bind FtsZ.3 We therefore assessed the effects of FzlA on protofilaments formed by CTL variants by TEM (Fig. 12, refer to supplemental Figs. 4 and 5 for larger images). Our laboratory recently determined that, although WT FtsZ robustly forms helices with His6-tagged FzlA at pH 7.2 (12), it more robustly forms helices with untagged FzlA at pH 6.5.3 We therefore first examined the structures formed by the CTL variants with untagged FzlA at lower pH. Under these conditions, we observed helical bundles only for WT FtsZ (Fig. 12, B and D, and supplemental Fig. 4, B and D). However, we did observe an apparent stabilizing effect of FzlA on ΔCTL and L14 polymers (Fig. 12, A and D, and supplemental Fig. 4). At 2.5 mm MgCl2 and 50 mm KCl concentrations, we did not observe many protofilaments of ΔCTL or L14 in the absence of FzlA (Fig. 12A and supplemental Fig. 4A). However, in the presence of FzlA, we readily observed WT-like single filaments for both proteins (Fig. 12B and supplemental Fig. 4B). Moreover, we also observed straight double- and triple-filament bundles for ΔCTL with FzlA under these conditions (Fig. 12B and supplemental Fig. 4B). It is unclear whether this is an effect on protofilament stability in solution or on binding EM grids. At 10 mm MgCl2 concentration, we observed thick bundles for ΔCTL that were unaffected by the presence of FzlA (Fig. 10, C and D, and supplemental Fig. 4, C and D). Despite the increased number of protofilaments observed for L14 in the presence of FzlA, we still failed to observe very large bundles like those seen for ΔCTL either at low or high MgCl2 concentrations (Fig. 12, B and D, and supplemental Fig. 4, B and D). The structures formed by HnCTL appeared similar in the presence or absence of untagged FzlA at pH 6.5 and either low or high magnesium concentrations (Fig. 12, A–D, and supplemental Fig. 4).
Figure 12.
FzlA-induced bundling of FtsZ protofilaments into helices is CTL-dependent. A–E, electron micrographs of polymers formed by 2 μm FtsZ CTL variants in the presence or absence of 2 μm FzlA at different pH (6.5 versus 7.2) with 2.5 or 10 mm MgCl2, 50 mm KCl, and 2 mm GTP spotted on grids 15 min after addition of nucleotide and stained with uranyl formate. F, electron micrographs of polymers formed by 2 μm FtsZ CTL variants in the presence of 2 μm glutathione S-transferase (GST) at pH 7.2 with 2.5 mm MgCl2, 50 mm KCl, and 2 mm GTP spotted on grids 15 min after addition of nucleotide and stained with uranyl formate. G, electron micrographs of polymers formed by 4 μm FtsZ CTL variants in the presence of 8 μm His6-FzlA at pH 7.2 with 2.5 mm MgCl2, 50 mm KCl and 2 mm GTP spotted on grids 15 min after addition of nucleotide and stained with uranyl formate. Scale bar, 100 nm. §, helical bundles of FtsZ (or CTL variant) with FzlA; @, spiral structures seen for ΔCTL with His6-FzlA.
We also assessed the effects of untagged FzlA on the structures of polymers formed by CTL variants at the presumably more physiological pH of 7.2 (Fig. 12E and supplemental Fig. 5). Again, we observed helical bundles only for WT FtsZ, and we observed a stabilizing effect of FzlA on ΔCTL and L14 (Fig. 12E and supplemental Fig. 5, A and B). This effect was specific to FzlA because glutathione S-transferase, which has structural homology to FzlA but does not bind FtsZ, did not affect polymer structure or density on grids under identical conditions (Fig. 12F).
