ABSTRACT
The genus Limnohabitans (Comamonadaceae, Betaproteobacteria) is a common and a highly active component of freshwater bacterioplanktonic communities. To date, the genus has been considered to contain only heterotrophic species. In this study, we detected the photosynthesis genes pufLM and bchY in 28 of 46 strains from three Limnohabitans lineages. The pufM sequences obtained are very closely related to environmental pufM sequences detected in various freshwater habitats, indicating the ubiquity and potential importance of photoheterotrophic Limnohabitans in nature. Additionally, we sequenced and analyzed the genomes of 5 potentially photoheterotrophic Limnohabitans strains, to gain further insights into their phototrophic capacity. The structure of the photosynthesis gene cluster turned out to be highly conserved within the genus Limnohabitans and also among all potentially photosynthetic Betaproteobacteria strains. The expression of photosynthetic complexes was detected in a culture of Limnohabitans planktonicus II-D5T using spectroscopic and pigment analyses. This was further verified by a novel combination of infrared microscopy and fluorescent in situ hybridization.
IMPORTANCE The data presented document that the capacity to perform anoxygenic photosynthesis is common among the members of the genus Limnohabitans, indicating that they may have a novel role in freshwater habitats.
KEYWORDS: FISH, IR microscopy, Limnohabitans, bacteriochlorophyll, bchY, freshwater Betaproteobacteria, photosynthetic bacteria, pufM
INTRODUCTION
Aerobic anoxygenic phototrophs (AAPs) are bacteria that supplement their mostly heterotrophic metabolism with light energy harvested using bacteriochlorophyll-containing reaction centers (1). AAPs constitute 1 to 11% of the total prokaryotes in the euphotic zone of the world's oceans (2–4), while they may represent up to 34% in more eutrophic environments such as shelf seas and estuaries (5, 6). AAPs also represent a common component of freshwater microbial communities. Large numbers of AAPs were reported for temperate lakes (7–9), peat bogs (10), and rivers (11), frequently correlating with the trophic state and temperature (12). Cultivation-independent approaches showed that AAPs represent a phylogenetically heterogeneous group that includes different clades of Alphaproteobacteria, Gammaproteobacteria, and Betaproteobacteria (13, 14). While the first two clades dominate the marine environment, Betaproteobacteria strains were documented in brackish and freshwater environmental samples. In particular, a large portion of pufM sequences were affiliated with a purple nonsulfur bacterium, Rhodoferax fermentans (Comamonadaceae), as the closest cultured relative (15–18). However, Rhodoferax spp. do not usually constitute an important fraction of freshwater bacterioplankton, in contrast to members of the closely related genus Limnohabitans (19).
The genus Limnohabitans represents an average of 12% of freshwater bacterioplankton, with global distribution in a broad range of habitats (20). Five lineages were differentiated within the genus Limnohabitans (21), but only four (called the “R-BT cluster”) can be targeted by fluorescence in situ hybridization (FISH) with the R-BT065 probe (22). The members of the R-BT cluster share a high growth potential, have relatively large mean cell volumes (0.05 to 0.16 μm3), compared to typical species of bacterioplankton, and are subject to high levels of grazing by protists (21, 23, 24). These ecophysiological traits underline their importance in carbon flow through freshwater food webs (25). The members of four lineages of the genus Limnohabitans are available as cultivated strains; however, due to the lack of any pigmentation, this genus has been considered to be heterotrophic (26–28).
Since the genus Limnohabitans was originally delineated from the genus Rhodoferax (it encompasses species formerly affiliated with the “Rhodoferax sp. BAL47” cluster) (29), we speculated that the environmental betaproteobacterial pufM sequences may actually originate from the genus Limnohabitans. To verify our hypothesis, we investigated the phototrophic potential in our collection of 46 Limnohabitans strains isolated from the epilimnion of various freshwater lakes (21).
RESULTS
Detection of photosynthesis genes in Limnohabitans strains.
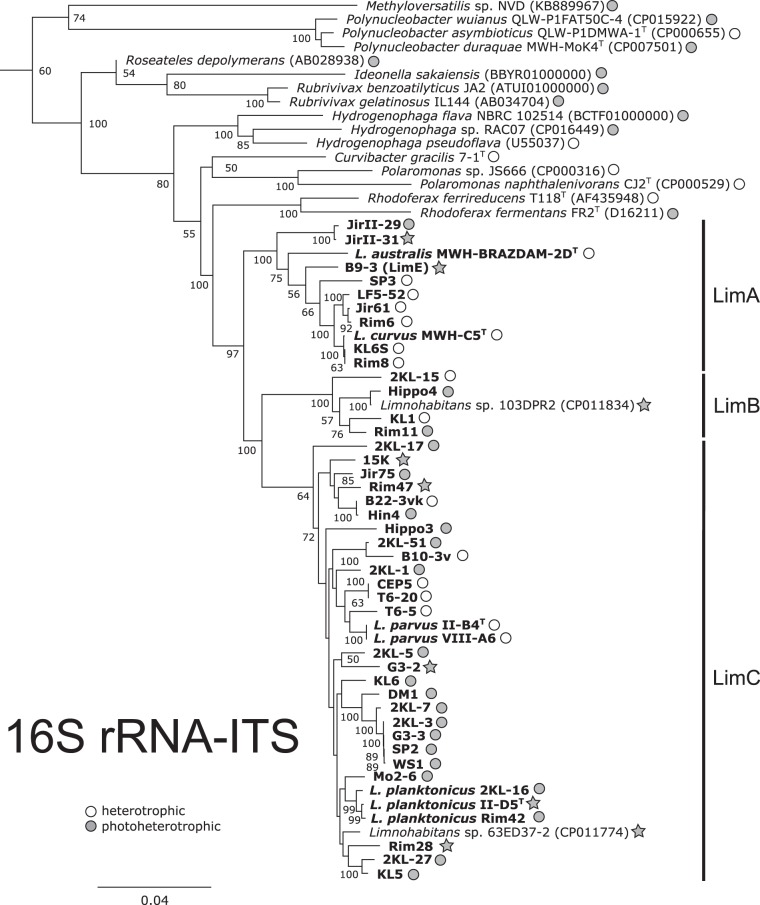
To identify potentially phototrophic strains, we screened our collection of 46 Limnohabitans strains for the presence of pufLM genes (encoding large and small subunits of the bacterial photosynthesis reaction centers) and the bchY gene (encoding a subunit of bacteriochlorophyll reductase Y), using PCR. The pufLM genes were detected in 28 of 46 examined strains from A, B, and C Limnohabitans lineages (Fig. 1), but amplification of the bacteriochlorophyll reductase gene bchY was positive for only 22 pufLM-positive strains. Since phototrophic organisms always contain the pufLM and bchY genes, this indicates that the bchY primers failed to amplify certain forms of bchY genes (see references 30 and 31).
FIG 1.
Phylogenetic tree with the metabolic diversification of the genus Limnohabitans and selected Betaproteobacteria species. The phylogenetic tree was calculated from sequences of the small ribosomal subunits and the internal transcribed spacer (ITS) region using Bayesian inference with 5 million generations. The metabolic type is indicated by color, as follows: gray, photoheterotrophy (pufLM presence); white, heterotrophy (none of the selected genes was detected). The stars indicate strains with available genomes.
