Abstract
Within the central nervous system, gene regulatory mechanisms are crucial regulators of cellular development and function, and dysregulation of these systems is commonly observed in major neuropsychiatric and neurological disorders. However, due to a lack of tools to specifically modulate the genome and epigenome in the central nervous system, many molecular and genetic mechanisms underlying cognitive function and behavior are still unknown. Although genome editing tools have been around for decades, the recent emergence of inexpensive, straightforward, and widely accessible CRISPR/Cas9 systems has led to a revolution in gene editing. The development of the catalytically dead Cas9 (dCas9) expanded this flexibility even further by acting as an anchoring system for fused effector proteins, structural scaffolds, and RNAs. Together, these advances have enabled robust, modular approaches for specific targeting and modification of the local chromatin environment at a single gene. This review highlights these advancements and how the combination of powerful modulatory tools paired with the versatility of CRISPR-Cas9-based systems offer great potential for understanding the underlying genetic and epigenetic contributions of neuronal function, behavior, and neurobiological diseases.
Keywords: CRISPR/Cas9, genome editing, neuron, epigenetics, neuroepigenetics, epigenetic editing
Introduction
The idea of editing a gene likely began with the first indication that a mutation within a gene can cause a disease. The concept is straightforward – correct a single mutation, such as the single nucleotide polymorphism (SNP) at the HBB gene observed in patients with sickle cell anemia, and a disease could be reversed. In practice, gene editing is a field that researchers have been struggling with since its inception. The discovery of gene editing systems such as zinc finger nucleases (ZFNs) [1] and transcription activator-like effector nucleases (TALENs) [2,3] provided the first demonstration of more efficient gene editing approaches, but both systems are cumbersome to use. The recent repurposing of the prokaryotic immune system clustered regularly interspaced short palindromic repeats (CRISPR) [4] for eukaryotic genome editing [5-7] has revitalized the field due to its modularity, precision, and accessibility. In addition to widespread use for gene editing, CRISPR/Cas9-based methods have expanded to recruit effector proteins and RNAs for gene activation, repression, and epigenetic reprogramming [8]. Within a few short years of being adapted for mammalian gene editing, CRISPR-related tools are now poised to enable robust and specific manipulation of the mammalian genome and epigenome.
This suite of tools to interrogate a single gene in unprecedented ways has the potential to push many areas of neuroscience towards more effective therapeutics, in part by enabling the generation of novel animal models with relevance to human disease. CRISPR-Cas9 genome editing has already decreased costs and time commitment required to generate transgenic mouse models, but it will also speed creation of transgenic species that have previously been more resilient to early gene editing techniques, such as the laboratory rat [9] or non-human primates [10]. Given that these species are frequently used as model systems for more complex behavioral and cognitive testing across neurosciences, this advance will be a major breakthrough for many subdisciplines. Moreover, the adaptation of these tools for epigenome editing has the potential to show how modulation of epigenetic marks at a single gene target can impact neuronal function and behavior [11-13]. Thus, these tools can provide novel insights into epigenetic aberrations that have been identified in numerous neuropsychiatric diseases, such as depression, schizophrenia, autism spectrum disorders, and addiction [14-21]. Taken together, these advances have generated tremendous momentum and excitement for more detailed genetic and epigenetic interrogation of the central nervous system. This review will summarize the current toolbox available to researchers interested in genetic and epigenetic editing, and highlight the benefits and pitfalls of using these techniques to understand how gene regulatory mechanisms contribute to neuronal function, behavior, and brain disease.
Evolution of the CRISPR/Cas9 Toolbox
CRISPR/Cas9
CRISPR was first discovered as an adaptive immune system in bacteria [4] and was later adapted for eukaryotic expression for gene editing [6,7,22]. There are two major components of the CRISPR-Cas9 gene editing system: the Cas9 nuclease protein and a chimeric single guide RNA (gRNA) (Figure 1a) [5,22]. The gRNA directs the Cas9 protein to its target sequence, where the DNA strands are separated and cleaved [5]. The gRNA sequence generally targets a 20bp nucleotide region immediately adjacent to a protospacer adjacent motif (PAM) sequence, which is specific to the species of Cas9 being used [5,23]. After DNA is cut, the cell attempts to repair this dsDNA break in one of two ways: homologous recombination (HR) or non-homologous end joining (NHEJ) [24]. HR requires a template and is the most common way to correct a mutation or knock-in a specific sequence. NHEJ is the cell’s emergency DNA repair system – it is an indel prone method that quickly repairs DNA without a template. This method is commonly used to induce a frameshift mutation in a gene, thus creating a functional knockout in the cell by introducing a premature stop codon. Alternatively, the Cas9 system targeted to multiple locations on a chromosome causes chromosomal rearrangements or deletions [25-27]. Overall, Cas9-based gene and chromosome editing both are widely employed methods in virtually any species to achieve specific alteration of a DNA sequence.
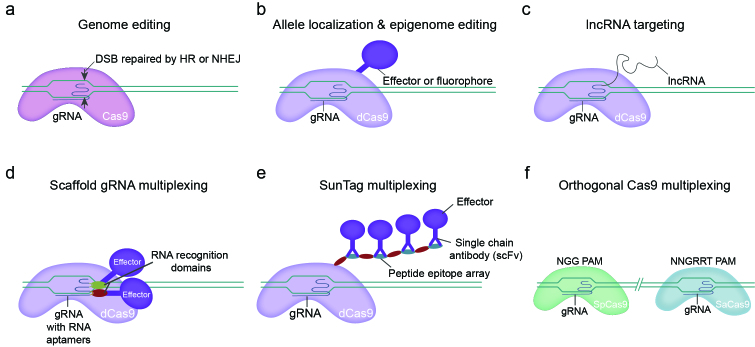
Figure 1.
CRISPR/Cas9-based gene and epigenetic editing and methods of multiplexing. a) The CRISPR gene editing system uses a guide RNA (gRNA) to recruit the Cas9 nuclease to a specific location on the genome. Cas9 induces a double stranded break (DSB) in DNA, resulting in activation of one of two DNA repair pathways—homologous recombination (HR) or non-homologous end joining (NHEJ). b) The catalytically dead Cas9 (dCas9) acts as an anchoring system for fusion proteins. Fluorescent proteins, transcriptional regulators, and epigenetic enzymes can be recruited with dCas9 in a gene-specific manner. c) The gRNA sequence tethered to a long non-coding RNA (lncRNA) permits the study of the localization and function of lncRNAs in transcriptional control. d) A gRNA fitted with RNA aptamers creates a scaffold for proteins with RNA recognition domains to bind, resulting in recruitment of effector proteins. Different aptamer/RNA recognition pairs can be used to recruit distinct modifiers either at the same locus or to coordinate complex control over many loci. e) SunTag multiplexing involves dCas9 fused with an array of peptide epitopes to achieve amplification of the recruited effector’s function. Effectors are fused with single chain antibodies (scFv) that are specific to the epitope fused to dCas9. f) Different species of Cas9 can be utilized to direct different effectors to distinct loci. The protospacer adjacent motif (PAM) sequence is species specific. For example, Streptococcus pyogenes (SpCas9) has a NGG PAM requirement, while Staphylococcus aureus (SaCas9) requires a NNGRRT PAM recognition sequence for gRNA binding.
