Abstract
Pituitary gonadotropin hormones are regulated by gonadotropin-releasing hormone (GnRH) via MAPK signaling pathways that stimulate gene transcription of the common α-subunit (Cga) and the hormone-specific β-subunits of gonadotropin. We have reported previously that GnRH-induced activities at these genes include various histone modifications, but we did not examine histone phosphorylation. This modification adds a negative charge to residues of the histone tails that interact with the negatively charged DNA, is associated with closed chromatin during mitosis, but is increased at certain genes for transcriptional activation. Thus, the functions of this modification are unclear. We initially hypothesized that GnRH might induce phosphorylation of Ser-10 in histone 3 (H3S10p) as part of its regulation of gonadotropin gene expression, possibly involving cross-talk with H3K9 acetylation. We found that GnRH increases the levels of both modifications around the Cga gene transcriptional start site and that JNK inhibition dramatically reduces H3S10p levels. However, this modification had only a minor effect on Cga expression and no effect on H3K9ac. GnRH also increased H3S28p and H3K27ac levels and also those of activated mitogen- and stress-activated protein kinase 1 (MSK1). MSK1 inhibition dramatically reduced H3S28p levels in untreated and GnRH-treated cells and also affected H3K27ac levels. Although not affecting basal Cga expression, MSK1/2 inhibition repressed GnRH activation of Cga expression. Moreover, ChIP analysis revealed that GnRH-activated MSK1 targets the first nucleosome just downstream from the TSS. Given that the elongating RNA polymerase II (RNAPII) stalls at this well positioned nucleosome, GnRH-induced H3S28p, possibly in association with H3K27ac, would facilitate the progression of RNAPII.
Keywords: chromatin, histone modification, mitogen-activated protein kinase (MAPK), phosphorylation, pituitary gland
Introduction
Chromatin structure often responds to external cues to determine gene expression through changing the accessibility of regulatory DNA. We have previously reported that the hypothalamic gonadotropin releasing hormone (GnRH)2 up-regulates expression of the gonadotropin hormones via various actions on the chromatin. In partially differentiated gonadotrope cells, GnRH was seen to induce the removal of histone deacetylases from the repressed hormone β-subunit gene promoters (1, 2), while also activating expression of these genes by targeting trimethylation of histone H3 lysine 4 (H3K4me3) to their promoters (3). There are several reports that GnRH induces recruitment of transcriptional coactivators with histone acetyltransferase (HAT) activity to these gene promoters (e.g. Refs. 4–6) indicating that the histones are also acetylated as part of the induction of expression of these genes, although the specific residues affected and their precise roles in transcription have not been reported.
In contrast to histone acetylation and methylation, whose functions in transcriptional regulation are relatively well understood, the role of histone phosphorylation is much less clear (7, 8). The better studied phosphorylation of H3 at serine 10 (H3S10p) has been implicated in transcriptional regulation of various induced genes (7, 9, 10), but is also involved in the DNA damage response, apoptosis, and mitosis/meiosis (11–13), and is associated both with chromatin condensation during mitosis and chromatin relaxation during gene expression (11, 12). Such opposing biological effects of H3 phosphorylation of the same residue indicate that the interpretation of this modification is highly context-dependent and that the individual residues cannot be considered in isolation.
Besides altering the charge of the histone that leads to a loosening of its interaction with the DNA, phosphorylation may serve as a recognition signal for other regulatory proteins, although the only proteins found to bind specifically to phosphorylated histones in association with gene transcription are the 14-3-3 family members that can recruit modifying enzymes such as HATs for subsequent histone acetylation (14–19). Indeed H3 phosphorylation and acetylation are often seen together at active promoters and both are crucial for the activation of certain genes, such as the immediate early genes following stimulation of MAPK pathways (10, 20, 21). Furthermore, HAT enzymes such as GCN5, which targets H3K9ac, were shown to acetylate preferentially H3S10p peptides over non-phosphorylated ones (22), providing a possible mechanism for such cross-talk between these modifications.
A number of kinases are reported to phosphorylate H3, and the mitogen-activated protein kinases (MAPKs) MSK and RSK have been implicated to mediate the response to various extracellular stimuli (12, 23–25). These kinases are downstream of the ERK and p38 MAPK that mediate many signaling pathways, and MSK1/2 knock-out mice were seen to have vastly reduced phospho-H3 levels (26). However, MSK1/2 also target various transcription factors (27, 28). Another MAPK, JNK which mediates part of the stress response, translocates to the nucleus following its activation and is also sometimes associated with chromatin (23), was shown to phosphorylate specifically H3S10 at its target genes during neuronal differentiation. This was shown to involve JNK recruitment to the chromatin via NF-Y, which was essential for gene expression (30). Other kinases are also capable of phosphorylating H3S10, although the specific and diverse roles and contributions of each of these events to the transcriptional regulation are not understood (12).
GnRH exerts its actions on the gonadotropes to induce gonadotropin gene expression through the activation of various MAPKs including JNK (31, 32), while the Cga gene promoter contains an NF-Y binding motif, suggesting a possible pathway for GnRH induction of HS10p, which might up-regulate Cga expression. We thus considered it highly likely that gonadotrope exposure to GnRH induces histone phosphorylation to activate this gene, possibly involving also cross-talk with H3 acetylation. We thus aimed to examine the GnRH-induced changes in histone acetylation and phosphorylation of the Cga gene and their role in Cga gene expression. This led us to understand that phosphorylation at neither H3S10 nor H3S28 is required for the on-going transcription of this gene, but H3S28p is elevated by GnRH particularly at the +1 nucleosome, where it likely helps transition through the first nucleosome, and is required for the full Cga transcriptional response to GnRH.
Results
The open chromatin at the Cga gene transcriptional start site (TSS) is enriched with phosphorylated H3S10 compared with the more closed chromatin at the β-subunit genes
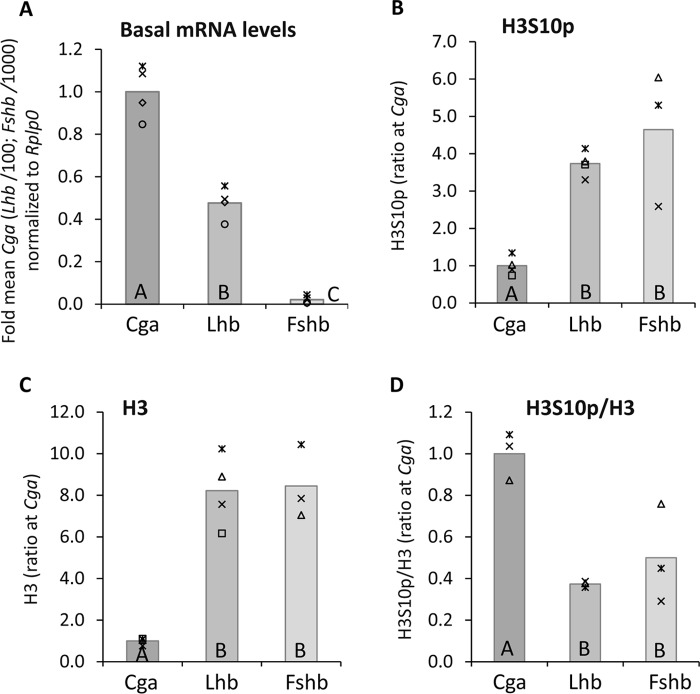
The αT3-1 cell line represents an immature partially-differentiated gonadotrope, and expresses high levels of the Cga gene, which encodes the common α subunit, but only low levels of the hormone-specific β-subunit genes, Lhb and Fshb (Fig. 1A). To examine whether the phosphorylation status of the histones around the TSSs of these genes correlates with their diverse expression levels, we first examined the levels of phosphorylation at H3S10 (H3S10p). Chromatin immunoprecipitation (ChIP) using antisera to H3S10p showed higher levels of this modification at the Lhb and Fshb genes as compared with the Cga gene (Fig. 1B). However H3 is found at much lower levels on this region of the Cga gene than on the β-subunit genes (Fig. 1C), and when the levels of this modification were normalized to those of total H3, the nucleosomes were clearly enriched with this modification at the Cga gene, correlating with the higher gene expression levels (Fig. 1D).