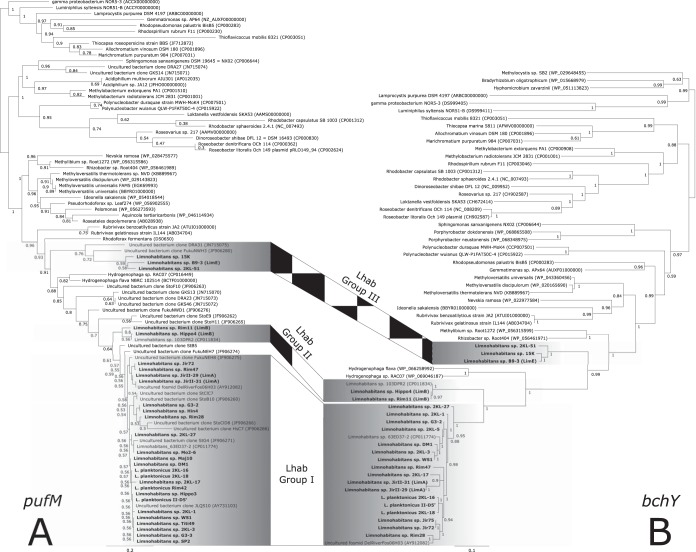
Phylogenetic analyses of the amino acid sequences of both the pufM (Fig. 2A) and bchY (Fig. 2B) genes showed a grouping of all Limnohabitans sequences close to betaproteobacterial sequences of Hydrogenophaga, Rhodoferax, Roseateles, Rubrivivax, and Methyloversatilis. In contrast, both sets of Limnohabitans sequences were found to be distant from two sequences of Polynucleobacter genus members, i.e., Polynucleobacter duraquae strain MWH-MoK4 and Polynucleobacter sp. strain QLW-P1FAT50C-4, and marine Alphaproteobacteria members, i.e., genera Dinoroseobacter, Rhodobacter, and Roseobacter.
FIG 2.
Phylogeny of phototrophy-related genes. The pufM gene (A) and bchY gene (B) phylogeny of the newly obtained Limnohabitans sequences (in bold) was determined with sequences retrieved from the NCBI RefSeq database. The pufM analysis allowed affiliation of environmental pufM clones from lakes in northern Germany (17), lakes in Austria (37), and the estuarine region of the East China Sea (67) with the genus Limnohabitans. The tree was calculated from amino acid alignments using the maximum likelihood approach with general time-reversible plus invariant plus γ distribution and χ2 statistics for branch support.
The studied Limnohabitans strains clustered in three phylogenetic groups (Fig. 2). All three groups were characterized by high within-group similarities of their sequences (>94% for the amino acid sequences and >83% for the nucleic acid sequences) for both pufM and bchY genes. Group I contained 24 strains from the LimC lineage (Fig. 1) (see reference 21) and 2 LimA strains (JirII-29 and JirII-31). Group II consisted of LimB members (strains Rim11, Hippo4, and 103DPR2), with <93% amino acid sequence similarities with group I. The most distant group, group III, included sequences from strains affiliated with LimE (strain B9-3) and LimC (strains 15K and 2KL-51) lineages. The amino acid sequence similarities of group III members with respect to both other groups were <81%. The phylogenetic distance of group III from the other two groups was supported by the fact that two Hydrogenophaga sequences separated Limnohabitans group III from groups I and II. The GC content of pufM sequences was highest for group I (57.6 to 63.3%) and lower for groups II and III (54.2 to 55% and 53.6 to 56.3%, respectively).
Phylogenetic analyses of pufM genes grouped 20 uncultured bacterial clones and 1 fosmid (all retrieved from public databases) within the genus Limnohabitans (Fig. 2A). The fosmid and 9 clones were placed directly within group I and 4 clones were closely related to group III, while none belonged directly to group II.
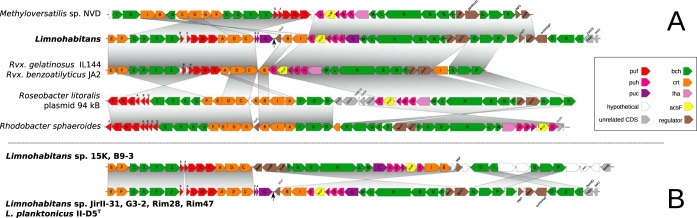
Conserved one-cluster photosynthesis gene organization.
We performed whole-genome sequencing of 5 selected strains, i.e., Limnohabitans planktonicus strain II-D5T and Limnohabitans sp. strains 15K, B9-3, JirII-31, and G3-2, to obtain more information about the photosynthesis gene organization of the pufM-positive strains (Table 1). We also analyzed previously sequenced genomes of Limnohabitans sp. strains Rim28 and Rim47. All of the sequenced species contained photosynthesis genes (from 42 to 47 genes) organized in a photosynthesis gene cluster (PGC). The main part of the PGC in Limnohabitans is formed by two conserved regions, with the first two regulatory genes being oriented in opposite directions (Fig. 3). The first conserved region contains the genes crtEF, bchCXYZ, pufBALMC, and crtADC and the second contains ppsR, ppaA, bchFNBHLM, pucC, puhABC, acsF, and puhE (Fig. 3). Both regions are also highly conserved in genomes of the Betaproteobacteria members Methyloversatilis sp. strain NVD, Rubrivivax gelatinosus, and Rubrivivax benzoatilyticus and even in the Alphaproteobacteria members Roseobacter litoralis and Rhodobacter sphaeroides. In contrast to the aforementioned genomes, the Limnohabitans PGC has a crtIB-bluF gene set located next to puhE. The whole PGC contained another three sets of genes, i.e., genes encoding bacteriochlorophyll synthase (bchGP) and bacteriochlorophyll reductase (bchIDO) and regulatory genes (tspO, ppaA, cvrA/nhaP, and F420), which are of variable order and composition (detailed in Table S1 in the supplemental material). Genomes of Limnohabitans sp. strains 15K and B9-3 have an opposite orientation of the second conserved region, compared to the genomes of strains II-D5, JirII-31, Rim28, Rim47, and G3-2. In strain II-D5T, the PGC is split into two parts (between pucC and bluF), separated by six genes related to arginine export.
TABLE 1.
Characteristics of Limnohabitans genomes and PGC clusters
| Characteristic | Strain JirII-31 | Strain 15K | Strain II-D5T | Strain G3-2 | Strain B9-3 |
|---|---|---|---|---|---|
| Limnohabitans lineage | LimA | LimC | LimC | LimC | LimE |
| Genome information | |||||
| Genome size (Mb) | 3.48 | 3.55 | 4.74 | 3.25 | 3.40 |
| Mean sequencing coverage (fold) | 200 | 210 | 60 | 250 | 220 |
| No. of contigs | 44 | 15 | 7 | 25 | 23 |
| G+C content (%) | 57.8 | 58.2 | 59.4 | 60.1 | 56.5 |
| No. of protein-coding genes | 3,368 | 3,342 | 4,273 | 3,025 | 3,174 |
| Coding region (% of bp) | 94.7 | 93.0 | 85.5 | 92.5 | 94.1 |
| No. of rRNA operons | 2 | 2 | 4 | 2 | 2 |
| No. of tRNA genes | 44 | 43 | 45 | 41 | 42 |
| Photosynthesis gene cluster | |||||
| PGC length (kb) | 48.0 | 54.2 | 48.7 | 45.6 | 51.1 |
| PGC type | I | II | I | I | II |
| Outer antenna (PucAB) | + | − | + | + | − |
| L-POR | − | + | − | − | − |
FIG 3.
