Abstract
Since its emergence in 2012, the genome editing technique known as CRISPR-Cas9 and its scientific use have rapidly expanded globally within a very short period of time. The technique consists of using an RNA guide molecule to bind to complementary DNA sequences, which simultaneously recruits the endonuclease Cas9 to introduce double-stranded breaks in the target DNA. The resulting double-stranded break is then repaired, allowing modification or removal of specific DNA bases. The technique has gained momentum in the laboratory because it is cheap, quick, and easy to use. Moreover, it is also being applied in vivo to generate more complex animal model systems. Such use of genome editing has proven to be highly effective and warrants a potential therapy for both genetic and non-genetic diseases. Although genome editing has the potential to be a transformative therapy for patients it is still in its infancy. Consequently, the legal and ethical frameworks are yet to be fully discussed and will be an increasingly important topic as the technology moves towards more contentious issues such as modification of the germline. Here, we review a number of scientific and ethical issues which may potentially influence the development of both the technology and its use in the clinical setting.
Keywords: CRISPR, Cas9, genome editing, bioethics
Introduction
In the era of “gene editing,” CRISPR-Cas9 has become hugely attractive for both the scientific community and the general public. The metaphorical use of the term “gene editing” gives the impression that genes as texts are static in nature and therefore can be corrected easily. However, a simplistic view of genes or editing may not be useful to appreciate the analogy of gene editing for it may easily dismiss collateral damage or adverse consequences.
This review considers science and ethics of CRISPR-Cas9 as two co-dependent factors required for better applications and effective ways of treating diseases. The first part of the review uncovers the science behind CRISPR-Cas9 and its applications. The second part elaborates the moral reasoning for the use of this rather attractive and useful technique for the betterment of humanity.
Development of the CRISPR-Cas9 System: A Historical Perspective
Since the emergence of CRISPR-Cas9 technology in 2012, techniques for making precise and targeted manipulations of DNA sequences—so called gene editing—in living cells have dominated the field of biology. However, genome editing is not a new concept as transgenic mice were successfully employed in research in the 1970s [1]. Transgenesis thereby became a powerful research tool to decipher the underlying biological mechanisms that exist in disease. Although this technique was largely employed to introduce a genetic component (transgene) into a cell it was unable to execute a targeted insertion into the genome. Further advances in the 1980s revealed directionality could be achieved using alterations in the genome of embryonic stem (ES) cells which retained their pluripotency to give rise to many other distinct cell types [2]. However, it was technically challenging and remained an inefficient method (success rate of less than 10 percent).
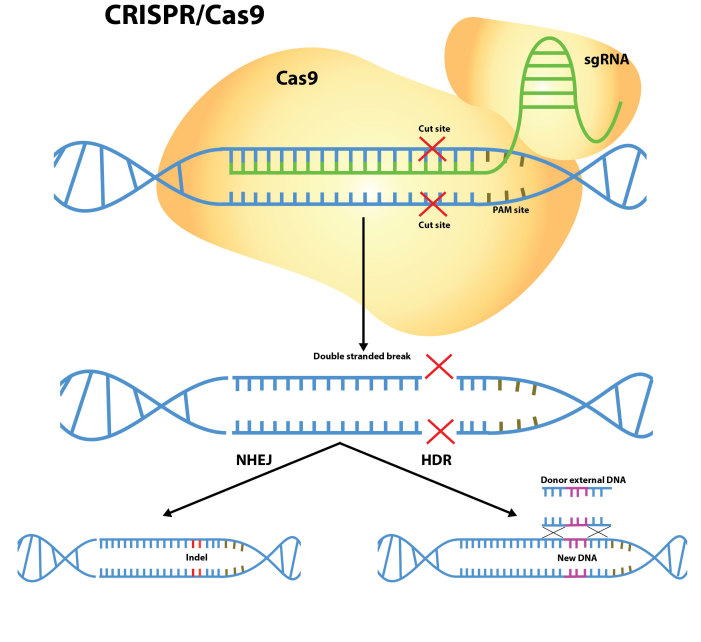
These limitations led to a concerted effort in developing alternative gene targeting technologies. In 2005, zinc finger nucleases (ZFNs) [3] and in 2010, Transcription activator-like effector nucleases (TALENs) [4,5], revealed the first systems that can be engineered to recognize specific sequences of DNA. These specific enzymes were able to recognize their target sequences on the genome to introduce double-stranded DNA breaks permitting a repair process by the cell to prevent lethality. This repair may occur in two ways (Figure 1). First, the ends of the DNA breaks are re-joined by the cell repair machinery, a process known as non-homologous end-joining (NHEJ) [6]. However, the imprecise nature of this repair process has the propensity to introduce insertions or deletions of DNA (Indels) that in turn may disrupt the translation of the targeted gene. Second, with the use of a complementary matching sequence of the DNA, a homology-directed repair (HDR) may occur to repair the double-stranded break in a targeted manner [7]. Although this technique can be used to add or remove specific DNA sequences at the site of the double-stranded break, it is found to be more inefficient when compared with NHEJ.
