Abstract
Fungi are ubiquitous microbes that are common in diverse environments including as commensal organisms on the human body. In addition to its obvious role as a digestive organ, the intestines have been further appreciated as important for the development, maintenance, and instruction of the immune system. The gut harbors many types of microorganisms including bacteria, archaea, fungi, and viruses, and many studies over the past couple of decades have documented an important role for intestinal bacteria in immunological function. Recent studies are now suggesting that intestinal fungi (the gut “mycobiome”) may similarly play important roles in host immunity and inflammation. This review will discuss recent studies that will influence our growing understanding of the role(s) of intestinal fungi in health and disease.
INTRODUCTION
Growing evidence documents that the mammalian gut is home to a diverse population of fungi that co-exist with other microbes in the intestinal microbiome including bacteria and viruses [1] (Fig. 1). Many of these fungi such as Candida spp. or Saccharomyces cerevisiae are well-known to most biologists, while most others such as Wallemia spp. or Cladosporium spp. are less well-known. Current definitions of the richness of the mammalian gut mycobiome mostly make use of culture-independent sequencing methods involving PCR amplification of internal transcribed spacer regions (ITS1 or ITS2) of fungal ribosomal DNA (rDNA) for species identification [2]. Characterizing fungal communities can be more challenging than bacterial communities due to uncertain methods of genomic DNA isolation, variations in the lengths of ITS regions in different fungi that influence PCR efficiencies, and insufficient reference databases. Further, we are still in early stages of understanding “normal” fungal diversity, time- and diet-based fluctuations, and the contributions of environmental exposures to definitions of the fungal microbiota [3]. Nevertheless, growing numbers of studies are suggesting that the intestinal mycobiome is skewed in the context of diseases including obesity, asthma, cirrhosis, and autism [4–6]. The potential for fungal dysbiosis to influence disease progression will be an active area of research in coming years.
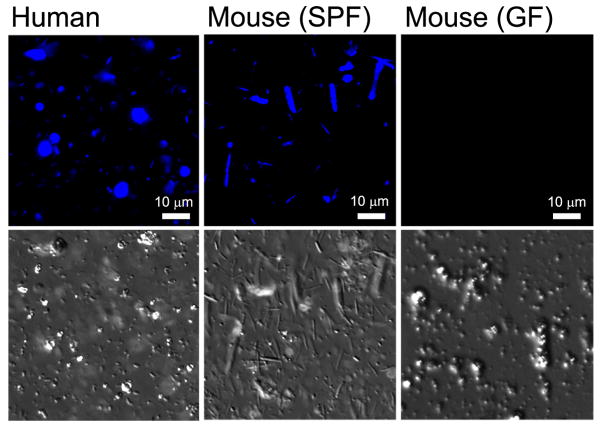
Figure 1. Fecal fungi.
Fungi are easily detectable in stool samples from humans and specific pathogen-free (SPF) mice, but not in germ-free (GF) mice. Stool was collected and stained fresh with calcofluor white, a fluorescent dye that binds to chitin in fungal cell walls and is commonly used to detect pathogenic fungi in histology sections. Fungi are readily recognizable by their size and are distinct from plant material, which can also stain with calcofluor white but are not visible in the images shown.
INTESTINAL FUNGAL MICROBIOTA
The intestinal mycobiome
A good balance of microbial communities in the gut and immune responses to intestinal microbes is essential for mammalian health. In most studies cataloging fungi found in the intestines of humans or mice, over 50 genera of fungi are commonly reported, although 10 or fewer typically account for vast majority of organisms detected [3,7]. The fungi most commonly reported in the intestines of mice and humans include Saccharomycetes including Candida and Saccharomyces spp., Eurotiomycetes including Aspergillus and Penicillium spp., Tremellomycetes including Cryptococcus and Trichosporon spp. as well as Cladosporium, Wallemia, and Malassezia spp. [4,7–15]. Some reports also include robust detection of fungi such as Phoma, Alternaria, Sclerotinia and others which are primarily plant pathogens and are likely to be carried by food or other environmental sources.