Despite the numerous challenges of expressing CRISPR/Cas9 constructs in neurons, many labs are starting to implement these tools to explore genetic mechanisms in neuronal systems. Multiple groups have successfully utilized Cas9 to achieve gene knockout in both cultured neurons and in vivo [28-32]. Use of the Cas9 system in the adult brain is advantageous for dissecting the function of a protein via genetic knockout. Traditionally, knockout approaches have required germline deletion, which is problematic as many proteins are required for development [33]. Even if an organism survives without the protein through development, the brain may compensate in other ways, obscuring the interpretation of the results. While conditional knockout animal models can help to circumvent this limitation, the advantage of the CRISPR system is the ability to achieve protein mutation at any point in development without the need for a transgenic animal. Indeed, this approach was validated for use in wild type C57BL/6J as well as Sprague-Dawley rats, where targeting of the NMDA receptor subunit GluN1 (Grin1 locus), which is embryonic lethal in a knockout animal, resulted in a diminished NMDA/AMPA current ratio [31]. Conversely, although gene editing relying on HR-mediated DNA repair will likely be less efficient in the adult CNS as post-mitotic neurons do not readily use this method of repair [34], HR-directed gene editing with CRISPR-Cas9 remains a valuable tool in generation of animal models or stem cell therapy.
CRISPR/dCas9 and the Recruitment of Effector Proteins
Precise and straightforward gene editing provides countless benefits to biomedical research. However, the development of the dCas9 (“dead” Cas9) anchoring system [35,36] has been equally impactful. Devoid of nuclease activity, dCas9 permits tethering of fusion proteins, which, like traditional Cas9, can be recruited to virtually any spot on the genome (as illustrated in Figure 1b). Both ZFP and TALE-based approaches also provide a scaffold for the site-specific recruitment of epigenetic modifiers, but the ability to use the same gRNAs with different effectors makes the CRISPR/dCas9 system more modular and straightforward to use. This subsection outlines many of the applications that are possible with dCas9, though many recent reviews have focused solely on this topic and are suggested for further reading [8,13,37,38].
Conventional methods to modulate gene expression have generally employed less specific genetic approaches. These methods generally include overexpression vectors and knockdown technologies such as RNA interference (RNAi) [39,40]. Although these methods offer evidence for the functional role of a gene, they ignore the endogenous gene locus. Likewise, interrogation of epigenetic mechanisms over the past two decades has largely been limited to genetic manipulation of the enzymes that create these modifications or global pharmacological inhibitors. Although many epigenetic marks correlate with gene expression changes or even behavioral experience, this limitation has meant that the causal links between epigenetic modifications and gene transcription have been elusive.
Use of the dCas9 fusion protein system has paved the way to understand how local regulatory modification affects chromatin state and conformation, transcriptional machinery, and gene expression. Gene activation using dCas9 fusion proteins is generally termed CRISPR activation (CRISPRa). For example, dCas9-based targeting of transactivators that recruit RNA Polymerase II (VP64, a concatemer of the herpes simplex viral protein VP16, and p65, a subunit of transcription factor NF-κB) results in robust, and largely specific, upregulation of targeted genes [41-46]. Likewise, combination of VP64, p65, and Rta (a gammaherpesvirus transactivator), termed VPR, also induces gene expression at targeted genes, and is significantly more effective than either VP64 or p65 alone [47]. This technology has already been employed to unravel some of the key mechanisms that control neuronal fate specification and function. For example, although it is well understood that distal enhancer elements in DNA are critical for expression of associated genes [48], until recently it has not been possible to activate enhancer in a precise way to study their individual roles in transcriptional regulation. Thus, this technology provided the means for a recent breakthrough, as CRISPR/dCas9-VP64 was used to drive genomic enhancer activity in neuronal systems for the first time [49]. Specifically targeting two development-dependent enhancers associated with Grin2c genewith dCas9-VP64 resulted in upregulation of Grin2c gene expression [49], providing insight into the importance of chromatin interactions for gene expression regulation.
Other fusion proteins that result in increased transcription actually target epigenetic modifications rather than recruitment of trans-activator proteins. For example, recruitment of p300, a histone acetyltransferase and transcriptional co-activator, results in robust gene expression by inducing transcriptionally permissive chromatin [50]. Similarly, dCas9-based targeting of Tet1, an enzyme involved in DNA hydroxymethylation (and, ultimately, cytosine demethylation), induces a methylation-repressed gene [51,52] by creating a local state of hypomethylation. In cultured cortical neurons, dCas9-Tet1 recruitment to the Bdnf promoter region IV induced Bdnf mRNA, enabling the function of this gene to be isolated from induction of other key plasticity and activity-regulated genes that are often regulated in parallel [52]. Additionally, the same study demonstrated that infusion of lentiviral constructs into the adult brain unsilenced a methylated reporter sequence. Taken together, these reports establish feasibility of using these systems in vitro and in vivo to alter transcriptional and epigenetic states.
On the other end of the spectrum, dCas9 fusion proteins aimed at repressing a specific locus have also been described. Generally, these constructs recruit histone modifying enzymes to condense the chromatin environment to decrease gene expression, while others covalently modify either DNA or histones to achieve their effects. For example, Krüppel-associated box (KRAB) targeting to a locus results in recruitment of chromatin condensing epigenetic enzymes linked to transcriptional silencing [41,53]. Enhancer-targeted dCas-KRAB has been used to understand the importance of different Fos (aka c-Fos) enhancers in cortical neurons [53], providing another instance in which dissecting enhancer function was not previously possible. While most of these repressing fusion proteins have been synthesized and validated for use with ZFP or TALE-based systems, many are now being repurposed for use with dCas9 [54]. For example, the DNA methyltransferase, DNMT, can be used to methylate specific regions and generally results in repression of gene expression [52,55,56]. Moreover, the impact on animal behavior of modulating the chromatin environment of a single gene in a single brain region has previously been explored with ZFN/TALEs. Specifically, histone methylation with ZFN-G9a targeted the Fosb locus in the nucleus accumbens resulted in blunted cocaine-induced locomotor sensitization [57]. This study highlights how the chromatin environment at a single gene can influence behavior, and open the door for future exploration into the role of epigenetic modifications in adaptive behavior and brain disease.
Tagging and Isolation
Antibody-based methods are powerful techniques to quantify protein levels present in a cell/tissue or isolate components bound to a protein. However, for many proteins, the lack of antibodies compatible with immunoprecipitation approaches severely limits understanding of protein localization and protein-protein or protein-nucleic acid interaction. The catalytically active Cas9 solves this problem by cutting DNA so that an exogenous epitope, such as a FLAG or HA tag, or fluorescent protein can be inserted into to the endogenous gene. Also, this method has been utilized in the mammalian CNS via in utero electroporation to tag proteins for high-resolution imaging to determine sub-cellular localization [58,59]. Likewise, DNA-binding proteins modulate many aspects of the transcriptional and epigenetic landscape. Identification of the proteins that are bound to a certain region imply the activity of that region, and assist with demarcation and functional roles of genetic elements like promoters and enhancers. This is typically characterized with chromatin immunoprecipitation (ChIP), which relies on the use of a protein-specific antibody to pull down DNA regions bound to the protein. Until recently, isolating proteins bound to a specific region of DNA has been more challenging. Development of engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) utilizes dCas proteins fused to an epitope tag [60]. Purification of the endogenous DNA region paired with mass-spectrometry identifies proteins bound to a specific region on the genome. Together, these powerful approaches help to solve current limitations in how protein-protein interactions and protein-DNA interactions are measured, and will be useful for examining gene regulation at a single locus.
