Abstract
The CRISPR/Cas9 system of genome editing has revolutionized molecular biology, offering a simple, and relatively inexpensive method of creating precise DNA edits. It has potential application in gene therapy treatment of retinal diseases providing targeted disruption, alteration, or transcriptional regulation of pathogenic genes. In vivo studies have demonstrated therapeutic benefit for a variety of diseases. Despite this, there are many challenges to clinical use of CRISPR/Cas9, including editing efficiency, off-target effects, and disease heterogeneity. This review details the mechanisms of the CRISPR/Cas9 system and the treatment strategies that can be applied to retinal diseases. It gives an overview of in vivo studies published to date and discusses the challenges and potential solutions to the wide-scale clinical use of CRISPR/Cas9 as a therapeutic intervention.
Keywords: CRISPR, Cas9, sgRNA, gene therapy, gene editing, retinal disease, NHEJ, HDR
Introduction
Inherited retinal diseases are an important cause of blindness and are estimated to affect 2 million people worldwide [1]. Despite this there is no available treatment for the majority of patients. Gene therapy could potentially offer a cure. By introducing, silencing, or editing genes involved in the pathogenesis of these diseases, progression can be halted or even reversed [2]. The eye has been at the forefront of gene therapy as it has several useful qualities. Firstly, the eye is immune-privileged meaning a higher tolerance of introduced antigens [3]. Secondly, the presence of the blood retina barrier reduces the likelihood that viral vectors introduced into the eye during gene therapy will migrate to other areas [4]. This lowers the risk associated with potential off-target effects of treatment. Thirdly, the target site for gene therapy is easily accessible via current ophthalmic techniques and only requires local anaesthetics. This access means that the amount of virus required for retinal transduction is minimal compared to systemic targets such as the liver [5].
Retinal gene therapy has traditionally involved gene replacement, a technique known as gene augmentation [6,7]. The targeted diseases are caused by a lack of wild type function of a gene, and therefore supplying the missing gene may revert the pathology towards a normal phenotype, particularly when applied before the onset of cell death. While gene augmentation offers a promising outcome for patients with loss-of-function mutations, it cannot be used to treat dominant gain-of-function mutations. In these cases, the pathogenic mutation will need to be silenced or corrected for normal cell function to return. This presents a greater challenge than gene augmentation because the introduction of molecular inhibition into a host cell brings the possibility of off-target effects. Prior to the CRISPR/Cas9 system, gene editing at the DNA level was directed by zinc finger nucleases (ZFNs) or transcription activator-like effector nucleases (TALENs). These techniques both have their disadvantages, with ZFNs having a low on-target efficiency and difficulty locating a potential target site [8,9], and TALENs being very large and therefore difficult to deliver to the cell [10]. The discovery of the CRISPR/Cas9 system has been revolutionary as it is simple to design and implement. Gene editing requires only three components: the presence of a short sequence, roughly 3 to 8 bp (the PAM site) adjacent to the target site, the endonuclease protein Cas9, and a customized piece of RNA which directs Cas9 to the target site for DNA cleavage [11-13].
CRISPR/Cas9 Mechanism
CRISPR/Cas9 is a naturally occurring system that has evolved in bacteria and archaea as a method of evading viral infection. The microorganisms store copies of short sections of viral DNA in their genome which are transcribed into RNA called CRISPR RNA (crRNA). The crRNA forms a complex with a second piece of RNA, the trans-activating crRNA (tracrRNA), and CRISPR-associated protein 9 (Cas9), which are all encoded in the bacterial genome. If this complex encounters viral (bacteriophage) DNA which is complementary to the crRNA sequence it will bind to it. Cas9 is an endonuclease which cleaves double-stranded DNA, slicing through the viral DNA and preventing transcription [11,13].
