Abstract
The recent discovery and use of CRISPR/Cas9 gene editing technology has provided new opportunities for scientific research in many fields of study including agriculture, genetic disorders, human disease, biotechnology, and basic biological research. The ability to precisely target DNA sequences and either remove, modify, or replace genetic sequences provides a new level of control in nearly all eukaryotic organisms, including budding yeast. Given the many discoveries made in Saccharomyces cerevisiae over the past decades spanning genetics, cell biology, and biochemistry, as well as the development of new technologies that have allowed high throughput screening, robotic automation, and a platform for synthetic genome engineering, the yeast community has also started to recognize the utility and complementary nature of CRISPR-based methodologies. Here we present and review a variety of recent uses of Cas9 in budding yeast—both nuclease dependent and independent applications spanning traditional gene editing and replacement, to transcriptional modulation, to novel uses including the development of living circuitry or robotic platforms for synthetic genome construction. Yeast continues to serve as a powerful model system, yet it can still benefit from use of CRISPR for basic research, industrial application, and innovation of new Cas9-based applications.
Keywords: Budding yeast, CRISPR/Cas9, review, S. cerevisiae, biotechnology, gene editing
Introduction
Saccharomyces cerevisiae (budding yeast) is one of the most well studied, genetically tractable organisms. As a model eukaryote, it has provided critical insight into the basic biology of the cell cycle [1], endomembrane vesicular trafficking [2], autophagy [3], and many other cellular systems. Part of the success for the tractability of yeast in both industry and basic research stems from the ability to rapidly edit and manipulate genomes. This has led to the development of genome-wide libraries [4-6], synthetic genetic array (SGA) technology [7], and markerless integration methods [8], to name only a few. The recent interest and explosion of research into CRISPR/Cas9-based editing across many model systems has also finally reached the yeast community.
CRISPR (clustered regularly interspaced palindromic repeats) has evolved as a primitive immune system in prokaryotes with the ability to precisely target and edit any genome [9-12]. Briefly, the Cas9 endonuclease of the Class II CRISPR system (typically from Streptococcus pyogenes) is first bound to a single-stranded piece of RNA (sgRNA; single guide) containing a short stretch of nucleotides that bind and recruit the Cas9/RNA complex to the corresponding sequence within a target genome that is also marked with a “PAM” (protospacer adjacent motif) 5’-NGG-3’ sequence [13]. The dual nuclease domains of Cas9 cause a double-stranded break (DSB) at the +3 position upstream of the PAM. Eukaryotes, including yeast, have evolved multiple DNA repair systems to handle the presence of DSBs, including repair by non-homologous end joining (NHEJ) [14] and homology directed repair (HDR) [15]. Introduction of DSBs at genomic position(s) by Cas9 has allowed for gene replacement, gene deletions, pathway construction, and single base editing in many eukaryotic organisms including humans [16-21]. This technology has obvious application to not only basic research, but industry, agriculture, biofuels, bioenergy, human pathogens, genetic disorders, and disease [22-25].
While the CRISPR system was first piloted in budding yeast in 2013 [26], a number of methodological and technical hurdles likely slowed its progression through the yeast community. First, we speculate that given the incredible efficiency by which S. cerevisiae already performs homologous recombination in vivo [27] sans any DSB, it seemed puzzling how the Cas9 nuclease might provide a significant advance from traditional molecular methodologies [5]. Second, and along these lines, a number of technical challenges including optimization of both expression and delivery of Cas9 and the sgRNA(s) had to first be overcome. However, recent efforts have provided a new suite of molecular tools using the CRISPR/Cas9 system that are being applied to a diverse array of methodologies in S. cerevisiae including multiplexed editing, markerless manipulation, chromosome splitting, transcriptional modulation, synthetic genome engineering, and gene drive technology.
Yeast Genome Manipulation using S. pyogenes Cas9
As the CRISPR/Cas9 gene editing system was tested in model systems, editing was also successfully demonstrated in S. cerevisiae using an inducible promoter to drive Cas9 expression and a high copy plasmid-driven RNA polymerase III regulated promoter (prSNR52) to express the sgRNA cassette [26]. Activation of the Cas9/sgRNA complex resulted in double-stranded break formation in > 99 percent of yeast and subsequent cell death. However, introduction of donor DNA (as an oligonucleotide) greatly increased repair of the DSB and resulted in successful editing; this process is also extremely efficient when using amplified PCR fragments even with limited homology [28].