Similarly, techniques such as fluorescence in situ hybridization (FISH) have provided an understanding of gene expression and chromatin organization by fluorescently labeling loci for visualization [61,62]. However, this technique requires that the design of specific probes and cells must be fixed, so visualization of a live cell is not possible. By fusing a fluorophore such as GFP to dCas9 and employing a specific gRNA to target a DNA region of interest, CRISPR-based approaches can deliver similar information without the need for FISH probes [63,64]. Moreover, this approach allows specific loci to be monitored in real time in live cells to understand the dynamics of chromatin looping, especially with neuronal activity [65]. Multiplexing loci with different fluorescent proteins is possible by utilizing different species of dCas9 [63] or a modified gRNA construct [64], which is described in more detail within the multiplexing subsection below (also, see Figure 2). Likewise, RNA localization is achievable with RNA-FISH [62] but faces the same limitations as DNA-FISH. By providing a mixed RNA/DNA oligo (PAMmer) with the dCas-GFP tagging system, RNA can also be visualized in real time [66]. These applications have the potential to markedly accelerate our understanding of RNA biology. For example, unique Bdnf transcripts arise from numerous distinct promoters, and the resulting transcript variants are induced by specific stimuli [67]. However, the localization and expression levels of these transcripts in live cells has been difficult to examine, and ultimately the function of different transcripts has remained elusive. Using dCas9 with a PAMmer allows for live imaging of Bdnf transcripts to understand the role of transcript dynamics and trafficking. Observing not only the localization of both DNA and RNA but also the dynamics of these systems in live cells is invaluable in neuroscience as chromosome looping and RNA trafficking are both implicated in basic neuronal function and disease [68].
Figure 2.
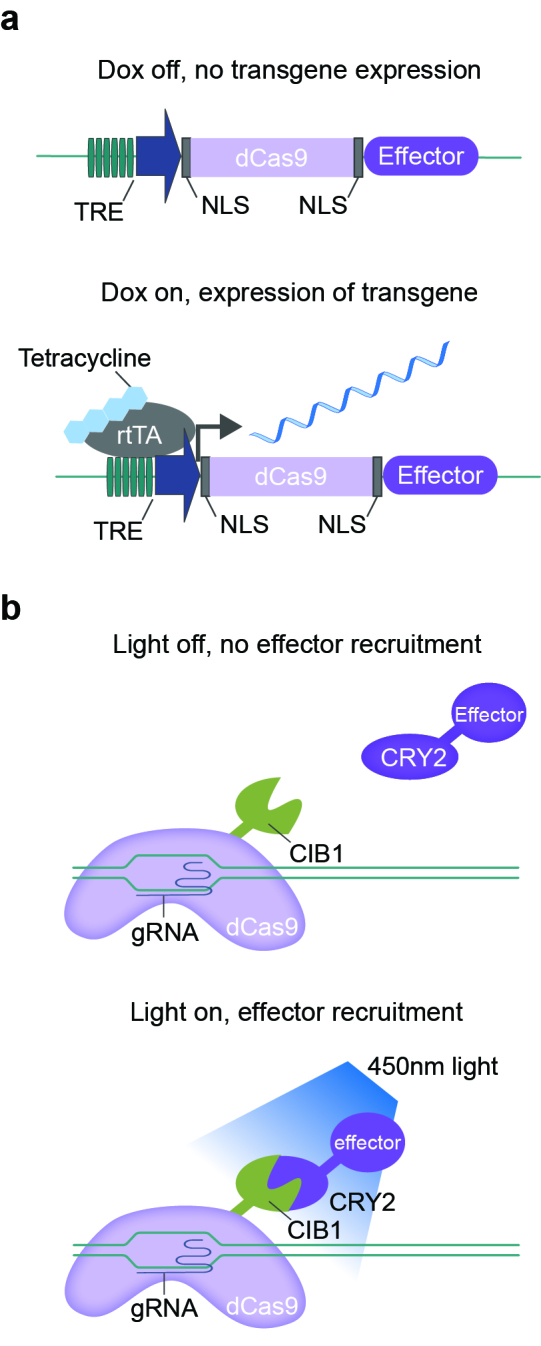
Chemical and optical methods to regulate the temporal expression of CRISPR-based techniques. a) The tetracycline-inducible system controls the expression of a transgene with administration of the common tetracycline, doxycycline (dox). The Tet-on system is illustrated here. At baseline, the dCas9-effector transgene, under the tetracycline response element (TRE), is not expressed at baseline. With dox treatment, rtTA is recruited to the TRE, which activates transgene transcription. NLS-nuclear localization sequence. b) Light induced CIB1/CRY2 binding partners regulate recruitment of an effector protein to dCas9 for temporal control. Exposure to 450nm blue light causes a confirmation change of CRY2, which is then able to bind to CIB1. When the light is removed, CRY2-CIBN interactions are reversed.
CRISPR-Display
Converging evidence from many different systems has implicated long non-coding RNAs in functional regulation of gene expression both cis and trans [69-71]. These RNA species contribute to epigenetic regulation and transcription alongside more well-known roles in imprinting, X-chromosome inactivation, and telomere function [71]. However, while lncRNA play critical roles in these processes, it has proven difficult to directly examine the importance lncRNA spatial localization due to technical limitations. Simply overexpressing a lncRNA does not guarantee that it will play the same role as an endogenous lncRNA, as many lncRNAs are thought to act through local interactions. To address this, another application of the dCas9 system is to “tether” a lncRNA of choice to the gRNA construct, allowing lncRNAs to be recruited in a spatially specific manner (Figure 1c) [72]. This approach, termed CRISPR-Display [72], has enabled experimental dissociation of sequence-based lncRNA effects from location-based lncRNA effects, and provides a powerful tool to understand the pleotropic molecular roles of lncRNA [73].
Multiplexing
Although the possibility of specifically targeting a single locus on the genome is a huge advancement to the field, multiplexing offers coordinated control over multiple loci on a single gene or concerted regulation of many different genes. Multiplexing is defined in multiple ways, either by the use of multiple guides or multiple dCas9 effector proteins. Multiplexed targeting of distinct transcriptional activators or repressors amplifies their individual effect [47,74]. In addition to providing insight into how single epigenetic modifications regulate gene expression, this technology enables more widespread epigenetic reprogramming at a single gene by multiplexing gRNAs or recruiting multiple copies of an epigenetic effector. For example, it is currently unclear whether DNA methylation can impact gene expression through methylation changes at single cytosines, or whether widespread alterations to a promoter or other genomic features are necessary to alter gene expression [75]. By enabling recruitment of DNA methylation machinery to highly specific locations or entire genomic elements, CRISPR-dCas9-based multiplexing strategies have the potential to bring much-needed resolution to this question.
