ABSTRACT
Cah is a calcium-binding autotransporter protein involved in autoaggregation and biofilm formation. Although cah is widespread in Shiga toxin-producing Escherichia coli (STEC), we detected mutations in cah at a frequency of 31.3% in this pathogen. In STEC O157:H7 supershedder strain SS17, a large deletion results in a smaller coding sequence, encoding a protein lacking the C-terminal 71 amino acids compared with Cah in STEC O157:H7 strain EDL933. We examined the function of Cah in biofilm formation and host colonization to better understand the selective pressures for cah mutations. EDL933-Cah played a conditional role in biofilm formation in vitro: it enhanced E. coli DH5α biofilm formation on glass surfaces under agitated culture conditions that prevented autoaggregation but inhibited biofilm formation under hydrostatic conditions that facilitated autoaggregation. This function appeared to be strain dependent since Cah-mediated biofilm formation was diminished when an EDL933 cah gene was expressed in SS17. Deletion of cah in EDL933 enhanced bacterial attachment to spinach leaves and altered the adherence pattern of EDL933 to bovine recto-anal junction squamous epithelial (RSE) cells. In contrast, in trans expression of EDL933 cah in SS17 increased its attachment to leaf surfaces, and in DH5α, it enhanced its adherence to RSE cells. Hence, the ecological function of Cah appears to be modulated by environmental conditions and other bacterial strain-specific properties. Considering the prevalence of cah in STEC and its role in attachment and biofilm formation, cah mutations might be selected in ecological niches in which inactivation of Cah would result in an increased fitness in STEC during colonization of plants or animal hosts.
IMPORTANCE Shiga toxin-producing Escherichia coli (STEC) harbors genes encoding diverse adhesins, and many of these are known to play an important role in bacterial attachment and host colonization. We demonstrated here that the autotransporter protein Cah confers on E. coli DH5α cells a strong autoaggregative phenotype that is inversely correlated with its ability to form biofilms and plays a strain-specific role in plant and animal colonization by STEC. Although cah is widespread in the STEC population, we detected a mutation rate of 31.3% in cah, which is similar to that reported for rpoS and fimH. The formation of cell aggregates due to increased bacterium-to-bacterium interactions may be disadvantageous to bacterial populations under conditions that favor a planktonic state in STEC. Therefore, a loss-of-function mutation in cah is likely a selective trait in STEC when autoaggregative properties become detrimental to bacterial cells and may contribute to the adaptability of STEC to fluctuating environments.
KEYWORDS: attachment, biofilms, adherence, autotransporter proteins, adhesins, enterohemorrhagic Escherichia coli (EHEC), Shiga toxin-producing Escherichia coli (STEC), adaptive mutations, produce, plant, epithelial cell
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC), one of the most important causal agents of foodborne illness linked to fresh leafy vegetables (1), is spread mainly from cattle into the environment by fecal shedding. Contamination of produce can occur either in pre- or postharvest environments, via contaminated water, manure, animals, or insects in the produce-growing fields (2–5) or in the water used for washing and processing produce, via workers, or by cross-contamination from other food at the postharvest level (6–8). Because produce is often consumed raw, contamination of produce by enteric pathogens poses a high health risk to consumers.
The formation of biofilm by enteric pathogens on vegetables and fruit enhances their persistence on plants. Biofilm-associated cells are more difficult to remove and more resistant to inactivation than planktonic cells (9). Furthermore, biofilm-associated cells as well as capsule-producing cells are more tolerant to desiccation (10). Therefore, biofilm formation by enteric pathogens on plants may confer protection against antimicrobial washes during the postharvest processing. Attachment is the first step to establish bacterial colonization on the plant surface. Enteric pathogens produce an array of adhesive structures and proteins for colonization of their animal hosts, many of which are important virulence factors and are involved in STEC colonization and biofilm formation on plants and abiotic surfaces; these include curli fimbriae, flagella, cellulose, lipopolysaccharide (LPS), colanic acid, and several outer membrane proteins (11–13). These overlapping functions may contribute to the fitness of STEC strains across animal hosts and secondary habitat environments and, consequently, to the occurrence of foodborne outbreaks.