In contrast to other eukaryotes, NHEJ is extremely inefficient in yeast and double-stranded DNA breaks are repaired by HR. This may represent one of the challenges in adopting the new technology in S. cerevisiae. Indeed, a number of studies have provided insight into optimizing use and application of the CRISPR technology in vivo. Early efforts have examined the mechanism of delivery and expression of the Cas9 nuclease—moderate, rather than high-expression is preferred [29], as well as sequential delivery of Cas9 prior to the sgRNA cassette [30]. Moreover, various strategies have been developed for expression and delivery of the sgRNA component including (i) the original [26] Pol III promoter (SNR52) and terminator (SUP4) pair, (ii) the “natural” split two-component crRNA/tracrRNA [31], (iii) an amplified linear guide cassette (sans any circular plasmid backbone) [30], or (iv) a modular sgRNA design with use of a self-cleaving ribozyme fused to the 5’ end of the guide sequence [29]. Finally, improvements to targeting multiple genomic loci (termed “multiplexing”) in a single transformation event by cloning and delivery of unique sgRNAs [30,32] have greatly expanded the possibilities for yeast strain creation.
The ability to manipulate the genome at multiple loci in a single editing event [30,31,33-37] presents a serious upgrade from conventional cloning methods (such as homologous recombination (HR)-dependent integration or mating yeast followed by sporulation) for several reasons. First, as previously mentioned, since Cas9-induced DSBs are poorly tolerated in yeast, general survival following editing can be utilized as a powerful selection tool without the need for any selectable markers [28,29,32,35,38-41]. The ability to manipulate genomic loci sans auxotrophic or drug resistance cassettes provides a serious advantage for all research areas in budding yeast. This allows for (i) the use of more plasmid-borne constructs with classic selectable markers, (ii) the manipulation of yeast strains that are lacking a variety of auxotrophic marker(s), and (iii) the use of stably integrated mutations at their endogenous loci instead of plasmid-driven versions that require selection, and may provide yeast with an opportunity to vary the plasmid copy number per cell. Second, this allows for introduction of precise genomic alterations including single point mutations [42] or editing of essential genes [28,36]. Third, DSB formation greatly aids in large-scale gene replacement, pathway integration, and modulation of existing (or new) biosynthetic pathways. Combining Cas9 editing with in vivo fragment assembly, Mans and colleagues reconstituted a six-gene pathway (pyruvate dehydrogenase complex) from E. faecalis at the ACS2 locus in a single step (Figure 1A) [43]. Other groups have also demonstrated the great utility of engineering entire pathways in vivo for both basic research and potential industrial application [29,35,36,38,40,44-46].
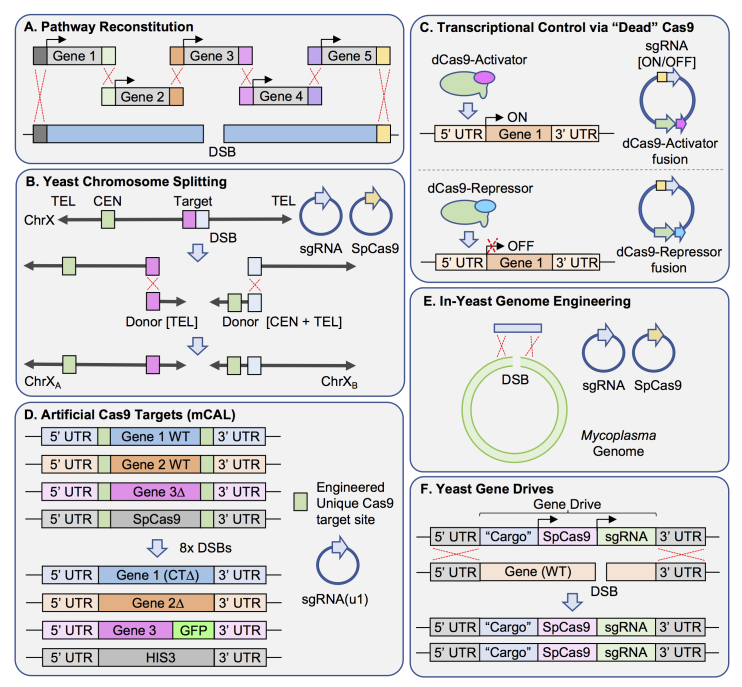
Figure 1.