Figure 1.
CRISPR-Cas9 mediated gene-editing mechanisms. A single guide RNA (sgRNA) recognizes a genomic region followed by 5’-NGG-3’ PAM sequence, which recruits the Cas9 DNA endonuclease. This introduces a double-stranded break that is repaired by (i) non-homologous end joining (NHEJ), an error prone pathway that can result in the creation of Indels that can disrupt the gene, or by (ii) homology directed repair (HDR) in the presence of a donor construct.
The role of ZFNs and TALENs in introducing double-stranded breaks is highly specific because of the requirement for two engineered proteins. However, to design this process, which involves the synthesis of proteins and optimization of the protocol to generate these breaks, requires considerable time and effort. These constraints led to limited adoption of this technology in the wider field of biology but sought for a simpler method for targeting double-stranded breaks in DNA. In 2012, it was discovered that the bacterium Streptococcus pyogenes contained a remarkable system of viral defense that could be adapted as a programmable system for genome editing [8]. This system is consisted of two parts, first, the “Clustered Regularly Interspaced Short Palindromic Repeat” (CRISPR) of RNA that acts as a guide for genome targeting and the second is the “CRISPR-associated protein 9” (Cas9) that acts as an endonuclease to enable double-stranded breaks. It was subsequently found that scientists could manipulate the CRISPR RNA molecule into a single guide RNA (sgRNA) that could be engineered to specifically target a genomic area of interest. The only requirement was that the sgRNA recognize a 17 to 22 bp genomic region that is followed by a 5’-NGG-3’ protospacer adjacent motif (PAM) site. Similar to ZFNs and TALENs, the endonuclease Cas9 can direct DNA cleavage to create double-stranded breaks that can be repaired through the NHEJ or HDR processes. Subsequently, this technology was adapted and shown to be capable of editing the mammalian genome with high efficiency and selectivity [9-11].
Technical Limitations of the CRISPR-Cas9 System
Given the rapid advancement of this technology, from a technical perspective, there are a number of questions that arise surrounding the potential limitations of this technology. One major concern is the potential off-target effects in areas of the genome that are sensitive to double-stranded breaks [12]. Moreover, the short length of an sgRNA (typically around 17 to 22 bp in length) combined with the CRISPR-Cas9 system is able to tolerate up to 1 to 3 mismatches between the sgRNA and the target site, which may further increase the chances of off-target effects. This problem can be reduced somewhat by evaluating the potential off-target effects computationally and only using sgRNAs that do not target other functional proteins or regions. However, given that the sgRNA may also have off-target effects in unknown long-range enhancer regions, such effects cannot be entirely eliminated. Despite this, scientists have engineered the Cas9 protein and sgRNA to increase specificity thereby lowering the off-target effects [13].
CRISPR-Cas9 technology is rapidly providing researchers with a viable method for dissecting the molecular mechanisms that underpin cellular function. However, if the technology is to progress towards the clinic, improvements in the delivery of the CRISPR-Cas9 must be considered. The use of transfection reagents that simultaneously deliver the sgRNA and Cas9 protein into target cells in the laboratory has proven its efficiency in a number of cell lines [14]. However, all or parts of the plasmids are often randomly integrated into the host genome [15,16]. It has been shown that plasmid DNA can also be inserted into both the on-target and off-target sites [17]. Furthermore, host immune responses can also be induced by these bacterial sequences, thereby dampening the efficiency of genome editing [18].