Cah was first identified in Escherichia coli O157:H7 strain EDL933 (14). The gene encoding Cah consists of a 2,850-bp open reading frame (ORF) and was named cah for calcium binding antigen 43 homolog (14). Cah shares high sequence similarity with Antigen 43 (Ag43), a surface-displayed autotransporter protein that confers cell autoaggregation due to its self-recognizing properties (15, 16), and with AIDA-1, an adhesin that mediates diffuse adherence to HeLa cells (17). Expression of EDL933 cah in E. coli DH5α conferred on bacterial cells an autoaggregative phenotype (14) and increased bacterial populations bound to alfalfa sprouts, suggesting a role for Cah in mediating cell-to-cell interaction and in attachment of bacteria to plant tissue (18). To date, a contribution of Cah to biofilm formation of STEC was demonstrated only in E. coli O157:H7 strain 86-24. Deletion of cah in this strain reduced its biofilm formation on an abiotic surface (14); however, it did not affect its ability to bind to alfalfa (18), implying diverse functions of Cah in biofilm formation and surface attachment.
We recently completed the genome sequence of E. coli O157:H7 supershedder strain SS17 (19). Comparative genomic analysis of SS17 placed this strain within the same cluster as the STEC O157:H7 strains linked to the 2006 spinach-associated outbreak, one of the largest leafy greens-associated outbreaks of STEC O157:H7 infection in the United States (http://www.cdc.gov/ecoli/2006/spinach-10-2006.html). Examination of putative adhesins in strain SS17 revealed that the coding region of cah in SS17 was truncated, resulting in a protein smaller than EDL933-Cah. Further examination of cah in other STEC strains revealed various mutations in the coding region of cah, including insertions and deletions. To gain insight into the biological role of natural mutations in STEC cah, we compared the functions of wild-type cah and truncated cah in three different systems: biofilm formation on abiotic surfaces, attachment to spinach leaf surfaces, and adherence to bovine recto-anal junction squamous epithelial (RSE) cells. Our results indicate a conditional contribution of Cah to biofilms on abiotic surfaces and a potential role of Cah in STEC colonization of both animal and plant hosts.
RESULTS
Genetic diversity and distribution of cah in STEC.
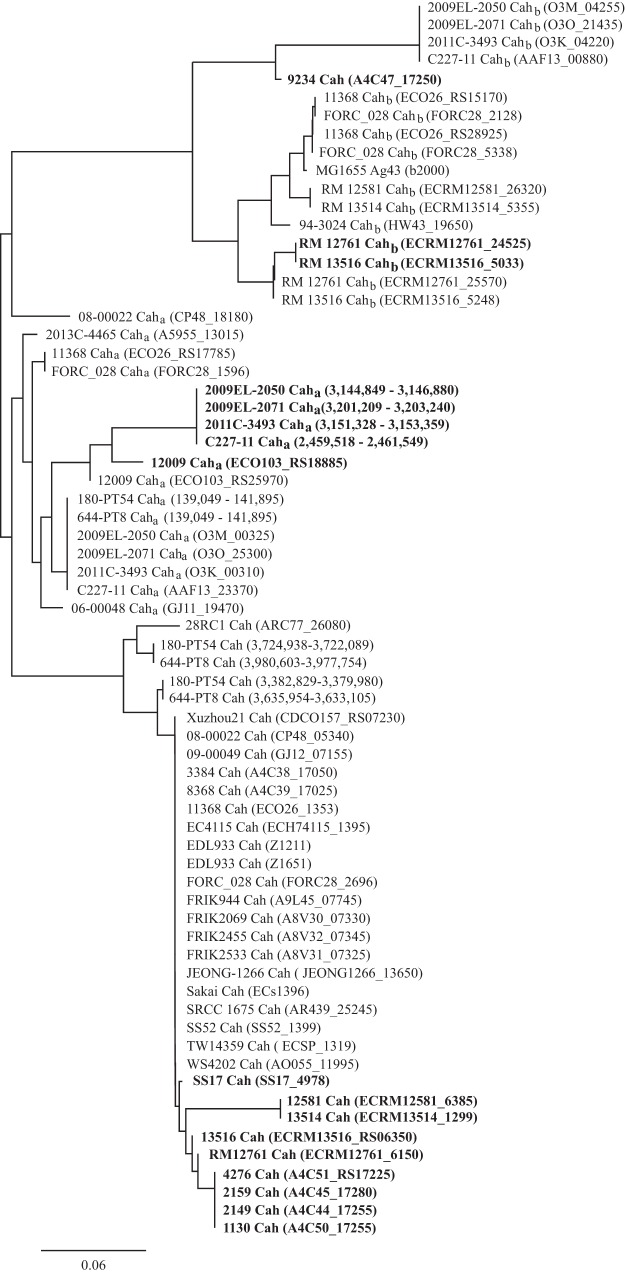