Photosynthesis gene cluster organization. (A) The PGCs in Limnohabitans, Methyloversatilis, and Rubrivivax (all Betaproteobacteria) and in Rhodobacter and Roseobacter (both Alphaproteobacteria) were compared. (B) The organization of PGC in Limnohabitans sp. strain 15K and B9-3 differed from that in other Limnohabitans genomes. Photosynthesis genes were annotated according to reference 13, and the full annotation is available in Table S1 in the supplemental material. Rvx., Rubrivivax; CDS, coding DNA sequence.
All sequenced Limnohabitans strains contain only an aerobic form of Mg-protoporphyrin cyclase (acsF gene product) in their genomes, whereas the anaerobic form (bchE gene product) is absent. The presence of the pufC gene in all strains indicates that Limnohabitans strains use anoxygenic reaction centers with a tetraheme cytochrome c subunit. In contrast, the pufX gene, which is frequently present among phototrophic Rhodobacterales strains, has not been found in any Limnohabitans strains. The PGC also contained the crtA gene, which encodes the spheroidene monooxygenase (Fig. 3) that converts spheroidene into spheroidenone in the final step of spheroidenone biosynthesis. Limnohabitans sp. strains 15K and B9-3 were the only strains lacking pucA and pucB genes, which encode the subunits of the outer light-harvesting complex that enlarge the optical cross section of photosynthesis reaction centers.
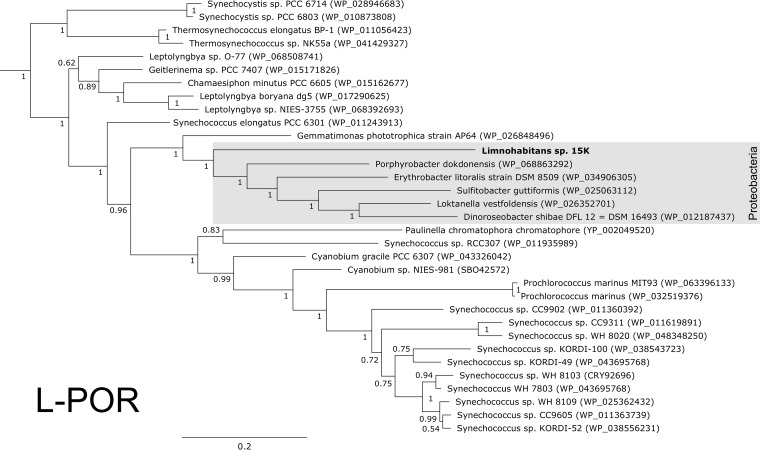
Light-dependent protochlorophyllide oxidoreductase in Limnohabitans.
A very interesting feature is the presence of light-dependent protochlorophyllide oxidoreductase (L-POR) in the genome of Limnohabitans sp. strain 15K. The percent identity between the Limnohabitans sp. strain 15K L-POR and the closest homologs was low, at 82.3%. The Limnohabitans L-POR sequence clustered together with sequences of the photoheterotrophic bacteria Dinoroseobacter shibae, Erythrobacter litoralis, and Gemmatimonas phototrophica and more distantly with the cyanobacteria Synechococcus sp. and Cyanobium sp. (Fig. 4).
FIG 4.
Phylogeny of the light-dependent protochlorophyllide reductase genes. Newly obtained sequences from two Limnohabitans genomes (in bold) clustered together with other proteobacterial L-POR sequences. The tree was calculated from an amino acid alignment using Bayesian inference, with posterior bootstrap support.
Bacteriochlorophyll evidence in Limnohabitans.
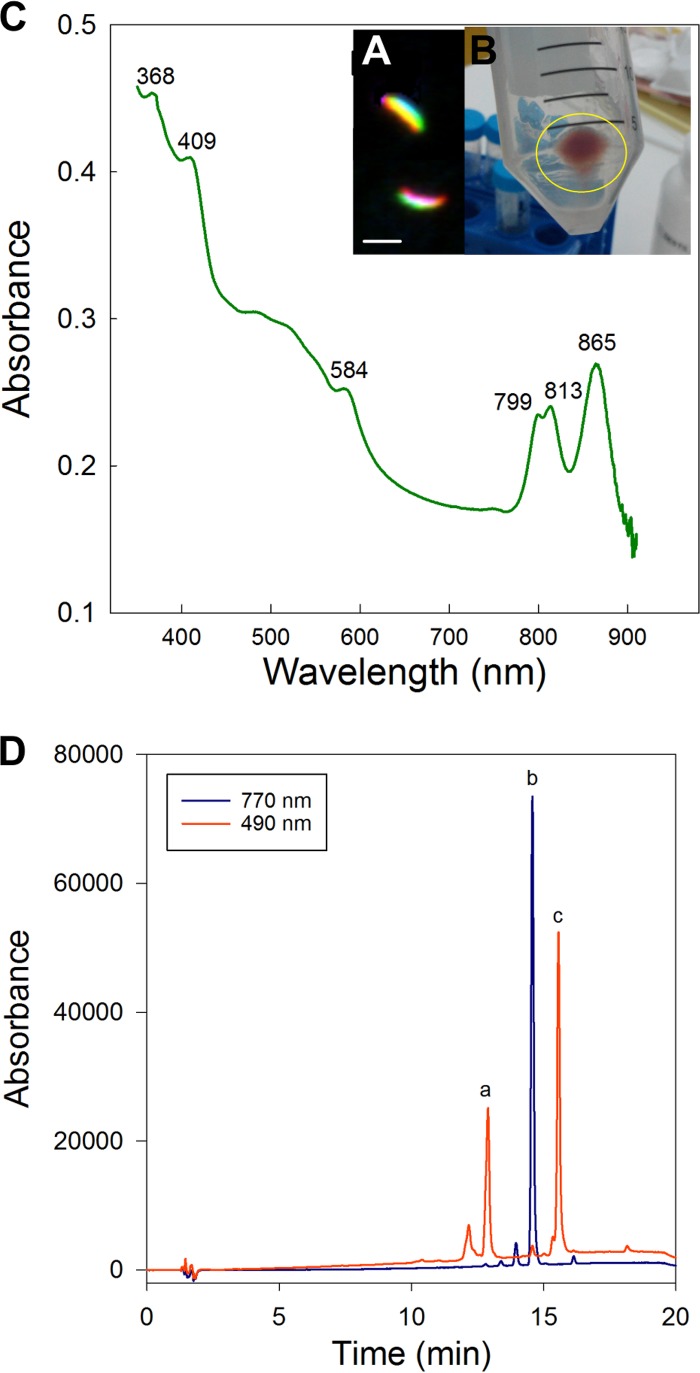
The expression of photosynthetic pigments was examined in batch cultures under different nutrient regimens without any detectable signal, except for L. planktonicus strain II-D5T grown in YST2 medium. The sedimented phototrophic cells of L. planktonicus II-D5T were pink (Fig. 5B). The recorded in vivo absorption spectrum documented the presence of three near-infrared (IR) peaks, at 799 nm, 813 nm, and 865 nm (Fig. 5C), originating from outer and inner light-harvesting complexes. High-performance liquid chromatography (HPLC) analysis revealed the presence of bacteriochlorophyll a (Bchl-a), hydroxyspheroidenone, and spheroidenone as the main carotenoids (Fig. 5D), which agrees with the prediction from the genomic information (crtA gene). Moreover, the expression of photosynthetic complexes of this strain was verified at the single-cell level using the optimized FISH protocol, which preserves Bchl-a autofluorescence in the near-infrared spectrum. Bchl-a autofluorescence was detectable at NaCl concentrations in the washing buffer below 135 mM (Table S2). This corresponds to the 20% formamide concentration in the hybridization buffer, and most probes, including R-BT065 targeting the B, C, D, and E lineages of the Limnohabitans genus, require much higher formamide concentrations (32). Experiments combining a high formamide concentration in the hybridization buffer with a high NaCl concentration in the washing buffer or a low formamide concentration in the hybridization buffer with a low NaCl concentration in the washing buffer (both not used in real FISH protocols) pointed to detrimental effects of high NaCl concentrations in the washing buffer. The new FISH-IR protocol allows determination of the phylogenetic affiliation of Bchl-a-containing bacteria. With this assay, we verified the purity of the cultures and confirmed that the cells showing bacteriochlorophyll autofluorescence in the near-infrared spectrum were indeed Limnohabitans (Fig. 5A).