To overcome the challenges of transfection delivery systems, the use of viral delivery systems have been evaluated. Viral vectors can enter a cell in large numbers efficiently under both in vitro and in vivo conditions. Presently, integrase-defective lentivirus vectors (IDLVs) [19], adenoviral vectors (AdVs) [20] and recombinant adeno-associated-viral vectors (rAAVs) [21] have been used to transfer CRISPR technology to mammalian cells. However, historically the use of viral vectors has been marred with controversy because of adverse events following clinical trials, leading to patient death [22,23]. Despite these early setbacks for gene therapy, a number of improvements in viral delivery systems are beginning to translate some encouraging preliminary results into the clinic [24-26]. As with CRISPR-Cas9 technology, delivery systems need further development to increase safety before they can be successfully used routinely.
Applications of the CRISPR-Cas9 System in Human Health
The CRISPR-Cas9 system has many potential biological research uses because of its ability to cleave the genome at a specifically desired location. Pertaining to genome editing it has been used to generate stable cell lines with specific functions to develop experimental models to understand the underlying disease pathology. CRISPR-Cas9 has been demonstrated to be effective in multiple organisms, including bacteria, fish, birds, fruit flies, and mammals [27,28]. In 2012, the application of this technology was successfully used in human cells by engineering a novel version of CRISPR-Cas9 [9,14]. Subsequently, the use of CRISPR as a novel system for investigating biological processes and pathologies began to expand to numerous fields. For example, it has been modified to program specific transcription factors to target and activate or silence specific genes [29,30]. Further adaptation of the Cas9, which enabled manipulation of methyl groups at specific positions on the DNA, has allowed researchers to evaluate how such changes affect gene expression [31]. More recently, CRISPR-Cas9 has been used to turn cells into programmable computers, where researchers have engineered molecular switches to control cell fate to enable them to program conditional behaviors [32]. Such examples demonstrate the versatility of the CRISPR-Cas9 system in generating basic research tools in vitro.
In addition to its in vitro application,CRISPR-Cas9 can also be used to generate in vivo animal models to study diseases more effectively. For example, mouse models have been developed to evaluate deleterious effects of mutations in cancer by using the system to introduce loss of function mutations in tumor suppressors or gain of function mutations in oncogenes [33]. Gene manipulation of mice at the germline level has enabled researchers to generate whole organism or conditional models that model human disease.
Germline editing is also a promising technique used for understanding early onset of human diseases by means of human embryos. However, research committees have been reluctant until recently to grant permission for such investigations. Recently, a group of researchers from China have carried out a systematic analysis of gene functions in modified human embryos in order to investigate the effectiveness of CRISPR-Cas9 for further research [34].
A major potential goal of developing CRISPR-Cas9 genome editing technology is its use in preventing or treating disease or disability. There is evidence that using CRISPR-Cas9 to target the genome of viruses such as Hepatitis B and HIV could control and ultimately cure patients of the disease [35]. For example, CRISPR-Cas9 has shown that introduction of Indels into HIV is lethal to the virus, however, it has also been shown that other modifications to the virus lead to increased virulence [36]. Recently, modifying the immune system to attack HIV has been gaining attraction as a promising therapeutic use of genome editing [37]. Similar strategies have been deployed for the treatment of leukemia and several other blood cancers [38]. Interestingly, cell-based therapies have shown significant advantages as cells can be removed, manipulated, expanded, and then reintroduced into the patient to enhance the desired therapeutic effect. However, for several other diseases such as solid tumor cancers or those that effect tissues or organs, CRISPR-Cas9 is unlikely to be effective given the present state of the technology. Despite the setbacks, there are currently active areas of research that are pursuing the application of CRISPR-Cas9 into editing CFTR gene in Cystic Fibrosis and dystrophin in Duchenne muscular dystrophy [39,40]. Despite these advancements, such research is currently in its infancy and the technology is faced with challenges related to delivery methods that were previously encountered by gene transfer technologies in the 1990s.