Alterations in the intestinal microbiome are associated with many diseases, and alterations (dysbiosis) of fungal communities may contribute to disease susceptibility or disease severity. Many factors can likely promote fungal dysbiosis including exposure to antibiotics, diet, or genetic diversity. Exposure to antibacterial antibiotics has long been known to promote overgrowth of Candida in the gut [16]. Similarly, treatment of mice with antifungal antibiotics reduces the prevalence of some fungi, while it increases the prevalence of others [10]. The genetics of the host can influence the fungal microbiota. Mice deficient in CARD9 or Dectin-1 have been reported to have alterations in bacterial and fungal communities including changes in overall fungal burden as well as specific alteration in Microbotryomycetes and Lactobacilli [17–19], although how reproducible this is in the context of diverse facilities with diverse baseline microbiota is not yet clear. We are only beginning to understand the factors that promote colonization and growth of certain fungi in the gastrointestinal tract.
Requirements for allowing Candida colonization
Candida albicans is probably the most well-known intestinal fungus, being both a commensal in healthy individuals and an opportunistic pathogen of the gastrointestinal tract as well as of other mucosal surfaces (e.g. skin and vagina) and the blood. Studies on the mechanisms by which Candida colonizes the gut may provide insights into how fungi establish themselves in the intestinal environment. Treatment of people or mice with oral antibiotics can permit expansion of intestinal Candida populations, and in mice this has been established as a model for GI Candida overgrowth and long-term colonization [20].
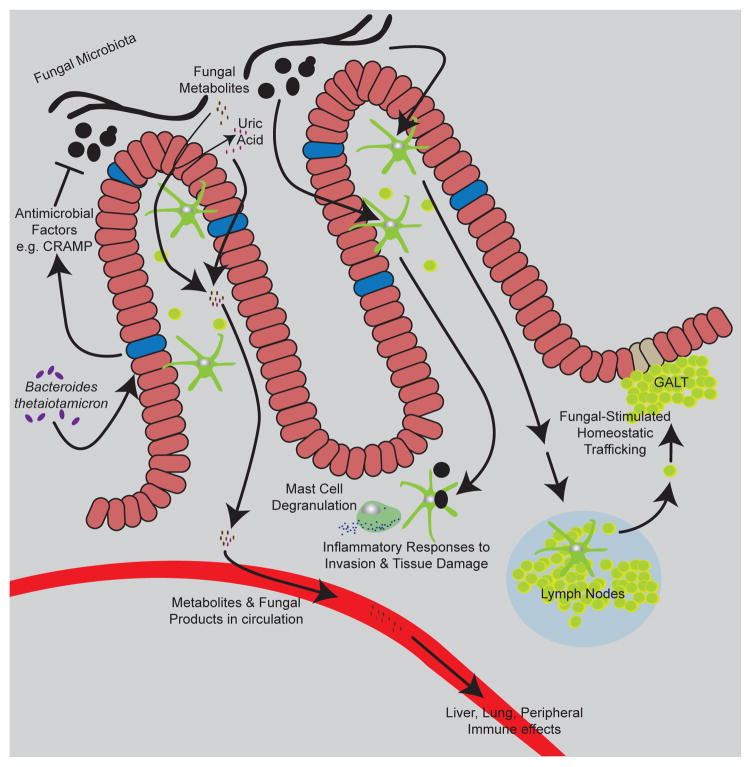
A recent study demonstrated that the mice colonized with Bacteroides thetaiotamicron are more resistant to C. albicans colonization. The authors proposed that B. thetaiotamicron stimulates colonocytes to express hypoxia-inducible factor-1α (HIF-1α), leading to production of antimicrobial peptide LL-37 (CRAMP in mice). Elevated levels of CRAMP contributed to killing of Candida and restricted GI colonization with the fungus [21] (Fig. 2). These findings suggest that inter-kingdom symbiosis between bacteria and fungi may be regulated, at least in part, by host immune responses to specific populations.
Figure 2. Diverse effects of commensal fungi.
