Abstract
NAC (NAM, no apical meristem; ATAF, Arabidopsis transcription activation factor and CUC, cup-shaped cotyledon) proteins are among the largest transcription factor (TF) families playing fundamental biological processes, including cell expansion and differentiation, and hormone signaling in response to biotic and abiotic stresses. In this study, 172 RsNACs comprising 17 membrane-bound members were identified from the whole radish genome. In total, 98 RsNAC genes were non-uniformly distributed across the nine radish chromosomes. In silico analysis revealed that expression patterns of several NAC genes were tissue-specific such as a preferential expression in roots and leaves. In addition, 21 representative NAC genes were selected to investigate their responses to heavy metals (HMs), salt, heat, drought and abscisic acid (ABA) stresses using real-time polymerase chain reaction (RT-qPCR). As a result, differential expressions among these genes were identified where RsNAC023 and RsNAC080 genes responded positively to all stresses except ABA, while RsNAC145 responded more actively to salt, heat and drought stresses compared with other genes. The results provides more valuable information and robust candidate genes for future functional analysis for improving abiotic stress tolerances in radish.
Keywords: Abiotic stress, Expression profiling, Radish, Genome-wide analysis, NAC, Transcription factor
Introduction
Radish (Raphanus sativus L.) is one of the most economically important root vegetables in Brassicaceae family. To date, several studies on the radish genome have been carried out in the past few years, which includes the recently released radish genome sequence of radish contributed significantly to further characterization of the genes in radish (Kitashiba et al., 2014; Mitsui et al., 2015). In the published sequence data, comprehensive analysis of a gene family can be performed to explore the phylogenetic evolution and expression patterns that will facilitate the understanding how NAC gene family controls traits at a genome-wide level. In radish, decreased yield and quality due to environmental stresses including HM, heat and high salt have been reported (Sun et al., 2016; Wang et al., 2015a; Xie et al., 2015). Therefore, plants tolerance to these stresses could contribute to higher yield and quality in radish production. Tolerance against stresses is regulated by several specific TFs, such as NACs (Vinocur & Altman, 2005). These TFs function together in regulating plant growth and development by activating or suppressing the transcriptional processes of their target genes (Udvardi et al., 2007). Recently, the NAC gene family has gained much attention due to its involvements in numerous functions such as the development of tolerance to environmental cues (Yang et al., 2011).
The NAC TF regarded as one of the largest TF families is composed of 117 in (Arabidopsis), 204 in Chinese cabbage (Brassica oleracea), 110 in potato (Solanum tuberosum), 104 in tomato (Solanum lycopersicum), 82 in melon (Cucumis melo), 74 in grape (Vitis vinifera), 151 in rice (Oryza sativa), 147 in foxtail millet (Setaria italica) and in 120 poplar (Populus Spp) NAC members (Hu et al., 2010; Liu et al., 2014; Nuruzzaman et al., 2010; Puranik et al., 2013; Singh et al., 2013; Wang et al., 2013a; Wei et al., 2016). In addition, several NAC proteins have been associated with plant defense against stress conditions. For example, high salinity (Zheng et al., 2009), drought (Jeong et al., 2010) and HMs (Wang et al., 2013b; Xu et al., 2013). Evidently, NAC family genes are critical regulator implicated in the survival of plants under various stress conditions. In Arabidopsis, expression levels of three NAC genes (ANAC072, ANAC055 and ANAC019) were found to be elevated under ABA, salt and drought stress conditions. Furthermore, overexpression of these genes increased abiotic stress tolerance as compared to the wild-type (Hu et al., 2006; Jensen et al., 2010). Similarly, transgenic ATAF1 showed improved tolerance to dehydration, ABA, salinity and oxidative stresses in Arabidopsis (Wu et al., 2009). Overexpression of OsNAC10, OsNAC6 and OsNAC5 genes significantly promoted the survival of the rice plants under high salinity and water deficit (Hu et al., 2006; Song et al., 2011).
NAC family members normally contain a highly conserved DNA-binding domain at the N-terminal composed of about 150–160 amino acid (aa) which is further subdivided into 5 (A–E) subdomains, but C-terminal is highly variable and consists of the transcriptional regulatory domain (TRD), which can either function as a transcriptional promoter or repressor (Kikuchi et al., 2000; Ooka et al., 2003). A special category of NAC members of NTL (NAC with transmembrane motif 1-like) has α-helical transmembrane motifs (TMs) at their C-terminal (Kim et al., 2010; Kim et al., 2007). These features facilitate inactivation or repression of various downstream genes controlling multiple cellular or molecular processes.
Because of the critical regulatory functions of NAC genes in plant responses to different stresses, the NAC gene family has been extensively studied in Arabidopsis thaliana and some other important plant species. To date, however, no genome-wide characterization of the NAC family has been conducted in radish. Therefore, it is important to comprehensively investigate the NAC TF family in radish to utilize them to develop stress tolerance cultivars in radish and other related root vegetables. The genome sequences of radish have been recently sequenced by Jeong et al. (2010), Kitashiba et al. (2014) and Mitsui et al. (2015) which allows for the in-depth analysis of the NAC TF family. In this study, 172 RsNAC genes were identified from the radish genome database and classified according to previously reported Arabidopsis and B. rapa NAC gene families. In addition, phylogenetic analysis, chromosome location and membrane-bound protein identification analysis were performed. Moreover, expression patterns of RsNAC genes in response to HMs, salt, heat, drought and ABA stresses were further investigated using qRT-PCR. These results will improve our understanding of the NAC gene family in radish, as well as contribute to screening of additional elite genes for future functional investigation of RsNACs under abiotic stresses.
Materials and Methods
Database search and detection of NAC transcription factors
The protein and nucleotide sequence datasets were obtained from the radish genome database for the detection of the RsNAC gene family (Mitsui et al., 2015). Radish protein sequences were retrieved using Hidden Markov Model profile (PF02365) generated from Pfam (Punta et al., 2012) with a cut-off E-value of 1e−3. To confirm the presence of conserved NAC domains, multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters. The data for Arabidopsis, B. rapa and rice NACs sequences were retrieved from the Arabidopsis Information Resource (Huala et al., 2001), Brassica Database (Cheng et al., 2011) and the Rice Genome Annotation Project (Kawahara et al., 2013), respectively. The sequences of all NAC TFs in other representative species were retrieved from the plant TF database (Jin et al., 2014). Additionally, TMHMM Server v. 2.0 (Krogh et al., 2001) was employed in discovering membrane-bound RsNAC proteins.
Phylogenetic analysis of RsNAC protein domains
Phylogenetic analysis was done using RsNAC domain sequences with previously grouped NAC domains from Arabidopsis and rice as reference sequences of eudicots and monocots, respectively (Abrouk et al., 2010). The phylogenetic tree was generated using MEGA6 program by the neighbor-joining method with 1,000 bootstrap replicates (Tamura et al., 2013).
Protein characterization and RsNAC sequence analyses
The Protparam program (Gasteiger et al., 2005) using ProtParam tool was employed to determine the molecular weight (MW) and isoelectric points (pI) using default settings. The online MEME program (Bailey et al., 2009) was used to search conserved motifs. The analysis was performed with default parameters except the following: number of repetitions, any; maximum number of motifs, 15; and optimum width of the motif, ≥50. The information for each RsNAC exon/intron organization was determined by mapping the CDS to DNA sequences using the Gene Structure Display Server 2.0 GSDS (Hu et al., 2014). To determine Arabidopsis orthologs, radish NAC proteins were used in BLASTP search against the Arabidopsis Information Resource, TAIR10 release (Huala et al., 2001). Reciprocal best BLAST hit blasting approach(Moreno-Hagelsieb & Latimer, 2008) was used and best hits were considered when bit-score of the compared proteins was more than 200, the sequence coverage value more than 50% and expected value E- value of ≤1e–3.