Outlining the Ethical Questions on CRISPR-Cas9 for Gene Editing
With the most recent advances in biological and medical research, CRISPR-Cas9 has become controversial because of its capacity to modify the plan of each cell in the human body. By altering the germline genome, many presume that it can even transfer not only the intended modifications but also other “unforeseeable alterations” to the offspring. Hence the question of “irreversible effects” on future generations is pronounced with trepidation. Recent discoveries from a Chinese group led by Junjiu Huang has created some real concerns and opened discussions regarding whether this technology should even be applied to pre-implantation embryos [34]. The research was published in Protein & Cell (following two rejections on ethical grounds by Nature and Science) with a significant backlash from the general public [34]. The story of CRISPR-Cas9, therefore, has awakened the potential power behind gene editing and the “nervousness” it generates among the public.
From a historical perspective, genetics and social engineering have had a profound toxic relationship, with the most notable example being such misuse in Nazi Germany [40]. Many well-respected scientists at the time, fearing the degeneration of the human race, attempted to promote policies that legally blocked the “breeding of inferior humans.” The rhetorical misuse of natural selection to promote eugenic policies led to widespread abominations, which included the mass-sterilization of thousands of individuals and the ethnic cleansing of millions of individuals. However, even though the views of eugenics held by the Nazis were later discredited, the sterilization of women with intellectual disabilities, on social and therapeutic grounds, was still legal practice in a number of European countries and in North America until the mid-twentieth century [41]. As a result of these misuses, there are significant ethical questions that arise for CRISPR-Cas9 use, the most pressing being whether its use may be harmful to society.
What is Ethically “Unacceptable” about CRISPR-Cas9?
As seen in the first part of this review CRISPR-Cas9 is still in its infancy of development. However, it is a rapidly evolving technology and with ever expanding applications it is also attracting private companies for investments despite certain ethical restrictions [42,43].
What is ethically repulsive about CRISPR-Cas9? Although bioethicists hold a wide range of opinions, the most compelling argument is about its ethical use in human germline manipulations [44]. The authors argue that the dearth of knowledge in the area of germ cell mutagenesis may cause uncertain future consequences. It appears that this argument from “potentiality” seems to incorporate not just the concerns for the safety of future generations susceptible of non-Mendelian (single gene) diseases, but how this technology can transform society in terms of societal values, its economy status, individuality, injustices, and accessibility [45]. It seems to appeal to the “transformative potential” of the technique which may have wider concerns for the overall moral and ethical texture of the society in which humans live.
Gene Editing with CRISPR-Cas9 and Maintaining Ethical Integrity
Given the complexity of the debate, a brief note about the current ethical stand may be necessary. There are several regulatory bodies including the WHO, UNESCO, and the Decleration on the Human Genome and Human Rights that are involved in the current debate on gene editing in view of formulating helpful guidelines. Although policies vary depending on the legal system, there are legally binding conventions such as “Oviedo Convention” (the Council of Europe Convention for the protection of Human Rights and Dignity of the Human Being with regard to the Application of Biology and Medicine: Convention on Human Rights and Biomedicine) and professional regulatory bodies (such as that of the HFEA in the U.K. and FDA in the U.S.) which are heavily involved in the deliberations. More recently, the U.S. National Academy of Sciences (NAS) and National Academy of Medicine (NAM) formulated a consensus report highlighting the importance of public engagement in this debate considering the benefits and risks of genome editing in humans [46].
There is a ban on human germline modification by many European countries [45] and in the United States, any edited embryos for pregnancy requires permission from the U.S. Food and Drug Administration. Similarly, in China, any clinical use of gene editing must be permitted by the Chinese Ministry of Health. The recent report by NAS/NAM classifies gene editing using CRISPR-Cas9 on the basis of purpose and heritability: purpose being therapeutic interventions (treat or prevent disease) or enhancement and heritability involving somatic or germline cells. From a public point of view, the report observes that the majority agrees with the use of genome editing in both somatic (64 percent) and germline cells (65 percent) for therapeutic purposes but not for enhancement applications [47]. However, it is important to note that basic research objectives in gene editing techniques are also closely tied up with therapeutic/enhancement interventions thereby adding a layer of complexity for the principle purpose of the technique.
