Abstract
Inherited metabolic diseases (IMDs) of the liver represent a vast and diverse group of rare genetic diseases characterized by the loss or dysfunction of enzymes or proteins essential for metabolic pathways in the liver. Conventional gene therapy involving adeno-associated virus (AAV) serotype 8 vectors provide therapeutically high levels of hepatic transgene expression facilitating the correction of the disease phenotype in pre-clinical studies and are currently being evaluated in clinical trials for multiple IMDs. However, insertional mutagenesis and immunogenicity risks as well as efficacy limitations represent major drawbacks for the AAV system. Genome editing tools, particularly the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system, offer multiple advantages over conventional gene transfer and have the potential to further advance the promises of gene therapy. Here, we provide a critical assessment of conventional gene therapy and genome editing approaches for therapeutic correction of the most investigated metabolic liver disorders, namely familial hypercholesterolemia, hemophilia, ornithine transcarbamylase deficiency, hereditary tyrosinemia type 1, and alpha-1 antitrypsin deficiency. In addition, we elaborate on the barriers and future directions for advancing novel nuclease mediated gene therapies for IMDs.
Keywords: gene therapy, genome editing, nucleases, inherited metabolic disease of the liver, inborn errors of metabolism, hepatocytes, therapy for rare disease
Introduction
Inherited metabolic diseases (IMDs) are a vast and diverse group of rare genetic disorders. Although individually rare, their collective incidence is substantial; occurring in roughly 1 in 800 live births [1]. IMDs are characterized by the aberrant synthesis or deficiency of enzymes or other proteins, such as receptors or transporters, involved in biochemical pathways essential for metabolism. The blockage of a metabolic pathway results in the accumulation of upstream toxic substrates leading to organ intoxication or failure to synthesize downstream essential nutrients [2,3]. In many of these diseases, the underlying cause of acute or chronic clinical symptoms are single gene mutations. In acute cases, patients may present severe symptoms, such as liver failure or thromboembolic complications, as early as the newborn period. Whereas chronic cases are associated with failure to thrive and developmental delays. Genetic mutations associated with metabolic liver diseases are typically inherited as autosomal recessive, but also occur through X-linked or co-dominant inheritance modes.
The liver is a critical organ for most metabolic pathways and is, thus, the target tissue for many IMDs. Replacement of the diseased liver with a liver allograft from a healthy donor in a liver transplant procedure represents the only definitive therapy available for IMDs of the liver with the majority of transplants for IMD patients occurring in the pediatric age group [4]. Liver transplantation is associated with a 5-year patient survival rate greater than 88 percent in children with IMDs; providing correction in enzymatic defects prior to presenting structural liver damage [5,6]. Liver allographs from heterozygous donors, such as obligate heterozygous parents of patients can be used for transplantation. Post-transplantation, the recipient will have the same phenotype as the donor with correction conferred by the functional gene copy and, in some cases, require additional medical therapy [7-9]. Despite the clinical benefits, liver transplantation is limited by a high mortality risk and post-transplant complications as well as side effects from life-long treatment with immunosuppressive therapy to prevent graft rejection [5,6]. Presently, there is a shortage of safe and effective therapeutic options for IMD patients.
The limitations of liver transplantation have provided the impetus to develop novel liver-based gene therapies for metabolic diseases. Gene therapy and the emerging precision genome editing tools have shown great promise as therapeutic options for IMDs. Here we describe the potential application of gene therapy and genome-editing based approaches for curing IMDs of the liver. This review focuses on select metabolic liver disorders investigated in preclinical and clinical gene therapy studies, and describes technical and clinical challenges as well as potential future directions for advancing a novel therapy for metabolic liver disease using each approach discussed.
Conventional Gene Therapy for Metabolic Liver Disease
The premise of gene therapy is to transfer and activate the forced expression of the functional copy of the aberrant gene within clinically relevant cells from the patient using viral vectors. For IMDs of the liver, the goal of gene therapy is to obtain high levels of therapeutic gene transfer and expression in the patient’s hepatocytes or in extrahepatic tissues, correcting the disease phenotype as an autologous approach. In the proceeding paragraphs, we provide a critical assessment of conventional gene therapy studies for the treatment of metabolic liver disorders while elaborating on critical limitations of gene therapy that impede its clinical application.
Familial Hypercholesterolemia
One type of inborn error of hepatocellular metabolism is familial hypercholesterolemia (FH), a life threatening autosomal co-dominant disorder. According to recent epidemiological data, heterozygous FH occurs in 1 in 200 people, and up to 1 in 160,000 people have homozygous FH [10-12]. The most common form of FH is caused by the inheritance of one or two copies of LDL receptor (LDLR) null mutant genes, resulting in up to 10 times higher LDL cholesterol (LDL-C) levels in the blood and 100 times enhanced risk for early atherosclerotic cardiovascular disease, mainly coronary artery and aortic valve disease, compared to the general population and death in the 20s when left untreated [10,11]. While patients homozygous for FH generally have the severe form of the disease, the severity also depends on the mutation-associated residual LDLR activity. Lifelong lipid lowering therapies, such as statins, are the standard treatment for patients with homozygous FH starting in early childhood. Although providing enhanced survival, lipid lowering therapy fails to achieve LDL-C target levels and is further limited by tolerability and efficacy issues [11,13]. LDL apheresis, a procedure involving plasma removal of LDL-C, is the standard treatment for homozygous FH patients with low tolerance for drug therapy, but levels of LDL-C rebound within days after treatment, necessitating weekly administration [14].
Homozygous FH was the first metabolic liver disorder treated in a gene therapy clinical trial, specifically using the gene therapy approach developed by Grossman et al., involving retroviral transduction of wild-type LDLR into the patient’s hepatocytes ex vivo [15]. The approach also involved the transplantation of transduced hepatocytes back into the patient. Although successful in a single patient study [16], the pilot clinical trial showed patient-specific variable levels of reduced serum LDL as well as failure to achieve normalized LDL-C levels and improve hypercholesterolemia likely due to low gene transfer and repopulation efficiency by transduced hepatocytes [15]. Compared to other viral vector systems, including retroviral vectors, the adeno-associated virus (AAV) is safer for gene therapy because they exist mainly as extrachromosomal DNA within the transduced cell with low levels of integration [17]. Recent preclinical studies using LDLR expressing AAV8 (serotype 8) vectors, selected for its strong hepatocyte tropism, has shown promising results in correcting cholesterol levels in vivo using humanized FH mouse models [18,19]. These studies showed significantly reduced total and non-high density lipoprotein plasma cholesterol levels within seven days of vector administration that was sustained for six months along with regression of atherosclerosis. The major limitation in these studies was the dose dependent long-term correction of FH, such that the most significant total cholesterol level reduction was observed for the highest vector dose. Additional limitations include dose dependent gene transfer and transgene expression in hepatocytes as well as unclear long-term safety of the vector doses required for metabolic response [18,19]. A phase 1/2 clinical trial involving 12 patients is currently ongoing to evaluate the safety of the AAV8 LDLR vector and the efficacy of the gene therapy (NCT02651675).