Chromosomal location of RsNAC genes
The sequences of RsNAC retrieved from the genomic sequences of the scaffolds were anchored to the integrated genetic map published byJeong et al. (2010). All the acquired potential RsNAC genes were searched against radish the chromosome sequence database Jeong et al. (2010) using BLASTN program. The sequences with a similarity of ≥99% and length coverage difference ≤5 base pairs (bp) were regarded as similar genes (Wang et al., 2015b). Subsequently, all the similar RsNAC gene locations (megabase, Mb) on the genetic map were annotated according to their corresponding pseudomolecules. All anchored genes were located on their specific genetic map against their location parameters using MapInspect software (http://www.plantbreeding.wur.nl/UK/software_mapinspect.html). Tandem repeats located within four predicted genes or within 50 kb from each other were identified manually and marked on the radish genetic map (Cannon et al., 2004).
Expression analysis of RsNAC genes based on the RNA-Seq data
The normalized RNA-Seq data were retrieved from Nodai Radish Genome Database (Mitsui et al., 2015), and then used for the expression profiling of the RsNACs in two tissues (roots and leaves) and six developmental stages (7, 14, 20, 40, 60 and 90 days after sowing, DAS) represented by RPKM (Reads Per Kilobase of transcript per Million mapped reads) value.
Plant growth, abiotic stress treatments and expression analysis
The ‘NAU-YH’ genotype seedlings were germinated on a wet filter paper in darkness for three days then transferred to the pots containing soil and peat media (1:1) and cultured in the growth chamber under 16-h (25 °C) and 8-h (18 °C) light and dark condition, respectively. Thirty days after transplanting, seedlings were subjected to the different treatments. For heat and salt treatments, seedlings were exposed to 42 °C and 200 mM NaCl conditions, respectively. For HMs treatments, seedlings were exposed to 20 mg L−1 CdCl2 ⋅2.5H2O and 100 mg L−1 Pb (NO3)2, respectively. For drought treatment, seedlings were treated with 20% polyethylene glycol 6000 (PEG) while 100 µM ABA was sprayed to the seedlings to induce ABA response. The samples from all the treatments were harvested after 24 h and frozen immediately in liquid nitrogen for RNA extraction. Total RNAs from triplicate samples were isolated using Trizol (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocol and then digested with RNase-free DNase. The quality of total RNA was determined by agarose gel electrophoresis . The total RNA (2 µg) was used to synthesize the first-strand cDNA using PrimeScript™ RT Reagent Kit (TaKaRa, Dalian, Liaoning Sheng, China), and the obtained cDNA products were further diluted 1:9 with nuclease-free water for RT-qPCR. The transcript levels of 21 randomly selected RsNAC genes were investigated with RT-qPCR according to the method reported by Xu et al. (2013). The radish Actin gene was used as the internal control. The Beacon Designer 7.7 was used to generate gene-specific primers from the non-conserved region of NAC sequences (Table S1).
Results
Identification of the RsNAC genes and comparative analysis in radish
In the determination of NAC genes in radish, a total of 181 NAC genes were obtained after searching the entire radish genome by the HMMER software. Furthermore, SMART and Pfam programs (Letunic, Doerks & Bork, 2012; Punta et al., 2012) were used to further screen the presence of the NAM domain in all the RsNAC sequences with a cut-off E-value of 1e–3. Finally, 172 RsNAC proteins were selected for subsequent analysis after exclusion of nine members with either no N-terminal NAM domain or with an E-value of >1e–3. For nomenclature, the family designation (NAC) was preceded by prefix ‘Rs’ for radish, and then followed by a number based on their generic order. The comprehensive list of RsNAC proteins analyzed in this study, the annotation of each sequence and their preatomic characteristics are shown in Table S2. The number of aa in RsNAC proteins ranged from 143 (RsNAC143) to 652 (RsNAC011) residues resulting in an average of ∼336 aa, whereas the pI ranged from 4.44 (RsNAC0730) to 9.63 (RsNAC173) with an average of ∼6.6. The results indicated that different NACs genes might function in diverse microenvironments (Kiraga et al., 2007). The RsNAC domains were aligned and observed by GeneDoc program. The RsNAC proteins possess an N-terminal NAM domain which is highly conserved compared to the diverse C-terminal (Figs. S1A–S1C). Moreover, the N-terminal NAM domains of RsNAC proteins were grouped into five distinct clusters (A–E) which were similar to that in A. thaliana and C. sativus (Tran et al., 2004; Zhang et al., 2017). However, among the 172 RsNACs, only the RsNAC155 lack the conserved subdomains A, B and C, while RsNAC161 and RsNAC127 do not contain subdomain A and C, respectively. Additionally, several proteins (RsNAC172, RsNAC171, RsNAC098, RsNAC100, RsNAC101, RsNAC156 and RsNAC143) lack subdomain E (Fig. S1C).
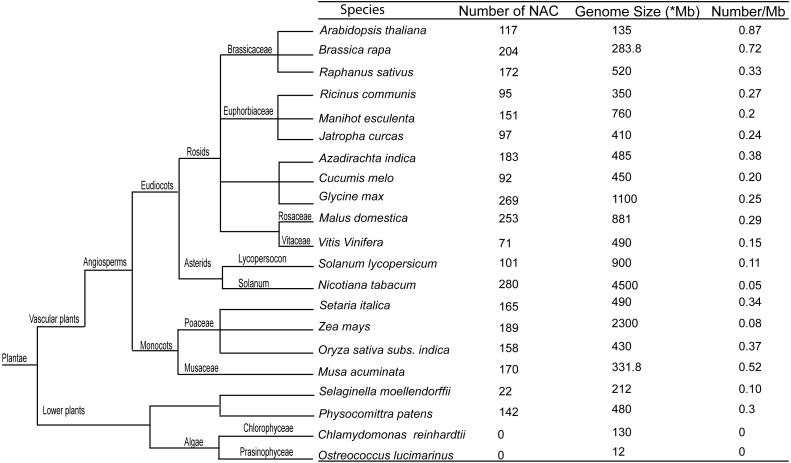
For comparative genomic analyses, we obtained NAC protein coding sequences for 21 representative plant species (Fig. 1), resulting to 2931 NAC TFs. Interestingly, all the vascular plants had NAC genes while in lower plants, two algae representatives had none. The phenomenon suggests that the NAC proteins might have expanded after the evolutionary split of the vascular plants from the lower plant species, and further indicate that this family is vascular plant-specific based on the missing NACs in algae. In conclusion, based on this study, the number of NAC in radish ranked among the members with high NAC representation in their genome. According to the density of NACs in the respective genomes, radish (0.33) ranged among the species with a high number of NAC in our analyses (Fig. 1). It was also noted that the densities of NAC TFs in most of the higher plant genomes were greater than those in all the lower plant genomes, revealing that NAC TFs increased considerably with the evolution of higher plants.
Figure 1. Comparative genomic analysis of NAC transcription factors of radish with other species.