Therefore, a sound bioethical reasoning may be required to clarify the purpose of CRISPR-Cas9, like techniques with current advancement in biotechnology. It also nurtures a healthy relationship between science and society and encourages public engagement in science for the betterment of living.
Arguments Towards Relevant Applications of CRISPR-Cas9: Separating Facts from Fiction
The consequentialist arguments, commonly used in medical situations, weigh both potential benefits and risks in ethical considerations. At the heart of such arguments would be to create some space for the moral obligation to promote medical intervention for preservation and promotion of life threatened by diseases. It does not necessarily mean that a significant risk for a serious genetic condition would invariably allow gene editing a reproductive option [45].
It is required to state that Pre-implantation Genetic Diagnosis (PGD) stands as the legal ethical stand in countries where secular bioethics discourse is permitted. However, there are few cases in which selection of unaffected embryos using PGD would not be possible and effective. In autosomal recessive diseases (e.g. cystic fibrosis, phenylketonuria) where both parents are homozygous and autosomal dominant diseases (e.g. Huntington’s disease, familial adenomatous polyposis) where at least one parent is homozygous, germline editing would be the only option for parents desiring a healthy child [45]. This makes CRISPR-Cas9 technology a potentially lucrative option in reproductive applications. It must be mentioned that PGD would be required to verify the success of the edits at least initially.
In general, the use of genome editing in somatic cells is ethically accepted because of its low risk to benefit ratio and the presence of informed consent [43]. Given safety concerns, germline applications for human embryos for implantation may have a high risk-to-benefit ratio as it may have an “unforeseeable” harming effects on future generations. This is justified given the nature of the technique and the level of currently available scientific evidence. Nevertheless, as a proof of concept, Ma et al., may argue that correcting MYBPC3 gene that causes hypertrophic cardiomyopathy may help better selection of corrected embryos for implantation as a remedy for monogenic inherited disorders [48]. However, PGD as a reproductive option is an effective and ethically acceptable mode of treatment for most of such diseases. Perhaps one may be able to justify the use of gene editing research for these genetic conditions as a way of developing research tools and conditions for more complex applications in the future. In this case it may be equally important to justify the use of human embryos in gene editing as monkey germline editing is not subjected to any particular restrictions [49].
Informed consent is another area of dispute concerning gene editing as a reproductive option [50]. It is a decision that may impact on the genetic traits of future generations whose informed consent cannot be obtained [51]. One might ask whether it is a logical oversimplification to expect an informed consent knowing the nature of the situation. In most countries, in vitro fertilization (IVF) is the standard way by which germline transmission of inherited diseases in human embryos can be screened. Obviously, the informed consent is given by a couple desiring the IVF who are properly informed of and conscious of their choice for the offspring. In contrast, it is not unfounded to reason that with CRISPR-Cas9 “the unforeseeable effects” may be greater than the actual level of genetic interference, thereby effectively making it impossible to derive an informed consent on behalf of the offspring. Moreover, because of the off-target effects of this technology on the germline, some of the potential side effects transmitted to future offspring may not be observed until several subsequent generations. In support of this some scientists are expressing a preference for a moratorium on the use of genome editing in human embryos, at least until the technology has developed to a stage that it is deemed mature [52,53]. Perhaps “deemed mature” may beg several questions including an assessment about the probability of successes/harmful effects of the application. Interestingly, in principle, this encourages an a priori condition for research to eliminate those factors hindering the success of gene editing.
Having a powerful tool in hand such as CRISPR-Cas9 to modify the human germline is constantly faced with criticisms about its potential future applications. It appears that the ethical dilemma of the technology is closely interwoven with potential but unforeseen harm or “abuses,” a secondary effect of the technology. At another layer those secondary effects may have lasting impact on human life because of their inherent capacity to be incorporated into the gene pool of human species. More recently, an international team of scientists showed improved efficiency and accuracy for CRISPR-Cas9 taking a significant step towards using the technology to correct debilitating birth defects [48]. We have learned from the past that advances in therapeutic treatment can be utilized for previously unintended cosmetic applications. For example, plastic and reconstructive surgery that was initially intended for the management of patients suffering from disfiguring conditions has been utilized for cosmetic purposes. However, it is understood that any discovery may have various unforeseen applications which may bring about beneficial or harmful effects. The discovery of radium sparked numerous benefits and several harmful consequences too. Should a benevolent technique such as CRISPR-Cas9 be penalized for the “unforeseen abuses?” In spite of sound moral reasoning, some argue that CRISPR-Cas9 may warrant “designer baby” technology and thereby may actively take part in societal change towards greater global inequality than that already seen today [54].