To use a more physiological pH while simultaneously favoring FzlA-induced bundling, we decided to use 8 μm His6-FzlA and 4 μm FtsZ or CTL variants at 2.5 mm MgCl2 concentration and pH 7.2. Under these conditions, all of the linker variants formed helical bundles similar to those observed for WT FtsZ (Fig. 12G and supplemental Fig. 5C). We observed fewer and shorter helical bundles for ΔCTL/His6-FzlA compared with the other CTL variants or WT. Surprisingly, we also observed flat spiral or coiled structures for ΔCTL/His6-FzlA (Fig. 12G, denoted by @, and supplemental Fig. 5C, inset). These structures resemble a highly-curved filament or filament bundle coiling onto itself. We hypothesize that these structures could result from increased self-interaction laterally between adjacent turns of helical double- or triple-filament bundles. Overall, our observations indicate that the CTL can affect FzlA-induced protofilament bundling and curvature. This is surprising because FzlA does not require the CTL for binding FtsZ3 and suggests that the CTL may influence the FzlA–FtsZ interaction through its intrinsic effects on the FtsZ assembly.
Discussion
In this study, we demonstrate a role for the intrinsically disordered C-terminal linker region of C. crescentus FtsZ in regulating polymer structure and dynamics in vitro. FtsZ variants lacking the CTL entirely (ΔCTL, ΔCTLC) exhibit increased propensity to bundle and reduced GTP hydrolysis rate and turnover compared with the other CTL variants or WT (Fig. 10). The exceptionally long and thick straight bundles of ΔCTL protofilaments form independently of GTP hydrolysis rates, because we see long, thick bundles only for ΔCTL even with the slowly hydrolyzed GTP analog GMPCPP (Figs. 2, B and C, and 3, B and C). Combining biochemistry, spectroscopy, and electron microscopy approaches, we confirmed that these bundles are not artifacts of observing a poorly polymerizing or depolymerizing FtsZ mutant under conditions that promote crowding (high MgCl2 or protein concentration). We confirmed that WT FtsZ and ΔCTL can copolymerize and observed that FtsZ/ΔCTL bundled copolymers appear much longer than FtsZ polymers (Fig. 9). We show that FzlC binding to the CTC domain can disrupt the ability of ΔCTL to form bundles (Fig. 11). This observation suggests a role for the membrane-anchoring protein FzlC in regulating FtsZ polymer structure and dynamics in a CTC-dependent manner. Additionally, it also suggests that ΔCTL has the ability to form gently curved single filaments given the right conditions and/or binding partners, but intrinsically it prefers to form bundles. Finally, we described a previously unidentified role for the CTL in facilitating FzlA to form helical bundles of FtsZ (Fig. 12 and supplemental Figs. 4 and 5). Overall, our characterization of the CTL variants and their interaction with FtsZ-binding proteins under different polymerizing conditions provide novel insights into the contributions of the CTL to FtsZ polymerization dynamics.
Based on our observations, we propose the following roles for the disordered linker in regulating lateral and longitudinal interactions between FtsZ monomers in protofilaments (Fig. 13). Following addition of GTP, WT FtsZ undergoes fast polymerization (activation, nucleation, and elongation), reaches a steady state characterized by dynamic turnover, annealing and fragmentation and transient lateral interactions, and eventually depolymerizes following exhaustion of GTP. The CTL likely plays a role in maintaining optimal interactions between protofilaments laterally, acting as flexible, charged “repulsive brushes” extending orthogonally around the protofilaments. Such a mechanism of regulating lateral interaction between polymers has been described for the intrinsically disordered C-terminal region of neurofilaments (34). Additionally, the CTL could influence longitudinal interactions between monomers through a similar electrostatic repulsion mechanism and thereby influence monomer on and off rates and/or fragmentation and annealing. In the case of ΔCTL, lateral interactions are more likely to form and less likely to break apart, with a strong propensity for the protofilaments to form long multifilament bundles. Moreover, ΔCTL might lack the repulsive interactions between adjacent monomers in a protofilament and therefore affect depolymerization and/or fragmentation. Together, these differences in lateral and longitudinal interactions would result in more stable polymers and slower turnover of monomers for ΔCTL.
Figure 13.
