Abstract
Moniliophthora perniciosa is the causative agent of witches' broom disease, which devastates cacao cultures in South America. This pathogenic fungus infects meristematic tissues and derives nutrients from the plant apoplast during an unusually long-lasting biotrophic stage. To survive, the fungus produces proteins to suppress the plant immune response. Proteins of the PR-1 (pathogenesis-related 1)/CAP superfamily have been implicated in fungal virulence and immune suppression. The genome of M. perniciosa encodes 11 homologues of plant PR-1 proteins, designated MpPR-1 proteins, but their precise mode of action is poorly understood. In this study, we expressed MpPR-1 proteins in a yeast model lacking endogenous CAP proteins. We show that some members of the MpPR-1 family bind and promote secretion of sterols, whereas others bind and promote secretion of fatty acids. Lipid binding by purified MpPR-1 occurs with micromolar affinity and is saturable in vitro. Sterol binding by MpPR-1 requires the presence of a flexible loop region containing aromatic amino acids, the caveolin-binding motif. Remarkably, MpPR-1 family members that do not bind sterols can be converted to sterol binders by a single point mutation in the caveolin-binding motif. We discuss the possible implications of the lipid-binding activity of MpPR-1 family members with regard to the mode of action of these proteins during M. perniciosa infections.
Keywords: fatty acid, fatty acid binding protein, infection, lipid-protein interaction, sterol
Introduction
Witches' broom disease of cacao (WBD)4 is one of the most devastating plant diseases in Southern America and a major threat to cacao production, resulting in extensive economic losses in affected countries (1, 2). WBD is caused by the basidiomycete fungus Moniliophthora perniciosa (3). This hemibiotrophic fungus has an atypical prolonged biotrophic stage of up to 90 days, during which it slowly grows in the apoplast of infected plants, inducing conspicuous morphological alterations that culminate with the formation of anomalous structures called “green brooms.” These chlorotic and swollen shoots are formed as a consequence of a hormonal imbalance and intense reprogramming of the plant metabolism induced by the fungal infection (4, 5). The necrotrophic phase of the disease begins upon the death of plant tissues, which are invaded by the proliferative mycelia. M. perniciosa also infects cacao flowers and fruits, causing swelling and abnormal ripening of the fruits and ultimately death of the infected tissues (1). A genome draft of M. perniciosa (6), a comprehensive transcriptomic analysis of the biotrophic stage of the interaction (4), and a series of functional analyses of M. perniciosa proteins provided insights into this intriguing disease (7–10).
Members of the CAP (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1) superfamily of proteins, also known as SCP/TAPS (sperm-coating protein/Tpx-1/Ag5/PR-1/Sc7) proteins, are present throughout the eukaryotic kingdom (11, 12). In plants, PR-1 (pathogenesis-related 1) superfamily members serve markers of an induced defense against pathogens (13). Interestingly, CAP proteins are abundantly expressed by human pathogenic nematodes such as Schistosoma mansoni and Necator americanus during infection and were speculated to function as immunomodulators to inhibit host defense (14). A CAP protein from a single isolate of Fusarium oxysporum f. sp. lycopersici, which can cause disease on both tomato plants and immunodepressed mice, is important in the infection of the animal model. However, this protein is not essential for virulence on tomato (15). Gr-Vap1, a CAP protein from the potato cyst nematode Globodera rostochiensis, on the other hand, induces the loss of basal immunity to unrelated pathogens when heterologously expressed in plants (16). In addition, the knockdown of Gr-Vap1 expression reduces the infectivity of nematode, indicating that this protein is important for pathogen-induced suppression of plant defense. An immunomodulatory function was also suggested for tablysin-15, a CAP protein from the horsefly Tabanus yao. Tablysin-15 binds leukotrienes and free fatty acids and thereby might inhibit the proinflammatory effects of leukotrienes that are released from mast cells in the vicinity of the bite (17).
The Saccharomyces cerevisiae CAP proteins, the Pry proteins (pathogen related in yeast) function to bind and export sterols and fatty acids out of the cells (18, 19). The sterol-binding activity is conserved among many CAP superfamily proteins, because expression of human CRISP2, parasitic SmVAL4, or plant PR-1 rescues the lack of Pry function in yeast, and these proteins bind cholesterol in vitro (18, 20, 21). Expression of tablysin-15, however, does not rescue the sterol export phenotype but rescues export of fatty acids (19). The sterol and fatty acid binding and export function of the yeast CAP proteins is confined to the CAP domain, because expression of the CAP domain alone is sufficient to rescue the sterol and fatty acid export phenotype of a pry1Δ pry2Δ double mutant, and the CAP domain of Pry1 alone binds sterols and fatty acids at distinct sites in vitro (19, 22, 23). Sterol binding by Pry proteins requires the displacement of a flexible loop containing aromatic amino acids, termed the caveolin-binding motif (CBM) within the CAP domain, because point mutations within this motif abolish sterol export and binding, whereas fatty acid binding takes place in the groove between two parallel running helices, α1 and α3 (19, 24). Taken together, these data indicate that CAP proteins may exert an immunomodulatory function through binding hydrophobic ligands, such as sterols or related small hydrophobic compounds and fatty acids and their derivatives such as leukotrienes (19, 23, 25).
The genome sequence of M. perniciosa indicates the presence of 11 CAP family members in this phytopathogen (6, 26). Furthermore, gene expression analyses revealed that some of M. perniciosa CAP genes, termed MpPR-1 genes, are highly and specifically expressed during the green broom stage of WBD and in germinating basidiospores (4, 26). These results suggest that some MpPR-1 proteins function during the M. perniciosa infective process.
In this work, we examine whether MpPR-1 proteins have lipid-binding activity in vivo, by complementing yeast Pry mutants, and in vitro, through sterol- and fatty acid-binding assays. Five of the ten MpPR-1 family members tested, MpPR-1c, MpPR-1d, MpPR-1g, MpPR-1j, and MpPR-1k, complemented the defect in sterol export of yeast mutants lacking Pry function. Consistent with their activity in vivo, MpPR-1d and MpPR-1k bound cholesterol in vitro. Interestingly, two of the MpPR-1 family members that did not bind sterols, MpPR-1e and MpPR-1i, bound fatty acids in vitro, and they also complemented the block in fatty acid export of mutant cells lacking their endogenous CAP function. Closer inspection of the sequence of the CBM of these four MpPR-1 proteins revealed that proteins that bound sterols have a conserved CBM, whereas those that did not bind harbored substitutions of an aromatic residue at position 3 of the CBM. Remarkably, single mutations within the CBM converted a sterol binder into a non-binder and vice versa. These data thus substantiate the importance of the CBM for sterol binding, and they suggest that the number of CAP proteins present in M. perniciosa and their sequence diversity might serve to bind a variety of small hydrophobic ligands to suppress the immune defense of its host.