The cah gene (2,850 bp) encodes an autoaggregative protein, Cah, of 949 amino acids (aa) in enterohemorrhagic E. coli O157:H7 strain EDL933. EDL933 possesses two copies of cah that are identical at the nucleotide level, one located on the O-island 43 (Z1211) and the other on the O-island 48 (Z1651). A BLAST search of EDL933 cah revealed that cah is widespread in the STEC population. Among 48 STEC strains examined, an identical or nearly identical Cah was identified in 32 strains, including 24 O157:H7 strains, 4 O145:H28 strains, 2 O26:H11 strains, 1 O168:H- strain, and 1 O136:H16 strain (Table 1). The majority of STEC strains examined in this study carry one copy of cah, except two O157:H7 strains, 644-PT8 and 180-PT54, which possess two copies of cah with slight sequence divergence; each encodes a protein sharing 98.5% and 95.2% identity with EDL933-Cah, respectively (Fig. 1). In several STEC strains, cah encodes a protein that differs in size from EDL933-Cah, such as in strain SS17 (782 aa), strains 1130, 2149, 2159, and 4276 (664 aa), strains RM13516 and RM12761 (432 aa), and strains RM13514 and RM12581 (136 aa) (Fig. 1 and Table 1). A BLAST search of EDL933-Cah failed to retrieve any homolog in O157:H7 strain 9234; however, a BLAST search of EDL933 cah retrieved a DNA fragment that aligned perfectly with the partial coding region of EDL933 cah encoding a peptide identical to the C-terminal 250 aa of EDL933-Cah (Fig. 1 and Table 1).
TABLE 1.
Distribution of cah and cah homologs in Shiga toxin-producing Escherichia colia
| Strain | Serotype | GenBank accession no. |
cah |
cah homologs |
||
|---|---|---|---|---|---|---|
| Gene (positions) or locus tag (corresponding protein length in aa) | % identity | Gene (locus tag or gene positions) (corresponding protein length in aa) | % identity | |||
| EDL933 | O157:H7 | CP008957 | Z1211 (949) | 100 | ND | ND |
| Z1651 (949) | 100 | |||||
| Sakai | O157:H7 | NC_002695 | ECs1396 (949) | 100 | ND | ND |
| TW14359 | O157:H7 | CP001368 | ECSP_1319 (949) | 100 | ND | ND |
| EC4115 | O157:H7 | CP001164 | ECH74115_1395 (949) | 100 | ND | ND |
| Xuzhou21 | O157:H7 | NC_017906 | CDCO157_RS07230 (949) | 100 | ND | ND |
| SS17 | O157:H7 | CP008805 | SS17_4978 (778) | 100 | ND | ND |
| SS52 | O157:H7 | NZ_CP010304 | SS52_1399 (949) | 100 | ND | ND |
| WS4202 | O157:H7 | NZ_CP012802 | AO055_11995 (949) | 100 | ND | ND |
| SRCC 1675 | O157:H7 | CP015023 | AR439_25245 (949) | 100 | ND | ND |
| JEONG-1266 | O157:H7 | CP014314 | JEONG1266_13650 (949) | 100 | ND | ND |
| FRIK944 | O157:H7 | CP016625 | A9L45_07745 (949) | 100 | ND | ND |
| FRIK2069 | O157:H7 | CP015846 | A8V30_07330 (949) | 100 | ND | ND |
| FRIK2455 | O157:H7 | CP015843 | A8V32_07345 (949) | 100 | ND | ND |