Considering the consequentialist arguments on modification of somatic cells by CRISPR-Cas9 does not have the same level of ethical impact as those raised by germline modification (Table 1). The ethical considerations for using CRISPR-Cas9 as a treatment for patients are focused on defining an appropriate risk-to-benefit ratio that favors beneficial outcome to the patient [55]. For CRISPR-Cas9 treatments, this balance will likely follow the same approach as other well-established medical treatments. It may depend on several factors such as disease type, disease progression, the type of cells/tissues treated, and the mode of therapeutic applications (e.g. enhancement vs. therapeutic applications). Importantly, risk-to-benefit ratio is likely to be affected significantly by the delivery methods used in the treatment. For example, a favored delivery method is a lentivirus approach because of its efficiency and stability. However, there had been adverse effects following gene therapy using lentiviral vectors in clinical trials [56]. With the development of safer delivery methods, some of these risks may be overcome.
Table 1. The potential risks associated with CRISPR-Cas9 gene editing technology.
| Specific CRISPR-Cas9-based applications in humans: Potential risks | ||||
| Technical | Social | |||
| Off-target Insertions and deletions (Indels) | Random integration of vector | Toxicity | Exacerbating social inequalities | |
| Germline editing | High - A potentially significant issue but screening of embryos prior to implantation could overcome this risk. | Medium - Random integration may result in inactivation/dysregulation of gene expression. Sequencing for the presence of integration would identify this. | Low | High – Dependency upon “enhancement” applications |
| Ex vivo delivery (cell transplants) | High - For clonally expanded cells, screening can be performed to identify off-target Indels. | Low | Low | Low |
| In vivo delivery (tissues & organs) | High - Poses the most significant risk because screening would be difficult to perform. | High – It would be very difficult to determine integration in vivo. If the integration occurs in a tumour suppressor or oncogene the development of cancer could be an issue. | High – The induction of inflammatory responses to vector components. | Low |
As indicated above, CRISPR-Cas9 may be used in somatic cells for modifying physical traits and capacities. These cosmetic applications may lead to “enhancement” of certain traits such as muscle development or intelligence. Some argue that enhancing applications should not be equated with treatments using CRISPR-Cas9 for muscular dystrophy or mental disability [45]. It may become clear in those cases the potential benefits may be unlikely to outweigh the risks involved. However, as noted by others, any proposed intervention by CRISPR-Cas9 for cosmetic applications will have to pass the well-established threshold that the potential risk-to-benefit ratio is acceptable, informed consent is given, and all regulatory approvals are obtained from the relevant medical regulatory bodies [55].
Do the Risks Outweigh the Benefits?
Any discussion about the use of CRISPR-Cas9 in the clinic needs prior assessment about the level of risks, since the technology has demonstrated to have potentially damaging off-target effects. It has been observed that CRISPR-Cas9 has a high propensity for off-target effects and single nucleotide variants in humans, due to the large number of repeats and highly homologous genome, and can cleave unintended sequences causing mutations that may have a likely effect on developing cancer like diseases. Therefore, further improvements are needed, especially for more precise modifications that will be required for therapeutic interventions [57]. In the drive towards using the technology in a clinical setting, it may not be practically possible to increase the biosafety in time for the first clinical trial, nonetheless reviewing the critical balance between risk and safety may help assess its impact prior to a clinical trial. Such safety reviews, performed at the Recombinant DNA Advisory Committee (RAC) at the U.S. National Institute of Health, are already at the stage of being granted [37,58]. The proposed 2-year trial will be conducted on 18 people with myeloma, sarcoma, or melanoma. Recruiting those that have stopped responding to conventional therapy has somewhat negated the risks associated with the technology. Following this approval, it is expected that other trials in different diseases will apply for permission. However, careful attention should be given by ethics committees to accurately evaluate the risks in different diseases, since the risk-to-benefit ratio changes with each disease.