Researchers soon realized that this system could be used for targeted gene editing in the genome of a chosen cell. By supplying a cell with Cas9, tracrRNA, and a crRNA specific to the target, Cas9 will cleave the region of interest. Since this discovery, the crRNA and tracrRNA have been artificially engineered into one single guide RNA (sgRNA) which can be customized to the target [13]. The final component required for CRISPR/Cas9-mediated gene editing is the presence of a protospacer adjacent motif (PAM) site adjacent to the target region [14]. A PAM site is a short DNA sequence (usually between 3 to 8 bp in length) that Cas9 binds to, inducing the double-stranded break approximately 3 bp upstream of the PAM [13]. These sites are naturally present on viral DNA and the exact sequence of the PAM is dependent on the species the Cas9 is isolated from. The most commonly used Cas9 to date has been SpCas9 from Streptococcus pyogenes, which requires a PAM of 5’-NGG-3’, where N is any nucleotide [15]. A number of bioinformatic programs are available which screen DNA sequences for PAM sites. Once a suitable PAM is selected, the sgRNA is designed to contain a region homologous to the sequence immediately upstream of the PAM (typically 18 to 25 bp long). By supplying a cell with the specific sgRNA and an active Cas9 protein, a double-stranded break will be induced at the site.
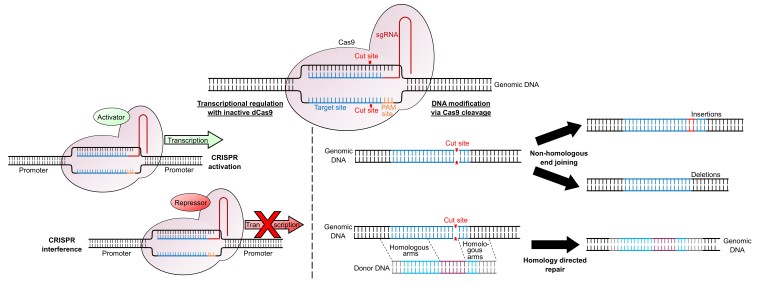
When a double-stranded break occurs in a host genome the cell will attempt to repair it using one of two processes: Non-Homologous End Joining (NHEJ) or Homology-Directed Repair (HDR) (Figure 1). In the absence of any homologous (identical complementary) DNA sequence, the cell will undergo NHEJ. Since there is no DNA template for repair, this process is somewhat random and hence extremely error prone, resulting in small insertions or deletions (indels) at the cleavage site. If these indels result in a frame-shift mutation in an exonic region, this can disrupt the gene, creating a non-functional protein. The HDR pathway allows researchers to perform precise edits of host DNA. Following a double-stranded break, any DNA molecules with a high level of homology to the target region will be substituted into the genome. By supplying a donor DNA molecule containing the desired mutation between two regions of homology to the target site (homologous arms), this increases the chances of the HDR pathway being used and the mutation being incorporated into the cell.
Figure 1.
Applications of CRISPR/Cas9 for transcriptional regulation and genomic modification. Following Cas9 binding, cleavage of both DNA strands allows DNA modification. In the absence of any homologous sequences, the cell will undergo non-homologous end joining, resulting in small insertions or deletions around the cut site. If donor DNA is supplied which has homologous arms matching the genomic DNA it will be incorporated into the genome via homology directed repair. Catalytically inactive dCas9 can be targeted to a promoter to alter transcriptional regulation. Fusing a transcriptional activator to dCas9 will upregulate gene expression (termed CRISPR activation) while fusing a transcriptional repressor to dCas9 will downregulate gene expression (termed CRISPR interference).
A third use of the CRISPR/Cas9 system involves introducing an artificially-engineered inactive version of Cas9 (dCas9) to alter the expression level of a gene at a transcriptional level, a technique called CRISPR interference (CRISPRi). dCas9 retains the ability to bind to DNA via the sgRNA and PAM site, but has no cleavage activity. By targeting dCas9 to regulatory elements of the target gene it can sterically inhibit transcriptional machinery such as transcription factors or RNA Polymerase, decreasing transcription of the gene [16]. This effect can be enhanced by fusing dCas9 with a transcriptional inhibitor, such as Krüppel associated box [17]. Alternatively, fusing dCas9 with a transcriptional activator can upregulate expression via CRISPR activation (CRISPRa) [18] (Figure 1).
Treatment Strategies
As knowledge of the CRISPR/Cas9 system increases, different applications of the technique are being developed. Many of these have already been applied in vivo for the treatment of retinal diseases, demonstrating its potential in gene therapy (Table 1).