CTL regulates lateral interactions between FtsZ protofilaments. Schematic describing polymerization dynamics of FtsZ and highlighting the proposed roles for the CTL in regulating longitudinal and lateral interactions. FtsZ monomers bind GTP, nucleate, and assemble into short protofilaments that can elongate further and/or anneal together. GTP-bound monomers within the protofilaments can undergo GTP hydrolysis leading to a conformational change that results in destabilization of longitudinal interactions. The CTL might play a role in regulating monomer–monomer interactions that contribute to this destabilization. This destabilization leads to fragmentation and/or depolymerization. At steady state, the forward reactions, nucleation, elongation, and annealing, balance out the reverse reactions, depolymerization and fragmentation, as long as GTP-bound FtsZ monomers are available. FtsZ protofilaments can also form lateral interactions that are most likely disrupted prior to fragmentation and/or depolymerization. We propose that the CTL plays a role in regulating this lateral interaction by functioning as a repulsive brush around FtsZ protofilaments. In the absence of the CTL (or both the CTL and the CTC), lateral interaction between protofilaments is stronger and/or less specific, thereby favoring the assembly of protofilaments into stable bundles. Furthermore, bundle formation could reduce protofilament turnover by stabilizing protofilaments against fragmentation/depolymerization.
We consistently observed reduced lateral interaction for HnCTL (165 aa CTL, Figs. 1A, 2, 3, and 12) protofilaments compared with WT (172 aa CTL, Fig. 1A), by EM, pelleting, and light scatter assays. It is interesting to note that although C. crescentus CTL has a net charge of −9 and pI of 4.66, HnCTL has a net charge of −18 and pI of 4.2. The reduction in lateral interaction for HnCTL, which has a more negatively charged CTL than WT, further supports our model that electrostatic repulsion between protofilaments mediated by the CTL is important for optimal lateral interaction and bundle formation. A similar observation has been reported earlier for B. subtilis FtsZ CTL variants; replacing the CTL (50 aa) of BsFtsZ with the CTL from E. coli (50 aa) or a 50-aa region of the CTL from Agrobacterium tumefaciens causes reduction in lateral interaction both by EM and light scatter (25). Whether the CTL sequence-dependent difference in protofilament bundling is due to CTL–CTL interactions or CTL–GTPase domain interactions between adjacent protofilaments needs further investigation, for example, by engineering CTL mutants to alter net charge and/or disorder and determining their assembly properties. An alternative explanation for the differences between WT and HnCTL could be that the CTL is in fact not disordered entirely but is capable of assuming secondary conformations that regulate interprotofilament interactions. However, the CTL sequence across species is predicted to result in intrinsically disordered peptides making this possibility less likely.
Across species of bacteria, many members of the division machinery, such as FzlA and the Zap family of proteins, have been implicated in promoting interactions between FtsZ protofilaments in vitro and/or regulating the density of protofilaments in the Z-ring (12, 13, 32, 33, 35, 36). Genetic manipulations of these proteins result in cell division defects, suggesting a role for regulating interfilament interactions in the cytokinetic function of FtsZ (13, 30).3 At least for the formation of helical bundles in vitro, untagged FzlA appears to require the CTL (Fig. 12). We have no evidence that FzlA interacts with the CTL. Thus, it is possible that the different intrinsic polymerization properties or lateral interactions of different CTL variants affect the curvature or polymer dynamics required for forming helical bundles upon FzlA binding. The His6-tagged variant of FzlA can form robust helices with all linker variants (Fig. 12G), which might be due to altered interactions with FtsZ. Interestingly, His6-FzlA also causes ΔCTL to form unique spiral or coiled structures that have never been observed for WT FtsZ with FzlA again suggesting that His6-FzlA might have non-canonical interactions with FtsZ.