Results
MpPR-1 proteins export sterols in vivo
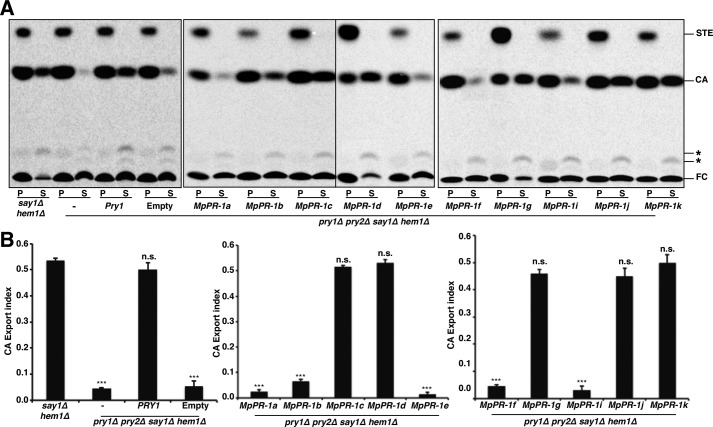
To determine whether the CAP proteins from the cacao pathogen M. perniciosa could bind sterols, we first tested whether they would complement a block in sterol export of yeast cells lacking their endogenous CAP family members Pry1 and Pry2. The yeast Pry proteins bind cholesterol in vitro and are required for the export of cholesteryl acetate into the culture supernatant in vivo (18). To test whether the expression of the MpPR-1s in pry1Δ pry2Δ double mutant cells rescued the defect in cholesterol export, hem1 and say1 (hem1Δ say1Δ) deficient cells of the indicated genotype were cultivated in the presence of radiolabeled [14C]cholesterol overnight. In these experiments, heme-deficient cells are being used because unlike wild-type cells, heme mutants can take up sterols form the media. In addition, these cells are deficient for the sterol deacetylase, Say1, and thus secrete cholesteryl acetate into the culture medium (27). The cells were then washed and diluted in fresh medium to allow the export of cholesterol and cholesteryl acetate. Lipids were extracted from the cell pellet and the culture supernatant, separated by TLC, visualized, and quantified by phosphorimaging. The relative percentages of cholesteryl acetate (CA) that was exported by the cells was plotted as an export index (ratio between extracellular CA and the sum of intracellular and extracellular CA). Mutant yeasts expressing MpPR-1c, MpPR-1d, MpPR-1g, MpPR-1j, and MpPR-1k exported high levels of CA into the culture supernatant, indicating that these MpPR-1 genes functionally complement the absence of yeast CAP proteins (pryΔ) in vivo (Fig. 1). The expression of MpPR-1a, MpPR-1b, MpPR-1e, MpPR-1f, or MpPR-1i, on the other hand, failed to rescue the block in sterol export. MpPR-1h could not be tested because the transformants died after initial growth, suggesting that this gene is toxic to yeast growth (data not shown). These results thus indicate that some but not all of the CAP proteins from M. perniciosa could bind sterols in vivo.
Figure 1.
Some MpPR-1 family members complement the sterol export defect of yeast mutants lacking Pry function. A, heme and Say1 deficient double mutant cells (hem1Δ say1Δ) containing either no plasmid (−), an empty plasmid (Empty), or a plasmid-borne copy of Pry1 or of the indicated MpPR-1 family member were cultivated in the presence of radiolabeled [14C]cholesterol. Lipids were extracted from the cell pellet (P) and the culture supernatant (S), separated by TLC, and visualized by phosphorimaging. The position of free cholesterol (FC), CA, steryl esters (STE) and unidentified lipids (*) are indicated to the right. The experiment was repeated five times with similar results. B, quantification of the export of CA. The export index indicates the relative levels of CA exported by the cells (the ratio between extracellular CA and the sum of intracellular and extracellular CA). The data represent the means ± S.D. of five independent experiments. Asterisks denote statistical significance of the export phenotype relative to hem1Δ say1Δ double mutant cells. ***, p < 0.0001. n.s., not significant.
Sequence analysis of the caveolin-binding motif of MpPR-1 proteins
To understand why some of the CAP proteins from M. perniciosa could bind sterols whereas others could not, we analyzed their sequence at the CBM. Our previous studies had revealed that the CBM is important for sterol binding of the yeast Pry1 protein (24). The hallmark of the CBM is the presence of aromatic amino acids in positions 1, 3, and 8 of the motif (øOXøOXXXXøO) (24, 28, 29). Although the first aromatic residue is conserved in all MpPR-1 proteins, the aromatic residues at positions 3 and 8 of the motif are only conserved in MpPR-1c, MpPR-1d, MpPR-1g, and MpPR-1j, as revealed by the sequence alignment shown in Fig. 2. MpPR-1k, has an alanine in position 3 of the CBM, which appears to be functional for sterol binding. Thus, MpPR-1 family members that bound sterols in vivo also had a conserved CBM. Conversely, in MpPR-1a, MpPR-1b, MpPR-1e, MpPR-1f, and MpPR-1i, which failed to complement the deficiency in the export of cholesteryl acetate, either only position 3 (MpPR-1a and MpPR-1e) or both positions (MpPR-1b, MpPR-1f, and MpPR-1i) within the CBM contain non-aromatic residues (Fig. 2). MpPR-1i is somewhat atypical because the loop region containing the CBM appears to be shortened, essentially resulting in a deletion of the residue in position 8 of the sequence motif (Fig. 2).
Figure 2.
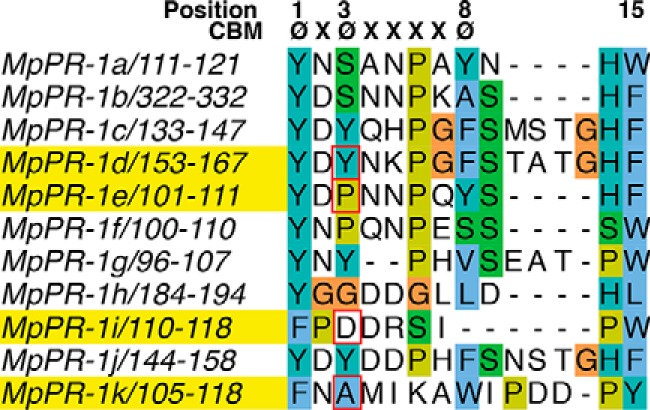
Comparison of the CBM of the 11 MpPR-1 family members from M. perniciosa. The positions of aromatic amino acids (ø) within the CBM are indicated at the top. Residues in red squares at position 3 of the motif mark the amino acids that were mutated. The aromatic amino acid in position 8 of the CBM is conserved only in few family members. Proteins selected for further analysis are highlighted in yellow: MpPR-1d, MpPR-1e, MpPR-1i, and MpPR-1k.