Diverse set of unique CRISPR/Cas9 methodologies employed in S. cerevisiae in recent years. (A) Traditional nuclease-based editing using Cas9 allows for the introduction of multiple non-native genes into the yeast genome in a single step. This study reconstituted the six genes (five illustrated) required for a pyruvate dehydrogenase complex (from E. faecalis) in vivo [43]. Each “Gene” represented contains both flanking UTR as well as unique 60-base pair segments (different colors used) to allow for homologous recombination. Following targeting of Cas9/sgRNA to the locus of interest, HR and subsequent integration of the entire six-gene cassette repairs the double-stranded break (DSB) and replaces the endogenous yeast gene. (B) Cas9 based method for chromosome splitting (CRISPR-PCS) [47]. A yeast chromosome (Arrow: telomere, Green box: centromere) is targeted for splitting into two or more smaller complete chromosomes by plasmid-expressed Cas9 and an sgRNA. Following introduction of the DSB, donor DNA is provided that will allow for repair of each fractured chromosome fragment. A homologous sequence (pink or blue) is included to link each unique donor DNA fragment to the appropriate chromosome segment. Left, the severed chromosome arm is lacking a telomere; the donor module includes a telomere seed sequence repeat. Right, the separated chromosome arm (now lacking a centromere) performs HR with the appropriate donor module to introduce both a capping telomere seed sequence and yeast centromere. This methodology allows for the generation of two (or more) functional chromosomes. (C) Transcriptional regulation of multiple yeast promoters using catalytic dead Cas9 (dCas9) fusions and an inducible sgRNA system [50]. The sgRNA cassette is under control of the TetO system (ON/OFF). Nuclease deficient Cas9 is fused to either a transcriptional activator (VPR; VP64-p65-RTA) or repressor (Mxi1). Expression of different sgRNAs recruits dCas9-A (activator) or dCas9-R (repressor) to the promoter element of interest (14 separate promoters tested with over 100 sgRNAs) to modulate transcription of the target gene(s). (D) Multiplexing using artificial Cas9 target sequences (mCAL) [28]. The introduction of non-native target sequences (20 bp target + 3 bp PAM) at multiple loci (illustrated as flanking three sample genes as well as an integrated copy of Cas9 at the HIS3 locus) allows for a single sgRNA construct (u1; unique sequence 1) to target this identical sequence at every position in the genome. Introduction of donor DNA with appropriate flanking sequence allows for HR-based integration of any version of each gene (full deletion, repair, domain deletions, point mutations, or tagged versions) as well as simultaneous excision of the Cas9-expressing cassette. (E) In-yeast genome engineering of a bacterial genome [59]. The combination of active Cas9, a targeting sgRNA (both on plasmids) as well as the entire Mycoplasma mycoides bacterial genome (1.2 Mb) was transformed into yeast. CRISPR-based DSB induction and subsequent HR-based repair (with a synthetic oligonucleotide) allowed for the deletion of a particular M. mycoides gene. (F) The study of gene drives using S. cerevisiae [67]. The Cas9-based “gene drive” consists of the following: (i) the Cas9 gene, (ii) the sgRNA-expressing cassette, and (iii) an optional “cargo” for a new or modified gene. In yeast, the sgRNA can be expressed from a plasmid or be integrated as the site of the drive. The entire drive is integrated into the genome and replaces (full or partial deletion) an endogenous gene. Activation of the gene drive system causes targeting of Cas9 to the homologous WT gene copy on the opposite chromosome (in a diploid yeast cell). Creation of the DSB induces HR-based repair using the entire flanking chromosomal sequence as donor DNA. Therefore, the entirety of the gene drive is copied and replaces the endogenous WT gene target. Illustrations are adapted from various sources.
The utility of Cas9 for DNA manipulation is continuing to expand beyond simple DSB formation and subsequent repair by HDR (Table 1). For instance, Sasano and colleagues have developed a modular toolkit for splitting and stable propagation of entire yeast chromosomes into two or more smaller chromosomes (Figure 1B). Following the DSB, HDR occurs on provided donor DNA modules that insert both a new centromere on the broken chromosome arm(s) as well as a telomere seed sequence repeat to the newly formatted chromosome ends [47]. Moreover, CRISPR has been used to delete large genomic fragments (> 30 Kb) [48] or to construct synthetic promoter elements in vivo [34].