The diagram depicts many of the effects of commensal fungi discussed in the text. Bacterial microbiota and the intestinal epithelium influence the ability of fungi to colonize the gut. Commensal fungi produce metabolites and products that influence immunity and inflammation at local and distal sites. Immune cells in the gut respond to commensal fungi to influence inflammation and immune homeostasis.
With respect to Candida, it is not clear what dictates the difference between commensal colonization and development of infectious disease. Candida albicans is a dimorphic fungus, meaning that it can grow in yeast and hyphal forms, and this ability to switch morphologies is linked to pathogenesis [22]. In addition, yeast can take on different morphologies including “opaque(a/α)” and “grey” as well as a morphology specifically associated with GI growth termed “gastrointestinally induced transition” (GUT) cells [23]. How these changes influence commensal colonization are still being worked out, but they likely reflect metabolic changes as well as changes in how the different types of cells interact with host tissues and immune cells. Until recently, Candida was not thought to produce lytic toxins targeting host cells, but a new study now reports that that the hyphal form of C. albicans secretes an amphiphilic cytolytic peptide, candidalysin, that disrupts epithelial cell membranes and promotes invasive disease [24].
INTESTINAL DISEASE
Inflammatory Bowel Disease
Crohn’s disease (CD) and ulcerative colitis (UC) are the major clinical forms of inflammatory bowel disease (IBD), a chronic inflammatory disease of the gastrointestinal tract [25]. The causes of IBD remain elusive, although data suggest that variations in environmental factors, host genetics, and gut microbiota, all contribute to susceptibility to disease. As the mycobiome has gained attention for its role in promoting both health and disease, several studies have begun to investigate whether there is evidence of fungal dysbiosis or altered immune responses to fungi in IBD patients [9,14,15,26,27].
Dectin-1 (CLEC7A), C-type lectin receptor, is a key innate immune receptor involved in coordinating host defense against fungi. It recognizes β-1,3-glucan, a major structural component of fungal cell walls [28]. Dectin-1 triggers phagocytosis of fungi and production of reactive oxygen species in macrophages and dendritic cells (DCs) and signals through CARD9 and NF-κB to direct production of inflammatory cytokines. Iliev et al. elucidated a central role of Dectin-1 in regulating the severity of colitis using the mouse DSS-induced colitis model [9]. Dectin-1 deficient mice were more susceptible to colitis, showing increased weight loss, pro-inflammatory cytokines, and delayed recovery from colitis (Fig. 2). Furthermore, they found that polymorphic variants in the CLEC7A gene are associated with increased severity of disease in ulcerative colitis patients, adding to previous studies showing a strong link between polymorphisms in CARD9 and IBD [9,29].
Recently Sokol et al. observed a decrease in α-diversity (species richness) of the fungal microbiota and an increase in the fungi-to-bacteria diversity ratio in fecal samples from over 200 IBD patients (mixed CD and UC) relative to a handful of healthy controls [14]. Ascomycota and Basidiomycota were the major phyla present in the fungal communities of both healthy subjects and IBD patients, and the Basidiomycota/Ascomycota ratio appeared to be elevated in IBD patients compared to controls. Specifically, the prevalence of Candida albicans was observed to be elevated in IBD patients, while Saccharomyces cerevisiae was reduced. Furthermore, correlation analysis revealed that the microbial network, including bacteria-fungi interactions, was altered in IBD patients compared to healthy subjects. The investigators noted stronger interactions between fungi and bacteria in UC patients whereas there was an uncoupling effect between the two in CD. These data suggest that fungi may differently influence how UC and CD develop.
Hoarau and coworkers take a slightly different approach in analyzing the micro- and mycobiome in IBD, specifically CD, by directly comparing CD patients and their healthy, non-CD relatives (NCDR) [15]. In this study, CD and NCDR samples did not differ in mycobiota diversity. Similar to Sokol et al., this study observed elevated Candida (although in this case Candida tropicalis) in CD patients. Anti-Saccharomyces cerevisiae antibodies (ASCA) are commonly found in the serum of IBD patients where these antibodies recognize mannan, a component in the fungal cell wall (including Candida spp.) [30]. Indeed, the authors observed elevated ASCA levels in the CD group, and they also noted a positive association between the presence of ASCA and C. tropicalis abundance in the CD group, suggesting immune responses to the intestinal fungi. In this group’s analysis of bacteria-fungal interactions, they specifically noted a positive association between C. tropicalis and Escherichia coli and Serratia marcescens. In vitro experiments revealed that together these three species generate a thicker biofilm than any of the species generate alone, leading the investigators to hypothesize that the organisms may synergize to establish a commensal niche. Investigating whether this triple-species biofilm forms in the gut and affects immune responses may provide novel insight about the pathogenesis of CD.