Hemophilia
Hemophilia is a debilitating X-linked recessive blood clotting disorder characterized by life threatening and spontaneous bleeding episodes and delayed blood clotting. Hemophilia A is caused by inherited deficiency of clotting factor VIII (FVIII) and occurs in 1 in 5,000 to 10,000 male births [20]. In hemophilia B, the molecular basis is the deficiency or loss of factor IX (FIX) and it has an incidence of 1 in 30,000 males globally [21]. The standard treatment for hemophilia is intravenous injection of recombinant FVIII or FIX. Because small increases in FIX expression levels ameliorates the disease phenotype, hemophilia B is a candidate for gene therapy. A recent gene therapy study for severe hemophilia B using AAV8 vectors showed promising results in a human trial (NCT00979238). After one year of therapy, 70 percent of participants discontinued intravenous recombinant FIX replacement therapy, whereas the frequency of recombinant therapy was reduced in 30 percent of patients [22]. While gene therapy has shown promise as a treatment for hemophilia type B, type A is more challenging to treat using this approach due to high risks of immunogenicity, a larger F8 transgene sequence that is difficult to package into a viral vector, and low protein expression levels following viral transduction [23]. FVIII stimulates the generation of neutralizing anti-FVIII antibody inhibitors and drug resistance in 25 to 30 percent of patients, underscoring the major barrier that immunogenicity presents to the advancement of a gene therapy for hemophilia A [23]. To address the issue caused by the large F8 transgene size and facilitate its packaging into AAV8 vectors, the FVIII B domain, not critical to the FVIII clotting function, was removed. The administration of AAV8 vector containing the human B domain deleted F8 resulted in improved therapy without induction of cellular stress response-mediated toxicity in hepatocytes at low vector doses in hemophilia A mice [23,24]. Although these pre-clinical results support the clinical advancement of AAV8-mediated delivery of FVIII for hemophilia A (NCT02576795), the safety of long-term FVIII expression should be further evaluated in larger animal models and clinical setting.
Ornithine Transcarbamylase Deficiency
Ornithine transcarbamylase (OTC) deficiency is an X-linked partially dominant urea cycle disorder with a prevalence of 1 in 14,000 [25,26]. Patients with OTC deficiency, predominantly hemizygous males, inherit one mutant OTC allele resulting in truncated or misshaped OTC enzymes with suboptimal functionality [27]. The OTC enzyme catalyzes reactions within the liver that convert nitrogen, generated from protein metabolism, to urea for excretion by the kidneys. The absence of functional OTC enzyme leads to ammonia accumulation within the body and a wide array of symptoms, including neurological abnormalities, progressive liver damage, abnormal lung function and body temperature, and seizures [28]. The treatment for the disorder involves dietary restrictions and drug therapy. In severe cases, liver transplantation is required. Recent advancements in gene therapy offer new hope for an alternative curative strategy for OTC deficiency. In the preclinical study by Wang et al., recombinant AAV2/8 vectors containing human OTC cDNA were administered into spfash mouse model and resulted in nearly a 100-fold increase in OTC expression comparable to healthy wild-type mice. In addition, the AAV-mediated delivery provided a 50 to 70 percent transduction efficiency and significantly reduced the OTC deficiency biomarker, urinary orotic aciduria [29]. Similar results were observed in a follow-up study using an OTC knock-out mouse model. Mice treated with AAV2/8 hOTC vectors showed correction of OTC deficiency with significant reduction in liver damage compared to controls [30]. Although these preclinical studies show promising results, clinical studies are needed to thoroughly evaluate the efficacy and safety, particularly immunogenicity, using these recently developed AAV8 vectors. Recently, a Phase 1/2 clinical study was initiated to assess the safety of AAV8-mediated gene transfer of OTC in adults with late-onset OTC deficiency (NCT02991144). In the first gene therapy clinical trial for OTC deficiency initiated in the late 1990s, the administration of adenovirus type 5 vector containing the human OTC cDNA resulted in a fatal inflammatory response [31,32]. This occurrence highlights the issues associated with transitioning from preclinical to clinical trials when advancing gene therapy as well as the risks associated with viral mediated gene transfer.
Hereditary Tyrosinemia Type 1
Hereditary tyrosinemia type 1 (HT1) is an autosomal recessive disorder characterized by severe liver and renal dysfunction and occurs in 1 in 100,000 individuals globally [33,34]. HT1 is caused by mutations in the fumarylacetoacetate hydrolase (FAH) gene resulting in non-functional FAH enzyme involved in the final step of the tyrosine catabolism pathway. The absence of FAH activity leads to the accumulation of toxic metabolites, oxidative damage in the liver that progresses to liver failure and hepatocarcinoma, injury in the kidneys, and neurological crisis as early as neonatal age [33]. The standard treatment for HT1 consists of protein-restricted diet with the drug 2-(2-nitro-4- trifluoromethylbenzoyl)-1,3 cyclohexane dione (NTBC). NTBC blocks the accumulation of toxic metabolites by inhibiting the 4-OH phenylpyruvate dioxygenase (HPD) enzyme upstream of FAH. Liver transplantation is the only curative therapy available for HT1 [35].
Preclinical studies for viral-mediated gene transfer for correction of HT1 have been completed over the past two decades. The study by Paulk et al. evaluated the use of AAV2 and 8 vector systems for targeted integration of wild-type murine Fah via homologous recombination in neonatal and adult FAH deficient mice. The results from the study showed correction efficiencies up to 0.1 percent for the AAV8 vector. Withdrawal of NTBC following administration of viral vectors and partial hepatectomy enabled the small population of repaired hepatocytes in AAV treated mice to have a selective proliferative advantage in the regenerating liver, resulting in FAH+ hepatocytes consisting of greater than 50 percent of the liver accompanied by correction of the disease phenotype [36]. In the preclinical study by Wang et al., AAV2 and 8 vector systems containing human FAH flanked by homology arms for 28S ribosomal repeat DNA recued FAH deficient mice at 10 to 30 times lower vector doses and higher transgene integration compared to FAH vectors without homologous sequences [37]. The major weakness of these studies is that low levels of integration events (2 to 4 percent in [37]) attributed to homologous recombination were observed, which indicates that non-specific vector integration is responsible for the stable integration of the transgene. Further studies are needed to characterize the non-specific integration sites. Ex vivo liver-directed gene therapy was also demonstrated in FAH deficient animal models [38]. With these studies in mind, gene therapy for repair of the FAH has been effective in animal models, mainly because corrected cells in HT1 have a natural competitive advantage for the repopulation of the liver.
Alpha-1 Antitrypsin Deficiency
Alpha-1 antitrypsin (AAT) deficiency is an autosomal co-dominant disorder occurring in 1 in 3,500 live births and is characterized by low or absence of AAT serum levels. AAT is a serine protease inhibitor, encoded by SERPINA1, primarily expressed and secreted by hepatocytes into the plasma where it protects against local connective tissue degradation by neutrophil elastase as well as other proteases [39]. Patients with AAT deficiency have a single point mutation (Glu34Lys; Z allele) in the SERPINA1 resulting in AAT protein polymerization in hepatocytes, manifesting as pediatric liver disease, including cirrhosis and hepatocellular carcinoma, and in adults as pulmonary disease [40]. Available therapies for AAT deficiency consists of weekly intravenous injections of plasma AAT, and lung augmentation and transplantation for severe cases of lung disease as well as liver transplantation [40,41]. After showing robust and persistent AAT serum levels in mouse [42] and non-human primate models [43], clinical trials evaluating the safety and efficacy of AAV2 (Phase I, NCT00377416) and AAV1 (Phase II, NCT01054339 and NCT00430768) vectors for intramuscular transduction of AAT transgene in AAT deficient patients were conducted. Overall, the clinical studies showed dose dependent, transiently detectable AAT levels persisting only with the highest vector doses [44-46]. The most robust transgene expression measured was only 3 percent of the therapeutic target level, and persisted for up to one year after vector injection, with the highest AAV2 vector dose [47]. Additional weaknesses of these studies are that they involved multiple intramuscular injections for transgene delivery; recipients showed rapid development and persistence of capsid specific T-cells and neutralizing antibodies in the peripheral blood, although without apparent toxicity; and target AAT expression levels were not met [44-47]. The activated immune response was suggested to be responsible for the limited efficacy in these studies. Additional serotypes of AAV vectors along with different delivery strategies were investigated. The preclinical study by Chiuchiolo et al. showed promising results obtained with the nonhuman primate-derived AAVrh.10 serotype vector administered intrapleurally [48], leading to the recent Phase 1/2 study in humans (NTC02168686).