It is important to bear in mind the drive towards commercialization of the technology and the conflicts that this creates when evaluating the safety aspects of any potential clinical trial. Filing a utility patent application in the United States or Europe generally gives the applicant protection for 20 years from the date the patent is filed. Therefore, there is a drive towards commercializing the technology quickly to recoup developmental investments. This sometimes conflicts with safety evaluations when clinical trials are not conducted effectively. For example, in 2004, the drug rofecobix (Vioxx) was withdrawn from the market over safety concerns after 88,000 to 140,000 people suffered heart attacks from taking it [59]. This was despite the pre-clinical finding that it had a protective effect. Therefore, if we are to avoid some of these failures when looking at the application of CRISPR-Cas9, we could reinvestigate the failure of gene therapy trials that were conducted in the 1990s to see if there are lessons that could be relevant to CRISPR-Cas9 trials.
As the genome-editing technology is rapidly progressing but still a way off from being adopted as a reliable technique, to modify somatic/germline cells in a clinical setting, discussions are required to debate the overall implications of the technique. If the technology advances to a stage that safety concerns are acceptable, then further discussions will be needed to consider the social, legal, and ethical implications of using this technology to avoid potential “abuses” of germline editing. As a matter of fact, although modification of human embryos for implantation is illegal in many countries, ethically informed discussions at public level may be necessary to evaluate the effects of gene editing techniques against a global picture, not just in the present but also into the future [60].
Conclusions
Genome editing techniques, particularly those related to CRISPR-Cas9, have lowered the cost and increased the output of genomic research, thereby allowing the genetic manipulation of cells and organisms that have traditionally been difficult or impossible to perform. The major direction the technology is moving towards is making it possible to alter gene function, rather than knocking out the function of the gene altogether. This approach will go some way towards making this technology applicable to treating a number of human diseases. However, there are some serious concerns regarding the ethical and moral aspects of the delivery of the technology, the creation of new variants on the human population and the potential negative impact on unintended consequences. Many researchers have urged a public discussion over the social, ethical, and legal implications of genome editing in the germline, however there are many other health concerns that also need to be discussed and debated. Although this technology is some way from being utilized in a clinically effective manner there is some urgency for these discussions to take place at a national, local, and governmental level.
Glossary
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeat
- Cas9
CRISPR-associated protein 9
- sgRNA
single guide RNA
- ZFNs
Zinc Finger Nucleases
- TALENs
Transcription Activator-Like Effector Nucleases
- NHEJ
Non-Homologous End-Joining
- HDR
Homology-directed repair
- RAC
Recombinant DNA Advisory Committee
- PAM
Protospacer Adjacent Motif
- Indels
Insertions or deletions of DNA
Author Contributions
APC wrote the manuscript and both authors were involved in the composition, reviewing, and editing the final manuscript. APC and SMWP contributed equally to the manuscript. No author received funding for their involvement.