Table 1. Overview of recent applications of CRISPR/Cas9 gene editing technology in S. cerevisiae.
| Category | Technology | Description | Reference | Additional Studies |
| Traditional Cas9-Based Gene Editing Methodologies | Di-CRISPR (Delta Integration CRISPR/Cas) | Multiplexing1 of Cas9 (markerless, single-step integration of biochemical pathways) by targeting repeated delta sites2 throughout yeast genome. | [38] | [30-32,37,41-45,70,71] |
| mCRISTAR (Multiplexed CRISPR Transformation-Associated Recombination) | Use of Cas9 to target and replace endogenous promoter elements. | [34] | ||
| Large Chromosomal Fragment Deletion | Generation of chromosomal deletions up to 30 Kb. | [48] | ||
| Cas9 Nickase | Use of a Cas9 nickase3 variant to edit bases distal (50+ bps) to the target site. | [72] | ||
| Novel Cas9-Based Applications | CRISPR-PCS (CRISPR PCR-Mediated Chromosome Splitting) | Use of Cas9 to split and generate new chromosomes complete with centromeres and telomere seed regions. | [47] | [73,74] |
| CRISPR-ChAP-MS (CRISPR-Based Chromatin Affinity Purification with Mass Spectrometry | Allows for dCas9-targeted purification of different chromatin regions coupled with protein and PTM identification via MS. | [55] | ||
| Transcriptional Regulation (via Dead Cas9) | CRISPRi (Genome Scale CRISPR Interference) | Use of nuclease deficient (“dead”) dCas9 for repression of gene expression of endogenous genes. | [49] | [52,53,75] |
| dCas9-Mediated Transcriptional Reprogramming | Use of either direct or indirect dCas9 constructs to transcriptional activator (VPR) or repressor (Mxi1) to modulate gene expression.4 | [50] | ||
| Graded Expression of Pathway Enzymes via dCas9 Positioning | Varied5 sgRNA targeting of dCas9 for tuned expression of metabolic pathway genes. | [51] | ||
| Synthetic Genome Engineering | Automated Multiplex Genome Engineering | High-throughput, robotic-based construction of overexpression or mutated alleles using Cas9 at repetitive genomic sequences. | [61] | |
| In-Yeast engineering of a Bacterial Genome | Engineering of a deletion mutant of the Mycoplasma bacterial genome (1.2 Mb) using Cas9. | [59] | ||
| CasHRA (Cas9-Facilitated Homologous Recombination Assembly) | Construction of the Minimal Escherichia coli genome (1.03 Mb) using large circular DNAs that are subsequently cleaved via Cas9 and assembled into the genome. | [60] | ||
| Synthetic Yeast Genome (SynV) Construction | De novo synthesis of the yeast Chromosome V (0.54 Mb) and replacement of the endogenous sequence using Cas9 and HR. | [58] | ||
| mCAL (Multiplexing of Cas9 at Artificial Loci) | Use of artificial6 Cas9 target sequences (20+3 bp PAM) to multiplex Cas9 with a single sgRNA to different loci. | [28] | ||
| Gene Drive | Gene Drive Safeguarding | Development and testing of a yeast Cas9-based gene drives7 to address safety concerns, positioning of the sgRNA (plasmid versus integrated), and fail safes to remove existing drives. | [67] |
1Multiplexing: Targeting of Cas9 to multiple genomic targets. This can be accomplished by a single sgRNA (to a repeated genomic sequence—telomeres, delta elements, etc.—or “engineered” target sites [28] placed throughout the genome). 2Delta sites: Repeated Ty retrotransposon delta sites within the yeast genome. 3Cas9 Nickase: a mutated enzyme variant that has one of the nuclease cleavage domains mutated—this causes a single-stranded break (a nick) rather than a double-stranded break. 4Two versions of transcriptional activator/repressor tethers were used to dCas9: (i) direct translational fusions to VPR (VP64-p65-Rta transcriptional activator) or Mxi1 (repressor) or (ii) indirect recruitment of a MCP-VPR or PCP-Mxi1 (both RNA scaffold binding protein fusions) to the scaffold-extended sgRNA sequences. 5Positions between +30 to +750 bps upstream of the TATA box were analyzed by sgRNA targeted dCas9. 6Artificial Cas9 target sequences: 20 base paid target sequences and a 3 bp 5’-NGG-3’ PAM sequence chosen to provide the maximum mismatch with the entire yeast genome were engineered and placed at several genomic loci. 7A Cas9 “gene drive” is defined as the placement of the Cas9 gene at a genetic locus (either deleting or modifying the native gene), accompanied by expression of an sgRNA that targets the Cas9 nuclease to the site of the WT endogenous gene on the opposite, homologous chromosome within a diploid cell. DSB formation causes the entire Cas9-containing drive to be copied to the second chromosome via homologous recombination.
