Abstract
Salvia miltiorrhiza is a traditional Chinese medicinal herb used for treating cardiovascular diseases. Depside salt from S. miltiorrhiza (DSSM) contains the following active components: magnesium lithospermate B, lithospermic acid, and rosmarinic acid. This study aimed to reveal the mechanisms of action of DSSM. After searching for DSSM-associated genes in GeneCards, Search Tool for Interacting Chemicals, SuperTarget, PubChem, and Comparative Toxicogenomics Database, they were subjected to enrichment analysis using Multifaceted Analysis Tool for Human Transcriptome. A protein-protein interaction (PPI) network was visualised; module analysis was conducted using the Cytoscape software. Finally, a transcriptional regulatory network was constructed using the TRRUST database and Cytoscape. Seventy-three DSSM-associated genes were identified. JUN, TNF, NFKB1, and FOS were hub nodes in the PPI network. Modules 1 and 2 were identified from the PPI network, with pathway enrichment analysis, showing that the presence of NFKB1 and BCL2 in module 1 was indicative of a particular association with the NF-κB signalling pathway. JUN, TNF, NFKB1, FOS, and BCL2 exhibited notable interactions among themselves in the PPI network. Several regulatory relationships (such as JUN → TNF/FOS, FOS → NFKB1 and NFKB1 → BCL2/TNF) were also found in the regulatory network. Thus, DSSM exerts effects against cardiovascular diseases by targeting JUN, TNF, NFKB1, FOS, and BCL2.
1. Introduction
Salvia miltiorrhiza (also named Danshen) is widely used as a traditional Chinese medicinal herb to prevent and treat vascular diseases [1, 2]. Depside salt from S. miltiorrhiza (DSSM) is a new medicine that contains the active components magnesium lithospermate B (MLB), lithospermic acid (LA), and rosmarinic acid (RA) [3]. MLB plays protective roles in relieving atherosclerosis and combating myocardial ischaemia-reperfusion injury [4]. RA and LA also have beneficial effects on cardiovascular diseases, such as atherosclerosis and neointimal hyperplasia [5, 6]. Thus, understanding the mechanisms of action of DSSM is important for its better utilisation in a clinical setting.
In patients with unstable angina, DSSM can suppress platelet activation and aggregation as well as matrix metallopeptidase 9 (MMP-9) expression and secretion [7]. MLB protects against diabetic atherosclerosis by inducing the nuclear factor erythroid 2-related factor-2-antioxidant responsive element-NAD(P)H: quinone oxidoreductase-1 pathway [8]. Du et al. reported that MLB can be used to treat ischaemic heart diseases as it specifically inhibits transforming growth factor β-activated protein kinase 1-binding protein 1-p38 apoptosis signalling [9]. Kim et al. also demonstrated that RA inhibits adriamycin-induced cardiotoxicity by suppressing reactive oxygen species generation as well as extracellular signal-regulated kinase and c-Jun N-terminal kinase (JNK) activation [10]. Moreover, LA inhibits foetal bovine serum-induced vascular smooth muscle cell (VSMC) proliferation by arresting cell cycle progression and suppressing cyclin D1 expression and lipopolysaccharide-induced VSMC migration by downregulating MMP-9 expression; thus, LA may be used for preventing neointimal hyperplasia, restenosis, and atherosclerosis [11]. However, no comprehensive survey of DSSM-associated genes has been reported.
In this study, we searched for DSSM-associated genes in several common databases. Subsequently, we applied multiple bioinformatic methods to identify further DSSM-associated key genes; these methods included enrichment analysis, protein-protein interaction (PPI) network and module analyses, and transcriptional regulatory network analysis. This study may provide a better understanding of the mechanisms of action of DSSM.
2. Methods
2.1. Search for DSSM-Associated Genes
Several databases were used in this study. The human genomic database GeneCards (http://www.genecards.org/) was used [12], which provides concise genome, transcriptome, proteome, and function data of all predicted and known human genes. Search Tool for Interacting Chemicals (STICH, version 5.0, http://stitch.embl.de/) was also sued [13], which is a database that integrates the interactions between chemicals and proteins. SuperTarget (http://insilico.charite.de/supertarget/) [14] was also used, which is a database that includes drug-associated information correlated with drug metabolism, adverse drug effects, medical indications, Gene Ontology (GO) terms, and pathways for target proteins. The PubChem (https://www.ncbi.nlm.nih.gov/pccompound/) [15] database was also used, which provides information on chemical substances and corresponding biological activities and is linked to the National Institutes of Health PubMed Entrez. Moreover, the study features the Comparative Toxicogenomics Database (CTD, http://ctdbase.org/) [16], which is a database associated with environmental chemical-gene product interactions as well as the transportation and accumulation of chemical substances in the human body. In GeneCards [12], STICH (parameters set as organism = human, score > 0.4) [13], SuperTarget [14], PubChem [15], and CTD [16] databases, “magnesium lithospermate B”, “lithospermic acid” and “rosmarinic acid” were used as keywords to search for genes associated with them (which were combined into a set of DSSM-associated genes).