Role of the CBM in sterol binding by M. perniciosa CAP proteins
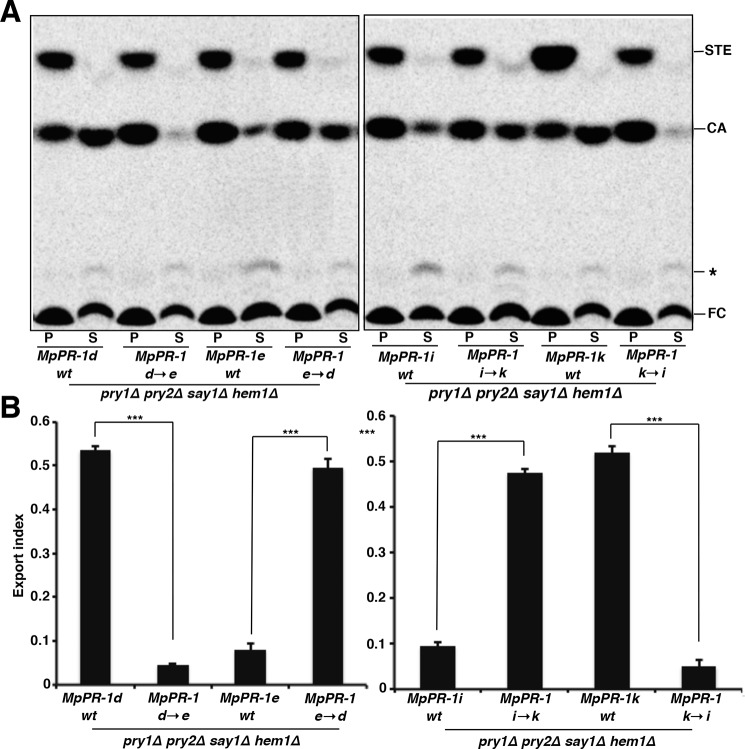
To evaluate the functional importance of the aromatic residues at position 3 of the CBM for sterol binding, we generated mutations in MpPR-d, MpPR-1e, MpPR-1i, and MpPR-1k. The rationale was to convert non-binding MpPR-1 family members into sterol-binding ones, and vice versa. To convert MpPR-1d into MpPR-1e, tyrosine at position 155 of MpPR-1d was mutated to proline (MpPR-1d→e; MpPR-1dY155P). To convert MpPR-1e to MpPR-1d, proline at position 103 of MpPR-1e was mutated to tyrosine (MpPR-1e→d; MpPR-1eP103Y). To convert MpPR-1k to MpPR-1i, alanine at position 107 of MpPR-1k was mutated to aspartic acid (MpPR-1k→i; MpPR-1kA107D), and to convert MpPR-1i to MpPR-1k, aspartic acid at position 112 was mutated to alanine (MpPR-1i→k; MpPR-1iD112A; see Fig. 2, amino acids boxed in red). The mutants were analyzed by the yeast complementation assay, and the results are depicted in Fig. 3. The conversion of MpPR-1d→e and that of MpPR-1k→i by exchanging the aromatic residues in position 3 of the CBM to either proline (MpPR-1d→e) or aspartic acid (MpPR-1k→i) resulted in a loss of sterol export. Conversely, the introduction of an aromatic residue at position 3 of the CBM of the non-binding MpPR-1e (MpPR-1e→d) and the removal of a negatively charged aspartic acid in MpPR-1i (MpPR-1i→k) resulted in a gain of sterol export phenotype. Thus, non-binding MpPR-1s can be converted to sterol binders by the introduction of an aromatic residue or alanine at position 3 of the CBM, whereas a sterol-binding MpPR-1 can be converted to a non-binder by substitution of the aromatic residues at position 3 of the CBM.
Figure 3.
Single point mutations in the CBM convert MpPR-1s that do not export sterols into sterol exporters. A, substitutions of aromatic amino acids in the third position of the CBM convert the sterol exporting MpPR-1d and MpPR-1k into non-exporting mutant versions (MpPR-1d→e and MpPR-1k→i). Conversely, introduction of an aromatic residue at position 3 of the CBM results in the conversion of the non-sterol exporting MpPR-1e and MpPR-1i into gain of function versions (MpPR-1e→d and MpPR-1i→k). hem1Δ say1Δ double mutant cells expressing the indicated wild-type (wt) or mutant version of MpPR-1 proteins were radiolabeled with [14C]cholesterol. Lipids were extracted from the cell pellet (P) and the culture supernatant (S) and separated by TLC. The position of free cholesterol (FC), CA, steryl esters (STE), and an unidentified lipid (*) are indicated to the right. The experiment was repeated three times with similar results. B, the export index was plotted as the ration between exported to total CA. The data represent the means ± S.D. of three independent experiments. Asterisks denote statistical significance of the export phenotype. ***, p < 0.0001.
MpPR-1 proteins bind cholesterol in vitro
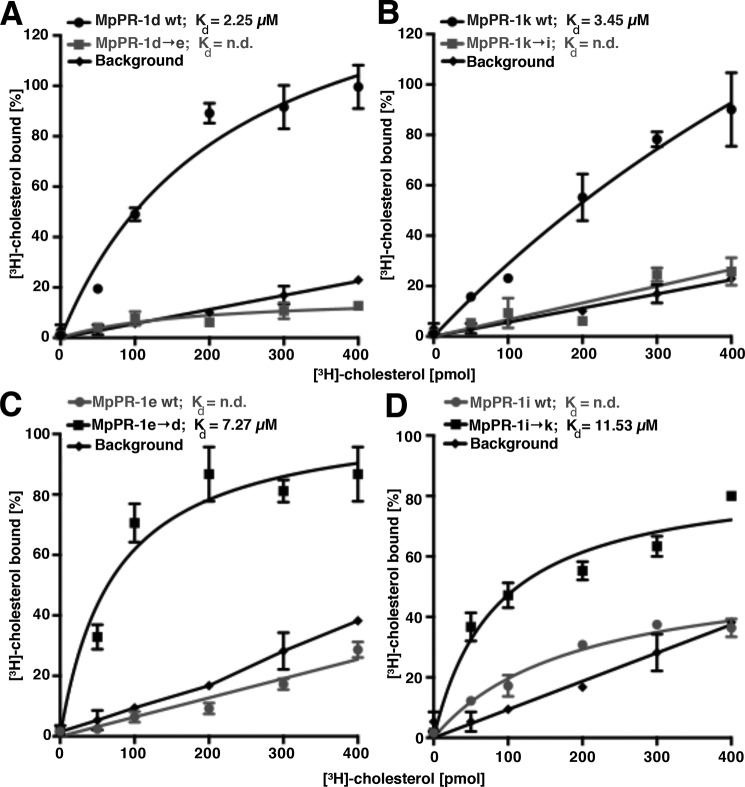
To test more directly whether the MpPR-1 proteins bind sterols in vitro, MpPR-1d and MpPR-1k were expressed as C-terminally polyhistidine-tagged versions in Escherichia coli and affinity-purified on nickel-agarose beads. Sterol binding was then assessed using increasing concentrations of the radioligand [3H]cholesterol (0–400 pmol) and a constant concentration of the purified proteins (100 pmol). These experiments revealed a concentration-dependent increase in radioligand binding, indicating that MpPR-1d and MpPR-1k bind cholesterol in vitro with both proteins displaying saturable binding (Fig. 4, A and B). In accordance with the observation that MpPR-1d→e and MpPR-1k→i mutant versions lost the capacity to export sterols in vivo, these mutant versions displayed poor cholesterol binding in vitro (Fig. 4, A and B). MpPR-1e and MpPR-1i, on the other hand, which did not complement the sterol export phenotype in vivo, also displayed poor binding when tested in vitro, thus confirming the results of the in vivo experiments (Fig. 4, C and D). Conversely, the corresponding gain-of-function mutants, MpPR-1e→d and MpPR-1i→k, displayed an increased affinity for sterols also in the in vitro assay. Taken together, these data indicate that the sterol export phenotype observed in the in vivo assay correlates well with the relative affinity of the respective MpPR-1 family member for sterols as determined in vitro and thus reinforce the importance of the aromatic residue in position 3 of the CBM for sterol binding.
Figure 4.
MpPR-1 bind cholesterol in vitro. A–D, MpPR-1 family members that export sterols in vivo also bind sterols in vitro. Sterol binding by the indicated MpPR-1 wild-type (wt) protein and the respective mutant versions was assessed using 100 pmol of purified protein and an increasing concentration of [3H]cholesterol (0–400 pmol). The protein was separated from unbound ligand by adsorption to an anion-exchange matrix, and bound radioligand was quantified by scintillation counting. Background binding was recorded by performing the binding assay in the absence of protein. The data represent the means ± S.D. of three independent experiments. n.d., not determined.