Irritable bowel syndrome (IBS)
Irritable bowel syndrome (IBS) is a gastrointestinal disorder causing cramping, abdominal pain, bloating, gas, diarrhea and constipation that is influenced by diet, lifestyle and stress [31]. Unlike IBD, IBS is not associated with overt inflammation or damage to intestinal tissues. However, a subset of IBS patients experiences visceral hypersensitivity, which is a lowered threshold for pain in the abdominal area. Although alterations in bacterial communities in IBS have been investigated, the potential role for fungi in IBS remains largely unexplored [32]. A provocative recent study now sheds light on fungal dysbiosis in IBS patients and how changes in the mycobiome may mediate visceral hypersensitivity [33]. The fecal mycobiome was compared amongst IBS patients that were categorized into hypersensitive and normally sensitive groups. Analysis of these groups revealed that the hypersensitive IBS patients have a different mycobiota compared to the normally-sensitive IBS patients. Utilizing a rat model of stress-induced IBS-like visceral hypersensitivity, the authors observed that treatment with an antifungal drug, high doses of soluble β-glucan (thought to inhibit the fungal β-glucan receptor Dectin-1), or a pharmacological Syk inhibitor (also aimed at inhibiting Dectin-1 signaling) could block development of visceral hypersensitivity. Additionally, gavage of cecum content from hypersensitive rats was able to restore visceral hypersensitivity in fungicide-treated recipient rats. Visceral hypersensitivity has been previously linked to mast cell degranulation [34], and the authors documented that particulate β-glucans (which can activate Dectin-1), can trigger histamine release from gut mucosal mast cells (Fig. 2). If this mechanism truly contributes to IBS visceral hypersensitivity, it will be important to understand how fungal antigens make their way from the gut lumen to mast cells in the mesentery and if activation of other antifungal immune receptors similarly affect development of visceral hypersensitivity.
Fungal influence on metabolites in the gut
Commensal fungi may directly or indirectly regulate gut homeostasis in many ways, and one recent hypothesis is that commensal fungi may influence the availability of specific biologically important metabolites [35]. Saccharomyces, commonly found in many foods, is a part of commensal fungi in the gut and is generally non-pathogenic in healthy individuals. However, even commensal fungi have detrimental effects if the relative burden is increased during fungal dysbiosis. Round and coworkers noted that intestinal S. cerevisiae could exacerbate colitis in mouse 2,4,6-trinitrobenzenesulfonic acid (TNBS) and DSS models, and they noted that monocolonization of germ-free mice with S. cerevisiae caused epithelial cells to produce increased levels of uric acid, promoting disruption of the intestinal barrier and increasing intestinal permeability [35] (Fig. 2). Uric acid was also produced during the S. cerevisiae-exacerbated disease in wild-type animals, and pharmacological inhibition of uric acid production ameliorated disease. Further, the investigators noted that uric acid and ASCA were positively correlated in human IBD patient sera. This and other fungal-influenced metabolite production may be good targets for clinical intervention in disease in the future.
ROLES FOR INTESTINAL FUNGI IN SITES BEYOND THE GUT
Influence of intestinal fungi on development of mucosal immunity
The gut microbiota regulates local and distal sites of the immune system [36]. It has a crucial role in developing secondary lymphoid organs during neonatal periods, and germ-free mice lacking microbes have immature lymph nodes and spleens [37]. Murine lymph nodes (LNs) undergo a maturation process that begins at birth and involves a shift in addressin expression from MAd-CAM1 to PNAd, allowing for recirculating lymphocytes to enter [38]. These changes in LN maturation are mediated in part by a subset of retinol dehydrogenase+ (RALDH) DCs [39]. Soon after birth, these RALDH+ DCs travel from the intestinal lamina propria to the peripheral and mesenteric lymph nodes (mLNs, pLNs) and promote LN maturation.