Table 1. In vivo experiments utilizing CRISPR/Cas9 for therapeutic interventions for retinal diseases.
| Reference | Disease | Gene Target | Methodology | Results | Cas9 species |
| Ruan et al. 2017 [25] | LCA10 | Removal of intron 25 of CEP290 | Subretinal injection of dual AAVs into mice | Successful gene editing in 7.5% to 26.4% of reads between treated eyes | SpCas9 |
| Kim et al. 2017 [33] | Wet age-related macular degeneration | Disruption of Hif1a | Subretinal injection of single AAV into CNV-induced mice | 20±4% reduction in CNV | CjCas9 |
| Courtney et al. 2016 [22] | Meesman’s epithelial corneal dystrophy | Allele-specific disruption of KRT12-L132P | Intrasomal injection of Cas9-GFP and gRNA in KRT12-L132P mice | KRT12-L132P disruption in 38% of isolated clones | SpCas9 |
| Suzuki et al. 2016 [32] | Retinitis pigmentosa | Insertion of missing Mertk exon 2 | Subretinal injection of dual AAVs into Royal College of Surgeons rat model | Increased Mertk mRNA and protein levels. Improved ONL preservation and ERG response | SpCas9 |
| Latella et al. 2016 [19] | Retinitis pigmentosa | Removal of 24 bp region of Rho at the P23H locus | Subretinal injection and electroporation of plasmid in Rho-/-P23HTg mice | 16% of reads contained the desired 24 bp deletion | SpCas9 |
| Yu et al. 2017 [37] | Retinitis pigmentosa | Disruption of Nrl | Subretinal injection of dual AAVs into Rhodopsin KO, Rd10, RHO-P347S mice | Delayed loss of rod function and prolonged cone survival | SpCas9 |
| Zhu et al. 2017 [38] | Retinitis pigmentosa | Disruption of Nrl or Nr2e3 | Subretinal injection of dual AAV into Rd10 and Rd1 mice | Significant rescue and restoration of both rod and cone function | SpCas9 |
| Bakondi et al. 2016 [23] | Retinitis pigmentosa | Allele-specific disruption of RhoS334 | Subretinal injection and electroporation of plasmid in S334ter-3 rats | Increased retinal preservation and 35% increased VA | Human codon optimized SpCas9 (hCas9) |
Silencing/Targeting the Pathogenic Mutation
The most common use of CRISPR/Cas9 in the treatment of eye diseases is the direct silencing of dominant negative pathogenic mutations via the NHEJ pathway. If the target gene mutation has a dominant negative effect (e.g. Rhodopsin), then disrupting the mutant allele will restore wild type functionality of the gene. In contrast, if the target gene is haploinsufficient (i.e. two wild type alleles are needed to prevent the disease phenotype e.g. PAX6), or the CRISPR/Cas9 strategy is not allele-specific (knocking out both the wild type and mutant copy of the gene), then additional wild type copies of the gene can be reintroduced to the cells via gene augmentation [19].
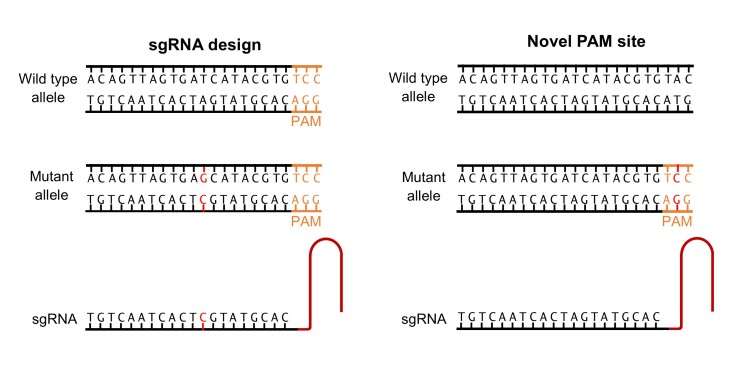
Allele-specific CRISPR/Cas9 binding can be achieved in two ways: sgRNA design or novel PAM sites (Figure 2). While the specificity of Cas9 is variable (see Off-target Effects), many papers have reported that a one bp difference in the DNA target sequence is sufficient to drive allele-specific cleavage. This allows just the mutant allele to be targeted by designing the sgRNA to the region containing the mutation [20,21]. Alternatively, some mutations will generate a novel PAM site that is not present on the wild type allele, ensuring the Cas9 only binds to the target strand. While it may seem unlikely that a novel PAM site will be generated, the wide range of Cas9 species and associated PAM sites available (see Table 2) makes this possible, with one researcher reporting that of 76 known missense mutations associated within four retinal dystrophy genes, 36 percent produced a novel PAM site for SpCas9 [22]. Both Courtney et al. (2016) and Bakondi et al. (2016) used novel PAM sites to target allele-specific disruption of their mutant gene in the treatment of Meesman’s epithelial corneal dystrophy and retinitis pigmentosa (RP), respectively [22,23]. Courtney et al. (2016) reported successful in vivo mutant gene disruptionin 5 of 13 clones (though this is too low to be therapeutically viable), while Bakondi et al. (2016) reported target cleavage efficiencies of 33 percent and 36 percent in two CRISPR-edited rats, corresponding with a partial phenotypic rescue. Neither paper reported a reduction in the wild type allele expression, demonstrating that allele-specific knock down is possible using CRISPR/Cas9.