Sterol-binding specificity of MpPR-1d
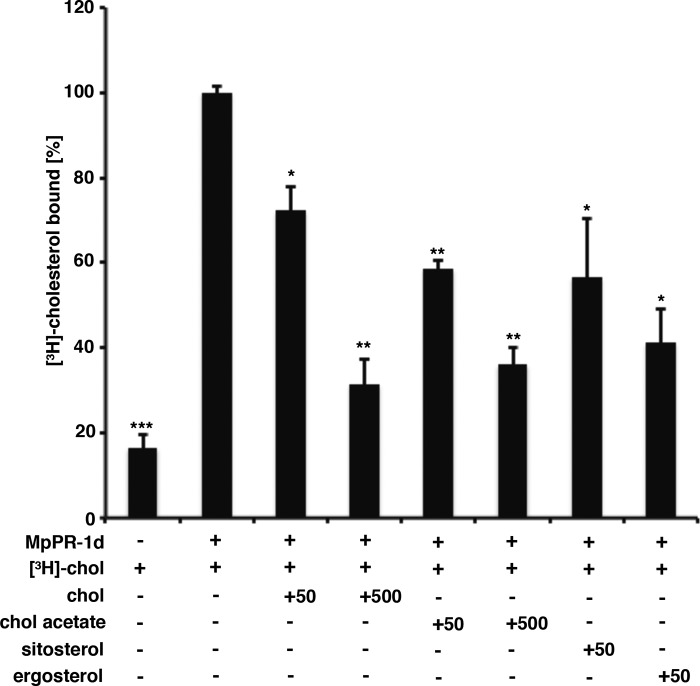
To examine the ligand specificity of MpPR-1d, the ability of various steroids to compete with radiolabeled cholesterol for binding to MpPR-1d, the M. perniciosa CAP family member with the highest affinity for cholesterol, was determined using a competitive binding assays (Fig. 5). The addition of unlabeled cholesterol or that of cholesteryl acetate (50 and 500 pmol) competed with the radiolabeled cholesterol for binding to MpPR-1d, indicating that sterol binding is specific. Additionally, both the plant sterol sitosterol and the fungal sterol ergosterol efficiently competed with the radiolabeled cholesterol for binding to MpPR-1d (Fig. 5). These results thus indicate that these MpPR-1 family members can bind different types of sterols including those from plants and fungi.
Figure 5.
MpPR-1d binds cholesteryl acetate, sitosterol, and ergosterol in vitro. Binding specificity of MpPR-1d was assessed in a competition binding assay in which an unlabeled sterol competes with [3H]cholesterol for binding to the protein. Purified MpPR-1d protein (100 pmol) was incubated with 50 pmol of [3H]cholesterol and either an equal concentration (50 pmol) or an excess (500 pmol) of the indicated sterol. The data represent the means ± S.D. of two independent experiments. Asterisks denote statistical significance relative to the control containing only the radiolabeled cholesterol and purified MpPR-1d. **, p < 0.001; *, p < 0.01. chol, cholesterol.
MpPR-1 proteins bind fatty acids in vivo and in vitro
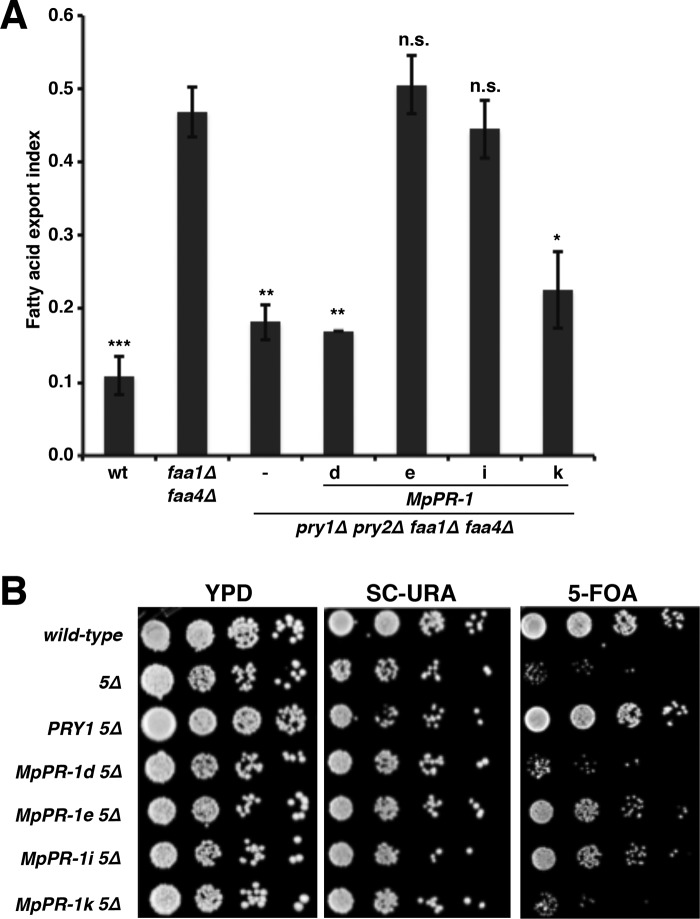
Given that the horsefly CAP protein, tablysin-15 and the yeast Pry1 bind fatty acids, we finally tested whether the four M. perniciosa CAP family members, which we analyzed in more detail for sterol binding, MpPR-1d, MpPR-1e, MpPR-1i, and MpPR-1k, would bind fatty acids as well (17, 19). Therefore, we first tested whether expression of these MpPR-1 proteins in yeast mutants lacking the two major acyl-CoA synthetases, Faa1 and Faa4, in addition to Pry1 and Pry2, would result in export of fatty acids. faa1Δ faa4Δ double mutant cells have previously been shown to export fatty acids into the culture media, and this fatty acid export requires the yeast CAP family members, the Pry proteins (19, 30). Quantification of fatty acids exported from pry1Δ pry2Δ faa1Δ faa4Δ mutant cells revealed that the expression of either MpPR-1e or MpPR-1i promoted fatty acid export, whereas export was reduced to basal levels in cells expressing either MpPR-1d or MpPR-1k (Fig. 6A). These data thus indicate that the MpPR-1 proteins that did not bind or export cholesterol, MpPR-1e and MpPR-1i, promoted fatty acid export, whereas those MpPR-1 proteins that bound and exported cholesterol did not export fatty acids, i.e. MpPR-1d and MpPR-1k.
Figure 6.
MpPR-1 family members that do not export sterols export fatty acids. A, wild-type (wt), faa1Δ faa4Δ double mutant and pry1Δ pry2Δ faa1Δ faa4Δ quadruple mutant cells expressing the indicated MpPR-1 family members were cultivated in minimal medium overnight at 30 °C. Lipids were then extracted from the cell pellet, and the culture supernatant and fatty acids were quantified by GC-MS. The values are plotted as export index representing the amount of exported fatty acids relative to total fatty acids present in the cell pellet and the supernatant. The data represent the means ± S.D. of three independent experiments. Asterisks denote statistical significance relative to the faa1Δ faa4Δ double mutant (***, p < 0.0001; **, p < 0.001; *, p < 0.01). n.s., not significant. B, MpPR-1 family members that do not export fatty acids do not provide the essential function of CAP proteins in cells that export fatty acids. Wild-type and quintuple mutant cells lacking Pry function and the acyl-CoA synthetases Faa1 and Faa4 expressing a plasmid borne copy of either Pry1 or the indicated MpPR-1 family member were serially diluted 10-fold and spotted on plates with the indicated medium. The URA3-marked plasmid-borne copy of FAA1 present in these cells cannot be lost on 5-fluoroorotic acid (5-FOA) medium in cells expressing MpPR-1d or MPPR-1k, the two MpPR-1 family members that do not export or bind fatty acids.