Challenges with Conventional Gene Therapy
AAV vectors have been applied for gene therapy strategies as a safer alternative compared to retroviral and adenoviral vectors that were linked to severe adverse events during early clinical gene therapy trials [49-51]. Although many of the recent preclinical and clinical studies using AAV vectors show promising results, the efficacy and safety of these vectors is still a major concern. Insertional mutagenesis by AAVs leading to a significant increase in hepatocellular carcinoma in neonatal mice has been observed [52-54]. Further, the study by Nault et al. observed clonal AAV insertions in cancer driver genes in 11 cases of hepatocellular carcinomas, suggesting potential oncogenicity of AAV vectors [55]. An additional issue with gene therapy concerns its pediatric application: as cells in developing livers or extrahepatic tissues proliferate following viral transduction, the non-integrated AAV vectors can become diluted out [56], limiting the efficacy of the therapy. Further, AAVs have associated immunogenicity issues as 38 percent of the western population have anti-AAV neutralizing antibodies, and thus would be precluded from gene therapy that use these vectors [57]. As observed in many of the preclinical studies discussed above, the development of anti-AAV antibodies results in the clearance of transduced cells containing the transgene and, thus necessitate potentially unsafe high viral doses.
Nuclease-Mediated Correction of Metabolic Liver Disease
Genome Editing Tools
Genome editing tools are tailorable nucleases with unprecedented potential to advance the goals of a gene therapy for IMDs. Unlike conventional gene therapy that only activates the expression of a recombinant transgene, therapeutic genome editing provides versatile strategies to precisely modify genes to correct an IMD-related deficiency while simultaneously engineering target cells to have a selective advantage for proliferation. The basis for genetic manipulation using nucleases is the activation of a double-stranded break (DSB) at a precise location in the genome for repair by two main pathways: non-homologous end joining (NHEJ) and homology directed repair (HDR). The error prone NHEJ pathway can be leveraged to disrupt a gene and knockdown its expression (Figure 1). Whereas the HDR pathway involves high fidelity repair of broken ends using endogenous or exogenous template sequences. By delivering a homologous donor template, either as a plasmid or short single-stranded DNA (ssDNA), along with a nuclease, the HDR pathway activates precise, site-specific gene correction or insertion (Figure 1).
Figure 1.
Gene modification outcomes using genome editing tools. After the nuclease has located the target sequence it induces a DSB. The DSB is resolved by endogenous DNA repair machinery via the HDR or NHEJ pathway. NHEJ-mediated repair (shown on the left) involves random insertion and deletion of base pairs at the break site and results in gene disruption. Alternatively, the HDR-mediated repair in the presence of a donor template DNA, either a short ssDNA (right) or long plasmid (center), results in a desired sequence to be incorporated for gene correction or insertion of a new gene.
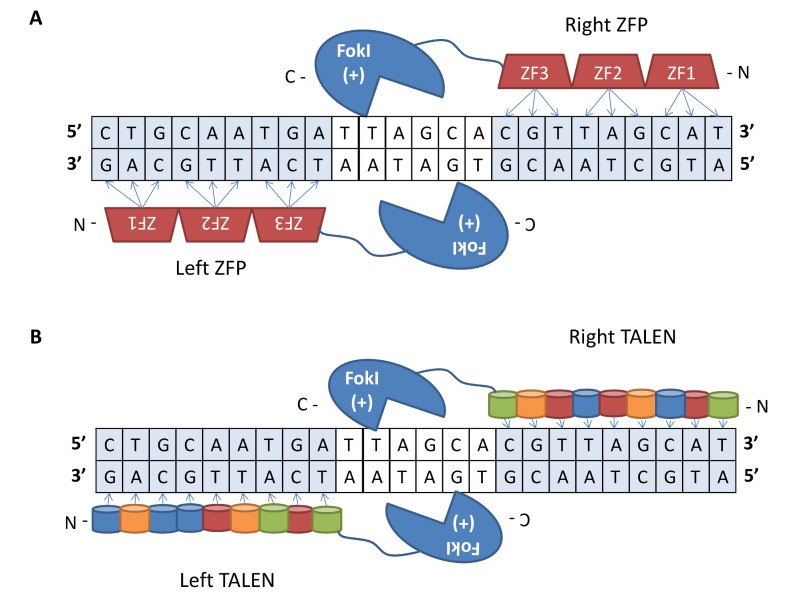
The first generation of genome editing tools are protein-based nucleases, including zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). ZFNs are hybrid proteins consisting of a tandem array of 3 to 6 Cys2-His2 zinc finger protein (ZFP) DNA-binding domains, each recognizing 3 base pairs in DNA, fused to a FokI restriction enzyme nuclease domain. A pair of ZFNs binding to neighboring target sequences in an inverted orientation separated by a spacer region to enable FokI dimerization results in the generation of a DSB at a precise location in the genome [58] (Figure 2A). TALENs consist of modular TALE DNA binding domains, each recognizing a specific nucleotide, fused to FokI. Similar to ZFNs, the generation of a DSB requires a TALEN pair, each binding to adjacent DNA segments separated by spacing for FokI dimerization [59] (Figure 2B). These tools are limited by complicated design and assembly procedures as well as narrow targeting ranges particularly for ZFNs [60].
Figure 2.
Protein-based programmable nucleases. (A) A ZFN pair consisting of a tandem array of ZFP domains, each binding to 3 nucleotides, fused to a FokI nuclease bound to adjacent DNA segments in an inverted orientation. Dimerization between the FokI domains activates a DSB at a location specified by the ZFP binding domains. (B) A TALEN pair bound to effector elements in the genome in a tail to tail orientation. Each TALEN consists of TALE repeat domains, each recognizing a single nucleotide, fused to a FokI nuclease domain. The effector domains from a TALEN pair bind to adjacent effector elements in a tail to tail orientation with optimized spacing. A DSB is induced by dimerized FokI domains upon binding of the TALE domains to the DNA target sites.