FIG 5.
Pigment analysis results for L. planktonicus II-D5T cells. (A) False-color photomicrographs of L. planktonicus II-D5T cells hybridized with the R-BT065 probe (green), showing bacteriochlorophyll a autofluorescence (red). The cells were hybridized using a probe for double labeling of oligonucleotides with a Cy-3 fluorochrome, but the signal is shown green (not orange) for clarity. Scale bar, 2 μm. (B) Pink coloration of L. planktonicus II-D5T cells. (C) In vivo absorption of the pigmented L. planktonicus II-D5T strain. (D) HPLC chromatograms of pigments extracted from L. planktonicus II-D5T. Peak a (at 490 nm) corresponds to hydroxyspheroidenone, peak b (at 770 nm) to bacteriochlorophyll a, and peak c (at 490 nm) to hydroxyspheroidenone.
DISCUSSION
Limnohabitans, new aerobic anoxygenic photoheterotrophs in freshwater.
We showed that the genus Limnohabitans, in addition to chemoheterotrophic strains, contains photoheterotrophic members. We found, by sequencing of two characteristic photosynthesis genes and an analysis of potentially photosynthetic genomes, that Limnohabitans strains possess the characteristics of AAPs, i.e., Bchl-a-containing reaction centers (33) and the presence of only the aerobic form of the oxidative cyclase (ascF gene). Photoheterotrophic Limnohabitans strains, like marine Alphaproteobacteria strains (34), have genes organized in a photosynthesis gene cluster, including the genes for spheroidene biosynthesis (35). We demonstrated that Limnohabitans planktonicus II-D5T produced bacteriochlorophyll a (Fig. 5); however, the synthesis of bacteriochlorophylls or carotenoids by Limnohabitans seems to be strongly downregulated under laboratory conditions. It is plausible that expression of the genes in the PGC can be regulated according to environmental conditions. We speculate that photoheterotrophy, which is very important in marine systems (33, 36), might be subjected to stronger regulations in freshwater systems, e.g., by the temperature (7, 8, 37). Further investigations into the regulation of sunlight energy utilization by Limnohabitans are needed and should be facilitated by the application of our optimized FISH-IR protocol.
Environmental significance for photoheterotrophic Limnohabitans.
Our phylogenetic analyses showed that a substantial part of the environmental betaproteobacterial pufM sequences retrieved from the epilimnia of freshwater lakes and rivers (15–18, 37) originated from Limnohabitans species (Fig. 2); for Lake Stechlin, Lake Stolp, and Lake Fuchskuhle northeast basin, they constituted 70 to 90% of all retrieved pufM clones (17). Moreover, several uncultured pufM clones and fosmids from the estuarine sites also were affiliated with Limnohabitans (4, 38). This is in concordance with the distribution of the R-BT cluster of Limnohabitans in a wide range of environmental conditions, including brackish waters (4, 20, 39). We postulate that it is photoheterotrophy combined with a broad range of carbon sources (21, 40) that give a significant advantage to the Limnohabitans genus over chemoheterotrophic bacteria and allows them to be regularly present within bacterioplanktonic communities, where they play a key role in the microbial food web (23). Still, the quantitative aspects of the occurrence of phototrophic Limnohabitans remain unknown. Here, we successfully combined two epifluorescence-based methods, i.e., FISH and IR microscopy (Fig. 5; also see Table S2 in the supplemental material). The newly developed approach should be easily applicable to environmental samples to advance our understanding of the ecology of phototrophic Limnohabitans and other AAPs.
Evolution of phototrophy in Betaproteobacteria.
The information on bacterial photosynthesis in Betaproteobacteria is increasing with every new genome-sequenced genus. Rubrivivax, Roseateles, Rhodoferax, Rhodocyclus, and Methyloversatilis were the only known genera within Betaproteobacteria for a long time (17, 37). Currently, other genera, including freshwater Polynucleobacter (41) and the genus Limnohabitans, have extended this list. Our data suggest that the inheritance of phototrophy in Betaproteobacteria was most likely vertical despite the large phylogenetic distances among the photosynthetic genera (Fig. 1). Surprisingly, most Betaproteobacteria members share very similar features of their PGCs, i.e., they are allocated on chromosomes, they have highly conserved gene orders, and phylogenetic analyses of the pufM and bchY genes indicated a common ancestry of these genes in Betaproteobacteria (Fig. 2), as was shown for the ascF gene (42). However, different PGC organizations were found in Limnohabitans sp. strains 15K and B9-3 (Fig. 3), suggesting that the PGCs in these strains had been acquired through horizontal gene transfer. Our hypothesis could be supported by (i) phylogenetic separation of their pufM and bchY genes (Fig. 2) and (ii) the presence of L-POR in strain 15K. This light-dependent protochlorophyllide oxidoreductase has long been thought to be specific to cyanobacteria, algae, and plants, while aerobic anoxygenic phototrophic bacteria usually employ the light-independent form. Recently, L-POR was confirmed also in the genomes of Gemmatimonas phototrophica and Alphaproteobacteria members such as Dinoroseobacter shibae and Erythrobacter litoralis (43).
Not all Limnohabitans strains have the potential for phototrophy, and many phototrophic strains are most closely related to a chemoheterotrophic bacterium (Fig. 1). This resembles the situation in the Roseobacter clade (44), where multiple regressive gene losses were suggested to explain the distribution of phototrophic genes (45). We suppose that such events, rather than multiple horizontal gene transfers, would predominantly explain the observed patchy distribution of phototrophy within Limnohabitans bacteria.
Limnohabitans, the freshwater Roseobacter?