Fatty acid export by Pry in faa1Δ faa4Δ mutant cells is essential as a quintuple mutant lacking both the acyl-CoA synthetases, and all three of the Pry proteins are lethal (pry1Δ pry2Δ pry3Δ faa1Δ faa4Δ) (19). To examine whether expression of either one of these MpPR-1 proteins would rescue the lethality of this quintuple mutant strain, we tested for loss of a URA3 marked plasmid containing FAA1. When cultivated on media containing (5-fluoroorotic acid), which counterselects for the presence of URA3, cells expressing MpPR-1e or MpPR-1i were able to grow but not those expressing MpPR-1d or MpPR-1k. This in vivo test thus confirms the results of the fatty acid export assay and indicates that expression of either MpPR-1e or MpPR-1i but not that of MpPR-1d or MpPR-1k complements the fatty acid export block imposed by the absence of the endogenous CAP proteins in yeast (Fig. 6B).
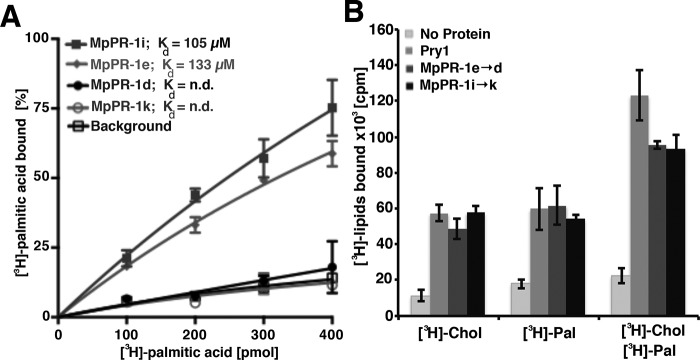
To test whether these MpPR-1 proteins would also bind fatty acid in vitro, polyhistidine-tagged versions were again expressed in E. coli and purified by affinity chromatography. They were then incubated with increasing concentration of [3H]palmitic acid, and binding of the radioligand was quantified. The results of this analysis are consistent with the in vivo data and indicate that both MpPR-1e and MpPR-1i bound fatty acids, whereas MpPR-1d and MpPR-1k displayed background binding (Fig. 7A). Thus, the MpPR-1 family members that export fatty acids in the in vivo assay also bound fatty acids when tested in vitro.
Figure 7.
MpPR-1 family members that export fatty acids in vivo also bind fatty acids in vitro. A, fatty acid binding to the indicated MpPR-1 family members was assessed by an in vitro binding assay with purified protein (100 pmol) and an increasing concentration of [3H]palmitic acid (0–400 pmol). The protein was separated from unbound ligand by adsorption to an anion-exchange matrix, and the bound radioligand was quantified by scintillation counting. Background binding was recorded by performing the binding assay in the absence of protein. B, CAP family members can bind both sterols and fatty acids simultaneously. Radioligand binding by yeast Pry1 and the respective gain of function versions of MpPR-1, MpPR-1e→d, and MpPR-1i→k was assessed using 100 pmol of purified protein and either [3H]cholesterol (50 pmol), [3H]palmitic acid (50 pmol), or both [3H]cholesterol and [3H]palmitic acid (each at 50 pmol) in the binding reaction. The data represent the means ± S.D. of three independent experiments.
To test whether the mutant versions of these proteins could now bind both ligands simultaneously, we performed in vitro binding assays with [3H]cholesterol (50 pmol) alone, with [3H]palmitic acid (50 pmol) alone and with both radioligands being present simultaneously (each at 50 pmol). The presence of both radioligands in the binding assay resulted in a duplication of the radioactivity associated with the proteins, indicating that all three proteins tested in this assay, Pry1, MpPR-1e→d, and MpPR-1i→k can bind both fatty acids and sterols simultaneously (Fig. 7B).
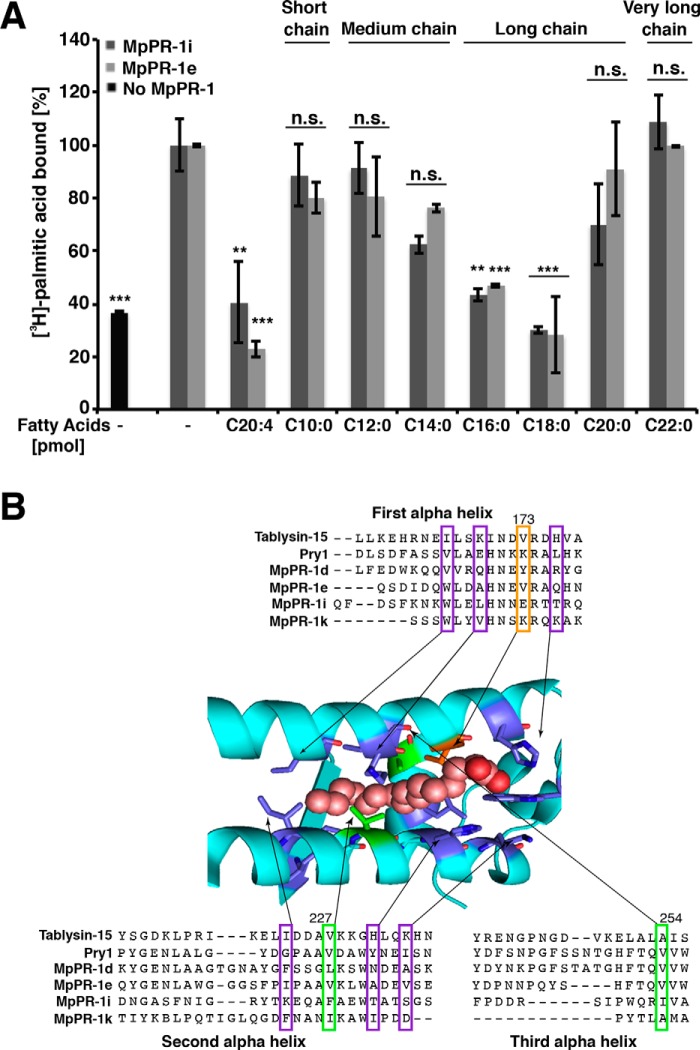
To assess the fatty acid chain length specificity of MpPR-1i and MpPR-1e, we performed competition binding experiments with unlabeled fatty acids of various chain length. The results of these experiments indicate that long chain fatty acids (C16, C18), but not C20, competed with [3H]palmitic acid for binding to these proteins (Fig. 8A). Short- (C10), medium- (C12, C14), and very long-chain (C22) fatty acids, on the other hand, failed to compete with the radioligand. The polyunsaturated arachidonic acid, the precursor to eicosanoids, however, bound to both MpPR-1i and MpPR-1e. The acyl chain substrate preference of MpPR-1i and MpPR-1e is thus comparable that that previously observed for tablysin-15 and Pry1 (17, 19).
Figure 8.
MpPR-1 family members bind saturated long chain and polyunsaturated fatty acids. A, fatty acid-binding specificity was assessed by a competition binding assay in which an equal concentration (50 pmol) of the unlabeled fatty acid of the indicated chain length and degree of unsaturation competes with the radiolabeled [3H]palmitic acid (50 pmol) for binding to the protein (100 pmol). The data represent the means ± S.D. of three independent experiments. Asterisks denote statistical significance relative to the control containing only the radiolabeled palmitic acid and purified MpPR-1i or MpPR-1e, respectively. ***, p < 0.0001; **, p < 0.001. n.s., not significant. B, sequence alignment around the fatty acid-binding pocket. The sequence of tablysin-15, yeast Pry1, and the M. perniciosa MpPR-1 family members analyzed in more detail are shown. The numbering on the top corresponds to the amino acid positions in Pry1. Valine at position 254 of Pry1 (boxed in green) is important for fatty acid binding (19). Key residues shown in colors are those forming the fatty acid binding pocket in tablysin-15 (17). B was adapted from Ref. 19.