2.2. Functional and Pathway Enrichment Analyses
The GO (http://www.geneontology.org) database describes the associations of gene products with the categories of biological process (BP), molecular function (MF), and cellular component (CC), as well as with more specific subcategories [17]. The Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.ad.jp/kegg) is a database used for annotating the functions of genes or other molecules [18]. The “BioCloud” online platform was developed for managing problems encountered in the analysis of high-throughput data. By applying the Multifaceted Analysis Tool for Human Transcriptome (MATHT, http://www.biocloudservice.com) in the “BioCloud” online platform, GO functional and KEGG pathway enrichment analyses were conducted for DSSM-associated genes, setting a threshold of <0.05 for the false discovery rate (FDR).
2.3. PPI Network and Module Analyses
On combining the Search Tool for the Retrieval of Interacting Genes (STRING, version 10.0, http://www.string-db.org/, combined score > 0.4) [19], Biological General Repository for Interaction Datasets (BioGRID, version 3.4, https://wiki.thebiogrid.org/) [20] and Human Protein Reference Database (release 9, http://www.hprd.org/) [21] interaction databases, PPI pairs among DSSM-associated genes were predicted. Subsequently, the PPI network was visualised for DSSM-associated genes using the Cytoscape software (http://www.cytoscape.org) [22]. Using the CytoNCA plug-in [23] (version 2.1.6, http://apps.cytoscape.org/apps/cytonca) in Cytoscape, degree centrality (DC), betweenness centrality (BC), and closeness centrality of the nodes were analysed to obtain the hub proteins in the PPI network [24]. The parameter was set as “without weight.”
Based on the MCODE plug-in [25] (version 1.4.2; http://apps.cytoscape.org/apps/mcode; parameters set as degree cut-off = 2, maximum depth = 100, node score cut-off = 0.2, and K-core = 2) in Cytoscape, module analysis was conducted for the PPI network. Subsequently, KEGG pathway enrichment analysis was performed for the nodes of significant modules, with FDR < 0.05 as the cut-off criterion.
2.4. Transcriptional Regulatory Network Construction
Using the transcriptional regulatory relationships unravelled by a sentence-based text-mining (TRRUST, http://www.grnpedia.org/trrust/) [26] database, transcription factors (TFs) among DSSM-associated genes were searched and then their targets were screened. Finally, a transcriptional regulatory network was constructed using Cytoscape [22].
3. Results
3.1. Search for DSSM-Associated Genes
The numbers of MLB-, RA-, and LA-associated genes identified from GeneCards, STICH, SuperTarget, PubChem, and CTD databases are listed in Table 1. The MLB-, RA-, and LA-associated genes were combined into a group of 73 DSSM-associated genes.
Table 1.
MLB-associated genes, RA-associated genes, and LA-associated genes searched from the GeneCards, STICH, SuperTarget, PubChem, and CTD databases.
| Database | MLB | RA | LA | |||
|---|---|---|---|---|---|---|
| Count | Symbol | Count | Symbol | Count | Symbol | |
| GeneCards | 6 | NOS2, NOS3, AKR1B1, XDH, PLOD1, PTPN1 | 33 | SLC16A1, PTGS2, TNF, ALOX5, IL1B, TAT, JUN, BCL2, CXCL8, CCL11, LCK, FOS, CCL2, CREB1, ERVK-6, PIK3CG, NFKB1, PLCG1, IKBKB, SHC1, MAP2K1, CCL3, RELA, GRB2, XIAP, EEF1A1, CCR3, CXCL2, BIRC2, ZAP70, BIRC3, ITK, MIR155 | 8 | NOS2, NOS3, AKR1B1, XDH, PLOD1, PTPN1, SPTLC1, SPTLC2 |
| STICH | 8 | IL15, IL2, IL4, KCNMA1, KCNU1, MAPK8, RRAGA, RRAGB | 7 | CCR3, FOS, IKBKB, IL2, LCK, PARG, PROCR | 0 | / |
| SuperTarget | 0 | / | 5 | ALDR, C1R, HYAL1, LCK, TYRO | 0 | / |
| PubChem | 0 | / | 10 | PROCR, PARG, TNF, ADAM17, SERPINE1, MAOB, MAOA, IL1B, DNMT1, COL1A1 | 0 | / |
| CTD | 7 | IFNG, IL1B, CASP3, NFE2L2, SIRT1, CAT, RELA | 21 | PROCR, ADAM17, IL1B, CXCL1, IL6, TNF, SERPINE1, AIFM1, CASP3, COL1A1, COMT, DNMT1, FN1, GPT, MAOA, MAOB, NOS2, NOS3, OGG1, PARG, RELA | 1 | XDH |
MLB, magnesium lithospermate B; RA, rosmarinic acid; LA, lithospermic acid, STICH, Search Tool for Interacting Chemicals; CTD, Comparative Toxicogenomics Database.