There were 17 vascular plants and four lower plants. *Megabases.
Phylogenetic analysis of NACs in radish
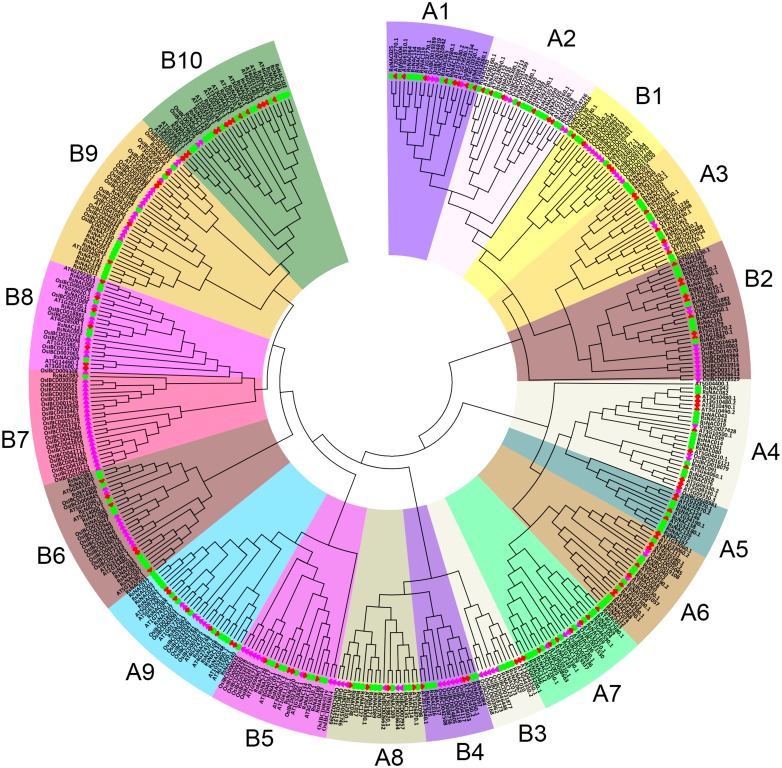
To gain further insights into the evolutionary relationship among NAC members from different plants, the NACs from radish were aligned against those in Arabidopsis and rice, and to plot an unrooted phylogenetic tree using MEGA 6 software (Fig. 2, Table S3). The RsNACs could be broadly categorized into two groups (group A and group B, respectively) based on their preferential homology to Arabidopsis and rice. As a result, group A and B showed high homology to Arabidopsis and rice, respectively. Additionally, 9 and 10 subgroups were obtained from group A (A1–A9) and B (B1–B10) in the phylogenetic analysis of the full-length NAC proteins of Arabidopsis, rice and radish based on similarity in NAC domain structures. Phylogenetic analysis revealed that most RsNAC members were closer to eudicot (AtNACs) than monocot (OsNACs) and preferentially clustered in group A. For example based on outer clades with a bootstrap value of >50, 12 RsNACs clustered with 10 AtNACs in A4 while 23 OsNACs clustered with only one RsNAC member (RsNAC085) in subgroup B7 (Fig. 2, Table S3).
Figure 2. Phylogenetic tree among the NAC TFs of radish, Arabidopsis, and rice.
Multiple sequence alignment of full-length NAC proteins was done using ClustalW and the phylogenetic tree was constructed using MEGA6 by the neighbor-joining method with bootstrap replicates, the tree was divided into two major groups (A and B). Group A was further subdivided in into nine phylogenetic subgroups designated A1–A9. Phylogenetic subgroups in-group B were designated as B1 to B10. The different sub-families are marked with different colored backgrounds. The colored shapes next to names represent radish (green circles), Red hexagons (Arabidopsis) and purple hexagons (rice).
Motif and gene structure analysis of the RsNAC genes
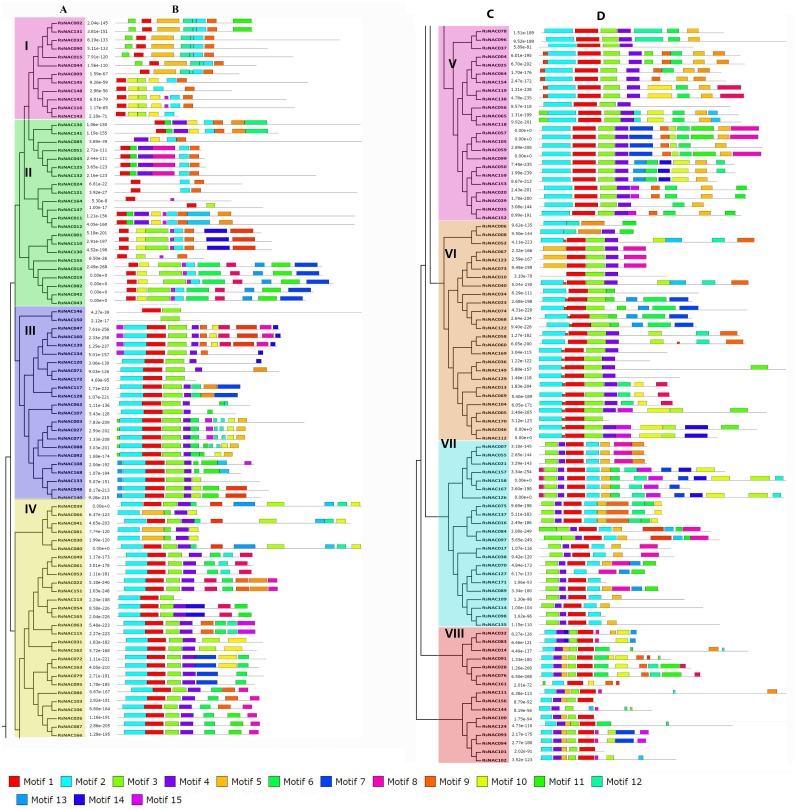
To explore the relationships among the RsNAC genes and predict their potential functions, the RsNAC proteins clustered into 9 subgroups based on the phylogenetic tree (Figs. 3A and 3C), named I to VIII. The gene number of each group ranged from 12 (subgroup I) to 28 (subgroup IV). RsNAC proteins belonging to a common cluster in the phylogenetic group (Figs. 3A and 3C) indicated a common motif distribution (Figs. 3B and 3D), suggesting functional similarities among members within the same subset. Fifteen (1–15)conserved motifs were analyzed in the RsNAC proteins (Figs. 3B and 3D). Notably, the majority of the discovered motifs were located in the N-terminal region of the NAC domain which could be an indication that these motifs are critical for the functioning of NACs. For example, subgroup I mainly contained six motifs (motifs 1, 2, 4, 5, 8 and 9), whereas subgroup II comprised of seven motifs (motifs 1, 2, 3, 4, 5, 7 and 13), which suggests that the RsNACs with similar functions tended to cluster into the same subgroups. Additionally, the structural features of the intron/exon distribution among RsNAC genes were analyzed based on their phylogenetic similarities (Figs. 4A–4D, Table S2). Exon/intron analysis depicted that introns distribution in RsNAC genes is diverse and varied from 0 to 6. Additionally, a total of 164 (∼95%) RsNAC genes had introns, while 8 (∼5%) RsNACs contained no intron (Figs. 4C and 4D). Among them, one RsNAC (RsNAC111) had six introns, two sets of 13 RsNAC members had five or four introns each and 109 RsNAC genes had two introns each. Particularly, the RsNACs with a maximum number of introns were located in subgroup VI with seven members having five introns, while the RsNACs without intron were located in subgroup I. These findings indicated a minimum diversity in gene structure among RsNAC genes belonging to a common group.