| FRIK2533 | O157:H7 | CP015842 | A8V31_07325 (949) | 100 | ND | ND |
| 644-PT8 | O157:H7 | CP015831 | cah (positions 3635954–3633105) (949) | 98.5 | caha (positions 141895–139049) (948) | 89.3 |
| cah (positions 3980603–3977754) (949) | 95.2 | |||||
| 180–PT54 | O157:H7 | CP015832 | cah (positions 3382829–3379980) (949) | 98.5 | caha (positions 141895–139049) (948) | 89.3 |
| cah (positions 3724938–3722089) (949) | 95.2 | |||||
| 28RC1 | O157:H7 | CP015020 | ARC77_26080 (949) | 93.8 | ND | ND |
| 1130 | O157:H7 | NZ_CP017434 | A4C50_17255 (650) | 100 | ND | ND |
| 2149 | O157:H7 | NZ_CP017436 | A4C44_17255 (650) | 100 | ND | ND |
| 2159 | O157:H7 | NZ_CP017438 | A4C45_17280 (650) | 100 | ND | ND |
| 3384 | O157:H7 | NZ_CP017440 | A4C38_17050 (949) | 100 | ND | ND |
| 4276 | O157:H7 | NZ_CP017442 | A4C51_17225 (650) | 100 | ND | ND |
| 8368 | O157:H7 | NZ_CP017444 | A4C39_17025 (949) | 100 | ND | ND |
| 9234 | O157:H7 | NZ_CP017446 | A4C47_17250/(250) | 100 | ND | ND |
| RM13514 | O145:H28 | CP006027 | ECRM13514_1299 (128) | 100 | cahb (ECRM13514_5355) (1,039) | 68.5 |
| RM12581 | O145:H28 | CP007136 | ECRM12581_6385 (128) | 100 | cahb (ECRM12581_26320) (1,039) | 68.5 |
| RM13516 | O145:H28 | CP006262 | ECRM13516_RS06350 (430) | 100 | cahb (ECRM13516_5248) (1,039) | 70.1 |
| cahb (ECRM13516_5033) (1,026) | 69.5 | |||||
| RM12761 | O145:H28 | CP007133 | ECRM12761_6150 (430) | 100 | cahb (ECRM12761_25570) (1,039) | 70.1 |
| cahb ECRM12761_24525 (1,026) | 69.5 | |||||
| 11368 | O26:H11 | NC_013361 | ECO26_1353 (949) | 100 | caha (ECO26_RS17785) (948) | 89.5 |
| cahb (ECO26_RS28925) (1,039) | 68.0 | |||||
| cahb (ECO26_RS15170) (1,039) | 67.9 | |||||
| FORC_028 | O26:H11 | CP012693 | FORC28_2696 (949) | 100 | caha (FORC28_1596) (948) | 89.7 |
| cahb (FORC28_5338) (1,039) | 68.0 | |||||
| cahb (FORC28_2128) (1,039) | 67.9 | |||||
| 08-00022 | O136:H16 | CP013662 | CP48_05340 (949) | 100 | caha (CP48_18180) (948) | 88.0 |
| 09-00049 | O168:H- | CP015228 | GJ12_07155 (949) | 100 | ND | ND |
| 12009 | O103:H2 | NC_013353 | ND | ND | caha (ECO103_RS25970) (948) | 88.4 |
| caha (ECO103_RS18885) (606) | 83.9 | |||||
| 2009EL-2050 | O104:H4 | CP003297 | ND | ND | caha (O3M_00325) (948) | 89.3 |
| cahb (O3M_04255) (1,039) | 68.7 | |||||
| caha (positions 3144849–3146880) (192) | 74.6 | |||||
| 2009EL-2071 | O104:H4 | CP003301 | ND | ND | caha (O3O_25300) (948) | 89.3 |
| cahb (O3O_21435) (1,039) | 68.7 | |||||
| caha (positions 3201209–3203240) (192) | 74.6 | |||||
| 2011C-3493 | O104:H4 | CP003289 | ND | ND | caha (O3K_00310) (948) | 89.3 |