3.2. Functional and Pathway Enrichment Analyses
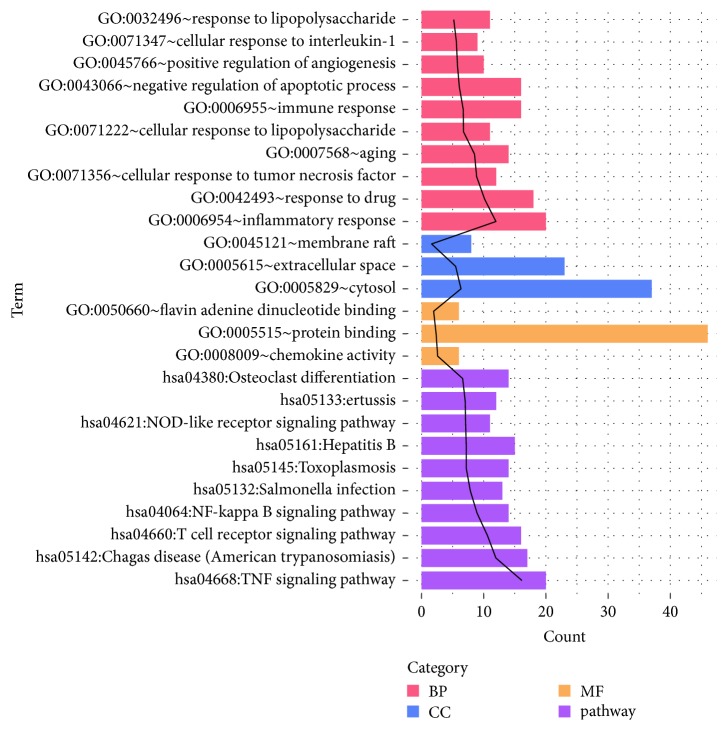
The main BP, CC, and MF terms, as well as KEGG pathways enriched for DSSM-associated genes, are shown in Figure 1. The significantly enriched terms included inflammatory response (BP), cytosol (CC), protein binding (MF), and tumour necrosis factor (TNF) signalling pathway (KEGG pathway).
Figure 1.
The main BP, CC, and MF terms as well as KEGG pathways enriched for DSSM-associated genes. BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; DSSM, depside salt from Salvia miltiorrhiza.
3.3. PPI Network and Module Analyses
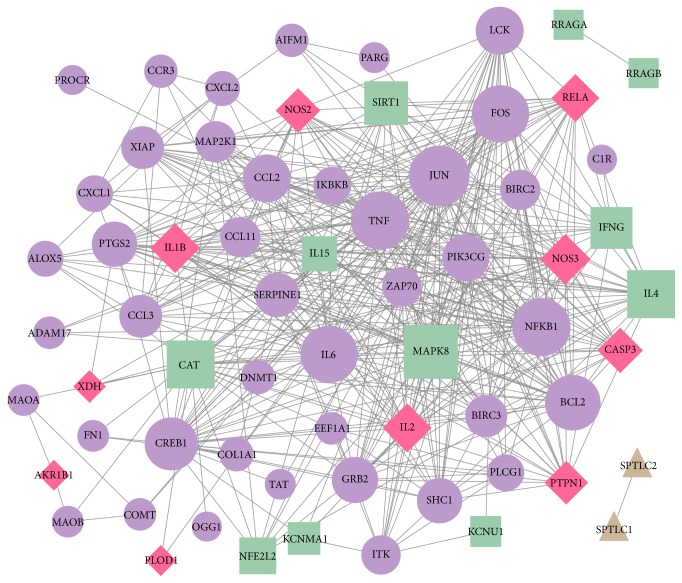
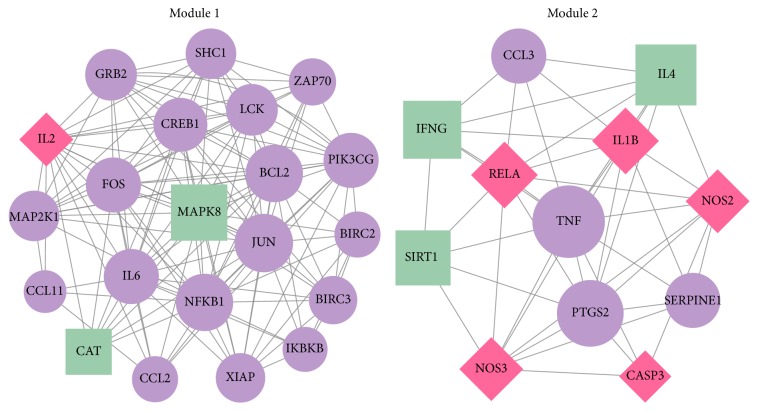
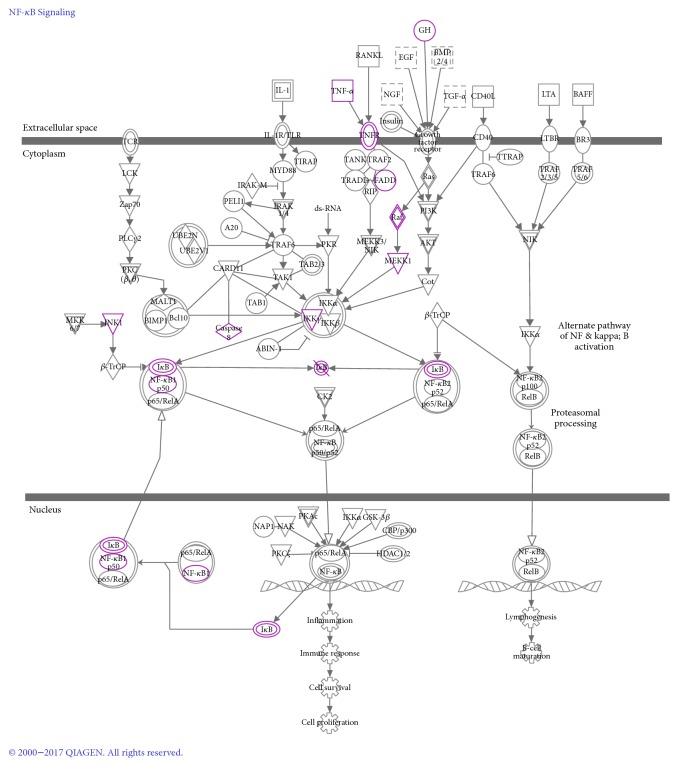
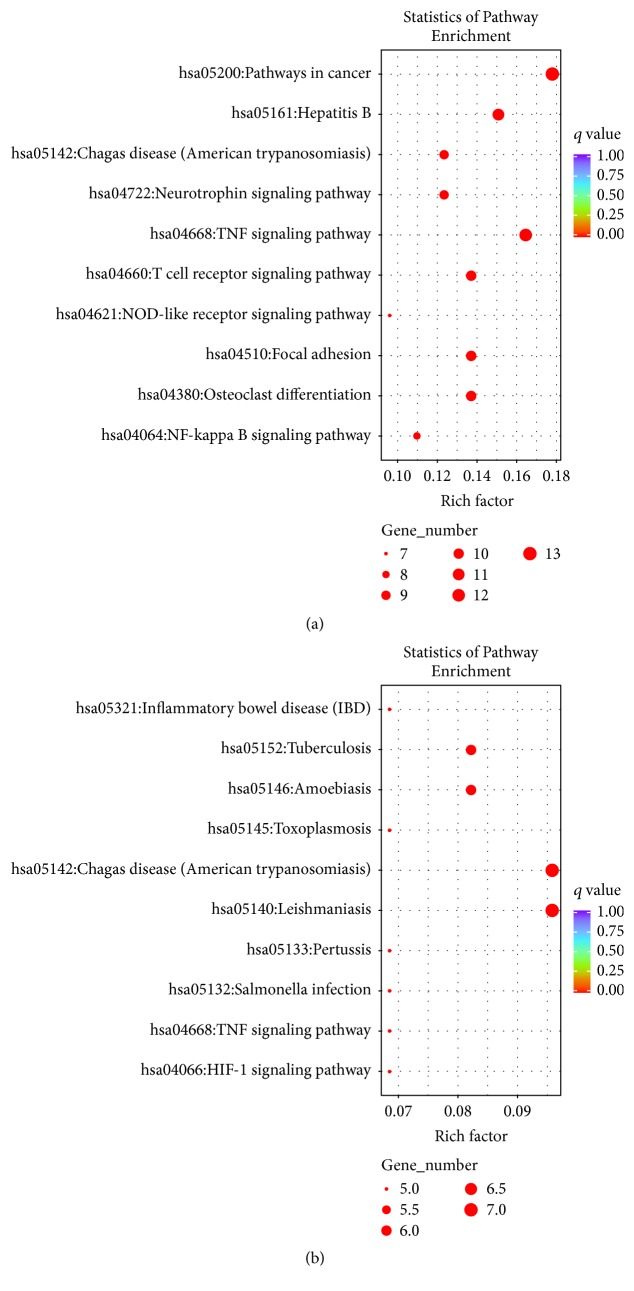
The PPI network for DSSM-associated genes comprised 65 nodes and 431 edges (Figure 2). The 65 nodes included 21 MLB-associated genes, 56 RA-associated genes, and eight LA-associated genes. Among them, two genes [nitric oxide synthase 2 (NOS2) and nitric oxide synthase 3 (NOS3)] were associated with all MLB, RA, and LA; four genes (interleukin 2, interleukin 1 beta, caspase 3, and v-rel avian reticuloendotheliosis viral oncogene homolog A) were associated with both MLB and RA; and four genes (xanthine dehydrogenase, aldo-keto reductase family 1 member B1, protein tyrosine phosphatase no-receptor type 1 and procollagen-lysine, and 2-oxoglutarate 5-dioxygenase) were associated with both MLB and LA. Jun protooncogene (JUN), TNF, nuclear factor kappa B subunit 1 (NFKB1), and Fos protooncogene (FOS) were noted as particular hub nodes according to their BC, closeness centrality, and DC scores (Table 2). Moreover, two modules were also identified from the PPI network: module 1 (21 nodes, score = 13.2) and module 2 (12 nodes, score = 7.8) (Figure 3). In addition, pathway enrichment analysis demonstrated that the nuclear factor kappa B (NF-κB) signalling pathway [Figure 4; p = 3.14E−06; involving NFKB1 and B-cell CLL/lymphoma 2 (BCL2)] and leishmaniasis (p = 4.72E−07) were particularly associated with the nodes in modules 1 and 2, respectively (Table 3 and Figure 5). JUN, TNF, NFKB1, FOS, and BCL2 also exhibited notable interactions among themselves in the PPI network.
Figure 2.