Freshwater and marine aquatic habitats differ greatly in nutrient dynamics and are inhabited by phylogenetically distinct lineages of microbes (46), although they seem to provide similar ecological constraints, particularly the organic carbon supply and the predation pressure by flagellates. The metabolic traits and ecophysiology of the investigated freshwater genus Limnohabitans (21) resemble those suggested for the marine Roseobacter clade (47). Both groups are actively growing organisms (48, 49) under high grazing pressure (50, 51). Both clades represent generalist species with larger genomes encoding multiple enzymatic activities (including phototrophy), allowing rapid growth. Other similarities between the two groups include the chessboard distribution of photosynthesis genes (44) and the poor expression of bacteriochlorophylls under laboratory conditions in some strains (12). Both groups are frequently found in algal cultures (52–54), which suggests a specific association between algae and bacteria. This life strategy may also explain the presence of the typically “algal” gene L-POR in Limnohabitans strains B9-3 and 15K. Interestingly, the presence of L-POR in Dinoroseobacter shibae (a Roseobacter clade member) has also been reported, but in that organism the gene is located on a plasmid (43). In light of the current data, we propose that the highly flexible and diverse genus of Limnohabitans represent a freshwater counterpart of the well-known marine Roseobacter clade.
MATERIALS AND METHODS
Bacterial strains, cultivation, and DNA extraction.
We used 46 Limnohabitans strains from our culture collection, including 8 novel strains (Limnohabitans sp. strains 2KL-51, DM1, JirII-29, JirII-31, Jir72, Jir75, Titi28, and Titi49). These strains were isolated, using the previously described procedure (21), from the Klíčava Reservoir (Czech Republic) sampled in 2008 (strain 2KL-51), from a Daphnia magna culture (University of Konstanz collection) sampled in 2009 (strain DM1), from the Jiřická Reservoir (Czech Republic) sampled in 2014 (strains JirII-29, JirII-31, Jir72, and Jir75), and from a sample collected at Lake Titicaca (Peru) during the summer of 2010 (strains Titi28 and Titi49). DNA was extracted by the phenol-chloroform-isoamyl alcohol method or using the UltraClean isolation kit (MoBio, Laboratories, Inc.).
Primer design and PCR product sequencing.
At least two PCRs, with the primer pairs targeting (i) photosynthetic reaction center L and M subunit (pufL and pufM) and (ii) bacteriochlorophyll reductase Y subunit (bchY) genes, were performed for each Limnohabitans strain (17, 30). The successfully amplified genes were subsequently sequenced to confirm their identity. We also designed new primers sets in order to obtain full-length sequences of the genes (for details, see Table 2). The optimal annealing temperatures for the new primer sets were determined by temperature gradient analysis (range of 49°C to 64°C, with steps of approximately 1°C). PCR products were sequenced by the Eurofins MWG Operon service.
TABLE 2.
Primer pairs used within this study
| Gene(s) | Forward primer |
Reverse primer |
Annealing temperature (°C)a | PCR product length (bp) | Reference(s) | ||
|---|---|---|---|---|---|---|---|
| Name | Sequence (5′ to 3′) | Name | Sequence (5′ to 3′) | ||||
| Photosynthetic reaction center (pufL and pufM) | pufL-64F | CTB TTC GAY TTC TGG RTS GG | pufM-754R | CCA TSG TCC AGC GCC AGA A | 55 | 1,600 | 13 |
| pufL | CTK TTC GAC TTC TGG GTS GG | pufM_uniR | CCA TSG TCC AGC GCC AGA A | 60 | 1,600 | 17 | |
| pufL-4F | GCC ATG CTG ARY TTT GAR AAA | pufM-1788R | CTT GAT GGC CCA SAG GTA | 50–61 (52) | 1,800 | This study | |
| Bacteriochlorophyll reductase (bchY) | bchY_fwd | CCN CAR WSN ATG TGY CCN GCN TTY | bchZ_rev2 | ART ABC CSC CNG CNC KRT CRW GRT | 52 | 1,500 | 30, 66 |
| bchY-22F | ACA CGC ACC ATC CCG ATT | bchY-1562R | CAT GWD CCC ATC YCC TCV GMT TT | 50–61 (52) | 1,500 | This study | |
The annealing temperature used for PCR is indicated in parentheses.
Genome sequencing and assembly.
Limnohabitans sp. strains JirII-31, B9-3, 15K, and G3-2 were sequenced using Illumina MiSeq v3 technology in the paired-end read module (2 by 300 bp). Reads were grouped by their names, and the low-quality regions (ends) were trimmed using the default settings in Geneious R8 (Biomatters Ltd.). Paired reads were assembled into contigs or scaffolds by using the Velvet assembler (55), the Spades assembler (56), or the assembler in Geneious R8. The assemblies obtained were compared with MUMmer 3.0 software (57) to improve the scaffolding. After genome annotation, additional scaffolding was performed manually based on conservative gene clustering. The reads were mapped onto the established scaffolds again to ensure that the scaffolding was correct.
The genome of Limnohabitans planktonicus II-D5T was sequenced by Beckman Coulter Genomics (France). Two libraries were constructed, i.e., a mate pair for 454 sequencing using GS FLX Titanium technology and a fragment library for Illumina GAII sequencing (100 bp). Subsequently, 331,000 Roche next-generation sequencing (NGS) reads (110.8 Mb) and 2,000,000 Illumina NGS reads (200 Mb) were produced, resulting in about 50-fold coverage of the genome. Reads were assembled with MIRA software in two modes, i.e., (i) both 454 and Illumina reads were used for elongation of contigs or (ii) Illumina reads were used mainly for polishing of the contigs established from 454 reads. Independent assembly was performed using Newbler software, with 454 reads being used for creation and elongation of contigs and Illumina reads being used for polishing. The assemblies obtained were manually curated and compared as described for the previous four genomes.
Genome annotation.
Draft genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (released in 2013) (http://www.ncbi.nlm.nih.gov/genome/annotation_prok) and using the SEED and Rapid Annotation using Subsystems Technology (RAST) systems (58). In addition to newly sequenced genomes, we included Limnohabitans sp. strains Rim28 and Rim47 (59) in further analyses. The automatic annotation of photosynthesis gene clusters (PGCs) obtained with the RAST algorithm was manually controlled using the known PGCs of Rhodoferax fermentans, Rubrivivax gelatinosus, and Rhodobacter sphaeroides and the environmental fosmid DelRiverFos06H03, obtained from the GenBank database. Moreover, we compared the annotations with a previous description of PGCs (60). In cases of incorrect automatic annotation, the genes were reanalyzed with the BLASTp or tBLASTx module within BLAST (http://blast.ncbi.nlm.nih.gov) and were reannotated using nonredundant Ref_Seq databases or a local database created from the correctly annotated strains. Similarly, nonrelated genes within the PGCs were analyzed and compared with each other to determine their conservative emergence in the Limnohabitans genomes.
Phylogenetic analyses.
Nucleotide sequences of pufLM or bchY genes from the Limnohabitans strains were compared with the NCBI nucleotide database (BLASTN) and translated nucleotide sequences with the translated nucleotide database (tBLASTx). Translated nucleotide sequences were aligned using the MUSCLE algorithm (61). Three independent algorithms (neighbor joining, maximum likelihood, and Bayesian inference) were used to determine the most appropriate phylogeny, based on the amino acid composition, using Geneious R8, PhyML 3.1 (62), and MrBayes (63) software, respectively. Phylogenetic trees were visualized with FigTree v1.4.1 (Andrew Rambaut, University of Edinburgh) (http://tree.bio.ed.ac.uk).
Pigment determination of L. planktonicus, microscopy, and optical measurements.