More recently, new strategies have emerged to allow targeting of multiple effectors with a single gRNA. One approach involves the addition of unique RNA-protein interaction domains engineered within the gRNA sequence to provide a scaffold for proteins with RNA recognition domains. This involves the modification of the secondary structure of the gRNA to include RNA aptamers, termed scaffold gRNA, creating stem loop structures with different RNA recognition motifs (Figure 1d). Many groups have utilized this technique with RNA aptamers that recruit bacteriophage MS2 coat protein [43,66,72,76,77], though it is possible with other RNA aptamers such as PP7 or Com [8,38,72]. These differing RNA binding motifs/binding partners can be used to recruit either multiple types of effectors to a single locus or to uniquely coordinate more complex control at multiple genes. Additionally, for epigenetic marks such as DNA methylation, this system is well suited to ensure that the region is targeted for DNA methylation/demethylation with multiple copies of an effector.
A second approach to recruit more effectors with only one gRNA is to use an array of peptide epitopes fused to dCas9. Termed SunTag [78], this strategy provides a protein scaffold for multiple binding sites of an effector protein (Figure 1e). The identity of the epitopes determines which effectors bind – a string of like epitopes recruit multiple effectors of the same type, while a variety of epitopes allow for the recruitment of different effectors. Recruitment of multiple activators or repressors may potentially produce a more robust effect over just one type of effector. This system has been used to induce robust gene expression with VP64, locus visualization with GFP [78], and induction of a repressed gene with Tet1 [51]. This type of approach may be essential for unsilencing of genes that are repressed by DNA methylation [51,78] or provide strong enough induction of gene expression to result in cell type conversion.
Though some processes involve the same modification at many genes, these tools could be even more powerful if they allowed for the coordinated, simultaneous activation and repression of distinct genes sets. Additionally, in some cases it would be useful to regulate the sequential recruitment of distinct individual factors (such as different epigenetic modifiers or transcription factors) to a single gene. Multiplexing with the use of multiple species of Cas9 is an attractive approach to coordinate more intricate control of a locus or many loci at once (Figure 1f) [6,23,79-81]. As mentioned before, the PAM sequence immediately adjacent to the crRNA site dictates which Cas species will bind. Therefore, if multiple species of Cas are utilized with their respective gRNA, then each species could be fused with a different effector protein. A potential caveat to this approach is that different PAM sequences may not be compatible with the desired targeting location. As an alternative strategy, the RNA aptamer system could be modified to include different RNA stem loop structures to facilitate the recruitment of different effectors to diverse loci while using the same species of dCas9.
Pairing CRISPR/Cas9 and Other Modulatory Technologies
Cell Type Specificity
The central nervous system is comprised of a heterogeneous population of cells, including different neuronal subtypes and glial cells. Due to this complexity, genetic manipulations that affect distinct populations of cells often yield different results. While stereotaxic surgery offers location specific control for basic research in animal models (Figure 3a), modern genetics has enabled even more precise cellular specificity by incorporating cell-type specific promoters to introduce transgenes into unique cellular populations. For example, excitatory forebrain neuronal populations are often targeted using the Camk2a promoter [82], whereas glial populations are often targeted in adult animals using the Gfap promoter [83]. Similarly, promoters that drive the expression of genes coding for specific neuronal receptors (e.g., Drd1, Drd2, Adora2a) [84,85], enzymes critical for neurotransmitter synthesis (e.g., Th, Chat, Tph2, Dbh) [86-90], or synaptic/vesicle proteins (e.g., Slc6a3, Slc1a3, and Syn1) [90,91] are often used to target more genetically defined neuronal populations. Finally, a number of activity-responsive promoters (e.g., Fos, Arc) [92] can be used to drive transgene expression in a manner that is sensitive to neuronal stimulation or activation in a given context. CRISPR-mediated gene editing in neurons has already utilized this strategy to achieve neuron-specific expression with the Syn1 promoter [30].
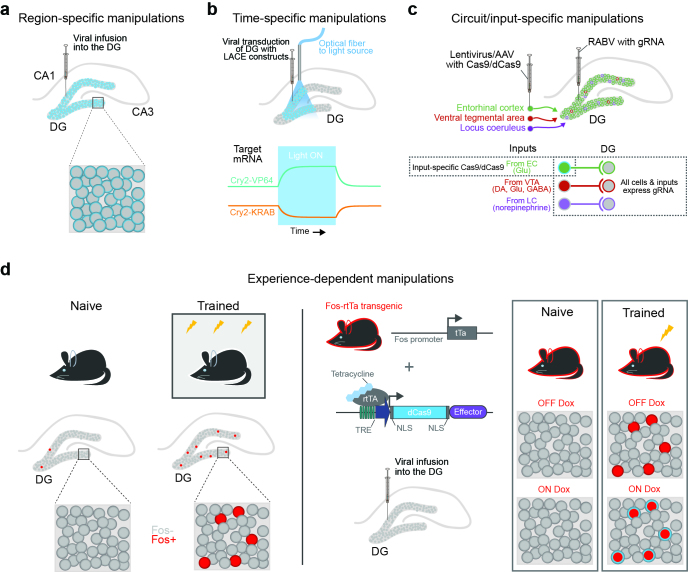
Figure 3.
The combination of CRISPR-based editing and neuronal manipulation techniques. a) Region-specific expression of CRISPR components is possible with stereotaxic surgery. In this example, the dentate gyrus (DG) of the hippocampus is targeted. With this approach, the promoter driving the transgene dictates which cells will express the construct in the region. b) (Top) Infusion of LACE viral constructs and implantation of an optical fiber allows temporally specific binding of effector proteins to dCas9-CIB1. (Bottom) Gene expression of a target gene can be bidirectionally controlled with different LACE systems. Cry2-VP64 induces transcription with exposure to light, while Cry2-KRAB represses expression. Removing light returns gene expression to baseline as the Cry-effectors are no longer recruited to the locus. c) (Top) Circuit specificity is achieved by infusing a monosynaptic retrograde rabies virus (RABV) containing the gRNA into the DG and a lentivirus or AAV carrying Cas9/dCas9 constructs into the entorhinal cortex (EC). (Bottom) Though there are multiple inputs into the DG, only the cells projecting from the EC to the DG receive both viruses and have a functional CRISPR system. d) (Left) Experience, such as fear conditioning, results in sparse coding in the DG, and cells participating in the encoding of the memory express the immediate early gene Fos. (Right) Use of a Fos promoter driving the Tet-on system (utilizing rtTA to activate the dCas9-effector transgene infused into the DG) can be used to express dCas9-effector in cells only activated by training. This is achieved by administration of doxycycline during training, providing a window in which active cells can express the transgene.