Figure 3. Phylogenetic classification and motif analysis of NAC transcription factors of radish.
Multiple sequence alignment of 172 full-length RsNAC proteins was done using ClustalW, and the phylogenetic tree was mapped using MEGA6 by the neighbor-joining method with 1,000 bootstrap replicates. MEME analysis explored conserved motifs of the RsNAC proteins. Gray lines illustrate the non-conserved sequences and each motif is showed by the colored boxes described at the bottom. The different color background indicated different groups. (A and C) multiple alignment, (B and D) conserved motifs.
Figure 4. Phylogenetic tree and Exon/Intron structure of 172 RsNAC genes.
Yellow boxes and black lines represent exons and introns, respectively, while the blue boxes represent the untranslated regions. (A and C) multiple alignment, (B and D) gene structure composition.
Membrane-associated RsNAC subfamily
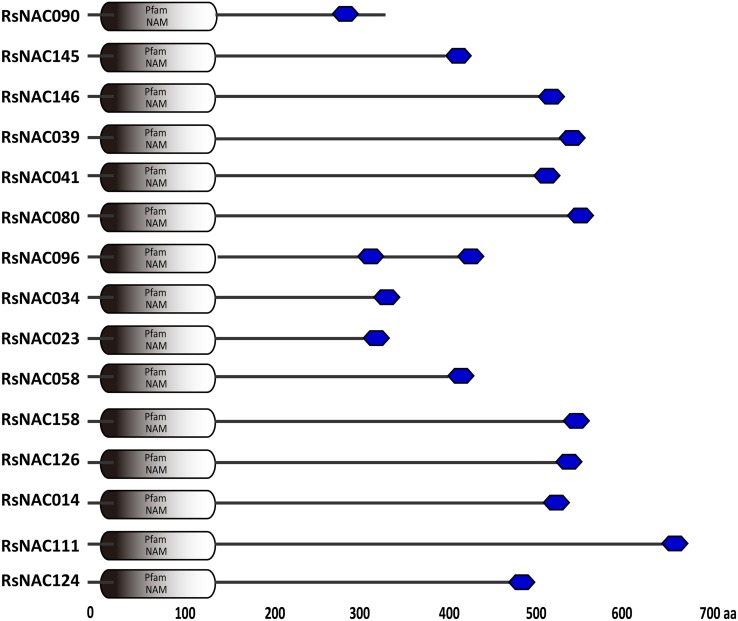
Fifteen RsNAC proteins (∼8.72%) were identified as membrane-associated RsNACs proteins (Fig. 5, Table 1). Classification of NTL proteins among the subgroups was discrete. Arabidopsis orthologues analysis indicated that 8 RsNAC NTLs were orthologs to previously characterized NTL from Arabidopsis (Table 1). The subgroups IV, VI and VIII contained three NTL proteins, while groups I and VII contained 2 NTLs, indicating that the functions of radish NTLs were complex.
Figure 5. Membrane-associated RsNAC subfamily.
Protein structure of membrane-bound NAC TFs poses indicating a conserved NAM domain located at the N-terminal of the proteins, TMs (transmembrane motifs) located at the N-terminal as shown by the blue hexagon.
Table 1. Description of the predicted membrane-bound NAC in radish.
| Protein | Length (aaa) | Tmb helix | Atc orthologue locus | At locus description |
|---|---|---|---|---|
| RsNAC090 | 339 | 281–303 | AT4G28530.1 | ANAC074 |
| RsNAC145 | 428 | 410–428 | AT5G14490.1 | ANAC085 |
| RsNAC146 | 546 | 517–539 | AT3G10480.1 | ANAC050 |
| RsNAC039 | 568 | 540–562 | AT3G15500.1 | ANAC055 |
| RsNAC041 | 535 | 510–532 | AT5G64530.1 | ANAC104 |
| RsNAC080 | 570 | 545–567 | AT5G64530.1 | ANAC104 |
| RsNAC096 | 444 | 309–331 | AT1G71930.1 | ANAC030 |
| RsNAC096 | 444 | 425–442 | AT1G71930.1 | ANAC030 |
| RsNAC034 | 349 | 323–345 | AT3G49530.1 | ANAC062 (NTL6) |
| RsNAC023 | 335 | 317–335 | AT2G27300.1 | ANAC040 (NTL8) |
| RsNAC058 | 440 | 408–430 | AT5G22290.1 | ANAC089 (NTL17) |
| RsNAC158 | 573 | 541–563 | AT1G34190.1 | ANAC017 (NTL7) |
| RsNAC126 | 565 | 534–556 | AT1G34190.1 | ANAC017 (NTL7) |
| RsNAC014 | 547 | 521–543 | AT5G04410.1 | ANAC078 (NTL11) |
| RsNAC111 | 652 | 612–634 | AT1G33060.1 | ANAC014 (NTL2) |
| RsNAC124 | 509 | 480–502 | AT1G32870.1 | ANAC013 (NTL1) |
Notes.
Amino acid.
Transmembrane segments predicted using online TMHMM server 2.0.
Arabidopsis thaliana.
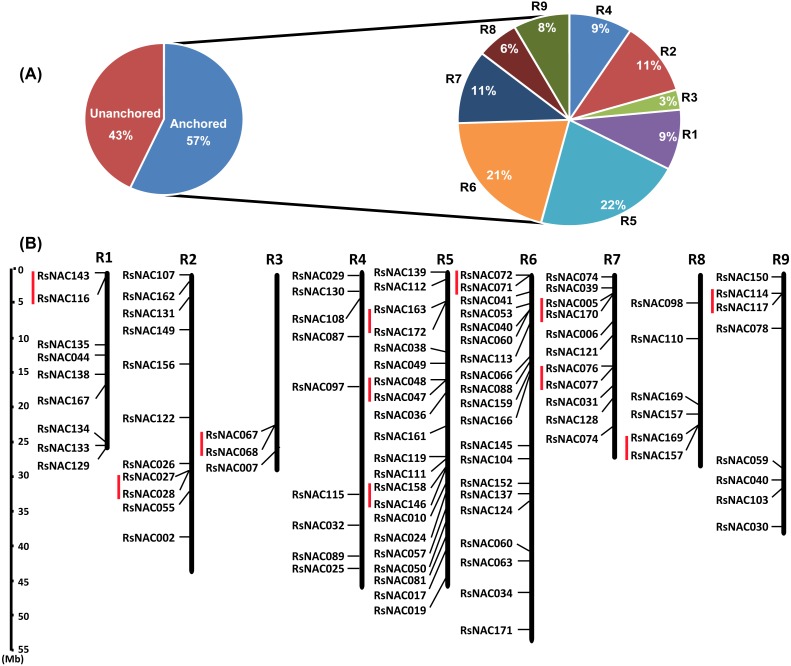
Chromosomal distribution of RsNAC genes in the radish genome
A total of 98 (∼57%) RsNAC genes were mapped onto 9 chromosomes (R1–R9) (Fig. 6, Table S4). For distribution and density of the RsNAC genes, R5 contained the maximum frequency (∼22%) of RsNAC genes followed by R6 (∼21%), while R3 had the lowest frequencies of RsNAC genes ∼3% (Fig. 6A). Thirteen RsNAC gene clusters containing 26 tandemly duplicated genes were identified in all chromosomes except chromosome R4 (Fig. 6B). Most tandem duplications were located on chromosome R5 with three gene clusters containing two genes each. The phenomenon indicated that some of the RsNAC genes have one or more paralogs, which could have been generated by whole-genome duplication (WGD) in radish. In general, majority of the RsNAC were located in the distal regions of the nine radish chromosomes while a few were found in proximal regions. Additionally, the RsNAC gene density in each chromosome ranged from 0.10 /Mb (R3) to 0.45 /Mb (R5). This indicated that the RsNAC genes were significantly non-random and unevenly distributed in the nine radish chromosomes.