Nuclease-dead Cas9 as a Targeting Scaffold
Aside from its traditional role as an endonuclease, Cas9 has also been engineered to separate its DNA targeting function from that of its DNA cleaving enzymatic function [49]. Mutation of only two residues (D10A and H840A) inactivates both nuclease domains, yet does not disrupt the ability of Cas9 to bind the sgRNA nor target the intended genomic loci. Termed “dead” Cas9 (dCas9), this serves as a molecular recruitment tool to ultimately deliver a secondary protein of interest to target regulatory sequences. In 2017, Jensen and colleagues demonstrated the ability of dCas9 to modulate gene expression in yeast using both (direct) translational fusions and appended sgRNA-RNA-binding domain (indirect) tethers to transcriptional activators (VP64) or repressors (Mxi1). This group screened various sgRNA sequences for ideal positioning of dCas9 complex to the site of 14 yeast promoters with the intent of modulating flux through two biosynthetic pathways (Figure 1C) [50]. Similar studies have also focused on sgRNA identity (and dCas9 positioning) in order to perturb metabolic pathways [51]. Work in yeast has also illustrated the use and development of more complex RNA binding scaffolds to recruit multiple RNA-binding proteins (fused to transcriptional activators) [52]. However, the utility of such transcriptional modulation is not limited to endogenous transcriptional regulation. A recent study has adopted the dCas9-Mxi1 fusion to develop digital “logic circuits” in yeast [53] using guide RNA switches genetically wired together. An engineered cell-to-cell communication system was also developed in yeast using CRISPR transcription factors [54]. The utility of dCas9 extends far beyond that of the nuclease active protein since a variety of additional DNA/chromatin-modifying enzymes can be routinely fused or recruited to Cas9 and would provide a powerful platform for genome-wide screening in yeast not only restricted to transcriptional modulation. One example of this has been use of a Protein A fusion to dCas9 to isolate and identify by mass spectrometry the “epiproteome” of a yeast promoter [55]. There is enormous potential to utilizing dCas9 as a programmable physical scaffold onto which other DNA (or epigenome) modifying enzymes can be targeted. An explosion of Cas9 protein fusions has provided an ever-expanding suite of options for inducible, chemically regulated, or even split nuclease systems [56].
Synthetic Genome Construction
Work in budding yeast has been instrumental in the field of synthetic genome engineering. The S. cerevisiae 2.0 project (Sc2.0) aims to create the entire yeast genome de novo with a variety of designed modifications (removal of introns, grouping of tRNAs, introduction of loxP recombination sites, telomere modifications, etc.) [57]. While this project has relied mainly on traditional HR-directed integration of artificial chromosomal segments in place of the native sequence, CRISPR/Cas9 allowed for repair of mutations found during this construction process [58]. Moreover, entire 1 Mb bacterial genomes (Mycoplasma and Escherichia) have been edited in vivo in yeast cells using CRISPR [59,60] (Figure 1E). Recent work in S. cerevisiae has also demonstrated the ability to utilize a combination of high-throughput automation, genome-scale engineering, and Cas9-based gene editing [61]. This group created standardized fragments of native yeast genes (either overexpression or knockdown using either sense or antisense transcripts in an RNAi active yeast background) and these constructs were all integrated at repetitive DNA sequences in the yeast genome using Cas9. However, this system is (currently) unable to target native loci; the development of genome-wide collections of sgRNAs would provide the option to manipulate endogenous genomic sites.
At the interface of engineered yeast genomes and CRISPR-based editing, the first use of introduced artificial Cas9 target sites into the budding yeast genome was recently performed [28]. This application will likely have great utility as the use of synthetic genes (and genomes) becomes more widespread. Pre-loading a yeast strain with identical DNA target sites (23 bps) allows for only a single guide RNA construct to be expressed to multiplex Cas9 across the genome (Figure 1D). In this way, all genes within a signaling pathway, or within a macromolecular complex, or evolved gene paralogs could be simultaneously targeted in any combination desired. A major concern of the CRISPR field has been to reduce and eliminate off-target effects—recruitment and unintentional editing of other genomic positions [62-64]. Use of artificial programmable genomic site(s) and the corresponding sgRNAs could aid in reducing off-target Cas9 editing in yeast and other organisms.