Many proteins that bind the CTC of FtsZ have been shown to affect Z-ring structure in vivo and/or affect FtsZ polymer structure in vitro (24, 37–39). It is unclear if and how these proteins regulate polymerization of FtsZ without interacting directly with the GTPase domain, because the CTC and the GTPase domains are linked by a long unstructured CTL. In the context of ΔCTL, the CTC does not obviously contribute to the regulation of protofilament structure or lateral interaction in C. crescentus FtsZ. However, FzlC binding to the CTC disrupts ΔCTL bundling. FzlC is conserved in α-proteobacteria (12). It acts as a membrane-anchoring protein for FtsZ in C. crescentus and may play a similar role in related species (24). Whether FzlC is relevant for regulating inter-protofilament spacing in WT FtsZ is unclear. It is curious, however, that whereas ZapA and FzlA, which bind the GTPase domain of FtsZ (13, 33),3 do not affect bundling of ΔCTL protofilaments, binding of FzlC to the CTC region disrupts bundling. It is possible that the binding of FzlC to the CTC of ΔCTL disrupts inter-protofilament lateral interactions at the GTPase domain by restricting the orientation of protofilaments. Alternatively, FzlC bound to the CTC of ΔCTL may perform a repulsive function to limit lateral interaction similar to the mechanism we propose for the CTL itself. Interestingly, the effect of FzlC on preventing ΔCTL bundles occurs in the absence of membrane, indicating that this ability of FzlC, if relevant in vivo, is independent from its function as a membrane anchor for FtsZ. Further in vitro studies on the interaction between FzlC and FtsZ would be required to understand the mechanism underlying this observation and to determine whether it extends to other factors that bind the CTC, such as FtsA. Differences in the effects of membrane-anchoring proteins such as FzlC or FtsA on FtsZ polymerization could contribute to the roles of these proteins in cell division, in addition to their abilities to recruit FtsZ protofilaments to the membrane.
How transient or dynamic are the bundles observed for ΔCTL? Does WT FtsZ also form similar bundles that dissociate more quickly and are difficult to capture for TEM? Our data from experiments using GMPCPP suggest that reducing turnover of FtsZ does not result in the same extent of bundling as observed for ΔCTL. Although TEM provides very high spatial resolution of protofilaments and polymer structure, it has two major limitations. First, we cannot observe changes in structures over time. Second, differences in staining and affinity for the carbon-coated grids could bias the observer toward large bundles and/or well spaced polymers. To overcome these limitations and resolve FtsZ polymerization dynamics, we would require high-resolution time-lapse microscopy and/or spectroscopy techniques.
Our characterization of the CTL variants and FtsZ mutants provides biochemical tools for resolving the roles of longitudinal and lateral interactions in regulating protofilament structures in vitro and in vivo. Similar studies of polymerizing proteins such as actin and microtubules in eukaryotes have potentiated a relatively thorough understanding of the regulation and function of their assembly properties. Our recent understanding that the Z-ring is more than a passive scaffold for the recruitment of cell wall enzymes and the role for its dynamics in regulating cytokinesis emphasizes the need for understanding its polymerization properties in molecular detail. Additionally, although our observations strengthen the correlation between polymerization dynamics in vitro and Z-ring structure and function in vivo, whether this correlation is suggestive of causation or the result of a confounding effect of the CTL on unrelated processes requires further investigation.
Experimental procedures
Purification of proteins
FtsZ and FtsZ variants (Fig. 1A), including FtsZL72W and ΔCTLL72W tryptophan mutants, were purified using the protocol described previously (28). Briefly, pET21 vectors (pMT219-FtsZ, pEG681-ΔCTL, pEG723-L14, pEG676-HnCTL, pEG765-ΔCTC, pEG678-ΔCTLC, and pEG948-FtsZL72W (29) and pEG1077-ΔCTLL72W) were used to express ftsZ or ftsZ variants in E. coli Rosetta(DE3)pLysS cells induced with 0.5 mm IPTG at 37 °C for 3 h when the A600 reached 1.0. Cells were pelleted, resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 50 mm KCl, 1 mm EDTA, 10% glycerol, DNase I, 1 mm β-mercaptoethanol, 2 mm PMSF with cOmplete mini, EDTA-free protease inhibitor tablet (Roche Applied Science)), and lysed using lysozyme treatment (1 mg/ml) followed by sonication. After anion-exchange chromatography (HiTrap Q HP 5 ml, GE Healthcare) and elution with a linear gradient of KCl, the fractions containing the FtsZ variant were pooled and subjected to ammonium sulfate precipitation. The ammonium sulfate precipitates (at 20–35% saturation, depending on the variant) were verified for each FtsZ variant by electrophoresis (SDS-PAGE) and Coomassie staining. The precipitate was resuspended in FtsZ storage buffer (50 mm HEPES-KOH, pH 7.2, 50 mm KCl, 0.1 mm EDTA, 1 mm β-mercaptoethanol, 10% glycerol) and purified further using size-exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare), snap-frozen in liquid nitrogen, and stored at −80 °C in FtsZ storage buffer.