Figure 6. Chromosomal distribution of RsNAC genes on the genetic map.
(A) A pie chart is indicating the percentage of the anchored and unanchored RsNAC genes and their abundant distribution on each radish chromosome with indication of percentages of RsNACs located on each chromosome. (B) Graphical representation of locations for putative RsNAC genes on each radish chromosome. The vertical red lines on the right of each chromosome indicate tandem duplicated genes. R1–R9 indicates the chromosome numbers. The scale is in megabase.
RNA-Seq-based expression analysis of RsNAC genes
For further investigation on the roles of RsNAC gene, the RNA-Seq based analysis was utilized to explore the expression patterns of the 172 RsNACs in roots and leaves at six different developmental stages (Fig. S2A–S2D, Table S5). As shown in Figs. S2A and S2C, there was no detectable expression of four RsNACs genes (RsNAC002, 131, 039 and 008). Moreover, tissue-specific expression patterns were observed in several NAC genes. These genes could be subdivided into three expression categories, which contained genes preferentially expressed in roots, leaves and both. For example, RsNAC002 (group I), RsNAC160 (group III), RsNAC165 (group IV), RsNAC069 (group V), RsNAC008 (group VI), RsNAC126 (group VII), and RsNAC124 (group VIII) were preferentially expressed in roots, while RsNAC097 and RsNAC017 (group VII) showed biased expression in leaves. However, some RsNACs exhibited multiple expressions in more than one organ and developmental stages. For example, RsNAC001, RsNAC027, RsNAC080 and RsNAC040 (group VI), RsNAC007 (group VII) and RsNAC028 (group VIII) were detected in roots and leaves at the six stages of development.
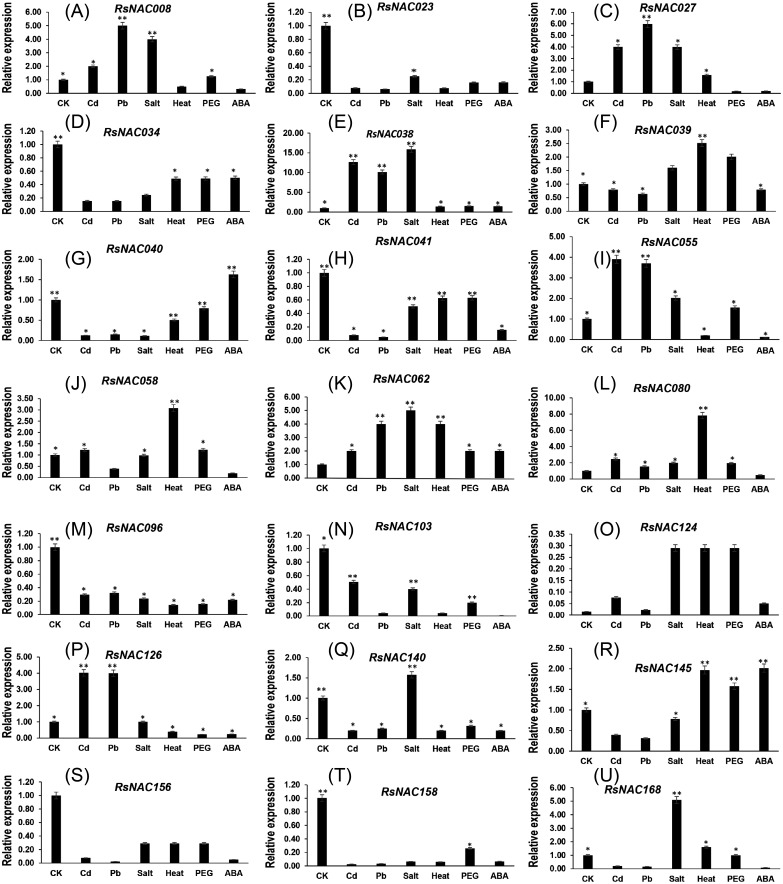
RsNAC genes expression profiling under various abiotic stresses and phytohormone treatment
To explore the expression patterns of RsNAC genes under various abiotic stresses and hormone treatments, 21 representative genes were identified for expression analysis under Cd, Pb, salt, PEG and ABA treatments. The results showed differential expression patterns of these genes under one or more treatments(Fig. 7, Table S6). Some genes (RsNAC027, RsNAC038 and RsNAC062) showed stable up-regulation under Cd, Pb and salt treatments. However, the other genes showed a weak expression level under Cd and Pb stress. Most RsNAC genes were preferentially down-regulated under HM stress whereas Pb stress up-regulated only a few of them, including RsNAC038, RsNAC008 and RsNAC062 (Fig. 7).
Figure 7. Expression patterns of RsNAC genes under abiotic stresses treatments.
RT-qPCR was used to analyze the expression profiles of 21 RsNAC genes under cadmium, lead, heat, salt, PEG, and ABA stress. CK is an untreated seedling. The Actin gene was used as an internal control. (A–U) represents expression patterns of the 21 RNAC genes in response to abiotic stresses, respectively. Error bars were obtained from three technical replicates. Asterisks indicate the genes significantly up-regulated or suppressed under abiotic stresses by t-test (* P < 0.05, ** P < 0.01).
Discussion
Characterization of RsNAC genes
NAC TFs are one of the largest plant-specific transcriptional regulators that play critical roles in numerous stress and developmental processes. However, there is still no detailed information available on NAC family in radish. To further explore the NAC information in radish, 172 putative RsNAC genes were identified from the radish genome. Remarkably, the number of the RsNAC members ranges among the largest NAC families. This finding is probably due to the selective pressure conferred by environmental cues to facilitate the growth and development of plants (Hughes & Friedman, 2003). In comparative genomic analysis among different plant species, N. tabacum was revealed harboring the highest numbers of NAC TFs (280). However, their NAC protein density (N. tabacum, 0.055) was lower than those in radish because of their large genome sizes. Additionally, stress responses of several RsNAC genes were detected based on their corresponding Arabidopsis homolog, which could be of benefit for further studies in functional characterization of NACs genes in root vegetables.