Gene Drive Technology
One of the most intriguing and powerful arrangements of the CRISPR/Cas9 technology is in a “gene drive.” Briefly, the endonuclease is integrated at the site of a target locus (typically replacing and deleting the endogenous gene) in addition to a guide RNA cassette (Figure 1F). The sgRNA targets Cas9 to a site present on the WT (wild type) copy of the gene on the homologous chromosome within a diploid cell. Double-stranded break formation and subsequent HR causes disruption of the WT allele on the homologous chromosome by the gene drive cassette. This mechanism bypasses the restrictions imposed by standard Mendelian genetics. This super-Mendelian arrangement can rapidly sweep through a population as nearly 100 percent of the progeny (from each generation) are homozygous diploid for the affected (or deleted) allele. Recent work has utilized this technology in flies and mosquitos with the intent of population control on a widespread level to restrict and eliminate the spread of insect-borne diseases [65,66]. Recent work in budding yeast has demonstrated the utility of this model organism for testing various gene drive arrangements with the intention of studying methods for (i) safely utilizing or (ii) halting active drives [67]. For example, the Church lab found that separation of the sgRNA-expressing cassette (on an unstable, high-copy plasmid) was a preferable safeguard to chromosomal integration adjacent to the Cas9 gene. Moreover, they piloted an experiment to illustrate the utility of a secondary gene drive-containing strain to actively target an original (theoretically “escaped”) drive. Given the severity of accidental (or intentional) release of an engineered, active gene drive-containing organism (of any type) [68,69], yeast can serve as a safe and useful model system to assay various Cas9 drive arrangements for future implementation in insects or other eukaryotic systems.
Future Perspectives
Given the profound contributions of the yeast community to many aspects of eukaryotic cell and molecular biology and the many recent technologies (SGA, genome wide imaging, genetic screening, automation, and synthetic and directed evolutionary biology) that are possible, it is critical that further exploration and innovation be performed in budding yeast using CRISPR/Cas9 editing. Recent studies have already begun to illustrate the utility of the traditional Cas9-based techniques over conventional cloning and integrating methods. Moreover, because the basic components (nuclease, guide RNA, target DNA, etc.) of the CRISPR system are highly similar (if not identical) in practice across different organisms, many of the findings (e.g., editing, Cas9 alterations, or guide specificity) can be directly applicable to the entire field and are not restricted to only Saccharomyces or fungi. The identification and use of dual Cas systems (Cas9 orthologs), genome-wide sgRNA collections, and new dCas9 translational fusions present exciting new platforms to merge with existing (or new) yeast technologies. Active research in budding yeast should continue to embrace the use of CRISPR/Cas9 to explore, innovate, and develop new molecular methodologies.
Glossary
- HR
homologous recombination
- HDR
homology directed repair
- PAM
protospacer adjacent motif
- CRISPR
clustered regularly interspaced short palindromic repeats
- SGA
synthetic genetic array
- sgRNA
single guide RNA
- DSB
double-stranded break
- dCas9
nuclease inactive (dead) Cas9
- PCR
polymerase chain reaction
- RNAi
RNA interference
- Mb
megabase, million base pairs
- Kb
kilobase, thousand base pairs
- TetO
tetracycline controlled transcriptional system
- WT
wild type
Author Contributions
RMG prepared figures, table, references, and wrote the manuscript. RMG was funded by Kansas State University, College of Arts & Sciences, Department of Biochemistry & Molecular Biophysics. GCF wrote the manuscript. This project was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103418 to GCF. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. This work was also supported by an Innovative Research Award (to GCF) from the Johnson Cancer Research Center at Kansas State University. The authors declare no conflict of interest.