Figure 1.
The open chromatin at the Cga gene is enriched with phosphorylated H3S10 compared with the more closed chromatin at the β-subunit genes. A, basal mRNA levels of Cga, Lhb, and Fshb in αT3-1 partially differentiated gonadotrope cells were analyzed by qPCR and normalized to those of Rplp0. The mRNA levels were measured against standard curves, comprising purified and quantitated cDNA of each gene, which was serially diluted. The corresponding Ct value was plotted against the log of the amount of DNA (repeated three times and averaged) and these values (normalized to Rplp0) are presented relative to the mean level of the Cga, with the gray bar indicating the mean. Analysis of variance followed by Bonferroni t test were used to determine significantly different means: those marked with the same letter are not significantly different (p > 0.05). B–D, ChIP analysis was carried out to determine the levels of (B) H3S10p and (C) total H3 at the promoter/5′ end of each of the three gonadotropin subunit genes. In D, levels of H3S10p are shown normalized to those of total H3 in the same sample. For all, an aliquot from the same cells before precipitation was designated as input, and the levels in input and precipitated DNA samples were quantitated by qPCR. The precipitated DNA was normalized against the respective input samples and these values were compared with the mean level at the Cga gene; presented and statistical analysis as above.
Basal levels of H3 histone acetylation at Lys-9, -18, and -27 also largely correlate with gene expression
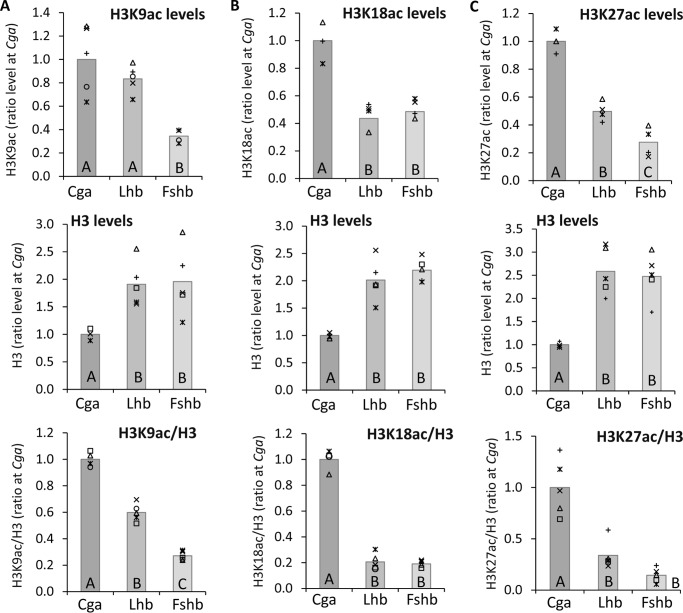
Histone acetylation is characteristically found at actively expressed genes, and levels of H3K9ac at these genes often correlate with those of H3S10p. Although we saw previously that the β-subunit genes are associated with histone deacetylases in these cells, the actual acetylation status of these genes, and how this compares with the acetylation levels at the more active Cga gene, are not known. We therefore measured, also by ChIP, the levels of acetylation of lysines 9, 18, and 27 of histone H3 at the same upstream regions of these three genes. The levels of acetylation at each of these lysine residues normalized to the level of total H3 in the same samples correlated with levels of expression, although K18ac and K27ac were at similarly low levels at the two β-subunit genes (Fig. 2, A–C).
Figure 2.
Basal states of H3 histone acetylation correlate similarly at Lys-9, Lys-18, and Lys-27. ChIP analysis was carried out as before to determine the levels of H3 acetylated at (A) Lys-9, (B) Lys-18, and (C) Lys-27 at the 5′ ends of each of the three genes. H3ac (top), total H3 (middle), and H3ac normalized to total H3 in the same sample (bottom) are shown as in Fig. 1, B–D, relative to the mean level at the Cga gene.
Exposure of these cells to GnRH elevates levels of H3S10p and H3 acetylation globally and at the Cga gene, correlating with an increase in its expression
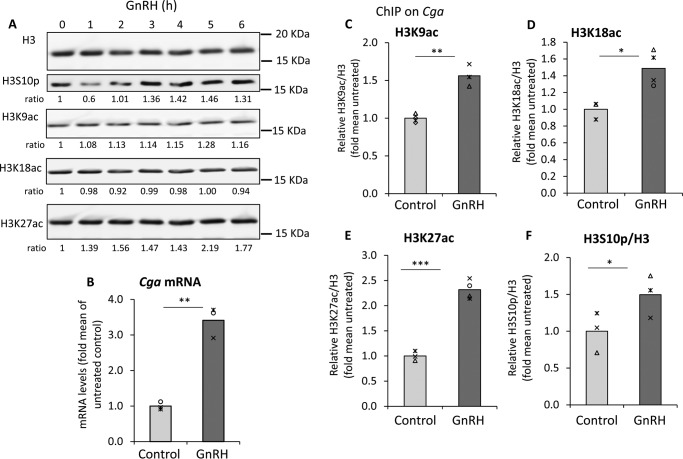
GnRH is a major regulator of gene expression in the gonadotropes as the activation of MAPK pathways target many genes, not just the gonadotropins, so we first assessed whether it had any global effects on histone acetylation and/or phosphorylation. Western analysis of histone extracts from cells exposed to 0–6 h GnRH revealed that although there was an initial drop in H3S10p levels, global levels of this modification and K27ac then increased, whereas the H3K9ac and K18ac appeared unaffected on a global scale (Fig. 3A).
Figure 3.
Exposure to GnRH elevates levels of H3S10p and H3ac globally and at the Cga gene, correlating with its increase in expression. A, cells treated with GnRH for 0–6 h were harvested at the same time, and Western blot analysis was carried out on the histone fraction using antibody to H3S10p, H3K9ac, H3K18ac, and H3K27ac, following stripping of the membrane after each reaction. Total histone H3 was used as the loading control. (The experiment was performed several times and blotting was performed in various sequences, but for the results shown here, the sequence was K27ac, H3, K18ac, S10p, and K9ac.) Relative levels of each modified protein signal are shown below the blot as fold over the level in untreated cells, after normalization to H3. B, serum-starved αT3-1 cells were treated with GnRH for 8 h after which total RNA was extracted for qPCR. Cga mRNA levels are shown after normalization with those of Rplp0, and relative to the mean in untreated control cells, as described in the legend to Fig. 1. Statistical analysis was Student's t test: **, p < 0.01. C–F, ChIP analysis at the Cga gene promoter/5′ end was carried out in untreated or GnRH-treated αT3–1 cells using antibodies for total H3 and: C, H3K9ac; D, H3K18ac; E, H3K27ac; or F, H3S10p. Levels of precipitated DNA were quantified by qPCR and analyzed as described in the legend to Fig. 1, presented relative to the mean levels in the untreated control; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Although expression of all three gonadotropin genes is induced by GnRH, these cells are considered the most appropriate model for study of the GnRH stimulation of the Cga gene, due to its higher expression levels and more predictable response. After confirming that GnRH elevates Cga mRNA levels in these cells (Fig. 3B), we went on to examine whether there was change in levels of H3S10p and H3ac at the Cga gene. ChIP was carried out with antisera to the particular modified histone and also total H3 for normalization. Acetylation of all three lysines was increased in the GnRH-treated cells but the response of K27ac was the most notable (Fig. 3, C–E). Phosphorylation of H3S10 was also significantly, albeit marginally, elevated after GnRH treatment (Fig. 3F).