L. planktonicus strain II-D5T was grown in YST2 medium (0.5 g liter−1 BD Bacto yeast extract, 0.5 g liter−1 BD Bacto casein acids, 0.5 g liter−1 potato flour [pH 7.2]) on an orbital shaker (150 rpm) at 24°C, with a 12-h/12-h dark/light cycle (Osram Dulux L 55W/865 light source). The presence of Bchl-a in the culture was analyzed by HPLC. Cells were harvested by centrifugation at 6,000 × g for 16 min (Sigma 2-16KL centrifuge), and the pellet was resuspended in 20 μl of deionized water. Pigments were extracted with 1 ml of acetone/methanol (7:2 [vol/vol]). The clear extract was analyzed with a Prominence-i HPLC system (LC-2030C; Shimadzu, Japan) equipped with a UV-visible diode array detector, as described previously (64). Carotenoids were detected at 490 nm and bacteriochlorophyll a at 770 nm. Pigments were identified based on their retention times and absorption spectra.
Bchl-a in individual bacterial cells was detected using infrared epifluorescence microscopy (65). Cells either were fixed in formaldehyde (final concentration of 2%) or remained unfixed for immediate analysis. The purity of the cultures was confirmed by using the catalyzed reporter deposition (CARD)-fluorescence in situ hybridization (FISH) method (32) with the R-BT065 probe (22) and by sequencing the pufM gene (13).
Optimization of fluorescence in situ hybridization for detection of Bchl-a in specific phylotypes at the single-cell level.
To prove that the observed autofluorescence in the infrared light came from L. planktonicus, we modified the FISH technique (32) to allow simultaneous detection of infrared and probe signals. The samples were fixed with fresh paraformaldehyde solution (final concentration of 2% [pH 7.5]) for 1 h at room temperature (about 21°C) or overnight at 4°C, filtered onto Nuclepore polycarbonate filters (pore size, 0.2 μm; Whatman, UK), and stored at −20°C. We decreased the hybridization temperature to 35°C and the washing temperature to 37°C, and we shortened the washing time to 20 min. The final rinsing of filters was performed in sterile deionized water and never in ethanol. Hybridized filter sections were mounted in Vectashield mounting medium.
We tested the effects of the hybridization conditions on Bchl-a autofluorescence with nine bacterial species, namely, Rubrivivax gelatinous, Rhodoferax antarcticus, Rhodoferax fermentans, Rhodospirillum rubrum, Rhodopseudomonas palustris, Methylobacter sp., Rhodobacter sphaeroides, Limnohabitans planktonicus, and Sphingomonas sp. We performed triplicate tests with formamide concentrations ranging from 0 to 70%, with steps of 10%. Additionally, we combined high formamide concentrations in the hybridization buffer with high NaCl concentrations in the washing buffer and low formamide concentrations in the hybridization buffer with low NaCl concentrations in the washing buffer to identify which step is critical for successful BChl-a detection upon FISH.
Upon optimization of the FISH-IR protocol, Bchl-a autofluorescence in L. planktonicus strain II-D5T was confirmed with the R-BT065 probe (with 55% formamide in the hybridization buffer) double-labeled with Cy3 dye (Biomers, Germany). Microscopic observation was performed using an AxioVision.Z2 microscope (Zeiss, Germany) with UV and blue excitation and emission, respectively, for 4′,6-diamidino-2-phenylindole (DAPI) staining, blue and green for CARD-FISH signals, green and orange for FISH signals, and white and infrared for Bchl-a autofluorescence.
Accession number(s).
The 5 genome-investigated strains were deposited in the Leibniz Institut-Deutsche Sammlung von Mikroorganismen und Zellkulturen, and their genome sequences are available as whole-genome shotgun projects at GenBank/ENA/DDBJ, with the following accession numbers: L. planktonicus strain II-D5T (DSM 21594), LFYT00000000; Limnohabitans sp. strain JirII-31 (DSM 104470), NESA00000000; Limnohabitans sp. strain G3-2 (DSM 104469), NESG00000000; Limnohabitans sp. strain B9-3 (DSM 104471), NESI00000000; and Limnohabitans sp. strain 15K (DSM 104472), NESM00000000. Novel gene sequences have the following GenBank accession numbers: pufLM, KM659099 to KM659120; bchY, KT948757 to KT948764; and 16S rRNA genes with internal transcribed spacer, KT899705 to KT899709.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Boldareva-Nuyanzina and Z. Cepáková for their help with the pufM sequences, M. W. Hahn for help with NGS, and K. Kopejtka and V. Selyanin for HPLC analysis. We are grateful to R. Dick (Eurofins Genomics) for carrying out all of the sequencing data analysis. We also thank R. Malá and M. Štojdlová for their excellent laboratory assistance and J. Dean for the language correction.
V.K. wrote the manuscript and provided Limnohabitans strains, PCR and sequencing, genome assembly and annotation, phylogenetic analyses, and project funding, Y.Z. provided sequencing and genome assembly and photosynthesis gene annotation, K.P. provided development of the FISH-IR protocol, procurement of photoheterotrophic L. planktonicus, and critical appraisal of the manuscript, K.Š. provided critical appraisal of the manuscript and project funding, H.K. provided DNA extraction, PCR, and sequencing, and M.K. provided the original idea, writing and critical appraisal of the manuscript, and project funding.
This study was largely supported by the European Social Fund (project CZ.1.07/2.3.00/30.0032) and the Czech Science Foundation under research grants 13-00243S (awarded to K.Š.) and 15-12197S (awarded to V.K. and K.P.). MŠMT project Algatech Plus (project LO1416) supported M.K. and K.P. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02116-17.