Recently, we have seen the advent of the second generation of programmable genome editing tools known as RNA-guided nucleases for genome editing in a wide variety of organisms [61-63]. This category of tools consists of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system from the bacterial adaptive immune system. In this system, the Cas9 nuclease from the type II CRISPR system in Streptococcus pyogenes is directed to a 20-nucleotide target sequence by a 100-nucleotide engineered single-guide RNA (sgRNA) that base pairs to the target site through Watson-Crick base-pairing rules. The Cas9 protein generates a DSB at the complementary target site specified by the sgRNA immediately 5’ of the NGG protospacer adjacent motif (PAM) sequence [64,65] (Figure 3A). The CRISPR-Cas9 system is an immensely powerful and versatile tool because of its facile design, such that changing the 20-nucleotide targeting sgRNA sequence redirects the system to a desired target; it is amenable to multiplex gene targeting; has a broad targeting range; and robust nuclease activity. The CRISPR-Cas9 system’s main disadvantage is it tolerates mismatches between the sgRNA target domain and gene sequences that facilitates high off-target activity and unwanted gene modifications [66,67]. Because of the high off-target activity, thorough screening of CRISPR activity at potential off-target sites is required during the design stage of a given sgRNA. This challenge inspired the characterization and development of new CRISPR-Cas systems and Cas9 orthologs. For example, a recently characterized RNA-guided nuclease is the putative type V CRISPR-Cas system, consisting of a Cas protein named Cpf1 and 42 nucleotide single-guide CRISPR RNA (crRNA). The Cpf1 nuclease from Acidaminococcus sp. BV3L6 and Lachnospiraceae bacterium ND2006 activates DSBs at complementary 23 nucleotide target sites 3’ of the TTTN PAM sequence [68] (Figure 3B). The CRISPR-Cpf1 system has been shown to provide higher specificity compared to the Cas9 system, although at the price of lower on-target activity [69]. The hybrid of the catalytically inactive dCas9 and FokI (fCas9) was recently developed to enhance the DNA cleavage specificity of the CRISPR system. In this system, a DSB is activated by the dimerization of two distinct fCas9 domains directed to adjacent target sites by two sgRNAs binding 15 or 25 bp apart [70] (Figure 3C). In the following paragraphs we will explore the development of therapeutic strategies involving genome editing tools for IMDs of the liver as summarized in Figure 4.
Figure 3.
RNA-guided nucleases. (A) An engineered 100 nucleotide sgRNA directs the Cas9 protein to a specific 20 nucleotide target sequence, which is found adjacent to the 5’ end of the PAM sequence. The 20 nucleotides base-pair with the target strand, which correctly positions the RuvC and HNH nuclease domains to generate a DSB at the complementary target site. (B) An engineered 42 nucleotide single-guide crRNA directs the Cpf1 protein to a specific 23 nucleotide target sequence, which is located adjacent to the 3’ end of the PAM sequence. The crRNA can then base-pair with the target strand, thereby positioning the two RuvC nuclease domains to generate a DSB. The RuvC domain on the target strand will generate a cut ~19 bp down from the PAM sequence, while the RuvC domain on the nontarget strand will generate a cut ~23 bp down from the PAM sequence, creating a DSB that has 5’ overhangs of ~5 nucleotides. (C) A programmed sgRNA directs a hybrid catalytically inactive dCas9 protein with a FokI nuclease called (fCas9) to the 5’ end of the PAM sequence. After the target nucleotide sequence base-pairs with the target strand, dimerization with an identical but inverted fCas9 allows the FokI nucleases to generate a DSB between the two fCas9 monomers bound ~15 or 25 bp apart.
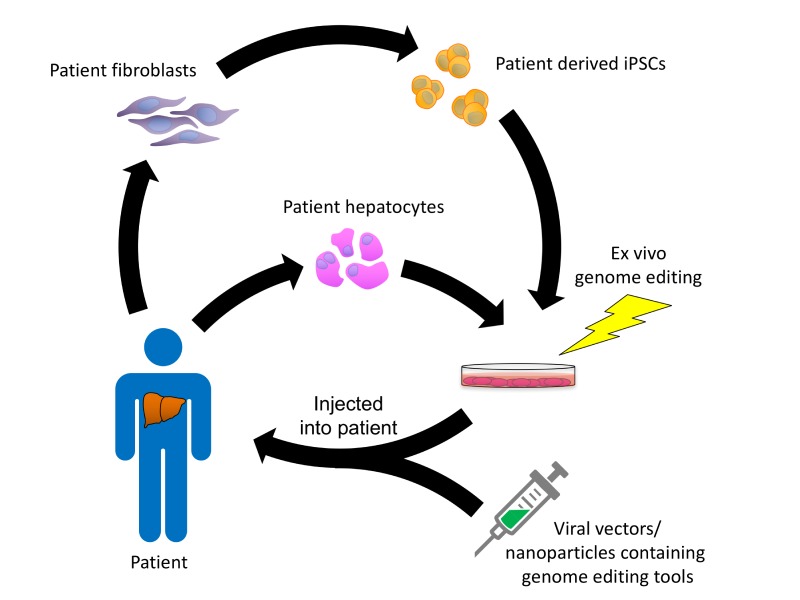
Figure 4.
Schematic of genome editing approaches for treating metabolic liver diseases. Hepatocytes are harvested from the patient and introduced with nucleases aiming the IMD target gene with or without donor template DNA ex vivo for in situ gene correction or target gene disruption. Alternatively, iPSCs derived from the patient can be used as target cells for ex vivo gene modification and subsequently differentiated into hepatocytes in vitro. The gene corrected hepatocytes or iPSC-derived hepatocyte-like cells are then perfused through the portal vein and engraft next to untreated hepatocytes in the liver. For in vivo gene targeting, viral particles or, alternatively, nanoparticles containing nucleases with or without donor template DNA are injected into the patient and are endocytosed by hepatocytes in the liver. Once in the liver, the gene editing tools mediate correction of the IMD in the patient’s cells in vivo.
Nuclease-mediated Gene Correction of IMD Mutations
One potential therapeutic strategy for IMDs involves the activation of HDR-mediated gene correction using a programmable nuclease and a donor template DNA containing a therapeutic transgene sequence. The nuclease, designed to target the mutant gene, facilitates HDR-mediated replacement of the mutant sequence in the patient’s genome with the therapeutic gene sequence from the donor template leading to in situ correction. The advantage of in situ correction is that it enables the expression of the corrected gene to remain under the control of endogenous regulatory elements. Alternatively, nucleases can be programmed to target a safe harbor site for HDR-mediated gene insertion of the therapeutic transgene with or without promoter elements from the donor template. The hallmark of both HDR-mediated gene modification approaches is that they involve precise, site-directed gene alterations, which provides improved safety compared to conventional gene therapy that has associated risks of random gene insertion.
An early study by Li et al. demonstrated the feasibility of using ZFNs and donor template DNA to induce HDR-mediated gene correction in vivo in a mouse model of hemophilia B [71]. The donor template DNA constructed with wild-type hF9 exons 2-8, spanning the site with 95 percent of F9 mutations, was delivered along with ZFNs using AAV8 vectors injected by intra-peritoneal injection into 2-day old neonatal mice. In this study, up to 3 percent HDR was observed in the extracted liver DNA accompanied by HDR-dependent plasma hFIX levels up to 7 percent of normal by 10 weeks of life. The HDR efficiency was sufficient to correct the coagulation time in the hemophilia B mice treated with ZFNs and donor template vectors. ZFN cleavage activity was observed in one off-target site at 1/10th the on-target activity. Further, compared to the conventional gene transfer strategy, genome editing provided higher and more persistent hFIX levels, whereas hFIX levels reduced dramatically with the episomal hF9 vector [71]. The loss of plasma hFIX levels after conventional viral mediated-gene transfer supports previous observations of AAV vectors, primarily existing as extra-chromosomal DNA, becoming lost in developing livers after a single vector administration [56]. In a separate study, AAV-mediated delivery of ZFNs and donor template vector was demonstrated in adult mice, resulting in long-term hFIX levels averaging 23 percent of normal and corrected clotting times [72], thus, providing evidence that in vivo HDR-mediated gene targeting is feasible at clinically meaningful levels in quiescent adult livers. As mentioned above, safe harbor sites represent an alternative targeting region for therapeutic transgene insertion by HDR. In the study by Sharma et al. AAV8 vectors containing ZFNs aiming the murine albumin gene and donor template encoding a promoter-free cassette of hF9 exons 2-8 injected into adult mice resulted in stable, corrective hFIX levels in the blood [73]. This study showed that intron 1 of the albumin gene is ideal as a safe harbor site because it provides high levels of functional transgene expression from an endogenous liver specific promoter with the use of a single ZFN design, providing the foundation for a versatile therapeutic strategy applicable to other IMDs of the liver.