JNK plays a crucial role in determining H3S10p levels, but this histone modification barely affects basal Cga gene expression and its absence does not influence H3K9ac
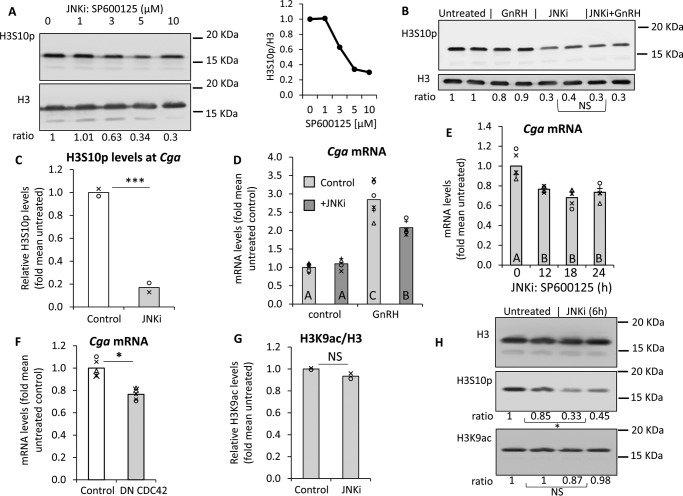
GnRH signaling to gonadotropin gene expression is through the activation of various MAPK phosphorylation cascades including JNK (31). JNK was reported to target H3S10p in neuronal cells, being recruited by the NF-Y transcription factor (30), which also has a binding motif on the Cga gene promoter (33). Therefore we examined whether the levels of H3S10p at the Cga gene might be regulated by JNK, first through exposure of the cells to a chemical JNK inhibitor, SP600125. This inhibitor dramatically reduced global levels H3S10p, which were not restored by exposure of the cells to GnRH (Fig. 4, A and B). This effect of the JNK inhibitor on H3S10p was apparent also specifically at the Cga gene in ChIP analysis, which revealed that the inhibitor reduced H3S10p levels to below 20% those in the control cells (Fig. 4C).
Figure 4.
JNK plays a crucial role in determining H3S10p levels, but this modification barely affects basal Cga gene expression and does not affect H3K9ac. A, αT3-1 cells were serum-starved overnight prior to treatment with 1–10 μm of the JNK inhibitor SP600125 (JNKi) for 6 h before isolation of the histone fraction and Western analysis using antibody to H3S10p, with H3 as loading control. Normalized values relative to the untreated control are shown below the blot, and presented graphically on the right. B, cells were prepared and treated similarly with GnRH and/or SP600125 added 30 min before GnRH, and H3S10p and H3 examined by Western blot analysis as above. NS, p > 0.05 for average H3S10p/H3 levels. C, following treatment with SP600125 (10 μm for 6 h) ChIP analysis was carried out, as described in the legend to Fig. 1, to determine the levels of H3S10p at the Cga promoter/5′ end. H3S10p levels are expressed as fold of the mean level in untreated cells; ***, p < 0.001. D, Cga mRNA levels were measured as described in the legend to Fig. 3B, in cells treated with GnRH for 7 h with or without SP600125 added 30 min beforehand. The mRNA levels are expressed as fold of the mean levels in untreated control cells. Statistical analysis was as described in the legend to Fig. 1. E, similarly qPCR was carried out on cells treated with SP600125 for 12–24 h, and Cga mRNA levels are expressed as fold of the mean levels in untreated cells. Statistical analysis was as described before. F, αT3–1 cells were transfected with control GFP plasmid or dominant-negative CDC42. Cells were lysed 48 h after transfection, and Cga mRNA levels measured and data presented as before. *, p < 0.05. G, ChIP analysis was also carried out for H3K9ac in cells treated with SP600125 for 6 h, and levels associated with the Cga gene are shown after normalization to H3, relative to the mean levels in untreated cells. NS, p > 0.05. H, Western blot analysis was also carried out on SP600125-treated cells, and the effects on global levels of both H3S10p and H3K9ac were evaluated using specific antisera with H3 as control. Average H3S10p/H3 or H3K9ac/H3 levels in JNKi-treated versus untreated cells are indicated: *, p < 0.05; NS, p > 0.05.
To determine the effect of this drop in H3S10p on Cga gene expression, we carried out quantitative PCR (qPCR) in serum-starved cells exposed to the inhibitor for 30 min before and during treatment with GnRH for 8 h. Surprisingly, the inhibitor had no apparent effect on the basal Cga mRNA levels, although the GnRH response was reduced somewhat (Fig. 4D), in line with previous reporter gene assays on the activity of this promoter (34). To determine whether an effect on basal gene expression might be seen during a longer time frame, we exposed cells to the inhibitor for up to 24 h; still there was only a minor effect on Cga mRNA levels that dropped, at the most, to around 70% that of the untreated controls (Fig. 4E). To confirm this result, we also transfected the cells with a construct to express a dominant-negative CDC42, which is upstream of JNK, and a similar effect was seen (Fig. 4F).
To investigate whether this drop in H3S10p is accompanied by a drop in the neighboring acetylation of H3K9, we carried out ChIP for this modification in the SP600125-treated cells. The levels of H3K9ac at the Cga gene promoter/TSS were unaffected by the JNK inhibition (Fig. 4G) and a similar lack of effect on H3K9ac was seen at a global level in Western analysis (Fig. 4H). Together, these results show clearly that JNK is responsible for H3S10p at the Cga gene in these cells, but this modification plays little role in determining the levels of Cga expression, and does not influence the levels of H3K9ac.