Discussion
Witches' broom disease of cacao is caused by M. perniciosa, a hemibiotrophic basidiomycete phytopathogen that contains 11 genes from CAP superfamily (26). Based on gene expression analysis, MpPR-1 genes could have roles in M. perniciosa pathogenicity as MpPR-1c, MpPR-1f, MpPR-1g, MpPR-1h, MpPR-1i, and MpPR-1k had highest expression in green broom stage, the hallmark of WBD progression (26). CAP homologues of S. cerevisiae are required for sterol and fatty acid export in yeast, and they bind cholesterol and fatty acids in vitro (18, 19). In the present study, we examined whether the MpPR-1 proteins could act as sterol and fatty acid binding and export proteins. Such studies are not possible in M. perniciosa, because transformation of this fungus is very inefficient, and they are unstable. In addition, M. perniciosa cannot be cultivated in vitro in its monokaryotic, biotrophic form because it rapidly converts to the dikaryotic, necrotrophic form, which is not of interest to understand its pathogenicity. We thus turned to yeast a model lacking endogenous CAP proteins for the expression and functional characterization of MpPR-1 family members. From the yeast complementation assay, two classes of MpPR-1 genes could be distinguished: the expression of MpPR-1c, MpPR-1d, MpPR-1g, MpPR-1j, and MpPR-1k cDNAs rescued the sterol export phenotype of yeast mutants lacking their endogenous CAP proteins. On the other hand, MpPR-1a, MpPR-1b, MpPR-1e, MpPR-1f, and MpPR-1i failed to complement the sterol export phenotype. However, expression of the sterol non-binding MpPR-1e and MpPR-1i complemented the fatty acid export defect of mutant cells lacking their endogenous CAP proteins, and they bound fatty acids in vitro. These data thus indicate that CAP proteins can bind various types of small hydrophobic compounds such as sterols or fatty acids, whereas the yeast Pry1 protein binds both of these ligands by two discrete, non-overlapping lipid-binding sites (19). The M. perniciosa PR-1 proteins, on the other hand, bind only one of the two ligands tested here. These MpPR-1 proteins, however, might bind as yet unidentified ligands, and this ligand binding and sequestration is likely important for the pathogenicity of the fungus under specific growth conditions.
However, we find no strict correlation between expression of a particular MpPR-1 family member during the fungal life cycle and its lipid ligand specificity. From the six MpPR-1 genes up-regulated in green broom stage (MpPR-1c, MpPR-1f, MpPR-1g, MpPR-1h, MpPR-1i, and MpPR-1k), three encode PR-1 proteins that bind sterols (MpPR-1c, MpPR-1g, and MpPR-1k) (4). Of the remaining, two encode PR-1 proteins that bind fatty acids (MpPR-1f and MpPR-1i), and one could not be tested because of its toxicity in yeast (MpPR-1h). From these data, it thus seems likely that binding of both sterols and fatty acids is important in WBD progression.
The data presented here are consistent with the proposition that CAP proteins constitute a class of secreted lipid-binding proteins and that they serve to sequester hydrophobic compounds that are secreted by the respective host to modulate its immune response. Such a possible mode of action of CAP proteins is supported by the observation that yeast mutants lacking Pry function are hypersensitive to eugenol, a member of the alkylbenzene class of compounds present in clove oil, nutmeg, cinnamon, and bay leaf, used as local antiseptic and anesthetic. Importantly, this eugenol sensitivity is proportional to expression levels of Pry proteins, and eugenol competes with cholesterol for binding to Pry in vitro, indicating that the flexible loop harboring the CBM can bind not only sterols but also other small hydrophobic compounds such as for example eugenol, thereby protecting cells from its toxicity (18, 23). A possible sequestration-based mode of action of CAP family members in plant innate immunity is supported by the fact that plant PR-1 binds sterols in vitro and addition of the purified PR-1 protein inhibits growth of the sterol-auxotrophic plant pathogenic oomycete Phytophthora (21). Thus, CAP family members secreted by both the host, and the pathogen may serve to sequester small hydrophobic compounds required for the survival of either one (31, 32).
M. perniciosa PR-1 proteins harbor a fold that is characteristic of all CAP proteins, an α-β-α sandwich fold (26, 33). The sterol-binding domain within the CAP domain is composed of a flexible loop containing aromatic amino acids (24). This caveolin-binding motif is conserved in the MpPR-1 members that binds sterols, but in members that do not bind cholesterol, the CBM contains substitutions of the important residues at positions 3 and 8 of the motif. Remarkably, MpPR-1 proteins that failed to bind sterols could be converted to sterol binders by the exchange of the amino acid in positon 3 of the CBM and vice versa.
In contrast to the sterol-binding CBM, the structural requirements for the fatty acid–binding pocket are less well-defined. The fatty acid-binding pocket of tablysin-15 is composed of two helices (α1 and α3) that run parallel to each other and a shorter third helix (α4) running perpendicular and forming the bottom closure of the fatty acid–binding pocket (17). Our analysis of fatty acid binding by Pry1 revealed that a conserved valine (position 254 of Pry1) at the bottom of the binding grove is important for binding fatty acids (19). This valine is conserved in MpPR-1e (position 114) and substituted by an isoleucine in MpPR-1i (position 121), both of which bind fatty acids. However, this valine is also conserved in MpPR-1d (position 170) and substituted by alanine in MpPR-1k (position 121), which both do not bind fatty acids (Fig. 8B). Thus, to define the sequence requirements of this second lipid-binding pocket, more structural information of different MpPR-1 family members will be required.
Taken together, the data presented here show that CAP family members from the cacao pathogenic fungus M. perniciosa display different lipid-binding properties. Five of the ten tested MpPR-1 family members (c, d, g, j, and k) exported cholesteryl acetate in vivo, and two of them (d and k) were tested and confirmed to bind cholesterol in vitro. In contrast, MpPR-1e and MpPR-1i, which did not export sterols, bound and exported fatty acids. Remarkably, both MpPR-1e and MpPR-1i could be converted to sterol binders by a single point mutation in their CBM, and the respective mutant proteins MpPR-1e→d and MpPR-1i→k could simultaneously bind both sterols and fatty acids in vitro. Thus, the diversification of the CAP family members in M. perniciosa may serve to bind a structurally diverse set of hydrophobic ligands at defined stages of the fungal life cycle.
Experimental procedures
Gene cloning and site-directed mutagenesis
cDNA was synthesized from 1 μg of total RNA using the SuperScript II reverse transcriptase (Invitrogen, Thermo Fisher Scientific), according to the manufacturer's instructions. Primers specifically designed for the amplification of each MpPR-1 gene, without the peptide signal sequence, were used in PCRs containing cDNA from the basidiomata (MpPR-1a, MpPR-1b, MpPR-1d, MpPR-1e, and MpPR-1j) or green broom stage of development (MpPR-1c, MpPR-1f, MpPR-1g, MpPR-1h, MpPR-1i, and MpPR-1k). Amplicons were subcloned into pGEM T-easy plasmid (Promega, Madison, WI), which was digested with appropriate restriction enzymes. Site-directed mutagenesis was performed by PCR using mismatch primers.
Yeast strains and growth conditions
Yeast strains were cultivated either in rich media (YPD; 1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose; US Biological, Swampscott, MA) or in minimal medium (containing 0.67% yeast nitrogen base without amino acids (US Biological), 0.73 g/liter amino acids, and 2% glucose). Media supplemented with sterols contained 0.05 mg/ml Tween 80 and 20 μg/ml cholesterol (Sigma). To bypass heme deficiency, cells were grown in medium supplemented with 10 μg/ml δ-aminolevulinic acid.