REFERENCES
- 1.Yurkov V, Csotonyi JT. 2009. New light on aerobic anoxygenic phototrophs, p 31–55. In Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed), Advances in photosynthesis and respiration, vol 28 The purple phototrophic bacteria. Springer, Dordrecht, Netherlands. [Google Scholar]
- 2.Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 3.Sieracki ME, Gilg IC, Thier EC, Poulton NJ, Goericke R. 2006. Distribution of planktonic aerobic anoxygenic photoheterotrophic bacteria in the northwest Atlantic. Limnol Oceanogr 51:38–46. doi: 10.4319/lo.2006.51.1.0038. [DOI] [Google Scholar]
- 4.Jiao N, Zhang Y, Zeng Y, Hong N, Liu R, Chen F, Wang P. 2007. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ Microbiol 9:3091–3099. doi: 10.1111/j.1462-2920.2007.01419.x. [DOI] [PubMed] [Google Scholar]
- 5.Cottrell MT, Waidner LA, Yu L, Kirchman DL. 2005. Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ Microbiol 7:1883–1895. doi: 10.1111/j.1462-2920.2005.00762.x. [DOI] [PubMed] [Google Scholar]
- 6.Mašín M, Zdun A, Stoń-Egiert J, Nausch M, Labrenz M, Moulisová V, Koblížek M. 2006. Seasonal changes and diversity of aerobic anoxygenic phototrophs in the Baltic Sea. Aquat Microb Ecol 45:247–254. doi: 10.3354/ame045247. [DOI] [Google Scholar]
- 7.Mašín M, Nedoma J, Pechar L, Koblížek M. 2008. Distribution of aerobic anoxygenic phototrophs in temperate freshwater systems. Environ Microbiol 10:1988–1996. doi: 10.1111/j.1462-2920.2008.01615.x. [DOI] [PubMed] [Google Scholar]
- 8.Masin M, Cuperova Z, Hojerova E, Salka I, Grossart H-P, Koblížek M. 2012. Distribution of aerobic anoxygenic phototrophic bacteria in glacial lakes of northern Europe. Aquat Microb Ecol 66:77–86. doi: 10.3354/ame01558. [DOI] [Google Scholar]
- 9.Fauteux L, Cottrell MT, Kirchman DL, Borrego CM, Garcia-Chaves MC, Del Giorgio PA. 2015. Patterns in abundance, cell size and pigment content of aerobic anoxygenic phototrophic bacteria along environmental gradients in northern lakes. PLoS One 10:e0124035. doi: 10.1371/journal.pone.0124035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew S, Koblížek M, Lew M, Medová H, Glińska-Lewczuk K, Owsianny PM. 2015. Seasonal changes of microbial communities in two shallow peat bog lakes. Folia Microbiol (Praha) 60:165–175. doi: 10.1007/s12223-014-0352-0. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-González C, Proia L, Ferrera I, Gasol JM, Sabater S. 2013. Effects of large river dam regulation on bacterioplankton community structure. FEMS Microbiol Ecol 84:316–331. doi: 10.1111/1574-6941.12063. [DOI] [PubMed] [Google Scholar]
- 12.Koblížek M. 2015. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 39:854–870. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 13.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 14.Yutin N, Suzuki MT, Teeling H, Weber M, Venter JC, Rusch DB, Béjà O. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling Expedition metagenomes. Environ Microbiol 9:1464–1475. doi: 10.1111/j.1462-2920.2007.01265.x. [DOI] [PubMed] [Google Scholar]
- 15.Waidner LA, Kirchman DL. 2008. Diversity and distribution of ecotypes of the aerobic anoxygenic phototrophy gene pufM in the Delaware estuary. Appl Environ Microbiol 74:4012–4021. doi: 10.1128/AEM.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Dong H, Yu B, Lv G, Deng S, Wu Y, Dai M, Jiao N. 2009. Abundance and diversity of aerobic anoxygenic phototrophic bacteria in saline lakes on the Tibetan plateau. FEMS Microbiol Ecol 67:268–278. doi: 10.1111/j.1574-6941.2008.00616.x. [DOI] [PubMed] [Google Scholar]
- 17.Salka I, Čuperová Z, Mašín M, Koblížek M, Grossart H-PP. 2011. Rhodoferax-related pufM gene cluster dominates the aerobic anoxygenic phototrophic communities in German freshwater lakes. Environ Microbiol 13:2865–2875. doi: 10.1111/j.1462-2920.2011.02562.x. [DOI] [PubMed] [Google Scholar]
- 18.Caliz J, Casamayor EO. 2014. Environmental controls and composition of anoxygenic photoheterotrophs in ultraoligotrophic high-altitude lakes (central Pyrenees). Environ Microbiol Rep 6:145–151. doi: 10.1111/1758-2229.12142. [DOI] [PubMed] [Google Scholar]
- 19.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Šimek K, Kasalický V, Jezbera J, Jezberová J, Hejzlar J, Hahn MW. 2010. Broad habitat range of the phylogenetically narrow R-BT065 cluster, representing a core group of the betaproteobacterial genus Limnohabitans. Appl Environ Microbiol 76:631–639. doi: 10.1128/AEM.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasalický V, Jezbera J, Hahn MW, Šimek K. 2013. The diversity of the Limnohabitans genus, an important group of freshwater bacterioplankton, by characterization of 35 isolated strains. PLoS One 8:e58209. doi: 10.1371/journal.pone.0058209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Šimek K, Pernthaler J, Weinbauer MG, Horňák K, Dolan JR, Nedoma J, Mašín M, Amann R. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl Environ Microbiol 67:2723–2733. doi: 10.1128/AEM.67.6.2723-2733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šimek K, Horňák K, Jezbera J, Nedoma J, Vrba J, Straškrábová V, Macek M, Dolan JR, Hahn MW. 2006. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ Microbiol 8:1613–1624. doi: 10.1111/j.1462-2920.2006.01053.x. [DOI] [PubMed] [Google Scholar]
- 24.Jezbera J, Horňák K, Šimek K. 2006. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol 8:1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 25.Šimek K, Kasalický V, Jezbera J, Horňák K, Nedoma J, Hahn MW, Bass D, Jost S, Boenigk J. 2013. Differential freshwater flagellate community response to bacterial food quality with a focus on Limnohabitans bacteria. ISME J 7:1519–1530. doi: 10.1038/ismej.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn MW, Kasalický V, Jezbera J, Brandt U, Jezberová J, Šimek K. 2010. Limnohabitans curvus gen. nov., sp. nov., a planktonic bacterium isolated from a freshwater lake. Int J Syst Evol Microbiol 60:1358–1365. doi: 10.1099/ijs.0.013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn MW, Kasalický V, Jezbera J, Brandt U, Šimek K. 2010. Limnohabitans australis sp. nov., isolated from a freshwater pond, and emended description of the genus Limnohabitans. Int J Syst Evol Microbiol 60:2946–2950. doi: 10.1099/ijs.0.022384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasalický V, Jezbera J, Šimek K, Hahn MW. 2010. Limnohabitans planktonicus sp. nov. and Limnohabitans parvus sp. nov., planktonic betaproteobacteria isolated from a freshwater reservoir, and emended description of the genus Limnohabitans. Int J Syst Evol Microbiol 60:2710–2714. doi: 10.1099/ijs.0.018952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han S-K. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155. doi: 10.3354/ame028141. [DOI] [Google Scholar]
- 30.Yutin N, Suzuki MT, Rosenberg M, Rotem D, Madigan MT, Süling J, Imhoff JF, Béjà O. 2009. BchY-based degenerate primers target all types of anoxygenic photosynthetic bacteria in a single PCR. Appl Environ Microbiol 75:7556–7559. doi: 10.1128/AEM.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imhoff J. 2016. New dimensions in microbial ecology: functional genes in studies to unravel the biodiversity and role of functional microbial groups in the environment. Microorganisms 4:19. doi: 10.3390/microorganisms4020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernthaler A, Pernthaler J, Amann R. 2004. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms, p 711–726. In Kowalchuk GA, de Bruijn F, Head IM, Van der Zijpp AJ, van Elsas JD (ed), Molecular microbial ecology manual, 2nd ed Springer, Dordrecht, Netherlands. [Google Scholar]