References
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6(6):507–12. [DOI] [PubMed] [Google Scholar]
- Bouabe H, Okkenhaug K. Gene targeting in mice: a review. Methods Mol Biol. 2013;1064:315–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–51. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma YM, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4(9):712–20. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337(6096):816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Lin G, Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2016;283(7):1218–31. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H. Toll meets bacterial CpG-DNA. Immunity. 2001;14(5):499–502. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33(2):175–8. [DOI] [PubMed] [Google Scholar]
- Holkers M, Maggio I, Henriques SF, Janssen JM, Cathomen T, Goncalves MA. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat Methods. 2014;11(10):1051–7. [DOI] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assessment of adenoviral vector safety and toxicity: report of the National Institutes of Health Recombinant DNA Advisory Committee. Hum Gene Ther. 2002;13(1):3–13. [DOI] [PubMed] [Google Scholar]
- Marshall E. Gene therapy death prompts review of adenovirus vector. Science. 1999;286(5448):2244–5. [DOI] [PubMed] [Google Scholar]
- Kay MA, Manno CS, Ragni MV, Larson PJ, Couto LB, McClelland A, et al. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 2000;24(3):257–61. [DOI] [PubMed] [Google Scholar]
- Khuri FR, Nemunaitis J, Ganly I, Arseneau J, Tannock IF, Romel L, et al. a controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6(8):879–85. [DOI] [PubMed] [Google Scholar]
- Finer M, Glorioso J. A brief account of viral vectors and their promise for gene therapy. Gene Ther. 2017;24(1):1–2. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports. 2013;4(1):220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon S. Welcome to the CRISPR zoo. Nature. 2016;531(7593):160–3. [DOI] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8(11):2180–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167(1):233-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry QR, Lyutova R, Fulga TA. Rational design of inducible CRISPR guide RNAs for de novo assembly of transcriptional programs. Nat Commun. 2017;8:14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15(7):387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, et al. CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate Viral Escape. Cell Reports. 2016;15(3):481–9. [DOI] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–8. [DOI] [PubMed] [Google Scholar]
- Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4(1):143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M. Sterilization, segregation and community care: ideology and solutions to the problem of mental deficiency in inter-war Britain. Hist Psychiatry. 1992;3(12):473–98. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- Baumann M. CRISPR/Cas9 genome editing – new and old ethical issues arising from a revolutionary technology. NanoEthics. 2016;10(2). [Google Scholar]
- Mulvihill JJ, Capps B, Joly Y, Lysaght T, Zwart HA, Chadwick R, et al. Ethical issues of CRISPR technology and gene editing through the lens of solidarity. Br Med Bull. 2017;122(1):17–29. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Caney S, Clancy T, Edwards J, Gallagher A, Greenfield A, et al. Nuffield Council on Bioethics Report:Genome editing. 2016. [Google Scholar]
- Scheufele DA. Science communication as political communication. Proc Natl Acad Sci USA. 2014;111 Suppl 4:13585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academy of Sciences and National Academy of Medicine Human Genome Editing: Science, Ethics, and Governance. Washington (DC): National Academies Press; 2017. [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–9. [DOI] [PubMed] [Google Scholar]
- Tu Z, Yang W, Yan S, Guo X, Li XJ. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyngell C. The Ethics of Human Life Extension: The Second Argument from Evolution. J Med Philos. 2015;40(6):696–713. [DOI] [PubMed] [Google Scholar]
- Billings PR, Hubbard R, Newman SA. Human germline gene modification: a dissent. Lancet. 1999;353(9167):1873–5. [DOI] [PubMed] [Google Scholar]
- Lanphier E, Urnov F, Haecker SE, Werner M, Smolenski J. Don’t edit the human germ line. Nature. 2015;519(7544):410–1. [DOI] [PubMed] [Google Scholar]
- Caplan AL, Parent B, Shen M, Plunkett C. No time to waste—the ethical challenges created by CRISPR: CRISPR/Cas, being an efficient, simple, and cheap technology to edit the genome of any organism, raises many ethical and regulatory issues beyond the use to manipulate human germ line cells. EMBO Rep. 2015;16(11):1421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokowski C, Pollack M, Pollack R. Cutting Eugenics Out of CRISPR-Cas9. Ethics Biol Eng Med. 2015;6(3-4):263–79. [Google Scholar]
- Kohn DB, Porteus MH, Scharenberg AM. Ethical and regulatory aspects of genome editing. Blood. 2016;127(21):2553–60. [DOI] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–58. [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhang L, Huang X. Genome modification by CRISPR/Cas9. FEBS J. 2014;281(23):5186–93. [DOI] [PubMed] [Google Scholar]
- Kaiser J. First proposed human test of CRISPR passes initial safety review. Science. 2016 [Google Scholar]
- Bhattacharya S. Up to 140,000 heart attacks linked to Vioxx. New Sci. 2005 [Google Scholar]
- Vasiliou SK, Diamandis EP, Church GM, Greely HT, Baylis F, Thompson C, et al. CRISPR-Cas9 System: opportunities and Concerns. Clin Chem. 2016;62(10):1304–11. [DOI] [PubMed] [Google Scholar]