According to multiple sequence alignment, all the identified RsNACs contain the expected NAM domain (Pfam PF02365) with approximately 160 aa apportioned into five distinct subdomains (A–E). However, several RsNACs have no typical NAC domain pattern. For example, RsNAC155 lacks the conserved domain A, B, C and D, while other 12 RsNACs lacked one or two sub-domains. Such variant NAC members are termed as NAC-like proteins as described in potato and rice NACs (Nuruzzaman et al., 2010; Singh et al., 2013). The C-terminal region of the RsNACs domains indicate a highly distinct pattern involved in biotic and abiotic stress processes (Mao et al., 2012). Previous studies revealed the involvement of NAC members in protein-binding activity which performs major functions in transcription regulation (Jensen et al., 2010; Nuruzzaman et al., 2010). For phylogenetic analysis, monocots specific phylogenetic clade was identified with only one RsNAC member. The results suggest the existence of evolutionary differences between NACs from monocots and dicots and a lineage-specific expansion after splitting from their common ancestor. Additionally, the majority of the monocots and dicots with diverse functions belonged to common clades indicating that greater expansion of the NAC family could have happened slightly before the splitting of monocots and dicots (Pereira-Santana et al., 2015). Based on the results of comparative genomic analysis, it was possible to predict the functions of several RsNAC genes in response to abiotic stress according to their Arabidopsis homologs, which could also be potentially utilized for further functional studies. It was found that ANAC019, ANAC055 and ANAC07 which are orthologs to RsNAC168, RsNAC140 and RsNAC105, respectively, have been implicated in positively responding to dehydration in Arabidopsis (Hu et al., 2006; Tran et al., 2004).
In this study, all the identified RsNAC NTLs proteins contained single TM at the C-terminal of their conserved domain except RsNAC096 with two TMs, which is similar to two soybean NAC proteins (GmNAC136 and GmNAC013) where each of them contained a pair of TMs (Le et al., 2011). Recently, NTLs were demonstrated to have critical functions in response to multiple stresses (Kim et al., 2007; Lee et al., 2014). Moreover, NTLs facilitates an efficient mechanism for gene regulation which is a survival feature that allows for timely responses to environmental cues (Kim et al., 2007). In Arabidopsis, it has been reported that four NTLs (At4g01540, At2g27300, At4g35580 and At3g49530) are involved in abiotic stress tolerance, which are membrane-related proteases located in the endoplasmic reticulum that initiates the activities of TFs in response to stresses (Chen, Slabaugh & Brandizzi, 2008; Kim et al., 2008; Seo, Kim & Park, 2008). It was found that the radish NTLs (RsNAC034, RsNAC023, RsNAC058, RsNAC158, RsNAC126, RsNAC014, RsNAC111 and RsNAC124) showed close phylogenetic relationship with AT3G49530.1, AT2G27300.1, AT5G22290.1, AT1G34190.1, AT1G34190.1, AT5G04410.1, AT1G33060.1 and AT1G32870.1, respectively. In Arabidopsis, these NAC were also identified as NAC NTLs (Kim et al., 2007). Thus, it is expected that some identified RsNAC NTLs could function critically in the adjustment of stress-responsive genes as a regulatory mechanism for the survival of radish under adverse growth conditions.
Chromosomal distribution and expression profiling of RsNAC genes
The vast number of genes in NAC family in radish confirms the occurrence of chromosome duplication events during evolution. In angiosperms, several genome duplications have occurred resulting in gene family enlargement (Cannon et al., 2004), this phenomenon occurred in radish about 5.1–8.4 and 12.8–21.4 million years ago (Shen et al., 2013). Genome duplication provides a critical evolutionary promoter in angiosperm genome enlargement (Van de Peer et al., 2009). Additionally, most studies have indicated that WGD is a key factor in angiosperm biology. During genome duplication processes, redundant genes are lost (fractionation), preserved, or fixed to perform a diverse task compared with the original genes. Therefore, gene family size could vary significantly among species due to WGD events that took place during their evolution (Abrouk et al., 2010; Cenci et al., 2014). In several TFs, genome duplication has been reported in plants including MYB and WRKY, which is the same in NAC (Karanja et al., 2017; Liu et al., 2014; Nuruzzaman et al., 2010). In this study, eleven clusters representing probable tandem duplication formed by RsNAC genes in the genome were identified to elucidate the mechanism behind a large number of the NAC family members in radish. In Arabidopsis, many gene families have enlarged due to the retention of original genes after WGD and transposition (Freeling et al., 2008; Rizzon, Ponger & Gaut, 2006). Each of these gene family expansions forms paralogs that potentially lead to genetic redundancy in addition to differential growth among different families (Pereira-Santana et al., 2015). This phenomenon could contribute to the expansion of the RsNAC TFs in radish, similar to B. rapa and M. domestica (Liu et al., 2014; Su et al., 2013). Tandem duplication also indicate the presence of redundant genes which according to existing theory cause loss or mutation of duplicate genes despite retention of several genes after WGD by functionalization (Lynch & Force, 2000).
In the present study, the publicly available RNA-Seq data was utilized in conducting in silico expression analysis of RsNAC genes. Transcript abundance of NAC genes in roots and leaves reveal a spatial, temporal and ubiquitous pattern of RsNAC genes of which RsNAC168 was preferentially expressed in leaves, which was similar to its corresponding orthologs in Arabidopsis (ANAC019) expressed in leaves for regulating drought and salinity responses (Jensen et al., 2010). Additionally, ANAC019 and ANAC055 (orthologs to RsNAC168 and RsNAC140, respectively) are involved in both ABA and JA-mediated regulation (Jensen et al., 2010). Notably, most expressed genes were responsive in more than one tissue and developmental stage, spatio-temporal expression pattern of the RsNACs could provide the specific usage of RsNACs in a transcriptional modification in any given tissue, while ubiquitously regulated RsNACs could control the transcription of a wide range genes. In rice, OsNAC10 is widely expressed in roots and flowers tissues in response to dehydration and salinization stresses (Jeong et al., 2010).
Differential expression of RsNAC genes under various abiotic stresses
Unfavorable environmental cues such as HMs, salinization, high temperature and dehydration could result in permanent and deleterious effects on the optimal survival of plants. In this study, the response of RsNAC genes to all the stress treatments indicate their integral role in regulating survival processes associated with stress response and signaling in radish and other related species. According to the previous report, NAC TFs were reported to coordinate stress by initiating stress-responsive genes (Jensen et al., 2010). Several RsNACs (including RsNAC008, RsNAC027, RsNAC038) were significantly upregulated in response to salt. In Arabidopsis, overexpression of ANAC072, ANAC055 (RsNAC039, RsNAC140) and ANAC019 (RsNAC168) showed improved stress tolerance under salt conditions than the wild-type (Hu et al., 2006; Jensen et al., 2010). The finding could suggest that up-regulating of these genes may have resulted in improved salt tolerance in radish, however, NAC genes were found to negatively control drought responses by up-regulating the expression of integral drought- responsive genes(Wang et al., 2013b). Some genes, such as RsNAC030, RsNAC041 and RsNAC145, showed diverse responses under different stresses, indicating that these may function as links between the various transduction pathways (Liu et al., 2014).