| cahb (O3K_04220) (1,039) | 68.7 | |||||
| caha (positions 3151328–3153359) (192) | 74.6 | |||||
| C227-11 | O104:H4 | CP011331 | ND | ND | caha (AAF13_23370) (948) | 89.3 |
| cahb (AAF13_00880) (1,039) | 68.7 | |||||
| caha (positions 2459518–2461549) (192) | 74.6 | |||||
| 94-3024 | O104:H21 | CP009106 | ND | ND | cahb (HW43_19650) (1,039) | 68.9 |
| 2013C-4465 | O55:H7 | CP015241 | ND | ND | caha (A5955_13015) (948) | 90.0 |
| 06-00048 | O36:H- | CP015229 | ND | ND | caha (GJ11_19470) (948) | 88.8 |
| CFSAN004176 | O145:H- | CP014583 | ND | ND | ND | ND |
| CFSAN004177 | O145:H- | CP014670 | ND | ND | ND | ND |
| 11128 | O111:H- | NC_013364 | ND | ND | ND | ND |
| RM9387 | O104:H7 | NZ_CP009104 | ND | ND | ND | ND |
| 2009C-3133 | O119:H4 | CP013025 | ND | ND | ND | ND |
| 2012C-4227 | O165:H25 | CP013029 | ND | ND | ND | ND |
| GB089 | O168:H- | CP013663 | ND | ND | ND | ND |
| 2011C-3911 | O-:H- | CP015240 | ND | ND | ND | ND |
The complete genomes of STEC that were available in GenBank as of September 2016 were used to create a database to search for cah or cah homologs. BLAST was performed in Geneious8.1.8 using EDL933 cah as a query. The retrieved cah and cah homologs were translated using the bacterial translation codon table. The locus tag of the coding DNA sequence (CDS) is based on the original genome annotation. If a locus tag is not available for a cah or cah homolog, the CDS is indicated by the name designated in this study, cah, caha, or cahb, followed by the genomics position of the corresponding gene in parentheses. Items in bold represent genes carrying a mutation in their coding region. The overall mutation rate was 31.3% for cah, 29.4% for caha, and 13.3% for cahb. ND, not detected by BLAST search.
FIG 1.
Clustering analysis of Cah in Shiga toxin-producing Escherichia coli strains. The cah gene in strain EDL933 is 2,850 bp in length, encoding a 949-aa protein. The DNA sequences of cah in STEC strains were retrieved by a BLAST search of a database containing all STEC strains for which a complete genome was deposited in GenBank as of September 2016. The sequences of Cah were aligned using the Geneious global alignment module in Geneious (Geneious8.1.8; Biomatters) using Blosum 62 as the distance matrix. A neighbor-joining tree was constructed in Geneious using the Jukes-Cantor model and resampled by bootstrap (10,000 replicates). The GenBank accession number for each genome is presented in Table 1. Strains carrying a different-size cah are shown in bold. The locus tag of Cah in each strain is shown in parentheses.