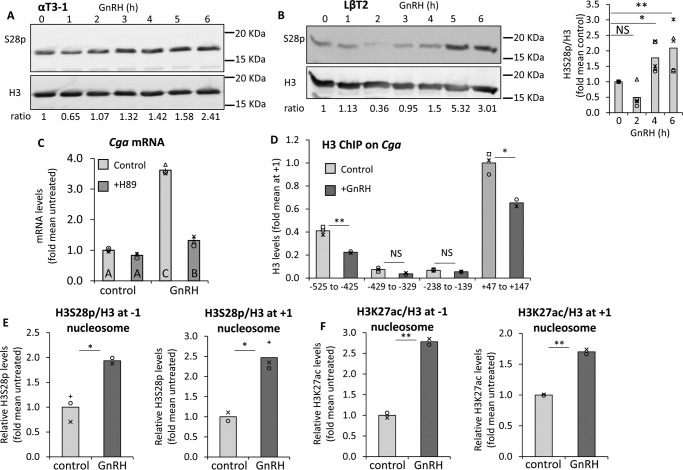
Phosphorylation at H3S28 also increases following GnRH, correlating with K27ac, and this plays a role in GnRH-induced gene expression
As H3S10p did not appear to play much of a role in determining Cga gene expression levels, we next examined phosphorylation at H3S28 (H3S28p), which is thought to be associated with transcriptional activation, but is much less studied than H3S10p. Initial global Western analysis in αT3-1 cells revealed that, like H3S10p, levels of this modification also dropped during the first hour after GnRH exposure, but it increased by 3 h of GnRH treatment (Fig. 5A). A similar effect was seen with another gonadotrope cell line, LβT2, although both the initial drop and increase above basal levels were somewhat delayed, the maximum response (3.12 ± 0.7; n = 5) was also reached after 4–6 h (Fig. 5B). H3S28p reportedly can be phosphorylated by MSK (9, 11, 13, 35), which is downstream of the ERK and p38 MAPK signaling pathways both of which are activated by GnRH, with ERK being particularly crucial to up-regulation of the Cga gene (31). We therefore tested the effects of a broad chemical inhibitor of MSK, H89, which was applied to the cells 30 min before and during exposure to GnRH. The inhibitor virtually abolished the GnRH stimulation of Cga gene expression, but did not affect the basal levels (Fig. 5C), suggesting a possible role for this pathway in the GnRH up-regulation of this gene.
Figure 5.
Phosphorylation at H3S28 increases following GnRH, as does K27ac, and this plays a role in GnRH-induced gene expression. A and B, levels of H3S28p were analyzed in GnRH-treated αT3-1 cells as in Fig. 3. In A, aT3–1 cells (same blot as in Fig 3A, hence same H3 panel; H328p was analyzed following stripping), or in B, LbT2 cells which were treated, analyzed, and presented similarly. Averages from repeated experiments in LβT2 cells are shown graphically; t test compared mean values with untreated controls, as described in the legend to Fig. 3. C, Cga mRNA levels were measured by qPCR as described in the legend to Fig. 3, in cells treated with GnRH (6 h) with or without H89 added 60 min prior to the GnRH. The mRNA levels are expressed as fold of the mean levels in untreated control cells and presented as described in the legend to Fig. 4D. D, high resolution ChIP was carried out for H3 in cells treated with GnRH (2 h); alternatively, for E, H3S28p or F, H3K27ac in untreated or GnRH-treated (5 h) cells to examine these modifications at the −1 and +1 nucleosomes on the Cga. Data were analyzed and are presented as described in the legend to Fig. 3. *, p < 0.05; **, p < 0.01.
We went on to examine whether H3S28p is induced by GnRH specifically at the Cga gene. For these experiments we utilized high resolution ChIP (36) which, through extensive sonication and multiple primer sets, is able to distinguish between the +1 nucleosome, which is the first one downstream of the TSS, and the −1 nucleosome, which is the closest of the promoter nucleosomes to the TSS, located 400 bp upstream (36). ChIP for H3 after this protocol is able to differentiate clearly between these two nucleosomes separated by a nucleosome-free region, and both nucleosomes are affected by GnRH (Fig. 5D). Subsequently, ChIP analysis for H3S28p revealed that the levels of phosphorylation at both nucleosomes are enriched following GnRH treatment, which was mimicked by the levels of H3K27ac (Fig. 5, E and F). Clearly actions on either one of these nucleosomes has distinct implications for the initiation and/or elongation mechanisms of regulating transcription.
GnRH activates MSK, which is required for S28p and also K27ac
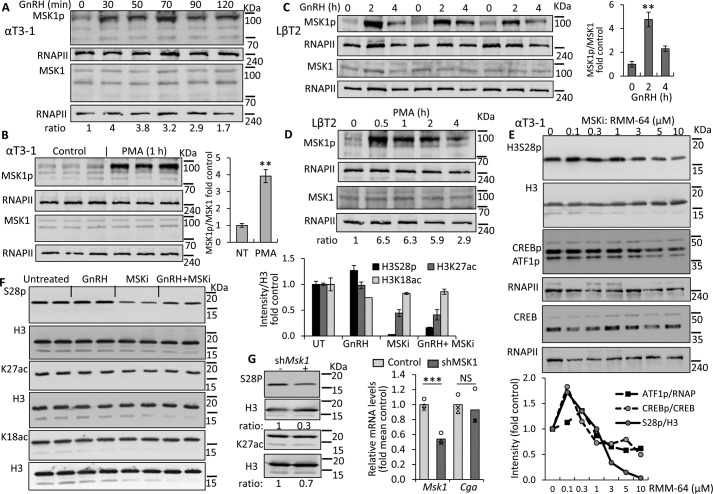
We next examined whether GnRH-activated MSK might be responsible for H3S28p at either of these nucleosomes. Western blotting of the active phosphorylated MSK1 (MSK1p) showed that its levels are indeed increased in αT3-1 cells within 30 min of GnRH treatment (Fig. 6A), and a similar effect was seen after exposure to PMA for 1 h (Fig. 6B). The LβT2 gonadotropes responded similarly to both GnRH (Fig. 6C) and PMA (Fig. 6D), with 4–6.5-fold increases over control levels. Moreover, we were able to reduce H3S28p levels dramatically through the use of the highly specific MSK-1 inhibitor, RMM-64 (Fig. 6E). The levels of phosphorylated H3S28, after exposure to 3–10 μm of the inhibitor, dropped even more than those of other known MSK1 targets, CREB and ATF1, which are both phosphorylated also by other kinases.
Figure 6.
GnRH activates MSK, which is required for S28p and also K27ac. A–D, Western blot analysis of nuclear extracts from (A and B) αT3-1 cells or (C and D) LβT2 cells exposed to (A and C) GnRH or (B and D) PMA was carried out using antibody for MSK1p and RNAPII as loading control, while different aliquots of the same sample were analyzed similarly for total MSK. Ratios of the MSK1p:MSK, each after normalization to RNAPII, are reported (A and D) under the gel, or (B and C) mean averages (±S.E.) shown graphically on the right of the gel; t test compared mean values with untreated controls, as described in the legend to Fig. 3. E, similarly, Western blot analysis was carried out for H3S28p, or using antibody to CREBp, which recognizes also ATF1p, in αT3-1 cells treated with various doses of the MSK1 inhibitor (MSKi), RMM-64 for 6 h with H3, RNAPII, or CREB as loading controls. Normalized ratios are shown graphically below. F, αT3-1 cells were serum-starved overnight prior to treatment with GnRH for 6 h and/or RMM-64, added 2 h beforehand. All cells were harvested at the same time, the histone fraction was isolated and Western blot analysis was carried out as described in the legend to Fig. 3; with normalized average (±S.E.) values relative to the untreated control shown graphically at the top right of the blot. G, cells were transfected with shRNA targeting MSK1 or control RNA and levels of H3S28p, H3K27ac, and H3 were assessed by Western blot analysis, as before or RNA extracted for qPCR for Msk1 and Cga; data were analyzed and presented as before. **, p < 0.01; ***, p < 0.001.
Subsequently we tested whether this inhibitor also affects the levels of H3K27ac, which might suggest cross-talk between these modifications. There was a clear drop in the levels of H3K27ac in cells exposed to the inhibitor, and neither this modification, nor the H3S28p could be rescued by GnRH, whereas there was no apparent effect on levels of H3K18ac (Fig. 6F). Confirmation of the role of MSK1 in H3S28p was then provided through shRNA-mediated knockdown (KD) of MSK1, which significantly reduced S28p levels, although it had less effect on K27ac. It is not clear whether this discrepancy is due to the different efficacy in reducing H3S28p levels, and/or might be due to nonspecific effects of the inhibitor on the K27ac. Notably, however, no effect was apparent on basal Cga mRNA levels (Fig. 6G).