Another powerful genetic system involves the combination of transgenic animals or viruses that express Cre recombinase in specific populations of cells with viruses containing strategically placed loxP sites [93]. Flanking loxP around transgene sites can be used to delete it when expression is no longer necessary. This could be used in multiple ways in the CNS. For example, deletion after Cas9-induced knockout would be advantageous to reduce the chance of off-target effects. Similarly, a floxed transgene can be flipped to become active, which is a common strategy for cell-type specificity or inducible expression [93]. This system is especially powerful in neuroscience as hundreds of Cre lines have been generated to drive expression in many different cell types [94,95]. Moreover, the Cre system is usable without the requirement of a transgenic animal by introduction of Cre alongside the transgene [96]. Taken together, the use of cell-type specific strategies to express CRISPR components will provide increased understanding to the complexity of the heterogeneous nature of the brain.
Inducible Expression
Inducible systems limit expression of a transgene to a specific period of time, which may be critical for exploration of temporally defined neuronal processes that underlie complex adaptive behavior, learning, and memory consolidation. Further, dynamic changes in gene expression and chromatin remodeling occur with experience [65,97]. Therefore, controlling the expression of CRISPR components in a way that more closely mimics natural regulation of gene expression will help to dissect essential contributions to behavior. There are two major systems used in neuroscience for temporal control of gene expression: the tetracycline inducible system and optogenetic approaches. The tetracycline inducible system (Tet-on and Tet-off) reversibly controls expression with exposure to a tetracycline, commonly doxycycline [98]. The presence of doxycycline either induces (Tet-on) or inhibits (Tet-off) expression of the transgene when the transgene is driven by a Tet operator (TetO), and an example of the Tet-on system is illustrated in Figure 2a.
Chemical induction of gene expression is a powerful tool, but it is not as temporally precise as optogenetic approaches, which operate on time scales more similar to neuronal activity and behavior. The use of optogenetics has revolutionized neuroscience as a way to temporally control neuronal activity and investigate circuit dynamics [99]. Traditionally, optogenetics involves light-sensitive channels or pumps (channelrhodopsins as well as halorhodopsin and archearhodopsin) [100], but the field has grown to include other light sensitive proteins. One notable example is the crypotochrome 2 (CRY2) protein from Arabidopsis thaliana [101]. CRY2 changes confirmation with blue light exposure and is then able dimerize with its binding partner, CIB1 (Figure 2b). This system has been integrated with CRISPR/dCas9 fused to VP64 or p65 transactivators (termed light-activated CRISPR effector, or LACE) to enable temporally controlled regulation of gene expression that is not possible with constitutive expression of dCas-VP64, [102,103]. While a TALE-based system has been validated in neurons [104], the CRISPR-based system is well suited for use both in vitro and in vivo and has been utilized in HEK cells to temporally induce gene expression [102,103]. While this system has only been utilized with CRISPRa with VP64 and p65, it is easily adaptable to other fusion proteins (Figure 3b).
Circuit-specific Gene and Epigenetic Editing
Retrograde tracers [105], channelrhodopsins, and designer receptors exclusively activated by designer drugs (DREADDs) [106] are critical tools for mapping circuits within the central nervous system, and all have helped to identify how genetically defined neuronal populations contribute to behavioral and cognitive processes. Nevertheless, it is also essential to identify the molecular and genetic mechanisms that regulate the function of specific brain regions and defined neuronal circuits. Combining CRISPR-based approaches with circuit-specific techniques enables this type of investigation via detailed interrogation of genetic and epigenetic mechanisms within specific neuronal pathways.
The most straightforward way to achieve circuit or pathway-specific modification using CRISPR approaches requires distinct types of viral vectors that carry transgenes for essential CRISPR components. For example, take the glutamatergic projection from the entorhinal cortex (EC) to the dentate gyrus (DG) (Figure 3c). To produce a knockout of a gene in these specific projections, a Cas9 nuclease could be packaged into a viral vector and infused into the EC. Likewise, the gRNA targeting the gene of interest could be packaged into a monosynaptic rabies virus and infused into the DG. The virus containing the gRNA construct would infect the terminals of the EC, resulting in only the EC neurons projecting to the DG obtaining all of the components necessary for gene-specific alteration. This type of projection-specific modulation has the potential to provide insight into the functional role of genes and epigenetic modifications in distinct circuits, much like optogenetics and DREADDs have defined the overall functional roles for these circuits. Circuit-specific targeting of specific receptors could indicate the importance of differential neuromodulatory signaling within a defined circuit rather than just activating or repressing the circuit such as the case with optogenetics and DREADDs. Additionally, any number of the dCas9 transcriptional or epigenetic modifications can be examined using this circuit specific arrangement.
Neuron-tagging Systems
Activity in a distinct subpopulation of cells within a region of the brain underlie many cognitive processes and behaviors. Tagging the neural ensembles that participate in an experience affords the ability to observe and even modulate this subpopulation. One notable example is the identification of an engram [107], which is difficult to detect due to sparse and temporal coding, notably in the dentate gyrus of the hippocampus (Figure 3a). Fortunately, neuron tagging systems permit direct observation of cells that are physiologically active during an experience. Traditionally, a tetracycline-inducible (TetTag) [108] or Cre system (targeted recombination in active populations, TRAP) [109] are driven by a promoter for a marker of neural activity, usually cFos. Therefore, cells that are active and inducing cFos mRNA will also produce the transcriptional induction protein used to later identify or manipulate this neuronal population [108,109]. Generally, expression of a fluorescent protein or a neuronal modulator such as ChR2 or DREADDs are used, but future studies could utilize CRISPR-based alterations within an active population (Figure 3d). In the example of learning and memory, it might be critical for a certain protein or epigenetic modification to occur in memory formation in a population of cells that are active with encoding of an experience. This system would lead to the dissection of what gene-specific alterations, either solely transcriptional or epigenetic, are responsible for different stages of memory.
Future Horizons of CRISPR in the CNS
Cell Reprogramming
Cellular reprogramming without the use of overexpression vectors is currently being explored by a number of groups utilizing CRISPRa. Here, the goal is to target genes identified to control cell fate to promote a more natural conversion of cell type as compared to overexpression vectors [74]. For cell type conversion to take place, transcription factors that determine cell fate must be highly expressed. While CRISPRa is being used for this type of work, generally just one VP64 unit fused to dCas is not enough to induce transcription to an appropriate level to modify cell type. Two different approaches have resolved this issue: two VP64 units fused to dCas or the use of dCas-VPR. In a recent report, human induced pluripotent stem cells transfected with dCas-VPR targeting two neuronal transcription factors transformed these cells into functional neurons [47]. Excitingly, these tools have also been demonstrated to allow direct trans-differentiation between cell types without initial reversion to a pluripotent state. Specifically, fibroblasts were directly converted to neuronal cells using VP64-dCas-VP64 by targeting BAM factors Brn2, Ascl1, and Myt1l [74]. Targeting these endogenous genes generated epigenetic remodeling and stable expression of neuronal genes, and resulted in converted cells with complex morphology and physiological properties that are characteristic of neurons. Efficient cellular reprogramming pushes the field towards personalized medicine. Patient-derived fibroblasts converted to functional neurons would allow for therapies to be tested for in vitro efficacy before being moved to the patient. This advancement would additionally pave the way for in vivo direct cellular conversion, which could be a potential therapeutic in many neurodegenerative diseases [110].