Figure 2.
Allele-specific Cas9 targeting via sgRNA design or novel PAM sites. Cas9 can be targeted to the mutant allele by designing the sgRNA to the region containing the mutation. The discrepancy between the sgRNA sequence and wild type may be sufficient to prevent binding. If the target mutation generates a novel PAM site this will allow Cas9 binding on the mutant but not the wild type strand.
Table 2. Cas9 species and their associated PAM site sequences.
| Species | Cas9 | PAM site | Reference |
| Streptococcus pyogenes | SpCas9 | NGG | Anders et al. 2014 [15] |
| Synthetically modified | SpCas9 VQR | NGAN or NGNG | Kleinstiver et al. 2015 [55] |
| Synthetically modified | SpCas9 EQR | NGAG | Kleinstiver et al. 2015 [55] |
| Synthetically modified | SpCas9 VRER | NGCG | Kleinstiver et al. 2015 [55] |
| Staphylococcus aureus | SaCas9 | NNGRRT | Ran et al. 2015 [60] |
| Campylobacter jejuni | CjCas9 | NNNNACAC | Kim et al. 2017 [33] |
| Neisseria meningitidis | NmCas9 | NNNNGATT | Hou et al. 2013 [61] |
| Streptococcus thermophilus | St1Cas9 | NNAGAAW | Müller et al. 2016 [62] |
| Streptococcus thermophilus | St3Cas9 | NGGNG | Müller et al. 2016 [62] |
Another method of silencing pathogenic mutations is the removal of sections of DNA using a pair of sgRNAs which bind to either side of the target region. Following excision, the genomic DNA is repaired using the NHEJ pathway [24]. Ruan et al. (2017) proposed this method as a treatment for Leber Congenital Amaurosis type 10 (LCA10) caused by an intronic mutation in intron 26 of CEP290 (IVS26 mutation) [25]. This mutation generates a premature stop codon in half of all transcripts due to aberrant splicing, resulting in reduced CEP290 activity [26]. This would normally be a viable candidate for gene augmentation but CEP290 has a large 7440 bp open reading frame which exceeds adeno-associated viral (AAV) packaging capacity, and CEP290 over-expression is cytotoxic to photoreceptors [27]. While there is no mouse model of human IVS26, mouse Cep290 intron 25 is homologous to human CEP290 intron 26 and was successfully removed from wild type mice retinal cells in vivo in 7.5 percent to 26.4 percent of reads. Since this region has no role in coding but simply directs splicing there is no need for a gRNA template for repair [25]. This dual sgRNA approach has also been used to excise a small 24 bp region surrounding the RHO P23H mutation with the aim to treat RP via gene silencing and augmentation [19].
Insertion of DNA
As well as inactivation of pathogenic mutations, CRISPR/Cas9 provides the opportunity to insert DNA at a specific locus to restore wild type functioning of a gene. Though this traditionally utilizes the HDR pathway [28-31], Suzuki et al. (2017) demonstrated a modified method of gene insertion via the NHEJ pathway, named Homology-Independent Targeting Integration [32]. Using this strategy, they successfully inserted the missing exon 2 into intron 1 of Mertk in the Royal College of Surgeons rat model of RP. This resulted in increased levels of Mertk mRNA and protein levels than the untreated animals, corresponding with improved outer nuclear layer (ONL) thickness and ERG responses indicating a partial rescue of the phenotype.