In recent years, NTLs have been extensively proven to play critical roles in cell division as well as abiotic stress responses (Lee et al., 2014). In selective breeding, genes with multiple stress responses possess high potential of stress tolerance manipulation in plants. However, an occasional overexpression of NAC genes may result in antagonistic responses such as delayed flowering and dwarfing in transgenic plants (Liu et al., 2011). Overexpression of ANAC019 and ANAC055 in Arabidopsis resulted in increased drought tolerance but also increased susceptibility to B. cinerea (Bu et al., 2008). Field evaluation and prudent choice of a suitable promoter can reduce the negative effects in the over-expression of some NAC genes (Shao, Wang & Tang, 2015). In this study, nine NTLs (RsNAC23, RsNAC34, RsNAC39, RsNAC041, RsNAC058, RsNAC080, RsNAC096, RsNAC124 and RsNAC145) were among the 21 responsive RsNAC genes that responded to multiple abiotic stresses. Among them, RsNAC023 and RsNAC080 responded to all stress treatments other than ABA, while RsNAC145 responded to salt, heat and drought only. Evidently, functional characterization of these genes could be of great interest in unraveling their potential for the development of radish cultivars tolerant to multiple abiotic stresses. Over-expression techniques will provide valuable insights into understanding the regulatory functions of NAC members in radish in response to abiotic stresses. Significantly, NTLs responded differently to various stress treatments, RsNAC058 was upregulated under Cd, heat and drought, but downregulated under Pb and ABA it was. The result implied that the NTL genes might be playing critical roles in plant response to abiotic stresses.
Conclusion
Characterization of NAC TF gene family in radish was performed with particular emphasis on their responses to abiotic stresses. In total, 172 RsNAC genes were identified and characterized in radish. The conserved motif, gene structure, chromosomal distribution and expression profiles of the RsNAC genes in response to abiotic stresses were analyzed. The expression profiles of RsNAC genes under Cd, Pb, heat, salt, PEG and ABA stress displayed differential expression pattern in response to one or more stresses. In this study, it was observed that several RsNAC genes could play key roles in conferring tolerance to different abiotic stresses. The findings could provide fundamental information for further functional studies of the NAC genes by unraveling potential NAC genes in the radish, and facilitate manipulating multiple stress tolerances in the radish and related root vegetables. The result could also facilitate developing agronomically superior traits for radish including enhanced tolerance to abiotic stresses.
Supplemental Information
Multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters.
Multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters.
Multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters.
The color scales for fold-change values are shown at the bottom such as elevated (red) and suppressed (green) genes. Several genes were not expressed (black) in any tissue.
Gene-specific primers generate from the non-conserved region of NAC sequences using Beacon Designer 7.7.
aTransmembrane motif predicted using the TMHMM server
aAmino acid cMolecular weight
dIsoelectric points
e Arabidopsis thaliana
Clustering of groups A and B based on the similarity in NAC domain structures, group A contain nine subgroups (A1–A9) mostly related to eudicot while group B is composed of ten subgroups (B1–B10) mostly related to monocot.
*Megabase
aReads Per Kilobase of transcript per Million mapped reads
bDays after sowing
Fold change values obtained from transcript levels of 21 randomly selected RsNAC genes using real-time polymerase chain reaction.
aPolyethylene glycol.
babscisic acid.
Acknowledgments
We extend our gratitude to Dr. Geofrey K. Gathungu of Chuka university and Mr. Henry K. Karanja of KEMRI Wellcome Trust Research Programme, Kenya, for their help with manuscript revision.
Funding Statement
This work was supported by grants from the Natural Science Foundation of China (31501759, 31601766), National Key Technology Research and Development Program of China (2016YFD0100204-25), Key Technology R & D Program of Jiangsu Province (BE2016379) and Jiangsu Agricultural Science and Technology Innovation Fund (JASTIF, CX (16) 1012). There was no additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Bernard Kinuthia Karanja conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables.
Liang Xu conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables.
Yan Wang, Everlyne M’mbone Muleke and Xianwen Zhu reviewed drafts of the paper.
Bashir Mohammed Jabir prepared figures and/or tables.
Yang Xie performed the experiments, reviewed drafts of the paper.
Wanwan Cheng performed the experiments.
Liwang Liu conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, attracted funding for the project and coordinated the work.
Data Availability
References
- Abrouk et al. (2010).Abrouk M, Murat F, Pont C, Messing J, Jackson S, Faraut T, Tannier E, Plomion C, Cooke R, Feuillet C. Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends in Plant Science. 2010;15:479–487. doi: 10.1016/j.tplants.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Bailey et al. (2009).Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu et al. (2008).Bu Q, Jiang H, Li C-B, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research. 2008;18:756–767. doi: 10.1038/cr.2008.53. [DOI] [PubMed] [Google Scholar]
- Cannon et al. (2004).Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004;4 doi: 10.1186/1471-2229-4-10. Article 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci et al. (2014).Cenci A, Guignon V, Roux N, Rouard M. Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Molecular Biology. 2014;85:63–80. doi: 10.1007/s11103-013-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Slabaugh & Brandizzi (2008).Chen Y-N, Slabaugh E, Brandizzi F. Membrane-tethered transcription factors in Arabidopsis thaliana: novel regulators in stress response and development. Current Opinion in Plant Biology. 2008;11:695–701. doi: 10.1016/j.pbi.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Cheng et al. (2011).Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biology. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling et al. (2008).Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Research. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger et al. (2005).Gasteiger E, Hoogland C, Gattiker A, Duvaud Se, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Humana Press; Totowa: 2005. pp. 571–607. [Google Scholar]
- Hu et al. (2014).Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2006).Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2010).Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in opulus trichocarpa. BMC Plant Biology. 2010;10:145. doi: 10.1186/1471-2229-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala et al. (2001).Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Research. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes & Friedman (2003).Hughes AL, Friedman R. Parallel evolution by gene duplication in the genomes of two unicellular fungi. Genome Research. 2003;13:794–799. doi: 10.1101/gr.714603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen et al. (2010).Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochemical Journal. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- Jeong et al. (2010).Jeong JS, Kim YS, Baek KH, Jung H, Ha S-H, Do Choi Y, Kim M, Reuzeau C, Kim J-K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiology. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin et al. (2014).Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research. 2014;42:D1182–D1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja et al. (2017).Karanja BK, Fan L, Xu L, Wang Y, Zhu X, Tang M, Wang R, Zhang F, Muleke EM, Liu L. Genome-wide characterization of the WRKY gene family in radish (Raphanus sativus L.) reveals its critical functions under different abiotic stresses. Plant Cell Reports. 2017;11:1757–1773. doi: 10.1007/s00299-017-2190-4. [DOI] [PubMed] [Google Scholar]