ZapA was purified as described previously (30). His6-SUMO-ZapA was produced from plasmid pEG620 in E. coli Rosetta(DE3)pLysS cells induced with 0.5 mm IPTG for 4 h at 37 °C. Cells were pelleted, resuspended in ZapA lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 20 mm imidazole, 10% glycerol, 2 mm PMSF with cOmplete mini, EDTA-free protease inhibitor tablet (Roche Applied Science)), and lysed with lysozyme treatment (1 mg/ml lysozyme, 2.5 mg/ml MgCl2, 1 mmCaCl2, and 2 units/ml DNase I) and sonication. His6-SUMO-ZapA was purified using a HisTrap FF 1-ml column (GE Healthcare) and eluted with 300 mm imidazole. The His6-SUMO tag was cleaved overnight during dialyses into ZapA lysis buffer using SUMO protease (His6-Ulp1) at a 100-fold molar excess. Cleaved His6-SUMO, uncleaved His6-SUMO-ZapA, and His6-Ulp1 were separated from untagged ZapA by passage over a HisTrap FF 1-ml column once again, this time collecting the unbound fraction. ZapA was further purified using anion exchange (HiTrap Q HP 1 ml, GE Healthcare) in ZapA QA buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% glycerol) with a linear gradient of NaCl and was dialyzed into FtsZ storage buffer before snap-freezing and storage at −80 °C.
His6-YFP-FzlC was purified as described previously (24). Briefly, His6-YFP-FzlC was produced in E. coli Rosetta (DE3)pLysS cells from plasmid pEG420 using 30 μm IPTG to induce expression overnight at 15 °C (24). Cells were pelleted and resuspended in FzlC lysis buffer (50 mm Tris-HCl, pH 8.0, 1 m KCl, 20 mm imidazole, 1 mm β-mercaptoethanol, and 20% glycerol, 2 mm PMSF with cOmplete mini, EDTA-free protease inhibitor tablet (Roche Applied Science)) and lysed using lysozyme treatment (with 1 mg/ml lysozyme, 2.5 mg/ml MgCl2, and 2 units/ml DNase I) and sonication. Lysate was supplemented with 3 mm ATP (to reduce DnaK co-purification), and His6-YFP-FzlC was purified using affinity chromatography (His-Trap FF 1-ml column, GE Healthcare) followed by gel filtration (Superdex 200 10/300 GL column, GE Healthcare) and was stored in FzlC storage buffer (50 mm Tris-HCl, pH 8.0, 300 mm KCl, 0.1 mm EDTA, 1 mm β-mercaptoethanol, and 10% glycerol) at −80 °C.
His6-FzlA was purified essentially as described previously (12). His6-FzlA was expressed from plasmid pEG994 in E. coli Rosetta(DE3)pLysS cells by induction with 0.5 mm IPTG for 4 h at 30 °C when A600 reached 0.5. Cells were pelleted, resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20 mm imidazole, 10% glycerol, 2 mm PMSF with cOmplete mini, EDTA-free protease inhibitor tablet (Roche Applied Science)), and lysed using lysozyme treatment (1 mg/ml lysozyme, 2 units/ml DNase I, 2.5 mm MgCl2) and sonication. His6-FzlA protein was isolated using affinity chromatography (HisTrap FF 1-ml column, GE Healthcare), eluted with 300 mm imidazole, and dialyzed into FzlA storage buffer (50 mm HEPES-KOH, pH 7.2, 300 mm KCl, pH 8.0, 10% glycerol).