Disease Variants
Cas9’s utility also extends to investigating the role of genetic variation, as SNPs are highly implicated in individual differences and susceptibility to disease. Not only have SNPs in coding regions been causally linked to a disorder, SNPs found in non-coding regions comprise the majority of genome-wide association study hits for disease phenotypes. These genetic variations can impact local chromatin environment or DNA-binding proteins. CRISPR-Cas9 provides a straightforward way to make specific alterations, such as introduction of a disease-linked SNP, in a known genetic background to characterize the functional result of the variant. For example, a non-coding SNP thought to be associated with a regulatory region for the alpha-synuclein (SNCA) gene, which is implicated in Parkinson’s disease, was specifically altered and found to contribute to SNCA’s expression [111]. Complex polygenic neuropsychiatric disorders such as depression [112,113], schizophrenia [114,115], addiction [116], and autism spectrum disorders [117] all have identified genetic variants, though the causality of each variant has not been explored. This approach is being adapted for use with patient-derived induced pluripotent stem cells (iPSCs) [111,118-120], which could be genetically altered with simple transfection approaches prior to differentiation into neurons. For example, a recent report utilized this approach with iPSCs to investigate a loss of function mutation in SCN1A, a subunit of the voltage-gated sodium channel Nav1.1 [119].
CRISPR could also revolutionize forward genetics approaches in neuronal systems. Forward genetic screens are comprehensive attempts to understand the underlying genetic contribution of a phenotype. However, mutagenesis through chemical agents or radiation are non-specific, making it cumbersome to identify the mutation of interest. One attractive characteristic of CRISPR-Cas9 systems is the ease of synthesizing the gRNAs. Therefore, a more cost-effective and high-throughput screen is possible with a gRNA library, allowing for identification of phenotype-causing alteration to be more readily identified [121].
Experience-dependent Expression and Epigenetic Changes
Temporal control of gene expression through CRISPRa/CRISPRi is beneficial when studying neuronal function and behavior, both of which occur in a temporally relevant fashion. Many groups have taken advantage of advancements in next-generation sequencing to characterize genome-wide changes in expression, epigenetic modifications, and chromatin looping within defined brain regions and during specific behavioral paradigms or manipulations. These data provide snapshots of the cellular environment that are correlated with behavior. For example, characterized changes in gene expression after an experience indicates that those genes differentially expressed have a role in the encoding of a memory or a behavior [122]. An inducible system combined with a multiplexed CRISPR-dCas9 system could mimic distinct “waves” of gene expression observed following specific experiences, allowing each wave to be characterized in relative isolation. Furthermore, due to the dynamic nature of epigenetic modifications with neuronal activity and experience, the prospect of understanding the permanence of epigenetic modifications in the brain is an exciting avenue of study.
Current Limitations
At present, the major limitation of using CRISPR in post-mitotic neurons is transgene expression efficiency. Typical transfection reagents have a very low efficiency in neurons, leaving transduction as one of the only viable ways to transfer CRISPR constructs into neurons with high efficiency. Depending on the application, a variety of viruses can be useful for neuronal delivery. However, genome capacity of the virus is a major issue. The Streptococcus Pyogenes Cas9 coding region alone is around 4.1kb [7], so some virus types will have difficulty packaging such a large transgene. The capacity of adeno-associated viruses, commonly used in neurons, is around 5kb, so packaging only the catalytically active Cas9 protein seems possible. The most promising viral type is the lentivirus because of its larger transgene capacity (~8 to 10kb) [123] and ability to infect non-dividing cells. However, in contrast to AAV viruses, it has a much lower spread and may require multiple injections if targeting in vivo. Additionally, achieving a high viral titer for long constructs can be difficult.
There are a few ways to minimize these problems. First, it is possible to use other species of Cas9, such as Staphylococcus aureus (SaCas9), which is approximately 1kb shorter than the commonly used Cas9 from S. pyogenes [23]. However, the drawback of using SaCas9 is that the PAM sequence (5’-NNGRRT-3’) is more complex, reducing the number of possible gRNA sequences in a given region of interest. Next, it is possible to break up CRISPR components into multiple vectors and perform a co-transduction. For example, many labs separate the gRNA and Cas9/dCas9 constructs, and there is one report of a split Cas9 [124]. Additionally, methods that segregate the dCas9 from its effector protein, such as SunTag [51], LACE [102,103], and the use of RNA aptamers [43,66,76] can reduce the load on a viral vector. The disadvantage to this approach is that cells must be co-transduced to receive all of the CRISPR components.
Methods that avoid the use of virus altogether involve infusion of recombinant protein and gRNA directly into the brain, biolistic transfection, and in utero electroporation. Infusion of the recombinant proteins largely depends on liposomal based approaches and has been validated in neuronal systems [32]. Alternatively, biolistic transfection of brain slices [29,31] and in utero electroporation has been successfully used to express CRISPR components in the brain [31], demonstrating its feasibility in neuronal systems. In summary, careful consideration is required to package and achieve expression in the central nervous system.
The major criticism of any gene editing systems is off-target effects, which include both similar genetic loci and global cellular responses. Great effort is being taken to accurately identify and minimize any off-target binding. The CRISPR system is RNA-guided, and the gRNA binds to DNA in a sequence-specific manner. Therefore, the possibility of binding in the presence of mismatches exists [125]. However, there is also a requirement for the presence of a PAM sequence, so off-targets that do not have a PAM sequence are not likely to bind to any significant extent [126]. The use of gRNA prediction software is critical to foresee potential off-targets in silico.
Experimentally, there are numerous ways to test for gRNA specificity. One straightforward assay is to conduct chromatin immunoprecipitation (ChIP) sequencing using antibodies for Cas9/dCas9 (or antibodies for fused tags) to determine the genomic location of CRISPR complex binding [127-129]. In one study, this assay demonstrated excellent specificity, as Cas9 binding was observed at only one off-target region of the genome, despite nearly 300 predicted potential off-target sites [127]. However, binding does not imply induction of a dsDNA break with Cas9. With full binding of the gRNA, a conformational change occurs that brings the nuclease domains into close contact with DNA. Therefore, if there are mismatches, the efficiency of a DSB is lower [130]. For dCas9 approaches, both ChIP and RNA-seq have been employed to characterize potential recruitment of fusion proteins with dCas9 to undesired loci. In a study utilizing dCas9-p300 and dCas9-VP64, RNA-seq was conducted to measure non-specific upregulations in gene expression [50], and no significant increases in gene expression were detected outside of the gRNA target for dCas-VP64, while just two other genes were significantly increased in dCas9-p300 transfected cells. Similar results were found with numerous other studies utilizing dCas9 [41,44,129,131,132]. Even with slight off-target effects in the dCas system, it is still a drastic improvement over constitutive overexpression vectors of epigenetic machinery.
Off-targets of Cas9 continue to be investigated by a number of groups. However, some recommendations exist to minimize the risk of off-target DSB induced by Cas9. This can be achieved through titrating Cas9, using a gene-inducible system [133], or introducing recombinant RNA/protein complexes directly to cells. It is important to limit the time Cas9 is expressed in cells since once the gene editing has occurred, the entire system is no longer needed. Secondly, use of Cas9 nickase will likely reduce off-target effects as recruitment of two CRISPR units is required for a dsDNA break. Finally, FokI, the nuclease commonly used in ZFN and TALENs, bound to dCas9 can produce more specific effects [134,135]. Overall, even with suspected off-target effects, careful expression and engineering of nucleases will likely provide safe and effective gene editing with CRISPR.