GnRH-induction of Cga expression requires MSK induced H3S28p at the +1 nucleosome
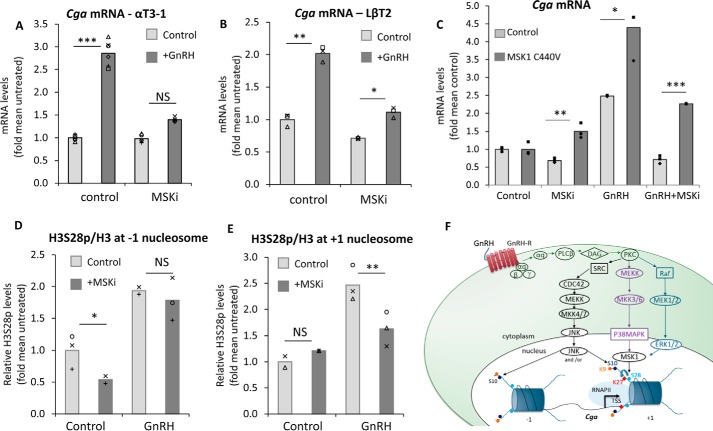
Having established that the H3S28p is regulated by MSK1, that MSK1 is activated following GnRH treatment, and that this can be blocked most effectively by the specific inhibitor, we sought to determine how this modification affects Cga transcription. The dramatic reduction in H3S28p levels by incubation of the cells with the MSK inhibitor, as after the MSK1 KD (Fig. 6G), did not appear to affect the basal levels of Cga expression, but clearly repressed GnRH-induced up-regulation of the gene in both gonadotrope cell lines (Fig. 7, A and B). Moreover, in confirmation of the specificity of the effect of the inhibitor, this repressive effect was significantly abated in cells expressing a RMM-64-insensitive MSK1 mutant (Fig. 7C).
Figure 7.
GnRH induction of Cga expression requires MSK induced S28p at the +1 nucleosome. A and C–E, αT3-1 cells or B, LβT2 cells were treated with GnRH and/or RMM-64 added 2 h prior to the GnRH, whereas some of the cells in C were transfected with a RMM-64 resistant MSK1 mutant, C440V, before (A–C) qPCR analysis of the Cga mRNA levels (GnRH 8 h); or (D and E) ChIP analysis (GnRH 5 h) to determine the levels of H3S28p at the nucleosomes upstream and downstream of the Cga TSS. The qPCR and ChIP were carried out and data presented as described in the legend to Fig. 1, with levels shown relative to mean values in untreated control cells; the control values with/without GnRH are from the experiment shown in Fig. 5E. F, schematic model showing the GnRH-induced pathways leading to phosphorylation of H3S28 at the +1 nucleosome of the Cga gene, which is required for the transcriptional response, possibly mediated also through H3K27ac both of which modifications would facilitate RNAPII transition of this well positioned nucleosome for transcriptional elongation. Although GnRH also activates JNK, which is essential for H3S10p, this modification does not affect H3K9ac and plays little effect in basal or GnRH-induced transcription of this gene. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
High resolution ChIP analysis of H3S28p revealed that the inhibition of MSK affected primarily the GnRH-induced levels of this modification at the +1 nucleosome, although the basal levels at the −1 nucleosome were also affected (Fig. 7, D and E). This suggests that phosphorylation of H3S28 at the upstream nucleosome is quite easily removed when MSK is not active, but that H3S28p at this nucleosome plays little role in basal rates of on-going transcription, as seen by the lack of effect on basal Cga expression (Fig. 7, A and B). In contrast, the GnRH-induced increase in H3S28p at the +1 nucleosome implies a role in promoter escape and/or the early steps in transcriptional elongation.
Discussion
GnRH is the major central regulator of the reproductive axis due to its role in stimulating production of the gonadotropin hormones, primarily via several MAPK pathways that activate gene-specific transcription factors (31, 32). Despite a crucial role for these kinases in increasing gonadotropin gene expression, there have not yet been any reports of a role for histone phosphorylation in the activation the GnRH target genes. In fact, although phosphorylation is a crucial and central player in intracellular signaling pathways in general, the role of histone phosphorylation as an end point in these pathways to activate gene transcription has remained understudied. We report here that, although H3S10p is found at all three gonadotropin genes in unstimulated gonadotropes, and is relatively enriched on the more highly expressed Cga gene, neither H3S10p nor H3S28p play much of a role in determining basal levels of Cga expression. Nevertheless, GnRH increases phosphorylation at both serines at the 5′ end of this gene, as well as increasing H3 acetylation. The GnRH-induced increase in H3S28p at the first nucleosome downstream of the TSS appears crucial for the GnRH stimulation of Cga expression, presumably facilitating RNAPII transition of this nucleosome and promoter escape (Fig. 7F).
Although JNK is activated by GnRH (31) and is clearly essential for the basal phosphorylation of H3S10, this modification surprisingly has very little effect on the on-going transcription of the Cga gene. This is in strong contrast with the situation in differentiating neuronal cells in which JNK-mediated phosphorylation of H3S10 is essential for expression of the target genes (30). The fact that the dramatic reduction in H3S10p levels following JNK inhibition also did not affect acetylation of the neighboring H3K9 indicates that this acetylation is JNK-independent and supports a previously suggested model that H3K9ac and H3S10p may be independent through spatially linked processes, and that phosphorylation is not necessarily a prerequisite for this acetylation (13, 35).
Reduction in the basal levels of H3S28p at the upstream nucleosome also had no apparent effect on the basal levels of Cga transcription, and was a transient response globally after GnRH exposure, presumably as a result of rapid phosphatase activation (32). However, after 3–6 h of GnRH, when increases in Cga expression are first noted, this modification was increased at both nucleosomes, as was H3K27ac. The link between H3S28p and H3 acetylation in stimulus-induced transcription is less well studied than for H3S10p, but cross-talk has been noted in several contexts, including at stress-responsive promoters where H3S28p is accompanied by increased histone acetylation (13), while targeting MSK1 to the α-globin promoter induced both H3K27ac and H3S28p (37). More recently it was demonstrated that H3S28p increases directly p300/CBP-dependent transcription in vitro, and MSK1 was seen to facilitate p300 recruitment to a chromatin template, leading to the proposition that H3S28p helps stabilize p300 association with the chromatin (25).
Our findings suggest a crucial role for MSK1 in the phosphorylation of H3S28 and in vitro kinase assays have previously shown that it is a direct target of this kinase (26, 38, 39). Moreover, MSK1 was shown to bind directly to the MLL1 complex (40), which we previously reported is found at the Cga gene TSS (3). We cannot exclude the possibility that the inhibitor also has other nonspecific effects; generally negligible effects on other kinases were reported (25), but the increase in S28p and CREBp at the lowest doses suggest a possible inhibition of the phosphatases that target these proteins although not ATF-1. In this study, however, we utilized the doses that were clearly inhibitory to the kinase activity. The MSK1/2 might also phosphorylate transcription factors that are responsible for recruiting the HAT enzymes to this gene, although CREB does not mediate the GnRH effect on this gene (41). It is also possible that MSK1/2 itself recruits HAT enzymes to the +1 nucleosome; MSK1 was shown previously to co-immunoprecipitate with multiple HATs including CBP and p300 both of which acetylate H3K27 (25, 42). However, in the context of these other findings, our results suggest that H3S28p facilitates H3K27ac at the 5′ end of the Cga gene as part of the GnRH-induced transcriptional response.