Shi and coworkers observed that RALDH+ DCs do not accumulate in the LNs of germ free (GF) mice, resulting in a lack of organized structure and cellularity within secondary lymphoid organs. Further, they observed a similar reduction of RALDH+ DCs in gut- and skin-draining LNs in wild-type mice treated with an anti-fungal cocktail but not in mice with an antibacterial treatment. Oral gavage with a mouse commensal fungal species, Candida tropicalis, was sufficient to enhance migration of gut RALDH+ DCs, to peripheral lymph nodes and promotes development of gut lymphoid tissues [40] (Fig. 2). This effect on RALDH+ DC migration and lymphoid tissue development was not efficiently promoted by two other fungi, Trichosporon asahii or Saccharomyces cerevisiae, suggesting that certain fungal species are specifically relevant in this context. We do not yet know if RALDH+ DCs must come in direct contact with commensal fungi to successfully migrate to the peripheral LNs or if they must phagocytose and present the commensal fungi to T cells to promote this development.
Influence of fungal dysbiosis on inflammation and immunity at distal sites
Huffnagle and coworkers have noted that high-level GI colonization with C. albicans in antibiotic-treated mice can exacerbate pathology in allergic airway disease models [41], and Wheeler at al. recently reported that inducing alterations in the existing fungal microbiome with antifungal drugs can similarly alter the course of house dust mite (HDM)-induced allergic airway disease [10]. Perturbing fungal homeostasis by oral treatment with the antifungal drug fluconazole reduced the prevalence of some intestinal fungi but allowed for increased growth of specific fungi including Aspergillus, Wallemia and Epicoccum. Anti-fungal drug-induced fungal dysbiosis or oral exposure to Aspergillus, Wallemia and Epicoccum caused increases in allergy-related antibodies (IgE and HDM IgG1), Th2 cytokine producing T cells (IL-4, IL-5, IL-10) and infiltration of eosinophils into the lungs upon HDM immunization [10].
Fujimura and coworkers investigated the influence of microbiome dysbiosis during the neonatal period in humans on developing multi-sensitized atopy and ultimately asthma [42]. By separating the neonates based upon their microbiome diversity, the authors defined three subgroups of neonatal gut microbiota (NGM1, NGM2, NGM3) and reported that NGM3 had the highest risk of developing atopy (heightened immune responses to allergens) by age 2 and asthma by age 4. Disruptions in the bacterial communities of NGM3 included a reduction in Bifidobacteria, Lactobacillus, Faecalibacterium and Akkermansia, coinciding with earlier studies showing atopy associated gut microbiome dysbiosis [43]. Important for this discussion, notable fungal changes associated with NGM3 included a decrease in Malassezia and an increase in Candida and Rhodotorula proportions. The specific functional consequences of these fungal changes have yet to be determined.
Cirrhosis is permanent scarring of the liver due to inflammation triggered by various factors such as abusive alcohol consumption, hepatitis infection, or autoimmune reaction. A major complication of liver cirrhosis is spontaneous bacterial peritonitis following treatments with antibiotics. As noted above, an unintended effect of antibiotic treatment can be fungal overgrowth and dysbiosis, and patients with cirrhosis are more susceptible to fungal infections [44]. Several years ago, investigators noted that patients with cirrhosis showed elevated serum ASCA levels and that ASCA levels correlated with disease severity [45]. More recently, a patient survey about the interaction between fungal and bacterial communities in cirrhosis suggested that the Bacteroidetes/Ascomycota ratio can predict future hospitalizations; a low ratio was associated with low hospitalizations [6]. Together the data suggest that an altered immune interaction with intestinal fungi may influence disease. Experimental evidence for this idea has been provided by Yang et al., observing that chronic alcohol feeding to mice resulted in increased intestinal fungal burden and that Candida is specifically expanded in the intestines of alcoholic patients [46]. These investigators noted that serum β-glucan levels were increased in the animals, and that development of liver disease was prevented in animals lacking Dectin-1. Specifically, the investigators propose that Kupffer cells, liver-resident macrophages that express Dectin-1, are activated by gut-derived β-glucan to secrete IL-1β which drives liver damage.