In situ gene correction of liver-based IMDs has been demonstrated using the CRISPR-Cas9 system and a variety of delivery strategies. In the study by Guan et al., CRISPR-Cas9 nucleases were used to develop a mouse model of hemophilia B recapitulating the genotype and severe disease phenotype observed in a family carrying a novel hF9 mutation [74]. Hydrodynamic tail vein injection of naked DNA encoding for CRISPR-Cas9 targeting the hF9 mutation site (in exon 8) along with donor template DNA, either short ssDNA or long plasmid DNA, in the patient-specific hemophilia mouse model resulted in 0.56 percent HDR, which was sufficient to normalize the coagulation times and dramatically enhance survival in a tail-clip challenge [74]. Although injection of naked DNA is not clinically feasible, the study reaffirms that modest levels of gene targeting is sufficient to ameliorate the hemophilia B disease phenotype.
The study by Yin et al. demonstrates the application of CRISPR-Cas9 nucleases for correction of the Fah mutation in a mouse model of HT1 [75]. Hydrodynamic injection of plasmids encoding for the Cas9 system and corrective short ssDNA donor template resulted in robust enrichment of gene corrected hepatocytes, initially only 0.4 percent and eventually increased to 33.3 percent of hepatocytes, due to NTBC withdrawal, enabling rescue from liver damage [75]. In a follow up study, Yin et al. used a dual delivery system involving delivery of Cas9 mRNA by lipid nanoparticles and sgRNA along with donor template by AAV2/8 vectors to achieve 6 percent in vivo gene correction of the Fah mutation in hepatocytes accompanied by correction of the HT1 disease phenotype in mice [76]. In contrast to other IMD disease models, gene correction of mutant Fah in HT1 provides hepatocytes with a competitive advantage over unmodified diseased cells, resulting in an enrichment of gene corrected cells in the liver, particularly when combined with cycles of NTBC withdrawal.
OTC deficiency is another disease model corrected using Cas9 nucleases in preclinical studies. The delivery of AAV8 vectors containing the Cas9 system and donor template in neonatal OTC mice resulted in 10 percent gene correction in hepatocytes along with a 40 percent reduction in blood ammonia levels as well as enhanced survival on a high protein diet [77]. In contrast to neonates, treated adult OTC mice had lower levels of gene correction (1.7 percent compared to 10 percent in neonates), which was insufficient to rescue OTC adults from severe ureagenesis likely attributed to large Cas9-induced indels detected only in the adult mice [77]. The study results suggest possible differences in DNA repair response between neonatal and adult hepatocytes. In reviewing the studies discussed so far, we learn that the low rate of in situ gene correction achieved in hepatocytes, particularly in adult cells, poses as a major challenge in advancing nucleases-mediated gene correction for clinical applications.
The low frequency of HDR can be addressed using donor templates containing selection cassettes for gene targeting in induced pluripotent stem cells (iPSCs). iPSCs are an attractive target cell for therapeutic genome editing of IMDs of the liver because they represent an unlimited source of human cells having the capacity to differentiate into hepatocyte-like cells. The seminal study by Yusa et al. was the first to provide proof-of-principle application of genome editing tools in human iPSCs for gene correction of an IMD [78]. In this study, ZFNs targeting the mutant z-allele and donor vector containing a puromycin cassette flanked by piggyBac repeats were transfected into iPSCs derived from patients with AAT deficiency. Following drug selection and removal of the selection cassette using piggyBac transposase, the iPSC lines showing correction in both z-alleles and unaltered genomes were differentiated into functional hepatocyte-like cells, capable of secreting normal enzymatically active monomeric AAT in vitro. Further, the corrected iPSC-derived hepatocyte-like cells were capable of integrating into the liver parenchyma in mice, providing strong evidence of in vivo functionality [78]. Correction of the z-allele in iPSC-derived hepatocyte-like cells from AAT deficient patients was also demonstrated using CRISPR-Cas9 nucleases [79]. Both studies showcase the potential to combine genome editing tools with iPSC technology as part of a cell-based therapeutic strategy.
Nuclease-induced Gene Disruption of IMD Target Genes
In mammalian cells, particularly quiescent hepatocytes in adult livers, the DNA repair pathway choice favors NHEJ over the HDR pathway, thus, making gene disruption a more likely genome editing outcome compared to in situ gene correction. An alternative strategy for the treatment of IMDs involves nuclease-induced loss of function mutations in a therapeutic target gene. The study by Ding et al. represents one of the first preclinical studies demonstrating this strategy [80]. In this study, adenoviral vectors containing the CRISPR-Cas9 system targeting the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene, an LDLR antagonist and therapeutic target for FH, injected into mice resulted in up to 50 percent cleavage activity in the PCSK9 without detectable off-target effects in hepatocytes accompanied by 35 to 40 percent lower plasma cholesterol compared to controls [80]. A similar study by Wang et al. demonstrated high levels of CRISPR-Cas9 cleavage efficiency and knockdown of the PCSK9 in human hepatocytes in vivo in a mouse model with chimeric humanized liver [81]. This study was critical because it provided a paradigm for preclinical evaluation of CRISPR-Cas9 systems targeting therapeutic genes in the human genome. A major weakness of these studies is that they involved the administration of adenoviral vectors, selected for testing because they provide high efficiency of delivery in mouse and human hepatocytes. Although not observed in [80,81], other studies have observed evidence of hepatic injury due to adenoviral vector-induced immunogenicity with high vector doses [74,82], indicating that safety of adenoviral vectors is a major issue. The AAV vector delivery system, selected because it is less immunogenic compared to other vectors, was used to confirm high levels of CRISPR-Cas9 disruption of PCSK9 results in dramatically reduced serum PCSK9 and total cholesterol levels in mice [83]. In this study, the smaller Cas9 ortholog from Staphylococcus aureus enabled packaging of the CRISPR-Cas9 system into a single AAV delivery vector to mediate in vivo genome editing with high specificity [83]. Recently, lipid-like nanoparticles were used to deliver PCSK9-aiming sgRNA and Cas9 mRNA into hepatocytes in vivo resulting in a 50 percent reduction in PCSK9 levels following two injections of nanoparticles [84]. Although this study represents the first to use a non-viral system to deliver the entire CRISPR-Cas9 system into hepatocytes in vivo, it did not investigate whether the efficiency of PSCK9 disruption was sufficient to correct an FH disease model. Further, this study did not evaluate whether the nanoparticle dose influenced the off-target cleavage activity. An additional therapeutic target proposed for FH is the Apoliporotein B (APOB) gene encoding the structural component in LDL particles. Jarrett et al. demonstrated that the co-injection of AAV-CRISPR vectors targeting Apob and Ldlr in miceresulted in an 82 percent reduction in plasma cholesterol and protection from atherosclerotic lesions in the aortae compared to FH disease mice treated with AAV-CRISPR-Ldlr vectors [85]. Interestingly, the concomitant disruption of Apob resulted in severe hepatic fat accumulation, which calls into question the safety of this strategy. Additional IMDs corrected in the literature using nucleases for NHEJ-mediated gene modification include hemophilia A [86] and HT1 [87].