References
- Nasmyth K. A prize for proliferation. Cell. 2001;107(6):689–701. [DOI] [PubMed] [Google Scholar]
- Mellman I, Emr SD. A Nobel Prize for membrane traffic: vesicles find their journey’s end. J Cell Biol. 2013;203(4):559–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A, Kainz K, Andryushkova A, Hofer S, Madeo F, Carmona-Gutierrez D. Autophagy: one more Nobel Prize for yeast. Microb Cell. 2016;3(12):579–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–91. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–91. [DOI] [PubMed] [Google Scholar]
- Douglas AC, Smith AM, Sharifpoor S, Yan Z, Durbic T, Heisler LE, et al. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda). 2012;2(10):1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353(6306):aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund PO, Davis TN. A high-efficiency method to replace essential genes with mutant alleles in yeast. Yeast. 2005;22(10):769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–66. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353(6299):aad5147. [DOI] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–51. [DOI] [PubMed] [Google Scholar]
- Coic E, Feldman T, Landman AS, Haber JE. Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Genetics. 2008;179(1):199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. [DOI] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6(6):2008–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci Y, Zhang B, Unver T. CRISPR/Cas9: an RNA-guided highly precise synthetic tool for plant genome editing [Epub ahead of print] J Cell Physiol. 2017 [DOI] [PubMed] [Google Scholar]
- Men K, Duan X, He Z, Yang Y, Yao S, Wei Y. CRISPR/Cas9-mediated correction of human genetic disease. Sci China Life Sci. 2017;60(5):447–57. [DOI] [PubMed] [Google Scholar]
- Estrela R, Cate JH. Energy biotechnology in the CRISPR-Cas9 era. Curr Opin Biotechnol. 2016;38:79–84. [DOI] [PubMed] [Google Scholar]
- Soppe JA, Lebbink RJ. Antiviral Goes Viral: Harnessing CRISPR/Cas9 to Combat Viruses in Humans. Trends Microbiol. 2017;25(10):833–50. [DOI] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiasen DP, Lisby M. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38(2):172–84. [DOI] [PubMed] [Google Scholar]
- Finnigan GC, Thorner J. mCAL: A New Approach for Versatile Multiplex Action of Cas9 Using One sgRNA and Loci Flanked by a Programmed Target Sequence. G3 (Bethesda). 2016;6(7):2147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife. 2014:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JM, Chandran SS, Horwitz AA. CRISPR-Cas-Assisted Multiplexing (CAM): Simple Same-Day Multi-Locus Engineering in Yeast. J Cell Physiol. 2016;231(12):2563–9. [DOI] [PubMed] [Google Scholar]
- Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, et al. Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4(5):585–94. [DOI] [PubMed] [Google Scholar]
- Jessop-Fabre MM, Jakociunas T, Stovicek V, Dai Z, Jensen MK, Keasling JD, et al. EasyClone-MarkerFree: A vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol J. 2016;11(8):1110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen LE, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–22. [DOI] [PubMed] [Google Scholar]
- Kang HS, Charlop-Powers Z, Brady SF. Multiplexed CRISPR/Cas9- and TAR-Mediated Promoter Engineering of Natural Product Biosynthetic Gene Clusters in Yeast. ACS Synth Biol. 2016;5(9):1002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Cate JH. Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol. 2014;546:473–89. [DOI] [PubMed] [Google Scholar]
- Horwitz Andrew A, Walter Jessica M, Schubert Max G, Kung Stephanie H, Hawkins K, Platt Darren M, et al. Efficient Multiplexed Integration of Synergistic Alleles and Metabolic Pathways in Yeasts via CRISPR-Cas. Cell Syst. 2015;1(1):88–96. [DOI] [PubMed] [Google Scholar]
- Ronda C, Maury J, Jakociunas T, Jacobsen SA, Germann SM, Harrison SJ, et al. CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Liang Y, Zhang MM, Ang EL, Zhao H. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng. 2016;33:19–27. [DOI] [PubMed] [Google Scholar]
- Ryan OW, Poddar S, Cate JH. CRISPR-Cas9 Genome Engineering in Saccharomyces cerevisiae Cells. Cold Spring Harb Protoc. 2016;2016(6):pdb.prot086827. [DOI] [PubMed] [Google Scholar]
- Tsai CS, Kong II, Lesmana A, Million G, Zhang GC, Kim SR, et al. Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR. Biotechnol Bioeng. 2015;112(11):2406–11. [DOI] [PubMed] [Google Scholar]
- Jakociunas T, Rajkumar AS, Zhang J, Arsovska D, Rodriguez A, Jendresen CB, et al. CasEMBLR: Cas9-Facilitated Multiloci Genomic Integration of in Vivo Assembled DNA Parts in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4(11):1226–34. [DOI] [PubMed] [Google Scholar]