For the purification of untagged FzlA, His6-SUMO-FzlA was expressed from plasmid pEG327 in E. coli Rosetta(DE3)pLysS cells by induction with 0.5 mm IPTG for 4 h at 30 °C. His6-SUMO-FzlA was purified and cleaved to FzlA using a similar protocol to ZapA purification from His6-SUMO-ZapA, with a few changes; FzlA lysis buffer (50 mm HEPES-KOH, pH 7.2, 300 mm KCl, 20 mm imidazole, 10% glycerol) was used for lysis. Affinity chromatography (HisTrap FF 1-ml column, GE Healthcare) followed by SUMO protease cleavage produced untagged FzlA that was further purified as the unbound fraction of another passage of a HisTrap FF 1-ml column (GE Healthcare). Untagged FzlA was dialyzed and stored in 50 mm HEPES-KOH, pH 7.2, 300 mm KCl, 10% glycerol.
GST was produced from pGEX4T1 in E. coli Rosetta (DE3)pLysS cells grown at 30 °C and induced at A600 of 0.5 with 0.5 mm IPTG for 3 h. Cells were resuspended in phosphate-buffered saline (PBS) with 300 mm KCl, lysed with 1 mg/ml lysozyme treatment and sonication, and purified by affinity chromatography (glutathione-Sepharose 4B column, GE Healthcare). Protein was eluted with 10 mm glutathione and dialyzed into GST storage buffer (50 mm HEPES-KOH, pH 7.2, 50 mm KCl, 0.1 mm EDTA) before snap freezing and storage at −80 °C. His6-FzlA, FzlA, and GST were buffer-exchanged into FtsZ storage buffer (50 mm HEPES-KOH, pH 7.2, 0.1 mm EDTA, 50 mm KCl, 10% glycerol), prior to addition to polymerization reactions with FtsZ.
Polymerization buffer and conditions
HEK50, pH 7.2 buffer (50 mm HEPES-KOH, pH 7.2, 0.1 mm EDTA, 50 mm KCl), was used for FtsZ polymerization assays, unless specified otherwise. 2.5 mm MgCl2 (low magnesium), 10 mm MgCl2 (high magnesium), or 300 mm KCl (high potassium) were used as mentioned under “Results” and figure legends. GTP or GMPCPP was used at 2 or 0.2 mm concentrations, respectively, unless specified otherwise. For experiments with FtsZ-CTL variants and FzlA at pH 6.5, MESK50 pH 6.5 buffer (50 mm MES-KOH, pH 6.5, 50 mm KCl) was used. All reactions were carried out at room temperature. When additional proteins were added, the same volume of the corresponding storage buffer was added to a corresponding control reaction with no added protein.
GTP hydrolysis rate measurement
GTP hydrolysis rates were measured using a malachite green dye-based reporter for inorganic phosphate as described previously (28). At the beginning of the reaction, 2 mm GTP was added to FtsZ or FtsZ variants in polymerization buffer at 2.5 or 10 mm MgCl2 concentration and 50 or 300 mm KCl concentration. Reaction was stopped using quench buffer (50 mm HEPES-KOH, pH 7.2, 21.3 mm EDTA, 50 mm KCl), and inorganic phosphate in the solution was measured using SensoLyte MG phosphate assay kit Colorimetric (AnaSpec, Inc., Fremont, CA).
Polymerization kinetic assays
FtsZ polymerization was measured using Fluoromax-3 spectrofluorometer (Jobin Yvon Inc.) to measure right angle light scatter (excitation and emission at 350 nm, 2-nm slits). Tryptophan fluorescence experiments using FtsZL72W and ΔCTLL72W mutants were performed using the same equipment, with excitation and emission at 295 and 344 nm, respectively, with 2-nm slits. Because GTP is fluorescent at these excitation/emission conditions, 50 μm GTP was used for the tryptophan assay (29). Similarly, 100 μm GMPCPP was used for the tryptophan fluorescence assay with increasing MgCl2 concentrations (Fig. 8, A and B). Note that addition of 100 μm GMPCPP does not cause significant increase in fluorescence signal at 344 nm by itself (Fig. 8, A and B). In both right angle light scatter and tryptophan fluorescence experiments, measurements were taken every 10 s.