CONCLUSION
The roles of intestinal fungi in health and disease have been underappreciated compared to commensal bacteria in the gut, and we are just beginning to develop an appreciation for the impact of intestinal fungi on diverse pathologies. Intestinal fungi may influence microbiota (bacterial, fungal, and others) community structure in the gut, may alter production and consumption of key metabolites in the gut, and may interact with host immune cells to influence immune development and homeostasis. All of these may influence host health and the development of disease in the gut and at peripheral sites. Future studies will help develop mechanistic understandings of these roles for commensal intestinal fungi and may lead to the development of novel therapeutic approaches to preventing or treating diverse diseases.
Highlights.
Fungi are commensal organisms in the mammalian gut.
Fungal dysbiosis is associated with diverse diseases.
Intestinal fungi shape development and maintenance of the immune system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES AND RECOMMENDED READING
- 1.Wheeler ML, Limon JJ, Underhill DM. Immunity to commensal fungi: Detente and disease. Annu Rev Pathol. 2017;12:359–385. doi: 10.1146/annurev-pathol-052016-100342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, Miller AN, et al. Nuclear ribosomal internal transcribed spacer (its) region as a universal DNA barcode marker for fungi. P Natl Acad Sci USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2015;12:77–87. doi: 10.1038/nrgastro.2014.188. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez MM, Perez D, Chaves FJ, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jove M, Pamplona R, Ricart W, Portero-Otin M, et al. Obesity changes the human gut mycobiome. Sci Rep-Uk. 2015:5. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabro A, De Filippo C. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017:5. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajaj JS, Liu EJ, Kheradman R, Fagan A, Heuman DM, White M, Gavis EA, Hylemon P, Sikaroodi M, Gillevet PM. Fungal dysbiosis in cirrhosis. Gut. 2017 doi: 10.1136/gutjnl-2016-313170. [DOI] [PubMed] [Google Scholar]
- 7.Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dollive S, Chen YY, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, Cuff C, Lewis JD, Wu GD, Bushman FD. Fungi of the murine gut: Episodic variation and proliferation during antibiotic treatment. PLoS One. 2013;8:e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, et al. Interactions between commensal fungi and the c-type lectin receptor dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, Arditi M, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. This study investigates in mouse model systems how fungal dysbiosis can influence development of immune responses in the gut and lung. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, Albenberg L, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric crohn’s disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, et al. Fungal microbiota dysbiosis in ibd. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial crohn’s disease. MBio. 2016:7. doi: 10.1128/mBio.01250-16. Together with the above reference, these studies provide interesting and contrasting descriptive looks at the fungal microbiome in fecal samples of adult patients with Crohn’s disease as assessed by culture-independent ITS rDNA sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donskey CJ. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis. 2004;39:219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 17••.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, et al. Card9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. This study suggests that CARD9, a signaling adaptor molecule involved in anti-fungal host-defense, may influence bacterial communities in the gut and how they alter intestinal metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol H, Conway KL, Zhang M, Choi M, Morin B, Cao Z, Villablanca EJ, Li C, Wijmenga C, Yun SH, Shi HN, et al. Card9 mediates intestinal epithelial cell restitution, t-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601. e593. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, Hattori M, Takeshita K, Kanai T, Saijo S, Ohno N, et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing lactobacillus-mediated regulatory t cell expansion in the intestine. Cell Host Microbe. 2015;18:183–197. doi: 10.1016/j.chom.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. Modulation of post-antibiotic bacterial community reassembly and host response by candida albicans. Sci Rep-Uk. 2013;3:2191. doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY. Activation of hif-1alpha and ll-37 by commensal bacteria inhibits candida albicans colonization. Nat Med. 2015;21:808–814. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moyes DL, Naglik JR. Mucosal immunity and candida albicans infection. Clin Dev Immunol. 2011;2011:346307. doi: 10.1155/2011/346307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 2017;15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Hofs S, Gratacap RL, Robbins J, Runglall M, Murciano C, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 2016;532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Souza HS, Fiocchi C. Immunopathogenesis of ibd: Current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 26.Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, Campieri M, et al. Fungal dysbiosis in mucosa-associated microbiota of crohn’s disease patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 28.Brown GD. Dectin-1: A signalling non-tlr pattern-recognition receptor. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 29.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, Green T, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Underhill DM, Iliev ID. The mycobiota: Interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 32.Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. 2015;9:318–331. doi: 10.5009/gnl14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Botschuijver S, Roeselers G, Levin E, Jonkers DM, Welting O, Heinsbroek SE, de Weerd HH, Boekhout T, Fornai M, Masclee AA, Schuren FHJ, et al. Intestinal fungal dysbiosis associates with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology. 2017;153:1026–1039. doi: 10.1053/j.gastro.2017.06.004. This study provides evidence suggesting that alterations in intestinal fungi may be responsible for pain sensitiviy in some patients with irritable bowel syndrome. [DOI] [PubMed] [Google Scholar]
- 34.Klooker TK, Braak B, Koopman KE, Welting O, Wouters MM, van der Heide S, Schemann M, Bischoff SC, van den Wijngaard RM, Boeckxstaens GE. The mast cell stabiliser ketotifen decreases visceral hypersensitivity and improves intestinal symptoms in patients with irritable bowel syndrome. Gut. 2010;59:1213–1221. doi: 10.1136/gut.2010.213108. [DOI] [PubMed] [Google Scholar]
- 35••.Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, Brown J, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017:9. doi: 10.1126/scitranslmed.aaf9044. This study provides evidence suggesting that intestinal fungi may directly alter production and metabolism of important metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: Shaping local and systemic immune responses. Semin Immunol. 2012;24:58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol. 1963;42:471–483. [PMC free article] [PubMed] [Google Scholar]
- 38.Mebius RE, Streeter PR, Michie S, Butcher EC, Weissman IL. A developmental switch in lymphocyte homing receptor and endothelial vascular addressin expression regulates lymphocyte homing and permits cd4+ cd3− cells to colonize lymph nodes. P Natl Acad Sci USA. 1996;93:11019–11024. doi: 10.1073/pnas.93.20.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Girard JP, Moussion C, Forster R. Hevs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol. 2012;12:762–773. doi: 10.1038/nri3298. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Li J, Zheng W, Zhao G, Zhang H, Wang X, Guo Y, Qin C, Shi Y. Peripheral lymphoid volume expansion and maintenance are controlled by gut microbiota via raldh+ dendritic cells. Immunity. 2016;44:330–342. doi: 10.1016/j.immuni.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and t cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 44.Lahmer T, Messer M, Mayr U, Saugel B, Noe S, Schultheiss C, Thies P, Spinner C, Nennstiel S, Schwerdtfeger C, Phillip V, et al. Fungal “colonisation” is associated with increased mortality in medical intensive care unit patients with liver cirrhosis. Mycopathologia. 2015;179:63–71. doi: 10.1007/s11046-014-9825-6. [DOI] [PubMed] [Google Scholar]
- 45.Papp M, Norman GL, Vitalis Z, Tornai I, Altorjay I, Foldi I, Udvardy M, Shums Z, Dinya T, Orosz P, Lombay B, Jr, et al. Presence of anti-microbial antibodies in liver cirrhosis--a tell-tale sign of compromised immunity? PLoS One. 2010;5:e12957. doi: 10.1371/journal.pone.0012957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Yang AM, Inamine T, Hochrath K, Chen P, Wang LR, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, Kisseleva T, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–2841. doi: 10.1172/JCI90562. This study provides provokative data in mouse and human studies implicating fungal dysbiosis and the Dectin-1 pathway in development of alcoholic liver disease. [DOI] [PMC free article] [PubMed] [Google Scholar]