Conclusions and Outlook
Genome editing tools have the potential to establish promising applications of gene therapy for IMDs of the liver while providing novel curative options (Figure 4). IMD correction could occur by several different approaches using programmable nucleases in a precise and site-specific manner, including in situ gene correction, insertion of a wild-type transgene, knockdown of a therapeutic target gene, or gene inversion in the patient’s liver or other extrahepatic organs in vivo. An alternative approach is ex vivo genome editing, particularly within the context of hepatocyte transplantation, involving isolation and gene targeting in the patient’s hepatocytes for subsequent infusion back into the patient for the repopulation of the liver by corrected cells. However, hepatocyte transplantation is experimental and requires further advancements prior to its clinical application [88]. Patient-derived iPSCs is a more attractive target cell compared to terminally differentiated hepatocytes because of their unmatched proliferative potential and capacity to differentiate into hepatocyte-like cells; thus, representing an exciting area warranting continued investigation for cell therapies and the generation of patient-specific disease models [78,89,90]. In comparison to conventional gene therapy, genome editing has the advantage of providing permanent and site-specific correction in the patient’s genome. AAVs will dilute out in a repopulating liver [56,71], requiring multiple infusions of viral particles and higher safety risks. The application of integrative viral vectors, such as retroviral vectors, greatly enhances insertional mutagenesis risks [49,91]. Further, genome editing provides the option for multiplexing, unmatched by conventional gene therapy, enabling new possibilities for engineering target cells, such that hepatocytes can be engineered to simultaneously correct the disease phenotype and have a competitive advantage for liver repopulation in a one-step process. For example, HPDknockdown has been shown to enhance liver repopulation by gene edited cells in the presence of drug inhibitors targeting the FAH enzyme, providing a system for the selection of gene edited hepatocytes [92]. Although nuclease-mediated gene therapy for IMDs is promising, there are several barriers to overcome for its clinical translation. Firstly, safe and efficient methods for delivery of functional nucleases and donor templates into target cells are lacking. Many preclinical studies performed to date have used viral vectors as well as hydrodynamic tail vein injection to deliver genome editing components into the liver [71,72,74,75,80]. The delivery of naked DNA is not clinically feasible. As a viral vector showing the best outcomes in clinical trials for liver disease, AAV8 vectors are the most practical as a delivery strategy for hepatic gene editing in humans, but immunogenicity and insertional mutagenesis risks are still a major concern. Therefore, future studies should focus on new recombinant AAV variants as described in [93-96] that eliminate immunogenicity risks and enhance potency as well as hepatocyte tropism. Lentiviral vectors are an alternative to AAVs. In contrast to AAVs, lentiviral vectors are associated with an absence of pre-existing immunity in patients. However, lentiviral vectors provide substantially lower transgene expression levels and higher proinflammatory risks compared to AAV8 vectors [97]. Non-viral delivery methods, such as lipid nanoparticles could deliver gene editing tools for targeting therapeutic genes in vivo as described in [84]. Future studies are needed to evaluate whether gene editing components delivered using lipid nanoparticles can correct an IMD disease model. Lipid nanoparticles are typically used to deliver nucleic acids, but modifications in nuclease proteins as described in [98] or the nanoparticle coating enable the possibility for nanoparticle delivery of safer and more potent forms of nucleases, such as Cas9 protein complexed with synthetic sgRNA. Further, lipid nanoparticles enable possible delivery of donor templates as asymmetrical ssDNA for high levels of HDR-mediated gene modification [99], which should be explored in future studies for therapeutic gene correction. However, developments in lipid nanoparticle modifications, delivery efficiency as well as specificity is warranted. Future studies should investigate other types of nanoparticles, including gold and polymer nanoparticles as delivery methods for genome editing components. Secondly, the optimization of genome editing tools to enhance their specificity is needed to ensure safety. To date, the CRISPR-Cas9 system is recognized as the most reliable and efficient tool available for therapeutic gene targeting applications, but it is associated with high frequency of off-target cleavage [100,101]. Recently, the class II CRISPR-Cas systems consisting of the Cpf1 nuclease has been shown to provide higher specificity compared to Cas9 nucleases, but at the cost of lower on-target activity [69,102]. Additional advantages of the Cpf1 system over Cas9 nucleases is the generation of 5’ overhangs that may be more favorable for HDR and non-HDR-mediated precise gene insertion or correction as well as shorter guide sequences for easier and cost-effective synthesis or packing into AAVs. Another CRISPR-Cas variant that addresses the off-target effects associated with the Cas9 system is the recently developed RNA-guided FokI nucleases, shown to provide high levels of gene targeting accompanied by enhanced specificity compared to the Cas9 system [103-105]. These new CRISPR variants should be tested in future preclinical studies for correction of an IMD. Thirdly, the production of humanized animal models is paramount to the clinical translation of genome editing strategies and is lacking for many IMDs of the liver. The CRISPR-Cas system enables fast and facile generation of mutation-specific disease models for IMDs that better recapitulate the human disease pathology and facilitate accurate assessment of therapeutic strategies in preclinical studies.
Glossary
- IMD
Inherited metabolic disease
- FH
familial hypercholesterolemia
- LDL
Low-Density Lipoprotein
- LDLR
LDL Receptor
- LDL-C
LDL cholesterol
- AAV
adeno-associated virus
- FVIII
factor VIII
- FIX
factor IX
- OTC
Ornithine transcarbamylase
- HT1
Hereditary tyrosinemia type 1
- FAH
fumarylacetoacetate hydrolase
- NTBC
2-(2-nitro-4- trifluoromethylbenzoyl)-1,3 cyclohexane dione
- HPD
4-OH phenylpyruvate dioxygenase
- AAT
Alpha-1 antitrypsin
- DSB
double-stranded break
- NHEJ
non-homologous end joining
- HDR
homology directed repair
- ZFN
zinc finger nuclease
- ZFP
zinc finger protein
- TALEN
transcription activator-like effector nuclease
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated protein 9
- sgRNA
single-guide RNA
- ssDNA
single-stranded DNA
- PAM
protospacer adjacent motif
- crRNA
CRISPR RNA
- iPSC
induced pluripotent stem cell
- PCSK9
proprotein convertase subtilisin/kexin type 9
- Apob
Apoliporotein B
Author Contributions
TEB and CMA contributed to writing the conventional gene therapy section. CMA and PHB prepared figures of genome editing outcomes and tools. RNC and TEB outlined the manuscript. RNC conceived the paper topics and scope, wrote multiple topics in the paper, helped to prepare figures, and supervised manuscript preparation and revisions. The NIH-funded SC INBRE Developmental Research Project (NIH 5P20GM103499-16) awarded to RNC.