The protein-protein interaction (PPI) network constructed for the depside salt from Salvia miltiorrhiza-associated genes. Green squares, purple circles, and brown triangles represent magnesium lithospermate B-associated genes, rosmarinic acid-associated genes, and lithospermic acid-associated genes, respectively. Red diamonds represent genes associated with more than one drug. The larger nodes indicate genes with higher degrees. The degree indicates the number of interactions with other proteins in the PPI network.
Table 2.
Top 10 nodes in the PPI network according to DC, BC, and CC scores.
| DC | BC | CC | |||
|---|---|---|---|---|---|
| Gene | Score | Gene | Score | Gene | Score |
| JUN | 37 | CAT | 577.2475 | JUN | 0.186047 |
| TNF | 34 | FOS | 309.3704 | TNF | 0.184438 |
| NFKB1 | 34 | TNF | 256.2388 | NFKB1 | 0.184438 |
| FOS | 33 | JUN | 251.4059 | FOS | 0.183908 |
| IL6 | 33 | CCL2 | 205.0727 | IL6 | 0.183381 |
| BCL2 | 31 | NFKB1 | 186.0261 | BCL2 | 0.181303 |
| MAPK8 | 29 | IL6 | 185.3155 | MAPK8 | 0.181303 |
| CREB1 | 28 | BCL2 | 166.0571 | CREB1 | 0.179775 |
| NOS3 | 25 | CREB1 | 164.3064 | NOS3 | 0.178771 |
| IL1B | 24 | CASP3 | 147.608 | IL1B | 0.178273 |
PPI, protein-protein interaction; DC, degree centrality; BC, betweenness centrality; CC, closeness centrality.
Figure 3.
Modules 1 and 2 identified from the protein-protein interaction network. Green squares and purple circles represent magnesium lithospermate B-associated genes and rosmarinic acid-associated genes, respectively. Red diamonds represent genes associated with more than one drug. The larger nodes indicate genes with higher degrees.
Figure 4.
The path diagram of the nuclear factor kappa B (NF-κB) signalling pathway. The highlighted genes represent heart failure-associated genes.
Table 3.
Pathways enriched for the genes separately involved in module 1 and module 2 (top 10 listed).
| Module | Term | Count | Genes | FDR |
|---|---|---|---|---|
| Module 1 | hsa04668:TNF signaling pathway | 12 | PIK3CG, FOS, IL6, CCL2, MAP2K1, JUN, CREB1, NFKB1, MAPK8, BIRC3, IKBKB, BIRC2 | 1.18E − 12 |
| hsa05161:Hepatitis B | 11 | PIK3CG, FOS, IL6, MAP2K1, GRB2, JUN, BCL2, CREB1, NFKB1, MAPK8, IKBKB | 2.20E − 09 | |
| hsa04660:T cell receptor signaling pathway | 10 | PIK3CG, FOS, MAP2K1, GRB2, JUN, LCK, ZAP70, NFKB1, IKBKB, IL2 | 4.40E − 09 | |
| hsa04380:Osteoclast differentiation | 10 | PIK3CG, FOS, MAP2K1, GRB2, JUN, CREB1, LCK, NFKB1, MAPK8, IKBKB | 3.98E − 08 | |
| hsa05200:Pathways in cancer | 13 | PIK3CG, FOS, IL6, XIAP, MAP2K1, GRB2, JUN, BCL2, NFKB1, MAPK8, BIRC3, IKBKB, BIRC2 | 9.52E − 08 | |
| hsa05142:Chagas disease (American trypanosomiasis) | 9 | PIK3CG, FOS, IL6, CCL2, JUN, NFKB1, MAPK8, IKBKB, IL2 | 2.56E − 07 | |
| hsa04722:Neurotrophin signaling pathway | 9 | PIK3CG, MAP2K1, GRB2, JUN, BCL2, NFKB1, MAPK8, SHC1, IKBKB | 8.15E − 07 | |
| hsa04510:Focal adhesion | 10 | PIK3CG, XIAP, MAP2K1, GRB2, JUN, BCL2, MAPK8, SHC1, BIRC3, BIRC2 | 2.33E − 06 | |
| hsa04064:NF-kappa B signaling pathway | 8 | XIAP, BCL2, LCK, ZAP70, NFKB1, BIRC3, IKBKB, BIRC2 | 3.14E − 06 | |
| hsa04621:NOD-like receptor signaling pathway | 7 | IL6, CCL2, NFKB1, MAPK8, BIRC3, IKBKB, BIRC2 | 8.05E − 06 | |
|
| ||||
| Module 2 | hsa05140:Leishmaniasis | 7 | IL4, TNF, PTGS2, RELA, IFNG, IL1B, NOS2 | 4.72E − 07 |
| hsa05142:Chagas disease (American trypanosomiasis) | 7 | CCL3, TNF, RELA, SERPINE1, IFNG, IL1B, NOS2 | 4.90E − 06 | |
| hsa05146:Amoebiasis | 6 | CASP3, TNF, RELA, IFNG, IL1B, NOS2 | 3.72E − 04 | |
| hsa05321:Inflammatory bowel disease (IBD) | 5 | IL4, TNF, RELA, IFNG, IL1B | 2.36E − 03 | |
| hsa05133:Pertussis | 5 | CASP3, TNF, RELA, IL1B, NOS2 | 4.47E − 03 | |
| hsa05152:Tuberculosis | 6 | CASP3, TNF, RELA, IFNG, IL1B, NOS2 | 4.76E − 03 | |
| hsa05132:Salmonella infection | 5 | CCL3, RELA, IFNG, IL1B, NOS2 | 6.71E − 03 | |
| hsa04066:HIF-1 signaling pathway | 5 | RELA, SERPINE1, IFNG, NOS3, NOS2 | 1.30E − 02 | |
| hsa04668:TNF signaling pathway | 5 | CASP3, TNF, PTGS2, RELA, IL1B | 1.78E − 02 | |
| hsa05145:Toxoplasmosis | 5 | CASP3, TNF, RELA, IFNG, NOS2 | 2.72E − 02 | |
Figure 5.