The precise roles of promoter H3S10p and S28p in basal Cga expression are not yet clear, but MSK was previously reported to cause displacement of repressive polycomb group proteins in the activation of a subset of genes via various signaling-response and neuronal differentiation pathways involving H3S28p (43). It is thus possible that these modifications are required primarily for the initial activation of closed chromatin, whereas in gonadotrope cells in which the Cga gene is already expressed, histone phosphorylation has little effect on the already open chromatin.
In these cells, the chromatin structure of the Cga, although it is a highly inducible gene, is more characteristic of a readily-expressed gene. In its basal state, the active proximal promoter is relatively free of nucleosomes, rendering it highly accessible for binding and activation by transcription factors. In contrast, the first nucleosome downstream of the TSS is well positioned, and provides an additional level of control by forming a strong barrier for early elongation (36). Such a barrier causes RNAPII stalling and backtracking, from which the RNAPII can proceed only after a recovery process involving spontaneous wrapping and unwrapping of the nucleosome (44–46). We have shown previously that incorporation of the H2A.Z histone variant allows greater mobility of the nucleosome to help overcome this barrier (36), whereas here we demonstrate that histone H3 phosphorylation, which reduces the histone-DNA charge interactions, likely also plays a role in the recovery from this backtracked state. Likewise, this barrier to elongation would be facilitated by the elevation in histone acetylation at H3K27, which removes a positive charge on the histone tail, and is reportedly facilitated by H3S28p-mediated HAT recruitment (25). Interestingly, a similar role has been shown in Drosophila for H3S10p, which is phosphorylated by JIL-1, a kinase essential for early transcriptional elongation; the 14-3-3 proteins recruited by H3S10p interact with the HAT Elongator protein 3, which acetylates H3K9 and this is also required for elongation to proceed (19).
The present study has thus demonstrated a specific role for H3S28 phosphorylation and reveals that it is regulated by a MAPK-activated pathway though MSK. Furthermore, this study emphasizes the importance of context in understanding the role of histone phosphorylation at specific nucleosomes. GnRH modulates the chromatin of the Cga gene to activate its expression primarily at the level of transcription elongation by modulating the +1 nucleosome barrier, so facilitating RNAPII progression. The GnRH induction of H3S28 phosphorylation by MSK1 thus plays a major role in regulating the Cga expression, by targeting this nucleosome.
These findings corroborate the recent report that H3S28p is a key event in stimulus-activated transcription in macrophages (25) and support the idea that this may be a widespread event in the induction of gene expression in response to specific stimuli, as a consequence of the activation of the commonly utilized MAPK signaling pathways. However, our study also places this in the context of the first nucleosome downstream of the TSS, with implications for promoter escape, linking the known activation of MAPK cascades as part of common signal transduction cascades to diverse downstream epigenetic effects at the target genes.
Experimental procedures
Cell culture
The murine gonadotrope αT3-1 and LβT2 cells (gifts from P. L. Mellon, University of California, San Diego) were cultured as reported (41), with serum removed (LβT2) or reduced to 0.5% (αT3-1) for 16 h before exposure to GnRH or PMA (both 100 nm), H89 (13.5 μm), SP600125 (all Sigma) or RMM-64 (both 10 μm; 25), with the inhibitors being added 30–120 min prior to the GnRH. Length of exposure to GnRH was characteristically 6–8 h for assessment of effects on gene expression, and 2–5 h when measuring histone modifications, according to the initial empirical evaluation of optimal response. After replacement of serum and antibiotics (30–60 min), cells at 70–80% confluence were transfected with plasmids expressing a dominant-negative CDC42 (a gift from BC Low, National University of Singapore) or MSK1 C440V (25), using PolyJet In Vitro DNA transfection reagent (Signagen) according to the manufacturer's instructions (3 μl reagent:1 μg of DNA). MSK1 was knocked down using the targeting sequence: AAAGTGAGCCTCCGTATCC in pSUPER, as previously described (41).
Quantitative PCR analysis
RNA was extracted using TRIzol (Invitrogen), total RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and real-time quantitative PCR (qPCR) carried out using PerfeCTa SYBR Green FastMix (Quanta Biosciences, Gaithersburg, MD) all as previously described (3). The primers for detection of Cga mRNA levels were, forward, ATGGATTACTACAGAAAATATGCAG and reverse, CCTGAATAATAAAGTCTCCATCAGG, and for Msk1, forward, TCCTTACTGTCAAGCACGAGC and reverse, AGGTGTTCCAGGGCAAGCACA. The normalizing gene was Rplp0, with forward primer, GCGACCTGGAAGTCCAACTA and reverse primer, ATCTGCTTGGAGCCCACAT. A standard curve, generated by 1:2 serial dilutions, was used to determine the levels in each sample. All reactions were repeated three times and averaged.
Protein isolation and purification
Cells were placed on ice and washed with ice-cold PBS before harvest in fresh PBS, transferred to a pre-cooled microcentrifuge tube, and centrifugation at 3000 rpm for 5 min at 4 °C. The supernatant was removed leaving the cell pellet on ice. For whole cell extract, cells were lysed in radioimmunoprecipitation assay buffer (RIPA: 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, 2 mm EDTA, 150 mm NaCl, 50 mm Tris-HCl, pH 8) containing protease and phosphatase inhibitors (2 μg/ml of aprotinin; 10 μg/ml of leupeptin; 1 μg/ml of pepstatin A, 1 mm PMSF, 10 mm NaF, and 1 mm Na3VO4) for 30 min. After centrifugation at 12,000 rpm for 20 min at 4 °C, the supernatant was collected and transferred into a fresh tube. For the isolation of cytoplasmic and nuclear proteins, the NE-PER nuclear and cytoplasmic extraction kit (Pierce) was used, as previously described (47). For the isolation of histones, cells were lysed in Triton extraction buffer (TEB: PBS containing 0.5% Triton X-100 (v/v)) containing the protease and phosphatase inhibitors (as above). After centrifugation at 2000 rpm for 10 min at 4 °C, the pellet was washed in one-half volume of TEB and centrifuged as before. The supernatant was removed, the pellet was resuspended in 0.2 m HCl, and the samples were incubated overnight at 4 °C on ice. The samples were then centrifuged at 2000 rpm for 10 min at 4 °C and the supernatant was collected and transferred into a fresh tube. The protein concentrations were determined via Bradford Protein Assay reagent (Bio-Rad) according to the manufacturer's instructions, following which the samples were used immediately or stored at −80 °C.
Western blot analysis was carried out as reported previously (47), after loading the same amounts of protein in each lane of a 10 or 12.5% SDS-polyacrylamide electrophoresis gel and with nitrocellulose membranes. The membrane was incubated with a primary antibody diluted to 1,000–15,000 overnight at 4 °C or for 2 h at room temperature, and after washing with 1× PBST, the membrane was incubated with horseradish peroxidase-conjugated anti-mouse, -rabbit, or -rat diluted to 1:10,000 in 1× PBST for 1 h at room temperature. The immunoreactive proteins were detected using the Super Signal Pico West chemiluminescent system (Pierce Chemical Co.) according to the manufacturer's instructions, followed by exposure in the Las-4000 image analyzer (Fujifilm) and analysis using the ImageJ software (NIH). The magnitude of each signal peak was measured as a percent of the total size of the measured peaks and this value was normalized against that of the loading control in the same sample. Loading controls for histone modifications were the total H3 levels, which even after stripping give a strong signal. Modified MSK1 and CREB were normalized first to the RNAPII loading control, which is not affected by the treatments in this study; the total/unmodified proteins in the same samples were assessed similarly in parallel due to their lower levels and weaker signals, after which these relative values were normalized.