Treating Disease Symptoms
Retinal diseases with complex genetic and environmental risk factors may be unsuitable for traditional gene therapy approaches. In these cases, genetic modification may be used to reduce the severity of the disease by targeting genes involved in its pathogenesis. This approach was taken by Kim et al. (2017) for the treatment of wet age-related macular degeneration (AMD) [33]. In wet AMD, choroidal neovascularisation (CNV) leads to deterioration of central vision [34]. Kim et al. (2017) used CRISPR/Cas9 to disrupt Hif1a (crucial to the development of CNV [35]) in wild type mice who later underwent laser-induced CNV. The AAV-Hif1a treated eyes demonstrated a 20±4 percent reduction in area of CNV compared to the uninjected control, with no deleterious effects on cone function, demonstrating its potential as a therapeutic intervention.
Cellular Reprogramming
Therapeutic cellular reprogramming involves converting cells which are sensitive to a mutation to a functionally related cell type that is less affected by the mutation in the hopes of reducing the disease severity. This technique has potential in the treatment of retinal dystrophies due to the high levels of heterogeneity existing between diseases. RP, for example, is characterized by loss of rod cells leading to secondary cone cell death, but to date there have been over 3000 causative mutations in over 60 genes identified for this disease. This makes it difficult to develop gene therapy strategies to treat large cohorts of patients [36]. Two papers published within two months of each other sought to utilize CRISPR/Cas9 to induce cellular reprogramming of mutationally-sensitive rod cells to cone cells within four models of RP [37,38]. Nrl and Nr2e3 are transcription factors involved in the differentiation and regulation of rod cells. Absence of either of these transcription factors is known to cause rod cells to develop a cone cell fate [39,40]. CRISPR/Cas9 caused targeted gene disruption of either Nrl or Nr2e3 via subretinal injection of mice between P7 and P14. Both researchers observed a downregulation of rod-specific genes and an up-regulation of some cone-specific genes. This occurred alongside significant rescue of photoreceptor functioning. Zhu et al. (2017) reported significantly improved cone and rod function with preserved ONL, and Yu et al. (2017) reported some functional cone and rod activity at 4 months of age, a month after rod death is usually complete in their tested mouse lines. Both authors conclude that cellular reprogramming of rods to cones is a promising area of further study for retinal disease treatment.
Stem Cells
The use of gene modification has promise as an early intervention strategy to prevent retinal degeneration, but for patients with advanced disease progression it offers little hope of a cure. CRISPR/Cas9 has the potential to correct pathogenic mutations in patient’s own induced pluripotent stem cells (iPSCs). These can be differentiated into the required cell type and re-implanted into the eye. This route has the potential to be tailored to rare mutations found in small populations or individuals. Bassuk et al. (2016) demonstrated this in iPSCs derived from two brothers carrying a novel RPGR mutation [41]. iPSCs were differentiated from fibroblasts and the HDR pathway used to correct the c.3070G>T mutation in 13 percent of cells, a higher rate than previous CRISPR/Cas9 iPSC correction studies [42]. The use of retinal grafts to integrate stem cell-derived cells has demonstrated cell survival, safety, and efficacy in clinical trials, offering a potential therapeutic route for CRISPR/Cas9-corrrected iPSC therapies [43,44].
Challenges to Clinical Application
Efficiency of Gene Editing
While in vitro gene editing for retinal diseases using CRISPR/Cas9 often reports high rates of gene disruption in vitro (up to 82 percent [19]), the rates of successful gene editing in vivo are lower (up to 33 percent [19]). This success rate is highly variable between studies and may be dependent upon the type of Cas9 used [25,33], the design of the sgRNA, the target cell type, the delivery method, and the in vivo model amongst other factors [37,38]. One study using AAV-delivered CRISPR/Cas9 found a 50 percent in vitro deletion rate in neuro-2a cells which was reduced to 7.5 percent to 26.4 percent in treated retinas in vivo [25]. Though these rates are quite low, some diseases would not require complete silencing of the mutant gene. Meesmann’s epithelial corneal dystrophy, for example, can be alleviated with a 50 percent reduction in mutant allele expression levels [22]. Other studies suggest that only a partial gene correction can result in a significant therapeutic impact [45]. Regardless of the rate of the CRISPR/Cas9 gene editing, in vivo studies are reporting improvements in disease phenotypes, which could have significant impacts on a patient’s quality of life [32,33,37,38].