- Kawahara et al. (2013).Kawahara Y, De la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6 doi: 10.1186/1939-8433-6-4. Article 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi et al. (2000).Kikuchi K, Ueguchi-Tanaka M, Yoshida K, Nagato Y, Matsusoka M, Hirano H-Y. Molecular analysis of the NAC gene family in rice. Molecular and General Genetics. 2000;262:1047–1051. doi: 10.1007/PL00008647. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2008).Kim SG, Lee AK, Yoon HK, Park CM. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. The Plant Journal. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2010).Kim S-G, Lee S, Seo PJ, Kim S-K, Kim J-K, Park C-M. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics. 2010;95:56–65. doi: 10.1016/j.ygeno.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2007).Kim S-Y, Kim S-G, Kim Y-S, Seo PJ, Bae M, Yoon H-K, Park C-M. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Research. 2007;35:203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraga et al. (2007).Kiraga J, Mackiewicz P, Mackiewicz D, Kowalczuk M, Biecek P, Polak N, Smolarczyk K, Dudek MR, Cebrat S. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. BMC Genomics. 2007;8:163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba et al. (2014).Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, Tonosaki K, Shirasawa S, Fukushima A, Yokoi S. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Research. 2014;21:481–490. doi: 10.1093/dnares/dsu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh et al. (2001).Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Le et al. (2011).Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Research. 2011;18:263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2014).Lee S, Lee H-J, Huh SU, Paek K-H, Ha J-H, Park C-M. The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Science. 2014;227:76–83. doi: 10.1016/j.plantsci.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Letunic, Doerks & Bork (2012).Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Research. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2014).Liu T, Song X, Duan W, Huang Z, Liu G, Li Y, Hou X. Genome-wide analysis and expression patterns of NAC transcription factor family under different developmental stages and abiotic stresses in Chinese cabbage. Plant Molecular Biology Reporter. 2014;32:1041–1056. doi: 10.1007/s11105-014-0712-6. [DOI] [Google Scholar]
- Liu et al. (2011).Liu X, Hong L, Li X-Y, Yao Y, Hu B, Li L. Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Bioscience, Biotechnology, and Biochemistry. 2011;75:443–445. doi: 10.1271/bbb.100614. [DOI] [PubMed] [Google Scholar]
- Lynch & Force (2000).Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao et al. (2012).Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. Journal of Experimental Botany. 2012;63:2933–2946. doi: 10.1093/jxb/err462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui et al. (2015).Mitsui Y, Shimomura M, Komatsu K, Namiki N, Shibata-Hatta M, Imai M, Katayose Y, Mukai Y, Kanamori H, Kurita K. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Scientific Reports. 2015;5:10835. doi: 10.1038/srep10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb & Latimer (2008).Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman et al. (2010).Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1007/s12298-017-0421-3. [DOI] [PubMed] [Google Scholar]
- Ooka et al. (2003).Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Pereira-Santana et al. (2015).Pereira-Santana A, Alcaraz LD, Castaño E, Sanchez-Calderon L, Sanchez-Teyer F, Rodriguez-Zapata L. Comparative genomics of NAC transcriptional factors in angiosperms: implications for the adaptation and diversification of flowering plants. PLOS ONE. 2015;10:e0141866. doi: 10.1371/journal.pone.0141866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta et al. (2012).Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Research. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik et al. (2013).Puranik S, Sahu PP, Mandal SN, Parida SK, Prasad M. Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.) PLOS ONE. 2013;8:e64594. doi: 10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon, Ponger & Gaut (2006).Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLOS Computational Biology. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, Kim & Park (2008).Seo PJ, Kim S-G, Park C-M. Membrane-bound transcription factors in plants. Trends in Plant Science. 2008;13:550–556. doi: 10.1016/j.tplants.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Shao, Wang & Tang (2015).Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Frontiers in Plant Science. 2015;6 doi: 10.3389/fpls.2015.00902. Article 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen et al. (2013).Shen D, Sun H, Huang M, Zheng Y, Qiu Y, Li X, Fei Z. Comprehensive analysis of expressed sequence tags from cultivated and wild radish (Raphanus spp.) BMC Genomics. 2013;14:721. doi: 10.1186/1471-2164-14-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh et al. (2013).Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.) DNA Research. 2013;20:403–423. doi: 10.1093/dnares/dst019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song et al. (2011).Song S-Y, Chen Y, Chen J, Dai XY, Zhang WH. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta. 2011;234:331–345. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- Su et al. (2013).Su H, Zhang S, Yuan X, Chen C, Wang XF, Hao YJ. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1, 2-CUC2 transcription factor family in apple. Plant Physiology and Biochemistry. 2013;71:11–21. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2016).Sun X, Xu L, Wang Y, Luo X, Zhu X, Kinuthia KB, Nie S, Feng H, Li C, Liu L. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.) Plant Cell Reports. 2016;35:329–346. doi: 10.1007/s00299-015-1887-5. [DOI] [PubMed] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran et al. (2004).Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi et al. (2007).Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, Zhang J-Y, Benedito V, Hofer JM, Chueng F. Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiology. 2007;144:538–549. doi: 10.1016/j.plaphy.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer et al. (2009).Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: a tale of duplications. Trends in Plant Science. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Vinocur & Altman (2005).Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology. 2005;16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2013a).Wang N, Zheng Y, Xin H, Fang L, Li S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Reports. 2013a;32:61–75. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2015a).Wang R, Xu L, Zhu X, Zhai L, Wang Y, Yu R, Gong Y, Limera C, Liu L. Transcriptome-wide characterization of novel and heat-stress-responsive microRNAs in radish (Raphanus sativus L.) using next-generation sequencing. Plant Molecular Biology Reporter. 2015a;33:867–880. doi: 10.1007/s11105-014-0786-1. [DOI] [Google Scholar]
- Wang et al. (2013b).Wang Y, Xu L, Chen Y, Shen H, Gong Y, Limera C, Liu L. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLOS ONE. 2013b;8:e66539. doi: 10.1371/journal.pone.0066539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2015b).Wang Z, Tang J, Hu R, Wu P, Hou XL, Song XM, Xiong AS. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics. 2015b;16:17. doi: 10.1186/s12864-015-1216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2016).Wei S, Gao L, Zhang Y, Zhang F, Yang X, Huang D. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Reports. 2016;35:1827–1839. doi: 10.1007/s00299-016-1997-8. [DOI] [PubMed] [Google Scholar]
- Wu et al. (2009).Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Research. 2009;19:1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- Xie et al. (2015).Xie Y, Ye S, Wang Y, Xu L, Zhu X, Yang J, Feng H, Yu R, Karanja B, Gong Y. Transcriptome-based gene profiling provides novel insights into the characteristics of radish root response to Cr stress with next-generation sequencing. Frontiers in Plant Science. 2015;6 doi: 10.3389/fpls.2015.00202. Article 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2013).Xu L, Wang Y, Zhai L, Xu Y, Wang L, Zhu X, Gong Y, Yu R, Limera C, Liu L. Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. Journal of Experimental Botany. 2013;64:4271–4287. doi: 10.1093/jxb/ert240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2011).Yang R, Deng C, Ouyang B, Ye Z. Molecular analysis of two salt-responsive NAC-family genes and their expression analysis in tomato. Molecular Biology Reports. 2011;38:857–863. doi: 10.1007/s11033-010-0177-0. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2017).Zhang XM, Yu HJ, Sun C, Deng J, Zhang X, Liu P, Li YY, Li Q, Jiang WJ. Genome-wide characterization and expression profiling of the NAC genes under abiotic stresses in Cucumis sativus. Plant Physiology and Biochemistry. 2017;113:98–109. doi: 10.1016/j.plaphy.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Zheng et al. (2009).Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochemical and Biophysical Research Communications. 2009;379:985–989. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters.
Multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters.
Multiple alignment of full-length RsNAC protein sequences was carried out using the clustalW program with default pairwise and multiple alignment parameters.
The color scales for fold-change values are shown at the bottom such as elevated (red) and suppressed (green) genes. Several genes were not expressed (black) in any tissue.
Gene-specific primers generate from the non-conserved region of NAC sequences using Beacon Designer 7.7.
aTransmembrane motif predicted using the TMHMM server
aAmino acid cMolecular weight
dIsoelectric points
e Arabidopsis thaliana
Clustering of groups A and B based on the similarity in NAC domain structures, group A contain nine subgroups (A1–A9) mostly related to eudicot while group B is composed of ten subgroups (B1–B10) mostly related to monocot.
*Megabase
aReads Per Kilobase of transcript per Million mapped reads
bDays after sowing
Fold change values obtained from transcript levels of 21 randomly selected RsNAC genes using real-time polymerase chain reaction.
aPolyethylene glycol.
babscisic acid.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as Supplemental Information in Tables S1–S6.