High-speed pelleting assay
Steady-state polymer mass was measured using high-speed pelleting assay as described previously (12, 28). Briefly, FtsZ or FtsZ variants in storage buffer was centrifuged at 250,000 × g for 15 min at 4 °C to pellet and remove nonspecific aggregates in the absence of GTP. The clarified FtsZ variant (from the supernatant) was then incubated for 15 min with 2 mm GTP and 10 mm MgCl2 in HEK50 pH 7.2 buffer with 0.05% Triton X-100. Polymers were then pelleted by ultracentrifugation at 250,000 × g for 15 min at 25 °C. The concentrations of protein in the supernatant and pellet were estimated using SDS-PAGE and Coomassie staining followed by gel analysis using ImageLab (Bio-Rad).
Transmission electron microscopy
Polymers formed by FtsZ or FtsZ variants (in the presence or absence of FtsZ-binding proteins) were visualized by TEM as described earlier (28). Reaction mixtures of the relevant proteins in FtsZ polymerization buffer were spotted onto glow-discharged carbon-coated copper grids (Electron Microscopy Sciences, Hatfield, PA) at least 15 min after addition of GTP except when noted otherwise, blotted, and stained twice with 0.75% uranyl formate for 2 min. The grids were dried and imaged using a Philips/FEI BioTwin CM120 TEM (operated at 80 kV) equipped with an AMT XR80 8 megapixel CCD camera (AMT Imaging).
Author contributions
K. S. and E. D. G. designed the study. K. S. performed the experiments. K. S. and E. D. G. analyzed the results, wrote the paper, and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Harold Erickson (Duke University) for providing the construct for the purification of FtsZL72W. We also thank the following members of the Goley Laboratory: Elizabeth Meier, P. J. Lariviere, and Selam Woldemeskel for guidance with optimizing assays with FzlC, FzlA, and ZapA, respectively; and Allison Daitch and Chris Mahone for useful discussions about the manuscript. We also thank Mike Delannoy, Barbara Smith, and the Microscopy Facility of Johns Hopkins School of Medicine for assistance with the electron microscopy imaging. Finally, we thank Jie Xiao and Anthony Vecchiarelli for providing useful insights on this work.
This work was supported by the National Institutes of Health Grant R01GM108640 (to E. D. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. 1–5.
Lariviere, P. J., Szwedziak, P., Mahone, C., Löwe, J., and Goley, E. D. (2017). FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol. Microbiol. 10.1111/mmi.13876.
- CTC
- C-terminal conserved peptide
- CTL
- C-terminal linker
- TEM
- transmission electron microscopy
- GMPCPP
- guanosine-5′-[(α,β)-methyleno]triphosphate
- aa
- amino acid
- IPTG
- isopropyl β-d-1-thiogalactopyranoside.
References
- 1. Cabeen M. T., and Jacobs-Wagner C. (2005) Bacterial cell shape. Nat. Rev. Microbiol. 3, 601–610 [DOI] [PubMed] [Google Scholar]
- 2. Typas A., Banzhaf M., Gross C. A., and Vollmer W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Erickson H. P., Anderson D. E., and Osawa M. (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meier E. L., and Goley E. D. (2014) Form and function of the bacterial cytokinetic ring. Curr. Opin. Cell Biol. 26, 19–27 [DOI] [PubMed] [Google Scholar]
- 5. Haeusser D. P., and Margolin W. (2016) Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 14, 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z., Trimble M. J., Brun Y. V., and Jensen G. J. (2007) The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu G., Huang T., Buss J., Coltharp C., Hensel Z., and Xiao J. (2010) In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5, e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang X., Lyu Z., Miguel A., McQuillen R., Huang K. C., and Xiao J. (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bisson-Filho A. W., Hsu Y.-P., Squyres G. R., Kuru E., Wu F., Jukes C., Sun Y., Dekker C., Holden S., VanNieuwenhze M. S., Brun Y. V., and Garner E. C. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang K. H., Durand-Heredia J., and Janakiraman A. (2013) FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wagstaff J. M., Tsim M., Oliva M. A., García-Sanchez A., Kureisaite-Ciziene D., Andreu J. M., and Löwe J. (2017) A polymerisation-associated conformational switch in FtsZ that enables treadmilling. mBio 8, e00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goley E. D., Dye N. A., Werner J. N., Gitai Z., and Shapiro L. (2010) Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galli E., and Gerdes K. (2012) FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 194, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buss J., Coltharp C., Shtengel G., Yang X., Hess H., and Xiao J. (2015) A multilayered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11, e1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Löwe J., and Amos L. A. (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 [DOI] [PubMed] [Google Scholar]