Off-target Effects
As with any genetic modification technique, there are concerns regarding the off-target effects of CRISPR/Cas9. Non-specific gene editing could remove essential genes or disrupt tumor suppressing genes causing cancers [10]. It is therefore vital that any genetic modification methods are specific and off-target effects are predictable. As Cas9 binding is dependent upon both the presence of a PAM site and upstream homology to the sgRNA sequence, bioinformatic programs can be used to predict off-target effects. Encouragingly, some papers reported that a one nucleotide discrepancy in the sgRNA binding region prevents indel formation [20,21] and most studies demonstrate no off-target effects at the most probable sites. In studies employing whole genome screening, off-target effects are rare (up to several hundred), with some studies reporting only one off-target effect within the entire genome [46-50].
Despite this, off-target effects have been varied across studies, with some papers reporting DNA editing with a 5-bp discrepancy between the sgRNA and target region [12,51]. The effect of base pair mismatch between the sgRNA and the target have been showed to be highly influenced by the distance between the mismatch and the PAM site, with PAM-distal mismatches being more tolerated than PAM-proximal mismatches [11-13,52]. There have also been incidents of reduced gene expression in a site with sgRNA homology but no PAM site [22]. The cleavage sites can also vary between in vitro and in vivo studies [53], making it difficult to accurately predict off-target effects.
Two artificial Cas9 molecules have been created with the aim of reducing off-target effects: nickases and high-fidelity Cas9 (SpCas9-HF1). Nickases are modified Cas9 molecules which can only cleave one strand of DNA. To induce a double-stranded break into the DNA for gene modification, one nickase must bind to either strand of DNA (each guided by an sgRNA) in close proximity. The use of nickases can reduce off-target effects by 50 to 1500 times compared to SpCas9 at the same site, and has been successfully used to disrupt genes in mouse embryos [54]. SpCas9-HF1 is a modified SpCas9 that has four altered amino acids [55]. These result in decreased non-specific binding and have been reported to have no detectable off-target effects while efficiency remains comparable to the wild-type SpCas9.
It has been proposed that reducing the temporal expression of Cas9 will reduce the off-target effects, and so the use of a self-limiting Cas9 construct has been explored. In a self-limiting Cas9, the Cas9 expression system carries at least one copy of the sgRNA target site. The sgRNA will guide the Cas9 to both the genomic target site and the Cas9 vector, inactivating it and halting Cas9 transcription. This has been shown to successfully reduce Cas9 expression to negligible levels while maintaining targeted gene editing in vitro [25]. While limiting transgene expression may have advantages from a regulatory approach, its usefulness is debated, with some papers reporting that long-term Cas9 expression is not associated with an increased risk of off-target effects [33].
Heterogeneity of Disease
Despite retinal dystrophies having many characteristics which make them a useful target for gene therapy, the heterogeneity of these diseases remain a challenge for widescale treatment therapies. Many diseases have hundreds of identified causative mutations (over 100 variants in over 20 loci for LCA [56,57]), with some mutations specific to singular families or even individuals [36,41].
Cellular reprogramming, targeting of disease symptoms, and iPSC-derived retinal grafts (described in more detail in Treatment Strategies), have been suggested as potential solutions to this problem. Cellular reprogramming and targeting disease symptoms via CRISPR/Cas9 can be applied to patients without an identified causative mutation, but these are still in early stages of research [37,38]. Stem cell derived retinal grafts, alternatively, have been used in multiple clinical trials and have demonstrated efficacy and safety [43,44]. Using patients own iPSC would allow the pathogenic mutation to be corrected in vitro before differentiation into photoreceptors. This would allow rare mutations to be targeted on a case-by-case basis and detailed screening of on and off-target effects to be conducted before transplantation into the eye.