References
- Pampols T. Inherited metabolic rare disease. Adv Exp Med Biol. 2010;686:397–431. [DOI] [PubMed] [Google Scholar]
- Chanprasert S, Scaglia F. Adult liver disorders caused by inborn errors of metabolism: review and update. Mol Genet Metab. 2015;114(1):1–10. [DOI] [PubMed] [Google Scholar]
- Saudubray JM, Sedel F, Walter JH. Clinical approach to treatable inborn metabolic diseases: an introduction. J Inherit Metab Dis. 2006;29(2-3):261–74. [DOI] [PubMed] [Google Scholar]
- Schilsky ML. Transplantation for inherited metabolic disorders of the liver. Transplant Proc. 2013;45(2):455–62. [DOI] [PubMed] [Google Scholar]
- Arnon R, Kerkar N, Davis MK, Anand R, Yin W, Gonzalez-Peralta RP. Liver transplantation in children with metabolic diseases: the studies of pediatric liver transplantation experience. Pediatr Transplant. 2010;14(6):796–805. [DOI] [PubMed] [Google Scholar]
- Maiorana A, Nobili V, Calandra S, Francalanci P, Bernabei S, El Hachem M, et al. Preemptive liver transplantation in a child with familial hypercholesterolemia. Pediatr Transplant. 2011;15(2):E25–9. [DOI] [PubMed] [Google Scholar]
- Kawagishi N, Satoh K, Akamatsu Y, Sekiguchi S, Ishigaki Y, Oikawa S, et al. Long-term outcome after living donor liver transplantation for two cases of homozygous familial hypercholesterolemia from a heterozygous donor. J Atheroscler Thromb. 2007;14(2):94–8. [DOI] [PubMed] [Google Scholar]
- Shirahata Y, Ohkohchi N, Kawagishi N, Syouji M, Tsukamoto S, Sekiguchi S, et al. Living-donor liver transplantation for homozygous familial hypercholesterolemia from a donor with heterozygous hypercholesterolemia. Transpl Int. 2003;16(4):276–9. [DOI] [PubMed] [Google Scholar]
- Yoshitoshi EY, Takada Y, Oike F, Sakamoto S, Ogawa K, Kanazawa H, et al. Long-term outcomes for 32 cases of Wilson’s disease after living-donor liver transplantation. Transplantation. 2009;87(2):261–7. [DOI] [PubMed] [Google Scholar]
- Santos RD, Gidding SS, Hegele RA, Cuchel MA, Barter PJ, Watts GF, et al. Defining severe familial hypercholesterolaemia and the implications for clinical management: a consensus statement from the International Atherosclerosis Society Severe Familial Hypercholesterolemia Panel. Lancet Diabetes Endocrinol. 2016;4(10):850–61. [DOI] [PubMed] [Google Scholar]
- Cuchel M, Bruckert E, Ginsberg HN, Raal FJ, Santos RD, Hegele RA, et al. Homozygous familial hypercholesterolaemia: new insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur Heart J. 2014;35(32):2146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjouke B, Kusters DM, Kindt I, Besseling J, Defesche JC, Sijbrands EJ, et al. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur Heart J. 2015;36(9):560–5. [DOI] [PubMed] [Google Scholar]
- Raal FJ, Pilcher GJ, Panz VR, van Deventer HE, Brice BC, Blom DJ, et al. Reduction in mortality in subjects with homozygous familial hypercholesterolemia associated with advances in lipid-lowering therapy. Circulation. 2011;124(20):2202–7. [DOI] [PubMed] [Google Scholar]
- Kroon AA, van’t Hof MA, Demacker PN, Stalenhoef AF. The rebound of lipoproteins after LDL-apheresis. Kinetics and estimation of mean lipoprotein levels. Atherosclerosis. 2000;152(2):519–26. [DOI] [PubMed] [Google Scholar]
- Grossman M, Rader DJ, Muller DW, Kolansky DM, Kozarsky K, Clark BJ, 3rd, et al. A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med. 1995;1(11):1148–54. [DOI] [PubMed] [Google Scholar]
- Grossman M, Raper SE, Kozarsky K, Stein EA, Engelhardt JF, Muller D, et al. Successful ex vivo gene therapy directed to liver in a patient with familial hypercholesterolaemia. Nat Genet. 1994;6(4):335–41. [DOI] [PubMed] [Google Scholar]
- Baruteau J, Waddington SN, Alexander IE, Gissen P. Gene therapy for monogenic liver diseases: clinical successes, current challenges and future prospects. J Inherit Metab Dis. 2017:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassim SH, Li H, Vandenberghe LH, Hinderer C, Bell P, Marchadier D, et al. Gene Therapy in a Humanized Mouse Model of Familial Hypercholesterolemia Leads to Marked Regression of Atherosclerosis. PLoS One. 2010;5(10):e13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassim SH, Li H, Bell P, Somanathan S, Lagor W, Jacobs F, et al. Adeno-Associated Virus Serotype 8 Gene Therapy Leads to Significant Lowering of Plasma Cholesterol Levels in Humanized Mouse Models of Homozygous and Heterozygous Familial Hypercholesterolemia. Hum Gene Ther. 2013;24(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi L, Lawler AM, Antonarakis SE, High KA, Gearhart JD, Kazazian HH., Jr Targeted disruption of the mouse factor VIII gene produces a model of haemophilia A. Nat Genet. 1995;10(1):119–21. [DOI] [PubMed] [Google Scholar]
- Thompson AR, Chen SH. Characterization of factor IX defects in hemophilia B patients. Methods Enzymol. 1993;222:143–69. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371(21):1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin I, Markusic DM, Palaschak B, Hoffman BE, Srikanthan MA, Herzog RW. Potential for cellular stress response to hepatic factor VIII expression from AAV vector. Mol Ther Methods Clin Dev. 2016;3:16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange AM, Altynova ES, Nguyen GN, Sabatino DE. Overexpression of factor VIII after AAV delivery is transiently associated with cellular stress in hemophilia A mice. Mol Ther Methods Clin Dev. 2016;3:16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusilow SW, Maestri NE. Urea cycle disorders: diagnosis, pathophysiology, and therapy. Adv Pediatr. 1996;43:127–70. [PubMed] [Google Scholar]
- Yamaguchi S, Brailey LL, Morizono H, Bale AE, Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum Mutat. 2006;27(7):626–32. [DOI] [PubMed] [Google Scholar]
- Tuchman M, Jaleel N, Morizono H, Sheehy L, Lynch MG. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum Mutat. 2002;19(2):93–107. [DOI] [PubMed] [Google Scholar]
- Maestri NE, Brusilow SW, Clissold DB, Bassett SS. Long-Term Treatment of Girls with Ornithine Transcarbamylase Deficiency. N Engl J Med. 1996;335(12):855–60. [DOI] [PubMed] [Google Scholar]
- Wang L, Morizono H, Lin J, Bell P, Jones D, McMenamin D, et al. Preclinical evaluation of a clinical candidate AAV8 vector for ornithine transcarbamylase (OTC) deficiency reveals functional enzyme from each persisting vector genome. Mol Genet Metab. 2012;105(2):203–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bell P, Morizono H, He Z, Pumbo E, Yu H, et al. AAV gene therapy corrects OTC deficiency and prevents liver fibrosis in aged OTC-knock out heterozygous mice. Mol Genet Metab. 2017;120(4):299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80(1-2):148–58. [DOI] [PubMed] [Google Scholar]
- Wilson JM. Lessons learned from the gene therapy trial for ornithine transcarbamylase deficiency. Mol Genet Metab. 2009;96(4):151–7. [DOI] [PubMed] [Google Scholar]
- Grompe M. The pathophysiology and treatment of hereditary tyrosinemia type 1. Semin Liver Dis. 2001;21(4):563–71. [DOI] [PubMed] [Google Scholar]
- Angileri F, Bergeron A, Morrow G, Lettre F, Gray G, Hutchin T, et al. Geographical and Ethnic Distribution of Mutations of the Fumarylacetoacetate Hydrolase Gene in Hereditary Tyrosinemia Type 1. JIMD Rep. 2015;19:43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dhalimy M, Overturf K, Finegold M, Grompe M. Long-term therapy with NTBC and tyrosine-restricted diet in a murine model of hereditary tyrosinemia type I. Mol Genet Metab. 2002;75(1):38–45. [DOI] [PubMed] [Google Scholar]
- Paulk NK, Wursthorn K, Wang Z, Finegold MJ, Kay MA, Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51(4):1200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lisowski L, Finegold MJ, Nakai H, Kay MA, Grompe M. AAV vectors containing rDNA homology display increased chromosomal integration and transgene persistence. Mol Ther. 2012;20(10):1902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shao Y, Li L, Tian F, Cen J, Chen X, et al. Efficient liver repopulation of transplanted hepatocyte prevents cirrhosis in a rat model of hereditary tyrosinemia type I. Sci Rep. 2016;6:31460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks KD, Tavill AS. Liver Disease in Alpha 1-Antitrypsin Deficiency: A Review. Am J Gastroenterol. 2008;103(8):2136–41. [DOI] [PubMed] [Google Scholar]