Expression and purification of MpPR-1 proteins
MpPR-1 genes and their mutant versions were cloned into the NcoI and XhoI sites of pET22b vector (Novagen, Merck), which contains a PelB signal sequence to direct the secretion of the expressed protein into the periplasmic space. The vector was transformed into E. coli BL21, and expression of the polyhistidine-tagged protein was induced by lactose at 24 °C overnight. The cells were harvested, lysed, and incubated with nickel-nitrilotriacetic acid beads (Qiagen). The beads were washed, and the proteins were eluted with imidazole and quantified. Protein concentration was determined by Lowry assay using folin reagent and BSA as standard.
In vivo sterol export assay
The yeast sterol export assay is based on the secretion of CA into the culture supernatant and was performed as previously described (27). Yeast mutants deficient in heme biosynthesis (hem1Δ) and lacking the sterol deacetylase Say1 (say1Δ) were cultivated overnight in the presence of 20 μg/ml cold cholesterol and 0.5% Tween 80. The cells were harvested by centrifugation, washed twice with synthetic complete (SC) medium, diluted to an A600 of 1.0 into fresh medium containing 0.025 μCi/ml [14C]cholesterol (American Radiolabeled Chemicals, Inc., St. Louis, MO), and grown overnight. The cells were washed with SC medium and cultivated for another day with non-radiolabeled cholesterol. The cells were centrifuged, and lipids were extracted from the cell pellet and the culture supernatant using chloroform/methanol (1:1, v/v). The samples were dried and separated by thin-layer chromatography on silica gel 60 plates (TLC; Merck) using the solvent system petroleum ether/diethyl ether/acetic acid (70:30:2, v/v/v). TLCs were exposed to phosphorimaging screens, and radiolabeled lipids were visualized and quantified using a phosphorimaging device (GE Healthcare). The export index was calculated as the ratio of extracellular CA to the sum of intracellular and extracellular CA. Export experiments were performed in triplicate, and the export index is given as the mean ± S.D. of three independent experiments.
Fatty acid export and quantification
Wild-type and mutant cells were grown in SC medium at 30 °C. The cells were washed once in cold water, and a fatty acid standard (C17:0; 50 μg; Sigma) was added. The cells were disrupted using glass beads, and lipids were extracted using chloroform/methanol (1:1; v/v) for cellular lipids and with chloroform/methanol/concentrated HCl (1:2:0.03; v/v/v) for extracellular fatty acids. Fatty acid methyl esters (FAMEs) were prepared from 5 A600 nm units of yeast cells using boron trifluoride at 100 °C for 45 min and were recovered by extraction with petrol ether. After evaporation, extracts were resuspended in hexane. FAMEs were separated with an Agilent 7890A gas chromatograph equipped with a DB-23 capillary column (30 m × 0.25 mm × 0.25 m) (Agilent Technologies, Santa Clara, CA). The temperature of the injection port was set to 250 °C, its pressure was set to 26.24 p.s.i. (average velocity, 48.17 cm/s), and the septum purge flow was set to 3 ml/min. Split injections occurred through an Agilent 7693A automated liquid sampler. The initial oven temperature (100 °C; held for 2 min) was increased to 160 °C at a rate of 25 °C/min and was then increased again to 250 °C at 8 °C/min. The final oven temperature was held for an additional 4 min. FAMEs were detected with a flame ionization detector (Agilent Technologies) set at 270 °C with H2, air, and helium flows set at 30, 400, and 27.7 ml/min, respectively. FAMEs were quantified relative to the internal standard, and the relative response factor for each FAME was determined from a four-level calibration curve (r2, 0.999).
In vitro lipid binding and competition assay
To determine lipid binding in vitro, a radioligand-binding assay was performed as described previously (18, 34, 35). Purified protein (100 pmol) in binding buffer (20 mm Tris, pH 7.5, 30 mm NaCl, 0.05% Triton X-100) was incubated with [3H]cholesterol or [3H]palmitic acid (0–400 pmol) for 1 h at 30 °C. The protein was then separated from the unbound ligand by adsorption to Q-Sepharose beads (GE Healthcare); beads were washed with washing buffer (20 mm Tris, pH 7.5), proteins were eluted (20 mm Tris, pH 7.5, 1 m NaCl), and the radioligand was quantified by scintillation counting. To determine nonspecific binding, the binding reaction was performed without the addition of protein into the binding assay. For competition binding assays, unlabeled sterols or fatty acids (Sigma) were included in the binding reaction together with an equal concentration to the respective radiolabeled ligand (50 pmol).
At least two independent experiments were performed under each experimental condition, and the data are reported as the means ± S.D. Statistical significance of data was analyzed by a multiple Student's t test (Prism; GraphPad Software, La Jolla, CA).
Author contributions
R. D. conducted most of the experiments, analyzed the results, and generated the figures. O. E. A. helped in fatty acid binding and export experiments. R. M. B. extracted fungal RNA and generated the cDNA clones. P. J. P. L. T., J. M. C. M., R. M. B., R. D., G. A. G. P., and R. S. conceived the idea for the project and analyzed the data. R. M. B., R. D., J. M. C. M., G. A. G. P., and R. S. wrote and edited the manuscript.
Acknowledgments
We thank Laurent Mène-Saffrané for fatty acid analysis and quantification, Laurent Falquet for bioinformatics support, members of the labs for helpful discussions, and Stéphanie Cottier for comments on the manuscript.
This work was supported by Swiss National Science Foundation Projects 31003A_153416 and 31003A_173003 (to R. S.) and São Paulo State Research Foundation Projects 2009/50119-9, 2010/51884-8, and 2012/07657-2. The authors declare that they have no conflicts of interest with the contents of this article.
- WBD
- witches' broom disease
- FAME
- fatty acid methyl ester
- CBM
- caveolin-binding motif
- CA
- cholesteryl acetate
- SC
- synthetic complete.