Rates of HDR in the Eye
Precise gene correction with CRISPR/Cas9 relies on the HDR pathway. While HDR occurs most frequently in the late S and G2 stages of the cell cycle phase, retinal cells are post-mitotic, and therefore the HDR rate is low. Some studies have concluded these HDR rates are currently too low to have a therapeutic value.
The use of small molecules to suppress the NHEJ pathway and force cells to repair the double-stranded break with HDR has demonstrated efficacy in increasing the rates of HDR. SCR7 is an inhibitor of DNA ligase and has been shown to increase rates of HDR by 5-fold and 19-fold in separate in vitro studies [31,58]. It was subsequently used to enhance CRISPR/Cas9 editing of the HSV-1 viral genome and found to increase HDR rates by over 10-fold [59].
In 2016, a new CRISPR/Cas9 technique was described: Homology-Independent Targeted Integration. This allows precise gene knock-in without the HDR pathway. In this, the DNA insert is flanked by Cas9 cleavage sites. Following cleavage of both the genomic DNA target site and the DNA insert, the insert will be ligated into the genomic target site via NHEJ. The DNA insert is designed in such a way that integration in the reverse orientation will generate a CRISPR/Cas9 cleavage site allowing it to be removed by Cas9 again until it is integrated in the correct orientation. This was found to have higher knock-in efficiency than HDR both in vitro in HEK293 cells and in vivo,where it was used to correct a rat model of RP [32].
Conclusion
CRISPR/Cas9 is an exciting area of study which is revolutionizing all aspects of genetics, from basic biology to potential medical interventions. Due to its favorable characteristics and track record in gene therapy, retinal diseases are likely to be one of the earliest targets of CRISPR/Cas9 mediated therapy. In vivo studies demonstrate a promising future for CRISPR/Cas9 therapies, with rescue effects seen across a range of diseases. Its versatility lends itself to a variety of approaches to tackle these debilitating diseases. Despite all this, there is still a long way to go. HDR rates are currently below the therapeutic range, although there are avenues around this being explored. Reduction of off-target effects remains a priority as if CRISPR/Cas9 is to have a future in gene therapy, its off-target effects must be predictable and minimized.
Glossary
- ZNFs
zinc finger nucleases
- TALENs
transcription activator-like effector nucleases
- crRNA
CRISPR RNA
- tracrRNA
trans-activating crRNA
- Cas9
CRISPR-associated protein 9
- PAM
protospacer adjacent motif
- sgRNA
single guide RNA
- NHEJ
non-homology end joining
- HDR
homology-directed repair
- Indels
insertions or deletions
- dCas9
inactive Cas9
- CRISPRi
CRISPR interference
- CRISPRa
CRISPR activation
- IVS26 mutations
intronic mutation in intron 26 of CEP290
- AAV
adeno-associated virus
- LCA10
Leber Congenital Amaurosis type 10
- ONL
outer nuclear layer
- AMD
age-related macular degeneration
- RP
retinitis pigmentosa
- CNV
choroidal neovascularisation
- iPSC
induced pluripotent stem cells
Author Contributions
CFP: Conception and design of review, research, and analysis of materials, drafting manuscript, critical revision of manuscript. REM: Review conception and design, critical review of manuscript.