Conclusions
The flexibility and accessibility of CRISPR tools are attractive to many areas of research, explaining its exponential growth in the realm of gene editing. The modular nature of dCas9 makes it an ideal tool to investigate RNA/DNA localization as well as transcriptional and epigenetic mechanisms. These tools demonstrate enormous promise for advancing our understanding of the genetic and epigenetic basis of neuronal plasticity, behavior, and disorders of the CNS. Moreover, the application of these tools to human diseases holds tremendous relevance for therapeutics. Overall, CRISPR based systems will likely prove to be an indispensable method in interrogation of neuronal function, plasticity, behavior, and neuropsychiatric disorders.
Acknowledgments
We thank all of the members of the Day lab for their thoughtful discussions on these topics.
Glossary
- ChIP
chromatin immunoprecipitation
- CRISPR
clustered regularly interspaced short palindromic repeats
- DG
dentate gyrus
- DREADD
designer receptors exclusively activated by designer drugs
- EC
entorhinal cortex
- FISH
fluorescence in situ hybridization
- gRNA
guide RNA
- HR
homologous recombination
- iPSCs
induced pluripotent stem cells
- KRAB
Krüppel-associated box
- LACE
light-activated CRISPR effector
- NHEJ
non-homologous end joining
- SNP
single nucleotide polymorphism
- PAM
protospacer adjacent motif
- TALEN
transcription activator-like effector nuclease
Author Contributions
KE Savell and JJ Day both conceived of the idea, wrote the manuscript, and designed the figures. This work is supported by NIH grants DA042514 (KES), DA039650 (JJD), and DA034681 (JJD).
References
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. Genetics. 2011. August;188(4):773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang F, Li S, Wang Y, Bai Y, Xu X. TALE: a tale of genome editing. Prog Biophys Mol Biol. 2014. January;114(1):25–32. [DOI] [PubMed] [Google Scholar]
- de Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012. November;40(21):10596–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007. March;315(5819):1709–12. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity [Internet] Science. 2012. August;337(6096):816–21. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems [Internet] Science. 2013. February;339(6121):819–23. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013. February;339(6121):823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016. January;17(1):5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Guan Y, Wang L, Qiu Z, Liu M, Chen Y, et al. CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat Protoc. 2014. October;9(10):2493–512. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014. February;156(4):836–43. [DOI] [PubMed] [Google Scholar]
- Heidenreich M, Zhang F. Applications of CRISPR–Cas systems in neuroscience. Nature. 2015. December;17(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci. 2014. December;17(12):1720–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ. New approaches to manipulating the epigenome. Dialogues Clin Neurosci. 2014. September;16(3):345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The Emerging Field of Neuroepigenetics. Neuron. 2013. October;80(3):624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76 Pt B:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014. September;17(9):1164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari-Javan S, Varbanov H, Halder R, Benito E, Kaurani L, Burkhardt S, et al. HDAC1 links early life stress to schizophrenia-like phenotypes. Proc Natl Acad Sci USA. 2017. June;114(23):E4686–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SE, Gupta S, Moes A, West AB, Arking DE. Exaggerated CpH methylation in the autism-affected brain. Mol Autism. 2017;8(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J, et al. Tcf4 Regulates Synaptic Plasticity, DNA Methylation, and Memory Function. Cell Reports. 2016. September;16(10):2666–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Meng L, Pei F, Zheng Y, Leng J. A review of DNA methylation in depression. J Clin Neurosci. 2017;43:39–46. [DOI] [PubMed] [Google Scholar]
- Renthal WR, Nestler EJ. Epigenetic Mechanisms in Drug Addiction. Trends Mol Med. 2008. August;14(8):341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013. November;8(11):2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015. April;520(7546):186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma I, Gent DC. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr. 2012. November;3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014. December;24(12):2059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Comms. 2014. April;5:3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013. August;41(14):e141–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR [Internet] Neuron. 2014. September;83(5):1051–7. 10.1016/j.neuron.2014.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, et al. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016. September;353(6304):1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015. January;33(1):102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS ONE. 2014;9(8):e105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol. 2017. May;35(5):431–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciotto MR, Wickman K. Using knockout and transgenic mice to study neurophysiology and behavior. Physiol Rev. 1998. October;78(4):1131–63. [DOI] [PubMed] [Google Scholar]
- Fishel ML, Vasko MR, Kelley MR. DNA repair in neurons: so if they don’t divide what’s to repair? Mutat Res. 2007. January;614(1-2):24–36. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013. February;152(5):1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013. August;41(15):7429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016. January;13(2):127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015. May;16(5):299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998. February;391(6669):806–11. [DOI] [PubMed] [Google Scholar]
- Prelich G. Gene overexpression: uses, mechanisms, and interpretation. Genetics. 2012. March;190(3):841–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013. July;154(2):442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013. October;10(10):977–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013. September;31(9):833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013. October;10(10):973–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013. October;23(10):1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013. October;2(10):604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle MP, Iyer E, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015. April;12(4):326–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Kim TK, West AE, Nord AS, Markenscoff-Papadimitriou E, Lomvardas S. Genomic Views of Transcriptional Enhancers: Essential Determinants of Cellular Identity and Activity-Dependent Responses in the CNS. J Neurosci. 2015. October;35(41):13819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, et al. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015. May;18(5):647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015. April;33(5):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol. 2016. October;34(10):1060–5. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016. September;167(1):233–235.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JY, Schaukowitch K, Farbiak L, Kilaru G, Kim TK. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nature. 2015. November;19(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015. May;12(5):401–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016. March;44(12):gkw159–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res. 2016. November;45(4):1703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci. 2014. October;17(12):1720–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell. 2016. June;165(7):1803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Mori T, Kurihara T, Kawase S, Koike R, Satoga M, et al. Fluorescent protein tagging of endogenous protein in brain neurons using CRISPR/Cas9-mediated knock-in and in utero electroporation techniques. Nature. 2016. October;6(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun. 2013. September;439(1):132–6. [DOI] [PubMed] [Google Scholar]
- Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci USA. 1982. July;79(14):4381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008. September;5(10):877–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA. 2015. March;112(10):3002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Zhang W, Hu H, Xue B, Qin J, Sun C, et al. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 2016. May;44(9):e86–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano-Fernández A, Barco A. Nuclear organization and 3D chromatin architecture in cognition and neuropsychiatric disorders. Mol Brain. 2016. September;9(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016. April;165(2):488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perovic M, Tesic V, Mladenovic Djordjevic A, Smiljanic K, Loncarevic-Vasiljkovic N, Ruzdijic S, et al. BDNF transcripts, proBDNF and proNGF, in the cortex and hippocampus throughout the life span of the rat. Age (Dordr). 2013. December;35(6):2057–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, et al. Local RNA Translation at the Synapse and in Disease. J Neurosci. 2011. November;31(45):16086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savell KE, Gallus NV, Simon RC, Brown JA, Revanna JS, Osborn MK, et al. Extra-coding RNAs regulate neuronal DNA methylation dynamics. Nat Comms. 2016;7:12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol Cell. 2014. October;56(1):29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. Long Noncoding RNAs: Past, Present, and Future. Genetics. 2013. March;193(3):651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015. June;12(7):664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JB, Adler AF, Wang HG, D’Ippolito AM, Hutchinson HA, Reddy TE, et al. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell. 2016. September;19(3):406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology. 2012. July;38(1):23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol. 2015. November;33(11):1159–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015. January;517(7536):583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014. October;159(3):635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Comms. 2017. February;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013. September;110(39):15644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013. November;10(11):1116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Deguchi S, Yamauchi T. Characterization of 5′ flanking region of alpha isoform of rat Ca2+/calmodulin-dependent protein kinase II gene and neuronal cell type specific promoter activity. Neurosci Lett. 2001. July;307(2):117–21. [DOI] [PubMed] [Google Scholar]
- Morelli AE, Larregina AT, Smith-Arica J, Dewey RA, Southgate TD, Ambar B, et al. Neuronal and glial cell type-specific promoters within adenovirus recombinants restrict the expression of the apoptosis-inducing molecule Fas ligand to predetermined brain cell types, and abolish peripheral liver toxicity. J Gen Virol. 1999;80 (Pt 3)(3):571–83. [DOI] [PubMed] [Google Scholar]
- Lee KW, Kim Y, Kim AM, Helmin K, Nairn AC, Greengard P. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc Natl Acad Sci USA. 2006. February;103(9):3399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006. February;9(3):443–52. [DOI] [PubMed] [Google Scholar]
- Calipari ES, Juarez B, Morel C, Walker DM, Cahill ME, Ribeiro E, et al. Dopaminergic dynamics underlying sex-specific cocaine reward. Nat Comms. 2017. January;8:13877–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–25. [DOI] [PubMed] [Google Scholar]
- Scott MM, Wylie CJ, Lerch JK, Murphy R, Lobur K, Herlitze S, et al. A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc Natl Acad Sci USA. 2005. November;102(45):16472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouroux A, Houhou L, Biguet NF, Serck-Hanssen G, Guibert B, Icard-Liepkalns C, et al. Analysis of the human dopamine beta-hydroxylase promoter: transcriptional induction by cyclic AMP. J Neurochem. 1993. January;60(1):364–7. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Masson J, Gingrich JA, Rayport S, Hen R. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J Neurosci Methods. 2005. April;143(1):27–32. [DOI] [PubMed] [Google Scholar]
- Schoch S, Cibelli G, Thiel G. Neuron-specific Gene Expression of Synapsin I. J Biol Chem. 1996. February;271(6):3317–23. [DOI] [PubMed] [Google Scholar]
- DeNardo L, Luo L. Genetic strategies to access activated neurons. Curr Opin Neurobiol. 2017. May;45:121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F. Cre Recombinase and Other Tyrosine Recombinases. Chem Rev. 2016. October;116(20):12785–820. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, et al. Subregion-and Cell Type- Restricted Gene Knockout in Mouse Brain. Cell. 1996. December;87(7):1317–26. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007. September;27(37):9817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed BY, Chakravarthy S, Eggers R, Hermens WT, Zhang JY, Niclou SP, et al. Efficient delivery of Cre-recombinase to neurons in vivo and stable transduction of neurons using adeno-associated and lentiviral vectors. BMC Neurosci. 2004. January;5(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011. August;14(10):1345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995. June;268(5218):1766–9. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005. September;8(9):1263–8. [DOI] [PubMed] [Google Scholar]
- Guru A, Post RJ, Ho YY, Warden MR. Making Sense of Optogenetics. Int J Neuropsychopharmacol. 2015. July;18(11):pyv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature. 1998. April;392(6677):720–3. [DOI] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015. March;11(3):198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem Biol. 2015. February;22(2):169–74. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015. January;517(7536):583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield BR. Retrograde axonal tracing with fluorescent markers. Curr Protoc Neurosci. 2008. ;Chapter 1:Unit1.17–1.17.24. [DOI] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007. March;104(12):5163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Köhler S, Frankland PW. Finding the engram. Nat Rev Neurosci. 2015. September;16(9):521–34. [DOI] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature. 2016. June;534(7605):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013. June;78(5):773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Marchetto MC, Bardy C, Gage FH. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat Rev Neurosci. 2016. July;17(7):424–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016. May;533(7601):95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S, Lear M, Shanley L, Hing B, Baizan-Edge A, Herwig A, et al. Differential activity by polymorphic variants of a remote enhancer that supports galanin expression in the hypothalamus and amygdala: implications for obesity, depression and alcoholism. Neuropsychopharmacology. 2011. October;36(11):2211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EC, Brown RC, Dai Y, Rosand J, Nugent NR, Amstadter AB, et al. Genetic determinants of depression: recent findings and future directions. Harv Rev Psychiatry. 2015. January;23(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, et al. A role for noncoding variation in schizophrenia. Cell Reports. 2014. November;9(4):1417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ. Recent genetic findings in schizophrenia and their therapeutic relevance [Internet] J Psychopharmacol (Oxford). 2015. February;29(2):85–96. Available from: http://journals.sagepub.com/doi/10.1177/0269881114553647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducci F, Goldman D. The genetic basis of addictive disorders. Psychiatr Clin North Am. 2012. June;35(2):495–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, Cheng L, Chu K, Lu Z, et al. Single nucleotide polymorphisms predict symptom severity of autism spectrum disorder. J Autism Dev Disord. 2012. June;42(6):971–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JJ, Robinson TM, Germain ND, Sirois CL, Bolduc KA, Ward AJ, et al. Disrupted neuronal maturation in Angelman syndrome-derived induced pluripotent stem cells. Nat Commun. 2017. April;8:15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gao C, Chen W, Ma W, Li X, Shi Y, et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry. 2016. January;6(1):e703–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SM, Hartley BJ, Flaherty E, Rajarajan P, Abdelaal R, Obiorah I, et al. Evaluating Synthetic Activation and Repression of Neuropsychiatric-Related Genes in hiPSC-Derived NPCs, Neurons, and Astrocytes. Stem Cell Reports. 2017. August;9(2):615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017. March;12(4):828–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD. Experience-dependent epigenomic reorganization in the hippocampus. Learn Mem. 2017. July;24(7):278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther. 2001. October;12(15):1893–905. [DOI] [PubMed] [Google Scholar]
- Ma D, Peng S, Xie Z. Integration and exchange of split dCas9 domains for transcriptional controls in mammalian cells. Nat Commun. 2016. September;7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014. February;507(7490):62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016. May;17(5):300–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014. July;32(7):670–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014. July;32(7):677–83. [DOI] [PubMed] [Google Scholar]
- O’Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015. March;43(6):3389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015. November;527(7576):110–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015. December;12(12):1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, et al. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015. August;25(8):1158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Fisher J, O’Rourke KP, Muley A, Kastenhuber ER, Livshits G, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015. April;33(4):390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015. July;26(7):425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014. June;32(6):569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]