Chromatin immunoprecipitation (ChIP)
Cells were grown to ∼70–80% confluence before treatment with GnRH and/or the inhibitors. Cross-linking was by addition of 1% formaldehyde in ice-cold PBS for 10 min at room temperature on a shaking platform and terminated by 0.125 m glycine for 5 min at room temperature. Cells were washed twice and harvested in ice-cold 1 × PBS, and ChIP was carried out essentially as previously reported (41). The sonication to obtain fragments averaging 500–700 bp was carried out using 10 pulses of 33% amplitude for 15 s, with 10-s intervals between each pulse. Alternatively, to obtain DNA fragments of 100–150 bp (high resolution ChIP), which is able to distinguish between the nucleosomes, 60 pulses of 33% amplitude for 15 s, with 10-s intervals between each pulse were used (as demonstrated in Ref. 36). The immunoprecipitated DNA was collected and purified using the PCR purification kit (Qiagen) and quantified by qPCR as above using primers to the proximal promoters described in Ref. 3, centering at −60 to −160 bp, or as follows for Cga: forward +47, GTGGTCACAAATATTTTACTCTTT; forward −429, GGAGCAATTGTTTTATTTTTCTGT; forward −525: CACACCTGGACATATCTACTGT; reverse +147, ATCACCTGCCCAGAACAC; R −139: TTGATCATATCACATTGCAACCC; reverse −329, ATTAGCTAAGTACCTGATATTTTCA; reverse −425, CAATTGCTCCAATTTCTTAAATTAAC.
Antibodies
Antibodies used were: anti-H3 (Abcam; ab1791), anti-H3K27ac (Cell Signaling; number 4729), anti-H3K18ac (Abcam; ab9675), anti-H3S10p (Abcam; ab14955; all with various lot numbers over several years during which experiments were repeated), anti-H3K9ac (Cell Signaling Technology; number 9649, lot 11 08/2013), anti-H3S28p (Sigma; number H9908, lot H9908), anti-phospho-CREB (Cell Signaling Technology; number 9198, lot 14 05/2017), anti-CREB (Cell Signaling Technology; number 9197, lot 16 05/2017), anti-phospho-MSK1 (Thr-581: Cell Signaling Technology; number 9595, lot 3 03/2017), anti-MSK1 (Santa Cruz; sc25417, lot G2205), anti-RNAPII (Santa Cruz; sc899, lot K1513), goat anti-mouse IgG-HRP (Santa Cruz; sc-2005, lot D0116), goat anti-rabbit IgG-HRP (Santa Cruz; sc-2004, lot B1315), goat anti-rat IgG-HRP (Santa Cruz; sc-2006, lot G2514). The specificity of antibodies to modified proteins was determined through differential responses from those of the total/non-modified protein, as shown in this and previous studies (e.g. Refs. 3, 29, and 48).
For Western blot analysis, antibodies for total H3 and histone modifications were diluted 1:5000, except for that against H3S28p, which was diluted 1:2000. The antibodies for MSK1 and phospho-MSK1 were diluted 1:1000 and 1:1500, respectively, for CREB and phospho-CREB at 1:2000, RNAPII at 1:5000, and IgG was diluted 1:10,000. For ChIP, they were diluted 1:280, except for the antibodies against H3 and H3S28p, which were used at 1:350 and 1:140 dilutions, respectively.
Statistical analysis
All experiments were repeated on at least two occasions, and representative individual experiments or compiled results are presented. One-way analysis of variance, followed by Bonferroni's t test or Student's t test were employed to determine statistically different means. Differences were considered significant when p < 0.05.
Author contributions
M. H. conception, design, and performing the research, analysis of results, writing of draft manuscript. A. W. conception, design, and performing the research and analysis of results. S. R. conception and experimental design. J. T. supplied reagents that he validated. L. P. contributed to experimental design, performed the research, and analysis of results. P. M. conceived the project, oversaw the work, analysis, and interpretation of data, wrote the final version of the paper.
Acknowledgment
We acknowledge the gift from Pamela Mellon (University of California, San Diego) of the gonadotrope cell lines.
This work was supported by Israel Science Foundation Grant 840/12 (to P. M.) and the European Framework program Marie Curie Actions. The authors declare that they have no conflicts of interest with the contents of this article.
- GnRH
- gonadotropin-releasing hormone
- PMA
- phorbol 12-myristate 13-acetate
- CREB
- cAMP-response element-binding protein
- qPCR
- quantitative PCR
- HAT
- histone acetyltransferase
- MSK1
- mitogen- and stress-activated protein kinase 1
- TSS
- transcriptional start site.
References
- 1. Lim S., Luo M., Koh M., Yang M., bin Abdul Kadir M. N., Tan J. H., Ye Z., Wang W., and Melamed P. (2007) Distinct mechanisms involving diverse histone deacetylases repress expression of the two gonadotropin β-subunit genes in immature gonadotropes, and their actions are overcome by gonadotropin-releasing hormone. Mol. Cell. Biol. 27, 4105–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Melamed P. (2008) Histone deacetylases and repression of the gonadotropin genes. Trends Endocrinol. Metab. 19, 25–31 [DOI] [PubMed] [Google Scholar]
- 3. Wijeweera A., Haj M., Feldman A., Pnueli L., Luo Z., and Melamed P. (2015) Gonadotropin gene transcription is activated by menin-mediated effects on the chromatin. Biochim. Biophys. Acta 1849, 328–341 [DOI] [PubMed] [Google Scholar]
- 4. Melamed P., Kadir M. N., Wijeweera A., and Seah S. (2006) Transcription of gonadotropin β subunit genes involves cross-talk between the transcription factors and co-regulators that mediate actions of the regulatory hormones. Mol. Cell. Endocrinol. 252, 167–183 [DOI] [PubMed] [Google Scholar]
- 5. Mouillet J. F., Sonnenberg-Hirche C., Yan X., and Sadovsky Y. (2004) p300 regulates the synergy of steroidogenic factor-1 and early growth response-1 in activating luteinizing hormone-β subunit gene. J. Biol. Chem. 279, 7832–7839 [DOI] [PubMed] [Google Scholar]
- 6. Miller R. S., Wolfe A., He L., Radovick S., and Wondisford F. E. (2012) CREB binding protein (CBP) activation is required for luteinizing hormone β expression and normal fertility in mice. Mol. Cell. Biol. 32, 2349–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baek S. H. (2011) When signaling kinases meet histones and histone modifiers in the nucleus. Mol. Cell. 42, 274–284 [DOI] [PubMed] [Google Scholar]
- 9. Banerjee T., and Chakravarti D. (2011) A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 31, 4858–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunn K. L., and Davie J. R. (2005) Stimulation of the Ras-MAPK pathway leads to independent phosphorylation of histone H3 on serine 10 and 28. Oncogene. 24, 3492–3502 [DOI] [PubMed] [Google Scholar]
- 11. Rossetto D., Avvakumov N., and Côté J. (2012) Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7, 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sawicka A., and Seiser C. (2012) Histone H3 phosphorylation: a versatile chromatin modification for different occasions. Biochimie 94, 2193–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sawicka A., and Seiser C. (2014) Sensing core histone phosphorylation: a matter of perfect timing. Biochim. Biophys. Acta 1839, 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walter W., Clynes D., Tang Y., Marmorstein R., Mellor J., and Berger S. L. (2008) 14-3-3 interaction with histone H3 involves a dual modification pattern of phosphoacetylation. Mol. Cell. Biol. 28, 2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winter S., Simboeck E., Fischle W., Zupkovitz G., Dohnal I., Mechtler K., Ammerer G., and Seiser C. (2008) 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 27, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., and Oliviero S. (2009) Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 138, 1122–1136 [DOI] [PubMed] [Google Scholar]