References
- Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29(5):335–75. [DOI] [PubMed] [Google Scholar]
- McClements ME, MacLaren RE. Gene therapy for retinal disease. Transl Res. 2013;161(4):241–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar I, London A, Schwartz M. The privileged immunity of immune privileged organs: the case of the eye. Front Immunol. 2012;3:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Caspi RR. Ocular immune privilege. F1000 Biol Rep. 2010;•••:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski DM, Thake M, MacLaren RE. Clinical applications of retinal gene therapy. Prog Retin Eye Res. 2013;32:22–47. [DOI] [PubMed] [Google Scholar]
- Boye SE, Boye SL, Lewin AS, Hauswirth WW. A comprehensive review of retinal gene therapy. Mol Ther. 2013;21(3):509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob SR, Finn A, Papakostas TD, Eliott D. Clinical Trials in Retinal Dystrophies. Middle East Afr J Ophthalmol. 2016;23(1):49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest. 2014;124(10):4154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SS, McCaughey T, Swann O, Pebay A, Hewitt AW. Genome engineering in ophthalmology: application of CRISPR/Cas to the treatment of eye disease. Prog Retin Eye Res. 2016;53:1–20. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–40. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41(15):7429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A, et al. In vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-based CRISPR/Cas9 in the Mouse Retina. Mol Ther Nucleic Acids. 2016;5(11):e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P, et al. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther. 2015;23(3):570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K, Kaneko T, Voigt B, Mashimo T. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun. 2014;5:4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Moore JE, Atkinson SD, Maurizi E, Allen EH, Pedrioli DM, et al. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23(1):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, et al. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol Ther. 2016;24(3):556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl C, Ortiz O, Rottig B, Wefers B, Wurst W, Kuhn R. Creation of targeted genomic deletions using TALEN or CRISPR/Cas nuclease pairs in one-cell mouse embryos. FEBS Open Bio. 2015;5:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan GX, Barry E, Yu D, Lukason M, Cheng SH, Scaria A. CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol Ther. 2017;25(2):331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79(3):556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E, Wang Q, Quiambao AB, Xu X, Qtaishat NM, Peachey NS, et al. The relationship between opsin overexpression and photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2001;42(3):589–600. [PubMed] [Google Scholar]
- Wang B, Li K, Wang A, Reiser M, Saunders T, Lockey RF, et al. Highly efficient CRISPR/HDR-mediated knock-in for mouse embryonic stem cells and zygotes. Biotechniques. 2015;59(4):201-2, 4, 6-8. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–9. [DOI] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–8. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540(7631):144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutto I, Lutty G. Understanding age-related macular degeneration (AMD): relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol Aspects Med. 2012;33(4):295–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre H, Tunik S, Aronsson M, Kvanta A. Hypoxia-Inducible Factor-1alpha Is Associated With Sprouting Angiogenesis in the Murine Laser-Induced Choroidal Neovascularization Model. Invest Ophthalmol Vis Sci. 2015;56(11):6591–604. [DOI] [PubMed] [Google Scholar]
- Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84(2):132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M, et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Ming C, Fu X, Duan Y, Hoang DA, Rutgard J, et al. Gene and mutation independent therapy via CRISPR-Cas9 mediated cellular reprogramming in rod photoreceptors. Cell Res. 2017;27(6):830–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana CL, Kolesnikov AV, Shen SQ, Myers CA, Kefalov VJ, Corbo JC. Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proc Natl Acad Sci USA. 2013;110(5):1732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13(15):1563–75. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Zheng A, Li Y, Tsang SH, Mahajan VB. Precision Medicine: Genetic Repair of Retinitis Pigmentosa in Patient-Derived Stem Cells. Sci Rep. 2016;6:19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41(19):9049–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SD, Regillo CD, Lam BL, Eliott D, Rosenfeld PJ, Gregori NZ, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015;385(9967):509–16. [DOI] [PubMed] [Google Scholar]
- Shirai H, Mandai M. Retinal regeneration by transplantation of retinal tissue derived from human embryonic or induced pluripotent stem cells. Inflamm Regen. 2016;36(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32(7):670–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, et al. Off-target mutations are rare in Cas9-modified mice. Nat Methods. 2015;12(6):479. [DOI] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237-43, 1 p following 43. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Sengillo JD, Justus S, Cabral T, Tsang SH, Mahajan VB. CRISPR-Cas Genome Surgery in Ophthalmology. Transl Vis Sci Technol. 2017;6(3):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI. Omics in Ophthalmology: Advances in Genomics and Precision Medicine for Leber Congenital Amaurosis and Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci. 2016;57(3):1378–87. [DOI] [PubMed] [Google Scholar]
- Hung SS, Chrysostomou V, Li F, Lim JK, Wang JH, Powell JE, et al. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Invest Ophthalmol Vis Sci. 2016;57(7):3470–6. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Li H, Hao M, Xiong D, Luo Y, Huang C, et al. Increasing the Efficiency of CRISPR/Cas9-mediated Precise Genome Editing of HSV-1 Virus in Human Cells. Sci Rep. 2016;6:34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110(39):15644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24(3):636–44. [DOI] [PMC free article] [PubMed] [Google Scholar]