Pathways enriched for the nodes involved in modules 1 (a) and 2 (b).
3.4. Transcriptional Regulatory Network Analysis
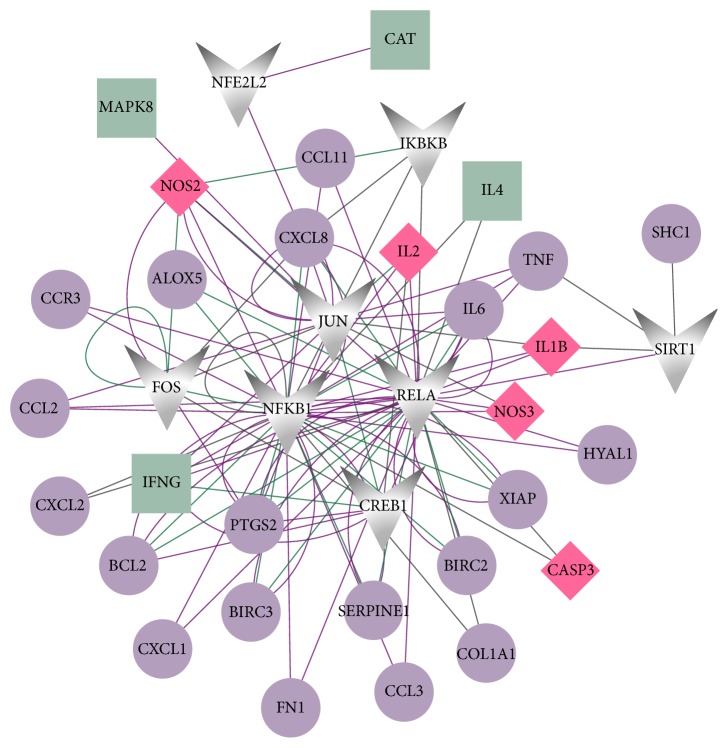
Based on the TRRUST database, TFs among DSSM-associated genes were searched for and their targets were screened. The constructed transcriptional regulatory network had 37 nodes (including eight TFs and 29 target genes) and 108 relationship pairs (such as JUN → TNF/FOS, FOS → NFKB1, and NFKB1 → BCL2/TNF) (Figure 6).
Figure 6.
The transcription factor- (TF-) target gene regulatory network. Green squares and purple circles represent magnesium lithospermate B-associated genes and rosmarinic acid-associated genes, respectively. Grey polygons represent TFs. Red diamonds represent genes associated with more than one drug. Larger nodes indicate genes with higher degrees. Purple lines, green lines, and grey lines represent activating regulatory, repressing regulatory, and unknown regulatory effects, respectively.
4. Discussion
In this study, a total of 73 DSSM-associated genes were identified from various databases. In the PPI network constructed on the basis of these genes and their interactions, JUN, TNF, NFKB1, and FOS were established as hub nodes according to their BC, closeness centrality, and DC scores. JUN, TNF, NFKB1, FOS, and BCL2 were also shown to interact among themselves in the PPI network. Moreover, two distinct modules (modules 1 and 2) of the PPI network were identified. In addition, several regulatory relationships (such as JUN → TNF/FOS, FOS → NFKB1, and NFKB1 → BCL2/TNF) were found to be involved in the transcriptional regulatory network.
Prolonged anti-TNF-α therapy has beneficial effects on the signs of subclinical cardiovascular disease in patients with severe psoriasis [27]. Studies have shown that, compared with nonbiologic disease-modifying antirheumatic drugs, TNF-α-blocking agents may contribute to the reduction of the risk of cardiovascular events in patients with rheumatoid arthritis [28, 29]. TNF-α plays a critical role in vascular dysfunction in cardiovascular diseases, which may be exploited to treat inflammation in a clinical setting [30]. TNF-α boosts atherosclerosis development by promoting the transcytosis of low-density lipoprotein (LDL) across endothelial cells, thus contributing to the reservoir of LDL in vascular walls [31]. These findings indicate that TNF may be associated with the functions of DSSM in cardiovascular diseases.
Blocking the JNK pathway may provide a new strategy for treating the cardiomyocyte death induced by myocardial ischaemia/reperfusion [32, 33]. Dominant negative c-Jun (DN-c-Jun) gene transfer inhibits VSMC proliferation, and JUN is associated with the intimal hyperplasia induced by balloon injury [34]. Saliques et al. considered that FOS is a novel factor that determines the severity and development of atherosclerosis and is thus involved in tobacco toxicity in coronary artery disease (CAD) patients [35]. Palomer et al. also demonstrated that miR-146a-targeting FOS is a potential tool for treating enhanced inflammation-associated cardiac disorders [36]. Thus, JUN and FOS may also be targets of DSSM in cardiovascular diseases.