- 33.Yurkov V, Beatty JT. 1998. Isolation of aerobic anoxygenic photosynthetic bacteria from black smoker plume waters of the Juan de Fuca Ridge in the Pacific Ocean. Appl Environ Microbiol 64:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Q, Koblížek M, Beatty JT, Jiao N. 2013. Evolutionary divergence of marine aerobic anoxygenic phototrophic bacteria as seen from diverse organisations of their photosynthesis gene clusters. Adv Bot Res 66:359–383. doi: 10.1016/B978-0-12-397923-0.00012-6. [DOI] [Google Scholar]
- 35.Zheng Q, Zhang R, Koblížek M, Boldareva EN, Yurkov V, Yan S, Jiao N. 2011. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One 6:e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomasch J, Gohl R, Bunk B, Diez MS, Wagner-Doebler I. 2011. Transcriptional response of the photoheterotrophic marine bacterium Dinoroseobacter shibae to changing light regimes. ISME J 5:1957–1968. doi: 10.1038/ismej.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Čuperová Z, Holzer E, Salka I, Sommarug R, Koblížek M. 2013. Temporal changes and altitudinal distribution of aerobic anoxygenic phototrophs in mountain lakes. Appl Environ Microbiol 79:6439–6446. doi: 10.1128/AEM.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waidner LA, Kirchman DL. 2005. Aerobic anoxygenic photosynthesis genes and operons in uncultured bacteria in the Delaware River. Environ Microbiol 7:1896–1908. doi: 10.1111/j.1462-2920.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- 39.Piwosz K, Salcher MM, Zeder M, Ameryk A, Pernthaler J. 2013. Seasonal dynamics and activity of typical freshwater bacteria in brackish waters of the Gulf of Gdansk. Limnol Oceanogr 58:817–826. doi: 10.4319/lo.2013.58.3.0817. [DOI] [Google Scholar]
- 40.Salcher MM, Posch T, Pernthaler J. 2013. In situ substrate preferences of abundant bacterioplankton populations in a prealpine freshwater lake. ISME J 7:896–907. doi: 10.1038/ismej.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn MW, Schmidt J, Pitt A, Taipale SJ, Lang E. 2016. Reclassification of four Polynucleobacter necessarius strains as representatives of Polynucleobacter asymbioticus comb. nov., Polynucleobacter duraquae sp. nov., Polynucleobacter yangtzensis sp. nov. and Polynucleobacter sinensis sp. nov., and emended description of Polynucleobacter necessarius. Int J Syst Evol Microbiol 66:2883–2892. doi: 10.1099/ijsem.0.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boldareva-Nuianzina EN, Bláhová Z, Sobotka R, Koblížek M. 2013. Distribution and origin of oxygen-dependent and oxygen-independent forms of Mg-protoporphyrin monomethylester cyclase among phototrophic proteobacteria. Appl Environ Microbiol 79:2596–2604. doi: 10.1128/AEM.00104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaschner M, Loeschcke A, Krause J, Minh BQ, Heck A, Endres S, Svensson V, Wirtz A, von Haeseler A, Jaeger KE, Drepper T, Krauss U. 2014. Discovery of the first light-dependent protochlorophyllide oxidoreductase in anoxygenic phototrophic bacteria. Mol Microbiol 93:1066–1078. doi: 10.1111/mmi.12719. [DOI] [PubMed] [Google Scholar]
- 44.Luo H, Moran MA. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78:573–587. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koblížek M, Zeng Y, Horák A, Oborník M, Beatty JT. 2013. Regressive evolution of photosynthesis in the Roseobacter clade. Adv Bot Res 66:385–405. doi: 10.1016/B978-0-12-397923-0.00013-8. [DOI] [Google Scholar]
- 46.Logares R, Bråte J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. 2010. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11). Mol Biol Evol 27:347–357. doi: 10.1093/molbev/msp239. [DOI] [PubMed] [Google Scholar]
- 47.Brinkhoff T, Giebel H-A, Simon M. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 48.Alonso C, Pernthaler J. 2006. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol 8:2022–2030. doi: 10.1111/j.1462-2920.2006.01082.x. [DOI] [PubMed] [Google Scholar]
- 49.Salcher MM, Pernthaler J, Posch T. 2010. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol Oceanogr 55:846–856. doi: 10.4319/lo.2010.55.2.0846. [DOI] [Google Scholar]
- 50.Simek K, Nedoma J, Znachor P, Kasalický V, Jezbera J, Hornák K, Sed'a J. 2014. A finely tuned symphony of factors modulates the microbial food web of a freshwater reservoir in spring. Limnol Oceanogr 59:1477–1492. doi: 10.4319/lo.2014.59.5.1477. [DOI] [Google Scholar]
- 51.Alonso-Sáez L, Unanue M, Latatu A, Azua I, Ayo B, Artolozaga I, Iriberri J. 2009. Changes in marine prokaryotic community induced by varying types of dissolved organic matter and subsequent grazing pressure. J Plankton Res 31:1373–1383. doi: 10.1093/plankt/fbp081. [DOI] [Google Scholar]
- 52.Allgaier M, Uphoff H, Felske A, Wagner-Döbler I. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl Environ Microbiol 69:5051–5059. doi: 10.1128/AEM.69.9.5051-5059.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Šimek K, Kasalický V, Zapomělová E, Horňák K. 2011. Alga-derived substrates select for distinct betaproteobacterial lineages and contribute to niche separation in Limnohabitans strains. Appl Environ Microbiol 77:7307–7315. doi: 10.1128/AEM.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, Cypionka H, Daniel R, Drepper T, Gerdts G, Hahnke S, Han C, Jahn D, Kalhoefer D, Kiss H, Klenk H-P, Kyrpides N, Liebl W, Liesegang H, Meincke L, Pati A, Petersen J, Piekarski T, Pommerenke C, Pradella S, Pukall R, Rabus R, Stackebrandt E, Thole S, Thompson L, Tielen P, Tomasch J, Von Jan M, Wanphrut N, Wichels A, Zech H, Simon M. 2010. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker's guide to life in the sea. ISME J 4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 55.Igarashi N, Harada J, Nagashima S, Matsuura K, Shimada K, Nagashima KVP. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J Mol Evol 52:333–341. doi: 10.1007/s002390010163. [DOI] [PubMed] [Google Scholar]
- 56.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of Microbial Genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng Y, Kasalický V, Šimek K, Koblížek M. 2012. Genome sequences of two freshwater betaproteobacterial isolates, Limnohabitans species strains Rim28 and Rim47, indicate their capabilities as both photoautotrophs and ammonia oxidizers. J Bacteriol 194:6302–6303. doi: 10.1128/JB.01481-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sander J, Dahl C. 2009. Metabolism of inorganic sulfur compounds in purple bacteria, p 595–622. In Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed), Advances in photosynthesis and respiration, vol 28 The purple phototrophic bacteria. Springer, Dordrecht, Netherlands. [Google Scholar]
- 61.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 63.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selyanin V, Hauruseu D, Koblížek M. 2016. The variability of light-harvesting complexes in aerobic anoxygenic phototrophs. Photosynth Res 128:35–43. doi: 10.1007/s11120-015-0197-7. [DOI] [PubMed] [Google Scholar]
- 65.Atamna-Ismaeel N, Finkel O, Glaser F, von Mering C, Vorholt JA, Koblížek M, Belkin S, Béjà O. 2012. Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ Microbiol Rep 4:209–216. doi: 10.1111/j.1758-2229.2011.00323.x. [DOI] [PubMed] [Google Scholar]
- 66.Koblížek M, Moulisová V, Muroňová M, Oborník M. 2015. Horizontal transfers of two types of puf operons among phototrophic members of the Roseobacter clade. Folia Microbiol (Praha) 60:37–43. doi: 10.1007/s12223-014-0337-z. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y, Zhang Y, Jiao N. 2011. Responses of aerobic anoxygenic phototrophic bacteria to algal blooms in the East China Sea. Hydrobiologia 661:435–443. doi: 10.1007/s10750-010-0553-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.