- Silverman EK, Sandhaus RA. Clinical practice. Alpha1-antitrypsin deficiency. N Engl J Med. 2009;360(26):2749–57. [DOI] [PubMed] [Google Scholar]
- Richmond RJ, Zellner KM. Alpha1-antitrypsin deficiency: incidence and implications. Dimens Crit Care Nurs. 2005;24(6):255–60. [DOI] [PubMed] [Google Scholar]
- Song S, Morgan M, Ellis T, Poirier A, Chesnut K, Wang J, et al. Sustained secretion of human alpha-1-antitrypsin from murine muscle transduced with adeno-associated virus vectors. Proc Natl Acad Sci USA. 1998;95(24):14384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Scott-Jorgensen M, Wang J, Poirier A, Crawford J, Campbell-Thompson M, et al. Intramuscular administration of recombinant adeno-associated virus 2 alpha-1 antitrypsin (rAAV-SERPINA1) vectors in a nonhuman primate model: safety and immunologic aspects. Mol Ther. 2002;6(3):329–35. [DOI] [PubMed] [Google Scholar]
- Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT, et al. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci USA. 2009;106(38):16363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantly ML, Spencer LT, Humphries M, Conlon TJ, Spencer CT, Poirier A, et al. Phase I trial of intramuscular injection of a recombinant adeno-associated virus serotype 2 alphal-antitrypsin (AAT) vector in AAT-deficient adults. Hum Gene Ther. 2006;17(12):1177–86. [DOI] [PubMed] [Google Scholar]
- Flotte TR, Trapnell BC, Humphries M, Carey B, Calcedo R, Rouhani F, et al. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing alpha1-antitrypsin: interim results. Hum Gene Ther. 2011;22(10):1239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, Chulay JD, Trapnell BC, Humphries M, Carey B, Sandhaus RA, et al. Human Treg responses allow sustained recombinant adeno-associated virus-mediated transgene expression. J Clin Invest. 2013;123(12):5310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De BP, Heguy A, Hackett NR, Ferris B, Leopold PL, Lee J, et al. High levels of persistent expression of alpha1-antitrypsin mediated by the nonhuman primate serotype rh.10 adeno-associated virus despite preexisting immunity to common human adeno-associated viruses. Mol Ther. 2006;13(1):67–76. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348(3):255–6. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffrat N, Leboulch P, et al. LMO2-associated clonal T-cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–9. [DOI] [PubMed] [Google Scholar]
- Somia N, Verma IM. Gene therapy: trials and tribulations. Nat Rev Genet. 2000;1(2):91–9. [DOI] [PubMed] [Google Scholar]
- Chandler RJ, LaFave MC, Varshney GK, Trivedi NS, Carrillo-Carrasco N, Senac JS, et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J Clin Invest. 2015;125(2):870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317(5837):477. [DOI] [PubMed] [Google Scholar]
- Wang PR, Xu M, Toffanin S, Li Y, Llovet JM, Russell DW. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc Natl Acad Sci USA. 2012;109(28):11264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47(10):1187–93. [DOI] [PubMed] [Google Scholar]
- Nakai H, Yant SR, Storm TA, Fuess S, Meuse L, Kay MA. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75(15):6969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21(6):704–12. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr Opin Biotechnol. 2012;23(5):644–50. [DOI] [PubMed] [Google Scholar]
- Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29(11):1717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013;41(20):e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–8. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima Si. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337(6096):816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013:9584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg Jonathan S, Abudayyeh Omar O, Slaymaker Ian M, Makarova Kira S, Essletzbichler P, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163(3):759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34(8):869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguela XM, Sharma R, Doyon Y, Miller JC, Li H, Haurigot V, et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122(19):3283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Anguela XM, Doyon Y, Wechsler T, DeKelver RC, Sproul S, et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126(15):1777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med. 2016;8(5):477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P, et al. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther. 2015;23(3):570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Raghavan A, Chen T, Qiao L, Zhang Y, Ding Q, et al. CRISPR-Cas9 Targeting of PCSK9 in Human Hepatocytes In Vivo-Brief Report. Arterioscler Thromb Vasc Biol. 2016;36(5):783–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Barnes MJ, Stillman IE, Libermann TA. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum Gene Ther. 1999;10(6):965–76. [DOI] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Mei M, Li B, Zhu X, Zu W, Tian Y, et al. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27(3):440–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta R, Zhang M, et al. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep. 2017;7:44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Kim J, Kweon J, Son JS, Lee JS, Yoo JE, et al. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc Natl Acad Sci USA. 2014;111(25):9253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankowicz FP, Barzi M, Legras X, Hubert L, Mi T, Tomolonis JA, et al. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun. 2016;7:12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan P. Liver transplantation and cell therapies for inborn errors of metabolism. J Inherit Metab Dis. 2013;36(4):675–80. [DOI] [PubMed] [Google Scholar]
- Lee PC, Truong B, Vega-Crespo A, Gilmore WB, Hermann K, Angarita SA, et al. Restoring Ureagenesis in Hepatocytes by CRISPR/Cas9-mediated Genomic Addition to Arginase-deficient Induced Pluripotent Stem Cells. Mol Ther Nucleic Acids. 2016;5(11):e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang Y, Zhou T, Wong LY, Tian XY, Hong X, et al. Generation of Human Liver Chimeric Mice with Hepatocytes from Familial Hypercholesterolemia Induced Pluripotent Stem Cells. Stem Cell Reports. 2017;8(3):605–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–9. [DOI] [PubMed] [Google Scholar]
- Nygaard S, Barzel A, Haft A, Major A, Finegold M, Kay MA, et al. A universal system to select gene-modified hepatocytes in vivo. Sci Transl Med. 2016;8(342):342ra79-ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530(7588):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch RC, Muth A, Muik A, Friedel T, Schmatz J, Dreier B, et al. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat Commun. 2015;6:6246. [DOI] [PubMed] [Google Scholar]
- Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506(7488):382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SR, Harris AF, Cabral DJ, Keeler AM, Sapp E, Ferreira JS, et al. Widespread Central Nervous System Gene Transfer and Silencing After Systemic Delivery of Novel AAV-AS Vector. Mol Ther. 2016;24(4):726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandendriessche T, Thorrez L, Acosta-Sanchez A, Petrus I, Wang L, Ma L, et al. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J Thromb Haemost. 2007;5(1):16–24. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–44. [DOI] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Cheong SA, Lee JG, Lee SW, Lee MS, Baek IJ, et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016; advance online publication [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015;26(7):425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlicek S, Shen Y, Alpagu Y, Bruntraeger MB, Zufir NB, Phuah ZY, et al. Re-engineered RNA-Guided FokI-Nucleases for Improved Genome Editing in Human Cells. Mol Ther. 2017;25(2):342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]