Studies involving Uygur and Han women in China have demonstrated that the DD genotype of NFKB1 polymorphism (rs28362491) may be a genetic marker of CAD [37, 38]. The NFKB1-94ins/del ATTG polymorphism may reduce the susceptibility to myocardial infarction by decreasing activated NF-κB, which is in turn correlated with the reduction of plasma inflammatory markers [39]. By increasing BCL2 expression, miR-21 promotes heart failure progression with preserved left ventricular ejection fraction and subsequently inhibits cardiac fibrosis [40]. Moreover, Tang et al. reported that miR-1 targets BCL2 functions in mediating cardiomyocyte apoptosis [41]. In this study, pathway enrichment analysis showing the presence of NFKB1 and BCL2 in module 1 indicated a particular association with the NF-κB signalling pathway, indicating that NFKB1 and BCL2 may be correlated with the effects of DSSM against cardiovascular diseases through this pathway.
5. Conclusion
In conclusion, a total of 73 DSSM-associated genes were identified by conducting a search of public databases. JUN, TNF, NFKB1, FOS, and BCL2 were also revealed as potential targets of DSSM for treating cardiovascular diseases. However, further experimental research should be performed to confirm these findings obtained by bioinformatic analysis.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (no. 81500229).
Contributor Information
Hua Li, Email: lihua199988@hotmail.com.
Hongying Liu, Email: liuhongying@green-valley.com.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Li X., Yu C., Lu Y., et al. Pharmacokinetics, tissue distribution, metabolism, and excretion of depside salts from Salvia miltiorrhiza in rats. Drug Metabolism and Disposition the Biological Fate of Chemicals. 2006;35(2):234–239. doi: 10.1124/dmd.106.013045. [DOI] [PubMed] [Google Scholar]
- 2.Wang B.-Q. Salvia miltiorrhiza chemical and pharmacological review of a medicinal plant. Journal of Medicinal Plants Research. 2010;425(25):2813–2820. [Google Scholar]
- 3.Zhou C. X., Luo H. W., Niwa M. Studies on isolation and identification of water-soluble constituents of Salvia miltiorrhiza. Journal of China Pharmaceutical University. 1999;30(6):411–416. [Google Scholar]
- 4.Li X., Yu C., Wang L., et al. Simultaneous determination of lithospermic acid B and its three metabolites by liquid chromatography/tandem mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2007;43(5):1864–1868. doi: 10.1016/j.jpba.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Huang S.-S., Zheng R.-L. Rosmarinic acid inhibits angiogenesis and its mechanism of action in vitro. Cancer Letters. 2006;239(2):271–280. doi: 10.1016/j.canlet.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 6.Wang L., Zhang Q., Li X., et al. Pharmacokinetics and metabolism of lithospermic acid by LC/MS/MS in rats. International Journal of Pharmaceutics. 2008;350(1-2):240–246. doi: 10.1016/j.ijpharm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Xinyu MU. Influence study of depside salt from Salvia Miltiorrhiza on blood platelet function and MMP-9 in patients with unstable angina. China Medical Herald. 2009;6(24):59–60. [Google Scholar]
- 8.Hur K. Y., Kim S. H., Choi M.-A., et al. Protective effects of magnesium lithospermate B against diabetic atherosclerosis via Nrf2-ARE-NQO1 transcriptional pathway. Atherosclerosis. 2010;211(1):69–76. doi: 10.1016/j.atherosclerosis.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 9.Du C.-S., Yang R.-F., Song S.-W., et al. Magnesium lithospermate B protects cardiomyocytes from ischemic injury via inhibition of TAB1-p38 apoptosis signaling. Frontiers in Pharmacology. 2010;AUG doi: 10.3389/fphar.2010.00111.Article 119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D.-S., Kim H.-R., Woo E.-R., Hong S.-T., Chae H.-J., Chae S.-W. Inhibitory effects of rosmarinic acid on adriamycin-induced apoptosis in H9c2 cardiac muscle cells by inhibiting reactive oxygen species and the activations of c-Jun N-terminal kinase and extracellular signal-regulated kinase. Biochemical Pharmacology. 2005;70(7):1066–1078. doi: 10.1016/j.bcp.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Chen L., Wang W.-Y., Wang Y.-P. Inhibitory effects of lithospermic acid on proliferation and migration of rat vascular smooth muscle cells. Acta Pharmacologica Sinica. 2009;30(9):1245–1252. doi: 10.1038/aps.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Safran M., Dalah I., Alexander J., et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010 doi: 10.1093/database/baq020.baq020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D., Santos A., Von Mering C., Jensen L. J., Bork P., Kuhn M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research. 2016;44(1):D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecker N., Ahmed J., von Eichborn J., et al. SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Research. 2012;40(1):D1113–D1117. doi: 10.1093/nar/gkr912.gkr912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S., Thiessen P. A., Bolton E. E., et al. PubChem substance and compound databases. Nucleic Acids Research. 2016;44(1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis A. P., Grondin C. J., Johnson R. J., et al. The Comparative Toxicogenomics Database: Update 2017. Nucleic Acids Research. 2017;45(1):D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tweedie S., Ashburner M., Falls K., et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Research. 