Two Cah homologs were detected in 17 STEC strains, including eight cah-negative strains (Table 1). The first homolog, Caha, is an autotransporter protein of 948 aa sharing 88 to 90% identify with EDL933-Cah. The second homolog, Cahb, also known as Ag43 or Flu in E. coli K-12 strains, is an autotransporter protein of 1,039 aa sharing 68 to 70% identity with EDL933-Cah. Genes encoding Cah homologs were detected in non-O157 strains more frequently than in O157 strains. Among the 24 non-O157 STEC strains, 15 strains were positive either for caha or cahb only or for both (Table 1). In contrast, only two O157:H7 strains were positive for caha among the 24 strains examined (Table 1). Certain STEC strains carry multiple copies of caha, such as O103:H2 strain 12009, or cahb, such as O26:H11 strains 11368 and FORC-028 (Table 1). Furthermore, several STEC strains carry partial coding sequences of caha or cahb. For example, there were two copies of caha in O103:H2 strain 12009, one encoding a full-length Caha (948 aa) and the other encoding a 650-aa peptide that shared 100% identity with the full-length Caha. Similarly, there were two copies of cahb in strains RM13516 and RM12761, one encoding a full-length Cahb (1,039 aa) and the other one encoding a peptide of 1,026 aa, which shared 98.2% identity with the same region of full-length Cahb (Table 1).
Natural mutation in STEC cah.
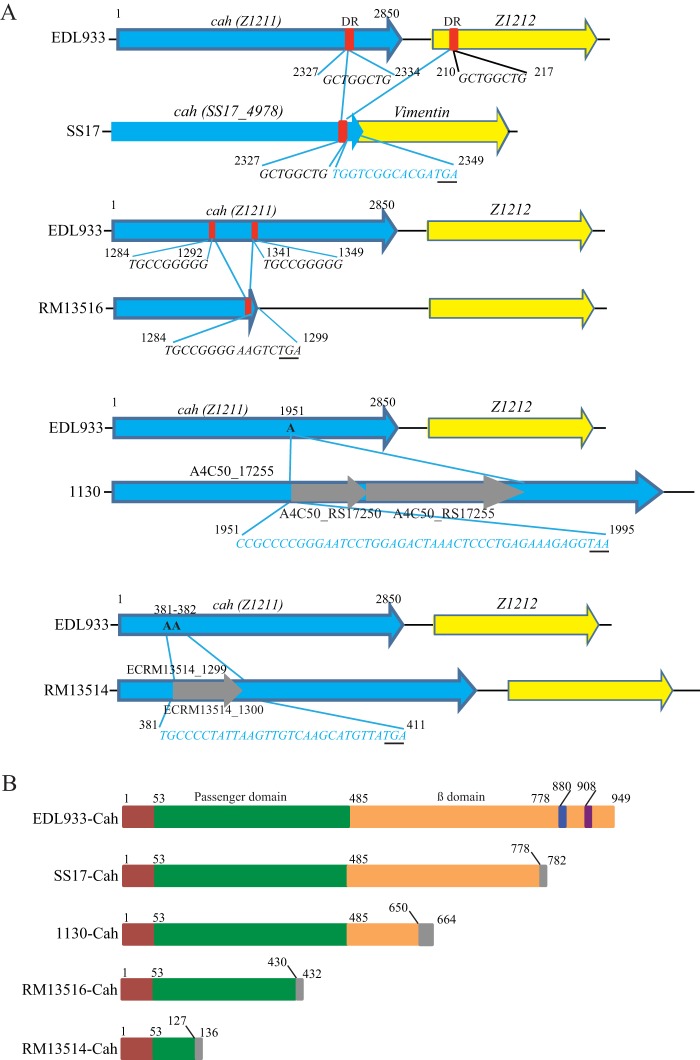
EDL933-Cah consists of a long signal peptide, a secreted passenger domain, and a β-barrel domain forming an integral outer membrane protein (14). We assessed the occurrence of natural mutations in cah in a database containing 48 complete STEC genomes. Among the 32 cah-positive STEC strains, various mutations were detected in the coding region of cah in 10 strains (31.3%), including several outbreak strains (Table 1). These mutations included mainly deletions (in strains SS17, RM13516, RM12761, and 9234) and insertions (in strains 1130, 2149, 2159, 4276, RM13514, and RM12581) (Fig. 2A and Table 1). The large deletion in strain SS17 was likely mediated by recombination between the two direct short repeats, GCTGGCTG, one located within the cah coding region (encoding aa 2327 to 2334 in EDL933-Cah) and the other immediately downstream of cah in Z1212 (aa 210 to 217) (Fig. 2A, SS17). This deletion eliminated an 843-bp DNA fragment that includes a partial cah coding sequence, the intergenic region between cah and Z1212, and a partial coding sequence of Z1212. Furthermore, this deletion joined the two ORFs, cah and Z1212, into an operon and created a stop codon within Z1212 (putative Vimentin gene) (Fig. 2A, SS17). Therefore, SS17-Cah appeared to be a hybrid protein, in which the first 778 aa at the N terminus are identical to those of EDL933-Cah, but the last 4 aa at its C terminus, WSAR, are unique to SS17-Cah (Fig. 2B, SS17-Cah). Similarly, the large deletion in strain RM13516 likely resulted from the recombination between the direct