- 17. Drobic B., Pérez-Cadahía B., Yu J., Kung S. K., and Davie J. R. (2010) Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Res. 38, 3196–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simboeck E., Sawicka A., Zupkovitz G., Senese S., Winter S., Dequiedt F., Ogris E., Di Croce L., Chiocca S., and Seiser C. (2010) A phosphorylation switch regulates the transcriptional activation of cell cycle regulator p21 by histone deacetylase inhibitors. J. Biol. Chem. 285, 41062–41073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karam C. S., Kellner W. A., Takenaka N., Clemmons A. W., and Corces V. G. (2010) 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet. 6, e1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheung P., Tanner K. G., Cheung W. L., Sassone-Corsi P., Denu J. M., and Allis C. D. (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 5, 905–915 [DOI] [PubMed] [Google Scholar]
- 21. Clayton A. L., Rose S., Barratt M. J., and Mahadevan L. C. (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19, 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo W. S., Trievel R. C., Rojas J. R., Duggan L., Hsu J. Y., Allis C. D., Marmorstein R., and Berger S. L. (2000) Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 5, 917–926 [DOI] [PubMed] [Google Scholar]
- 23. Klein A. M., Zaganjor E., and Cobb M. H. (2013) Chromatin-tethered MAPKs. Curr. Opin. Cell Biol. 25, 272–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S. H., Sharrocks A. D., and Whitmarsh A. J. (2013) MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13 [DOI] [PubMed] [Google Scholar]
- 25. Josefowicz S. Z., Shimada M., Armache A., Li C. H., Miller R. M., Lin S., Yang A., Dill B. D., Molina H., Park H. S., Garcia B. A., Taunton J., Roeder R. G., and Allis C. D. (2016) Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol. Cell. 64, 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., and Arthur J. S. (2003) MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vermeulen L., Vanden Berghe W., Beck I. M., De Bosscher K., and Haegeman G. (2009) The versatile role of MSKs in transcriptional regulation. Trends Biochem. Sci. 34, 311–318 [DOI] [PubMed] [Google Scholar]
- 28. Reyskens K. M., and Arthur J. S. (2016) Emerging roles of the mitogen and stress activated kinases MSK1 and MSK2. Front. Cell Dev. Biol. 4, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yosefzon Y., David C., Tsukerman A., Pnueli L., Qiao S., Boehm U., and Melamed P. (2017) An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis. Proc. Natl. Acad. Sci. U.S.A. 114, 10131–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tiwari V. K., Stadler M. B., Wirbelauer C., Paro R., Schübeler D., and Beisel C. (2011) A chromatin-modifying function of JNK during stem cell differentiation. Nat. Genet. 44, 94–100 [DOI] [PubMed] [Google Scholar]
- 31. Naor Z. (2009) Signaling by G-protein-coupled receptor (GPCR): studies on the GnRH receptor. Front. Neuroendocrinol. 30, 10–29 [DOI] [PubMed] [Google Scholar]
- 32. Lim S., Pnueli L., Tan J. H., Naor Z., Rajagopal G., and Melamed P. (2009) Negative feedback governs gonadotrope frequency-decoding of gonadotropin releasing hormone pulse-frequency. PLoS ONE 4, e7244 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jorgensen J. S., Quirk C. C., and Nilson J. H. (2004) Multiple and overlapping combinatorial codes orchestrate hormonal responsiveness and dictate cell-specific expression of the genes encoding luteinizing hormone. Endocr. Rev. 25, 521–542 [DOI] [PubMed] [Google Scholar]
- 34. Xie J., Bliss S. P., Nett T. M., Ebersole B. J., Sealfon S. C., and Roberson M. S. (2005) Transcript profiling of immediate early genes reveals a unique role for activating transcription factor 3 in mediating activation of the glycoprotein hormone alpha-subunit promoter by gonadotropin-releasing hormone. Mol. Endocrinol. 19, 2624–2638 [DOI] [PubMed] [Google Scholar]
- 35. Sawicka A., Hartl D., Goiser M., Pusch O., Stocsits R. R., Tamir I. M., Mechtler K., and Seiser C. (2014) H3S28 phosphorylation is a hallmark of the transcriptional response to cellular stress. Genome Res. 24, 1808–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rudnizky S., Bavly A., Malik O., Pnueli L., Melamed P., and Kaplan A. (2016) H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 7, 12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lau P. N., and Cheung P. (2011) Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl. Acad. Sci. U.S.A. 108, 2801–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dyson M. H., Thomson S., Inagaki M., Goto H., Arthur S. J., Nightingale K., Iborra F. J., and Mahadevan L. C. (2005) MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J. Cell Sci. 118, 2247–2259 [DOI] [PubMed] [Google Scholar]
- 39. Zhong S., Jansen C., She Q. B., Goto H., Inagaki M., Bode A. M., Ma W. Y., and Dong Z. (2001) Ultraviolet B-induced phosphorylation of histone H3 at serine 28 is mediated by MSK1. J. Biol. Chem. 276, 33213–33219 [DOI] [PubMed] [Google Scholar]
- 40. Wiersma M., Bussiere M., Halsall J. A., Turan N., Slany R., Turner B. M., and Nightingale K. P. (2016) Protein kinase MSK1 physically and functionally interacts with the KMT2A/MLL1 methyltransferase complex and contributes to the regulation of multiple target genes. Epigenetics Chromatin 9, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pnueli L., Luo M., Wang S., Naor Z., and Melamed P. (2011) Calcineurin mediates the gonadotropin-releasing hormone effect on expression of both subunits of the follicle-stimulating hormone through distinct mechanisms. Mol. Cell. Biol. 31, 5023–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janknecht R. (2003) Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1). Oncogene 22, 746–755 [DOI] [PubMed] [Google Scholar]
- 43. Gehani S. S., Agrawal-Singh S., Dietrich N., Christophersen N. S., Helin K., and Hansen K. (2010) Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3 S28 phosphorylation. Mol. Cell. 39, 886–900 [DOI] [PubMed] [Google Scholar]
- 44. Hodges C., Bintu L., Lubkowska L., Kashlev M., and Bustamante C. (2009) Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kireeva M. L., Hancock B., Cremona G. H., Walter W., Studitsky V. M., and Kashlev M. (2005) Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 18, 97–108 [DOI] [PubMed] [Google Scholar]
- 46. Petesch S. J., and Lis J. T. (2012) Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 28, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feng J., Lawson M. A., and Melamed P. (2008) A proteomic comparison of immature and mature mouse gonadotrophs reveals novel differentially expressed nuclear proteins that regulate gonadotropin gene transcription and RNA splicing. Biol. Reprod. 79, 546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pnueli L., Rudnizky S., Yosefzon Y., and Melamed P. (2015) RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin α-subunit gene. Proc. Natl. Acad. Sci. U.S.A. 112, 4369–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]