- Biot-Pelletier D, Martin VJ. Seamless site-directed mutagenesis of the Saccharomyces cerevisiae genome using CRISPR-Cas9. J Biol Eng. 2016;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NG, van den Broek M, et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15(2):fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YG, Jin YS, Cha YL, Seo JH. Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour Technol. 2017;228:355–61. [DOI] [PubMed] [Google Scholar]
- Vanegas KG, Lehka BJ, Mortensen UH. SWITCH: a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb Cell Fact. 2017;16(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EauClaire SF Zhang J, Rivera CG, Huang LL. Combinatorial metabolic pathway assembly in the yeast genome with RNA-guided Cas9. J Ind Microbiol Biotechnol. 2016;43(7):1001–15. [DOI] [PubMed] [Google Scholar]
- Sasano Y, Nagasawa K, Kaboli S, Sugiyama M, Harashima S. CRISPR-PCS: a powerful new approach to inducing multiple chromosome splitting in Saccharomyces cerevisiae. Sci Rep. 2016;6:30278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Wang X, Jia H, Yu M, Zhang X, Tang H, et al. Large fragment deletion using a CRISPR/Cas9 system in Saccharomyces cerevisiae. Anal Biochem. 2016;509:118–23. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ED, Ferreira R, Jakociunas T, Arsovska D, Zhang J, Ding L, et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb Cell Fact. 2017;16(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaner M, Alper HS. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab Eng. 2017;40:14–22. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1-2):339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gander MW, Vrana JD, Voje WE, Carothers JM, Klavins E. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat Commun. 2017;8:15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar A, Bolten NJ, Nemhauser J, Klavins E. Cell-Cell Communication in Yeast Using Auxin Biosynthesis and Auxin Responsive CRISPR Transcription Factors. ACS Synth Biol. 2016;5(4):279–86. [DOI] [PubMed] [Google Scholar]
- Waldrip ZJ, Byrum SD, Storey AJ, Gao J, Byrd AK, Mackintosh SG, et al. A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics. 2014;9(9):1207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Harrington LB, Doudna JA. Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem Biol. 2016;11(3):681–8. [DOI] [PubMed] [Google Scholar]
- Shen Y, Wang Y, Chen T, Gao F, Gong J, Abramczyk D, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355(6329):eaaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZX, Li BZ, Mitchell LA, Wu Y, Qi X, Jin Z, et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017;355(6329):eaaf4704. [DOI] [PubMed] [Google Scholar]
- Tsarmpopoulos I, Gourgues G, Blanchard A, Vashee S, Jores J, Lartigue C, et al. In-Yeast Engineering of a Bacterial Genome Using CRISPR/Cas9. ACS Synth Biol. 2016;5(1):104–9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wu R, Xue X, Qin Z. CasHRA (Cas9-facilitated Homologous Recombination Assembly) method of constructing megabase-sized DNA. Nucleic Acids Res. 2016;44(14):e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si T, Chao R, Min Y, Wu Y, Ren W, Zhao H. Automated multiplex genome-scale engineering in yeast. Nat Commun. 2017;8:15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal M, Khan FA, Da L, Habib Z, Dai J, Cao G. Keeping CRISPR/Cas on-Target. Curr Issues Mol Biol. 2016;20:1–12. [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112(49):E6736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34(1):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33(12):1250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari OS, Bellen HJ, Bier E, Bullock SL, Burt A, Church GM, et al. BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science. 2015;349(6251):927–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber BL, Raghu S, Edwards OR. Opinion: is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc Natl Acad Sci USA. 2015;112(34):10565–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generoso WC, Gottardi M, Oreb M, Boles E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J Microbiol Methods. 2016;127:203–5. [DOI] [PubMed] [Google Scholar]
- Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, et al. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast. 2015;32(12):711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satomura A, Nishioka R, Mori H, Sato K, Kuroda K, Ueda M. Precise genome-wide base editing by the CRISPR Nickase system in yeast. Sci Rep. 2017;7(1):2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhu MJ, Bloom JS, Day L, Kruglyak L. CRISPR-directed mitotic recombination enables genetic mapping without crosses. Science. 2016;352(6289):1113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NC, Larionov V, Kouprina N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015;43(8):e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Suresh S, Schlecht U, Wu M, Wagih O, Peltz G, et al. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]