- 16. Egelman E. H. (1998) Tubulin family: kinship of key proteins across phylogenetic domains. Curr. Biol. 8, R288–R290 [DOI] [PubMed] [Google Scholar]
- 17. Mukherjee A., and Lutkenhaus J. (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scheffers D. J., den Blaauwen T., and Driessen A. J. (2000) Non-hydrolysable GTP-γ-S stabilizes the FtsZ polymer in a GDP-bound state. Mol. Microbiol. 35, 1211–1219 [DOI] [PubMed] [Google Scholar]
- 19. Mukherjee A., and Lutkenhaus J. (1999) Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hou S., Wieczorek S. A., Kaminski T. S., Ziebacz N., Tabaka M., Sorto N. A., Foss M. H., Shaw J. T., Thanbichler M., Weibel D. B., Nieznanski K., Holyst R., and Garstecki P. (2012) Characterization of Caulobacter crescentus FtsZ protein using dynamic light scattering. J. Biol. Chem. 287, 23878–23886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaughan S., Wickstead B., Gull K., and Addinall S. G. (2004) Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58, 19–29 [DOI] [PubMed] [Google Scholar]
- 22. Din N., Quardokus E. M., Sackett M. J., and Brun Y. V. (1998) Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 27, 1051–1063 [DOI] [PubMed] [Google Scholar]
- 23. Pichoff S., and Lutkenhaus J. (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55, 1722–1734 [DOI] [PubMed] [Google Scholar]
- 24. Meier E. L., Razavi S., Inoue T., and Goley E. D. (2016) A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol. Microbiol. 101, 265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buske P. J., and Levin P. A. (2013) A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gardner K. A., Moore D. A., and Erickson H. P. (2013) The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol. 89, 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buske P. J., Mittal A., Pappu R. V., and Levin P. A. (2015) An intrinsically disordered linker plays a critical role in bacterial cell division. Semin. Cell Dev. Biol. 37, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sundararajan K., Miguel A., Desmarais S. M., Meier E. L., Casey Huang K., and Goley E. D. (2015) The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 6, 7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Milam S. L., and Erickson H. P. (2013) Rapid in vitro assembly of Caulobacter crescentus FtsZ protein at pH 6.5 and 7.2. J. Biol. Chem. 288, 23675–23679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Woldemeskel S. A., McQuillen R., Hessel A. M., Xiao J., and Goley E. D. (2017) A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus. Mol. Microbiol. 105, 721–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X., Huang J., Mukherjee A., Cao C., and Lutkenhaus J. (1997) Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179, 5551–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gueiros-Filho F. J., and Losick R. (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mohammadi T., Ploeger G. E., Verheul J., Comvalius A. D., Martos A., Alfonso C., van Marle J., Rivas G., and den Blaauwen T. (2009) The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48, 11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoh J. H. (1998) Functional protein domains from the thermally driven motion of polypeptide chains: a proposal. Proteins 32, 223–228 [PubMed] [Google Scholar]
- 35. Durand-Heredia J., Rivkin E., Fan G., Morales J., and Janakiraman A. (2012) Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buss J., Coltharp C., Huang T., Pohlmeyer C., Wang S.-C., Hatem C., and Xiao J. (2013) In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 89, 1099–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loose M., and Mitchison T. J. (2014) The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schumacher M. A., Huang K.-H., Zeng W., and Janakiraman A. (2017) Structure of the Z ring-associated protein, ZapD, bound to the C-terminal domain of the tubulin-like protein, FtsZ, suggests mechanism of Z ring stabilization through FtsZ cross-linking. J. Biol. Chem. 292, 3740–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y., Huang H., Osawa M., and Erickson H. P. (2017) ZipA and FtsA* stabilize FtsZ-GDP miniring structures. Sci. Rep. 7, 3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.