2009;37(1):D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatr-Aryamontri A., Breitkreutz B.-J., Oughtred R., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Research. 2015;43(1):D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keshava Prasad T. S., Goel R., Kandasamy K., et al. Human protein reference database—2009 update. Nucleic Acids Research. 2009;37(1):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito R., Smoot M. E., Ono K., et al. A travel guide to Cytoscape plugins. Nature Methods. 2012;9(11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y., Li M., Wang J., Pan Y., Wu F.-X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. BioSystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 24.He X., Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genetics. 2006;2(6):p. e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bader G. D., Hogue C. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4 doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han H., Shim H., Shin D., et al. TRRUST: A reference database of human transcriptional regulatory interactions. Scientific Reports. 2015;5 doi: 10.1038/srep11432.11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herédi E., Végh J., Pogácsás L., et al. Subclinical cardiovascular disease and it's improvement after long-term TNF-α inhibitor therapy in severe psoriatic patients. Journal of the European Academy of Dermatology and Venereology. 2016;30(9):1531–1536. doi: 10.1111/jdv.13649. [DOI] [PubMed] [Google Scholar]
- 28.Solomon D. H., Curtis J. R., Saag K. G., et al. Cardiovascular risk in rheumatoid arthritis: comparing tnf-α blockade with nonbiologic DMARDs. American Journal of Medicine. 2013;126(8):730.e9–730.e17. doi: 10.1016/j.amjmed.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westlake S. L., Colebatch A. N., Baird J., et al. Tumour necrosis factor antagonists and the risk of cardiovascular disease in patients with rheumatoid arthritis: a systematic literature review. Rheumatology. 2011;50(3):518–531. doi: 10.1093/rheumatology/keq316. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Park Y., Wu J., et al. Role of TNF-α in vascular dysfunction. Clinical Science. 2009;116(3):219–230. doi: 10.1042/CS20080196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Yang X., Bian F., et al. TNF-α promotes early atherosclerosis by increasing transcytosis of LDL across endothelial cells: Crosstalk between NF-κB and PPAR-γ. Journal of Molecular and Cellular Cardiology. 2014;72:85–94. doi: 10.1016/j.yjmcc.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Ferrandi C., Ballerio R., Gaillard P., et al. Inhibition of c-Jun N-terminal kinase decreases cardiomyocyte apoptosis and infarct size after myocardial ischemia and reperfusion in anaesthetized rats. British Journal of Pharmacology. 2004;142(6):953–960. doi: 10.1038/sj.bjp.0705873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milano G., Morel S., Bonny C., et al. A peptide inhibitor of c-Jun NH2-terminal kinase reduces myocardial ischemia-reperfusion injury and infarct size in vivo. American Lournal of Physiology, Heart and Circulatory Physiology. 2007;292(4):H1828–H1835. doi: 10.1152/ajpheart.01117.2006. [DOI] [PubMed] [Google Scholar]
- 34.Yasumoto H., Kim S., Zhan Y., et al. Dominant negative c-Jun gene transfer inhibits vascular smooth muscle cell proliferation and neointimal hyperplasia in rats. Gene Therapy. 2001;8(22):1682–1689. doi: 10.1038/sj.gt.3301590. [DOI] [PubMed] [Google Scholar]
- 35.Saliques S., Teyssier J.-R., Vergely C., et al. Smoking and FOS expression from blood leukocyte transcripts in patients with coronary artery disease. Atherosclerosis. 2011;219(2):931–936. doi: 10.1016/j.atherosclerosis.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 36.Palomer X., Capdevila-Busquets E., Botteri G., et al. miR-146a targets Fos expression in human cardiac cells. Disease Models and Mechanisms. 2015;8(9):1081–1091. doi: 10.1242/dmm.020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y. N., Zhang J. Y., Ma Y. T. GW25-e3074 94 ATTG Insertion/Deletion Polymorphism of the NFKB1 Gene Is Associated with Coronary Artery Disease in Han and Uygur Women in China. Genetic Testing and Molecular Biomarkers. 2014;18(6):430–438. doi: 10.1089/gtmb.2013.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai H., Chen Q., Li X., et al. Association between genetic polymorphism in NFKB1 and NFKBIA and coronary artery disease in a Chinese Han population. International Journal of Clinical and Experimental Medicine. 2015;8(11):21487–21496. [PMC free article] [PubMed] [Google Scholar]
- 39.Boccardi V., Rizzo M. R., Marfella R., et al. -94 ins/del ATTG NFKB1 gene variant is associated with lower susceptibility to myocardial infarction. Nutrition, Metabolism and Cardiovascular Diseases. 2011;21(9):679–684. doi: 10.1016/j.numecd.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Dong S., Ma W., Hao B., et al. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. International Journal of Clinical and Experimental Pathology. 2014;7(2):565–574. [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Y., Zheng J., Sun Y., Wu Z., Liu Z., Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. International Heart Journal. 2009;50(3):377–387. doi: 10.1536/ihj.50.377. [DOI] [PubMed] [Google Scholar]