References
- 1. Meinhardt L. W., Rincones J., Bailey B. A., Aime M. C., Griffith G. W., Zhang D., and Pereira G. A. (2008) Moniliophthora perniciosa, the causal agent of witches' broom disease of cacao: what's new from this old foe. Mol. Plant Pathol. 9, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Teixeira P. J., Thomazella D. P., and Pereira G. A. (2015) Time for chocolate: current understanding and new perspectives on cacao witches' broom disease research. PLoS Pathog. 11, e1005130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aime M. C., and Phillips-Mora W. (2005) The causal agents of witches' broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia 97, 1012–1022 [DOI] [PubMed] [Google Scholar]
- 4. Teixeira P. J., Thomazella D. P., Reis O., do Prado P. F., do Rio M. C., Fiorin G. L., José J., Costa G. G., Negri V. A., Mondego J. M., Mieczkowski P., and Pereira G. A. (2014) High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell 26, 4245–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barau J., Grandis A., Carvalho V. M., Teixeira G. S., Zaparoli G. H., do Rio M. C., Rincones J., Buckeridge M. S., and Pereira G. A. (2015) Apoplastic and intracellular plant sugars regulate developmental transitions in witches' broom disease of cacao. J. Exp. Bot. 66, 1325–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mondego J. M., Carazzolle M. F., Costa G. G., Formighieri E. F., Parizzi L. P., Rincones J., Cotomacci C., Carraro D. M., Cunha A. F., Carrer H., Vidal R. O., Estrela R. C., García O., Thomazella D. P., de Oliveira B. V., et al. (2008) A genome survey of Moniliophthora perniciosa gives new insights into witches' broom disease of cacao. BMC Genomics 9, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de O Barsottini M. R, de Oliveira J. F., Adamoski D., Teixeira P. J., do Prado P. F., Tiezzi H. O., Sforça M. L., Cassago A., Portugal R. V., de Oliveira P. S., de M Zeri A. C, Dias S. M., Pereira G. A., and Ambrosio A. L. (2013) Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization. Mol. Plant Microbe Interact. 26, 1281–1293 [DOI] [PubMed] [Google Scholar]
- 8. de Oliveira G. A., Pereira E. G., Dias C. V., Souza T. L., Ferretti G. D., Cordeiro Y., Camillo L. R., Cascardo J., Almeida F. C., Valente A. P., and Silva J. L. (2012) Moniliophthora perniciosa necrosis- and ethylene-inducing protein 2 (MpNep2) as a metastable dimer in solution: structural and functional implications. PLoS One 7, e45620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomazella D. P., Teixeira P. J., Oliveira H. C., Saviani E. E., Rincones J., Toni I. M., Reis O., Garcia O., Meinhardt L. W., Salgado I., and Pereira G. A. (2012) The hemibiotrophic cacao pathogen Moniliophthora perniciosa depends on a mitochondrial alternative oxidase for biotrophic development. New Phytol. 194, 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaparoli G., Barsottini M. R., de Oliveira J. F., Dyszy F., Teixeira P. J., Barau J. G., Garcia O., Costa-Filho A. J., Ambrosio A. L., Pereira G. A., and Dias S. M. (2011) The crystal structure of necrosis- and ethylene-inducing protein 2 from the causal agent of cacao's witches' broom disease reveals key elements for its activity. Biochemistry 50, 9901–9910 [DOI] [PubMed] [Google Scholar]
- 11. Gibbs G. M., Roelants K., and O'Bryan M. K. (2008) The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins: roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897 [DOI] [PubMed] [Google Scholar]
- 12. Cantacessi C., Campbell B. E., Visser A., Geldhof P., Nolan M. J., Nisbet A. J., Matthews J. B., Loukas A., Hofmann A., Otranto D., Sternberg P. W., and Gasser R. B. (2009) A portrait of the “SCP/TAPS” proteins of eukaryotes: developing a framework for fundamental research and biotechnological outcomes. Biotechnol Adv. 27, 376–388 [DOI] [PubMed] [Google Scholar]
- 13. van Loon L. C., Rep M., and Pieterse C. M. (2006) Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 [DOI] [PubMed] [Google Scholar]
- 14. Chalmers I. W., McArdle A. J., Coulson R. M., Wagner M. A., Schmid R., Hirai H., and Hoffmann K. F. (2008) Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics 9, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prados-Rosales R. C., Roldán-Rodríguez R., Serena C., López-Berges M. S., Guarro J., Martínez-del-Pozo Á., and Di Pietro A. (2012) A PR-1-like protein of Fusarium oxysporum functions in virulence on mammalian hosts. J. Biol. Chem. 287, 21970–21979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lozano-Torres J. L., Wilbers R. H., Warmerdam S., Finkers-Tomczak A., Diaz-Granados A., van Schaik C. C., Helder J., Bakker J., Goverse A., Schots A., and Smant G. (2014) Apoplastic venom allergen-like proteins of cyst nematodes modulate the activation of basal plant innate immunity by cell surface receptors. PLoS Pathog. 10, e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu X., Francischetti I. M., Lai R., Ribeiro J. M., and Andersen J. F. (2012) Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain. J. Biol. Chem. 287, 10967–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choudhary V., and Schneiter R. (2012) Pathogen-related yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 16882–16887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darwiche R., Mène-Saffrané L., Gfeller D., Asojo O. A., and Schneiter R. (2017) The pathogen-related yeast protein Pry1, a member of the CAP protein superfamily, is a fatty acid-binding protein. J. Biol. Chem. 292, 8304–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelleher A., Darwiche R., Rezende W. C., Farias L. P., Leite L. C., Schneiter R., and Asojo O. A. (2014) Schistosoma mansoni venom allergen-like protein 4 (SmVAL4) is a novel lipid-binding SCP/TAPS protein that lacks the prototypical CAP motifs. Acta Crystallogr. D Biol. Crystallogr. 70, 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gamir J., Darwiche R., Van't Hof P., Choudhary V., Stumpe M., Schneiter R., and Mauch F. (2017) The sterol-binding activity of pathogenesis-related protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509 [DOI] [PubMed] [Google Scholar]
- 22. Darwiche R., Kelleher A., Hudspeth E. M., Schneiter R., and Asojo O. A. (2016) Structural and functional characterization of the CAP domain of pathogen-related yeast 1 (Pry1) protein. Sci. Rep. 6, 28838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darwiche R., and Schneiter R. (2016) Cholesterol-binding by the yeast CAP faimily member, Pry1, requires the presence of an aliphatic side chain on cholesterol. J Steroids Hormon. Sci. 7, 2 [Google Scholar]
- 24. Choudhary V., Darwiche R., Gfeller D., Zoete V., Michielin O., and Schneiter R. (2014) The caveolin-binding motif of the pathogen-related yeast protein Pry1, a member of the CAP protein superfamily, is required for in vivo export of cholesteryl acetate. J. Lipid Res. 55, 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schneiter R., and Di Pietro A. (2013) The CAP protein superfamily: function in sterol export and fungal virulence. Biomol. Concepts 4, 519–525 [DOI] [PubMed] [Google Scholar]
- 26. Teixeira P. J., Thomazella D. P., Vidal R. O., do Prado P. F., Reis O., Baroni R. M., Franco S. F., Mieczkowski P., Pereira G. A., and Mondego J. M. (2012) The fungal pathogen Moniliophthora perniciosa has genes similar to plant PR-1 that are highly expressed during its interaction with cacao. PLoS One 7, e45929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tiwari R., Köffel R., and Schneiter R. (2007) An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae. EMBO J. 26, 5109–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Byrne D. P., Dart C., and Rigden D. J. (2012) Evaluating caveolin interactions: do proteins interact with the caveolin scaffolding domain through a widespread aromatic residue-rich motif? PLoS One 7, e44879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eberle H. B., Serrano R. L., Füllekrug J., Schlosser A., Lehmann W. D., Lottspeich F., Kaloyanova D., Wieland F. T., and Helms J. B. (2002) Identification and characterization of a novel human plant pathogenesis-related protein that localizes to lipid-enriched microdomains in the Golgi complex. J. Cell Sci. 115, 827–838 [DOI] [PubMed] [Google Scholar]
- 30. Scharnewski M., Pongdontri P., Mora G., Hoppert M., and Fulda M. (2008) Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 275, 2765–2778 [DOI] [PubMed] [Google Scholar]
- 31. Kazan K., and Gardiner D. M. (2017) Targeting pathogen sterols: defence and counterdefence. PLoS Pathog. 13, e1006297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Breen S., Williams S. J., Outram M., Kobe B., and Solomon P. S. (2017) Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879 [DOI] [PubMed] [Google Scholar]
- 33. Baroni R. M., Luo Z., Darwiche R., Hudspeth E. M., Schneiter R., Pereira G. A. G., Mondego J. M. C., and Asojo O. A. (2017) Crystal structure of MpPR-1i, a SCP/TAPS protein from Moniliophthora perniciosa, the fungus that causes witches' broom disease of cacao. Sci. Rep 7, 7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Im Y. J., Raychaudhuri S., Prinz W. A., and Hurley J. H. (2005) Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Darwiche R., and Schneiter R. (2017) A ligand-binding assay to measure the affinity and specificity of sterol-binding proteins in vitro. Methods Mol. Biol. 1645, 361–368 [DOI] [PubMed] [Google Scholar]