repeat, TGCCGGGGG, in the coding region of EDL933 cah, one spanning positions 1284 to 1292 and the other spanning positions 1341 to 1349 (Fig. 2A, RM13516). The deletion resulted in a premature stop codon, generating a truncated Cah with the first 430 aa identical to those of EDL933-Cah (Fig. 2B, RM13516-Cah). The same deletion in cah was detected in O145 strain RM12761, which was associated with the same outbreak as strain RM13516 (20, 21). The cah gene in O157 strains 1130, 2149, 2159, and 4276 was disrupted by an insertion mutation: the adenine (A) at position 1951 corresponding to the EDL933 cah was replaced with a DNA fragment containing coding regions for two transposases (Fig. 2A, 1130). Consequently, a premature stop codon was introduced, resulting in a truncated Cah with the first 650 aa identical to those of EDL933-Cah (Fig. 2B, 1130-Cah). In strain RM13514, cah was disrupted in a fashion similar to the one that we observed in strain 1130. The nucleotides AA at positions 381 and 382 corresponding to the EDL933 cah were replaced with a gene encoding a transposon-related mobile element (ECRM13514_1300). Also, a premature stop codon was introduced due to the insertion, resulting in a 136-aa protein with the first 127 aa at the N terminus identical to those of EDL933-Cah (Fig. 2B, 13514-Cah).
FIG 2.
Sequence analysis of mutations in cah and the corresponding protein in Shiga toxin-producing Escherichia coli. (A) Pairwise alignment of SS17 cah, RM13516 cah, 1130 cah, and RM13514 cah with EDL933 cah. The EDL933 cah was used as a reference; thus, deletion or insertion in cah of each STEC strain described here was the result of the comparison with EDL933 cah. In both SS17 and RM13516, the positions of deletion mutations were mapped to the corresponding position in EDL933 cah. The direct repeats in each gene are indicated by red boxes. Sequence alignment of EDL933 cah and SS17 cah revealed a deletion of a 516-bp DNA fragment spanning nucleic acids corresponding to positions 2334 to 2850 in EDL933 cah. The stop codon in SS17 cah is underlined. Insertion mutation in strains 1130 and RM13514 occurred by replacing one or two adenines within the coding region of cah. The premature stop codon is underlined. Nucleotides in blue indicate that sequences were not from cah. (B) Schematic representation of EDL933-Cah and truncated Cah in STEC strains. The putative signal sequence, passenger domain, and β-domain described in EDL933-Cah previously (14) are indicated by red, green, and orange boxes, respectively. The blue box represents the RGD motif, and the purple box represents the QAGLEA domain that is often found in the HlyD family of secretion protein. The truncated Cah proteins in STEC strains SS17, 1130, RM13516, and RM13514 retain peptides of 778 aa, 650 aa, 430 aa, and 127 aa, respectively, which are identical to the corresponding regions in the N terminus of EDL933-Cah.
Conditional contribution of Cah in E. coli biofilm formation.
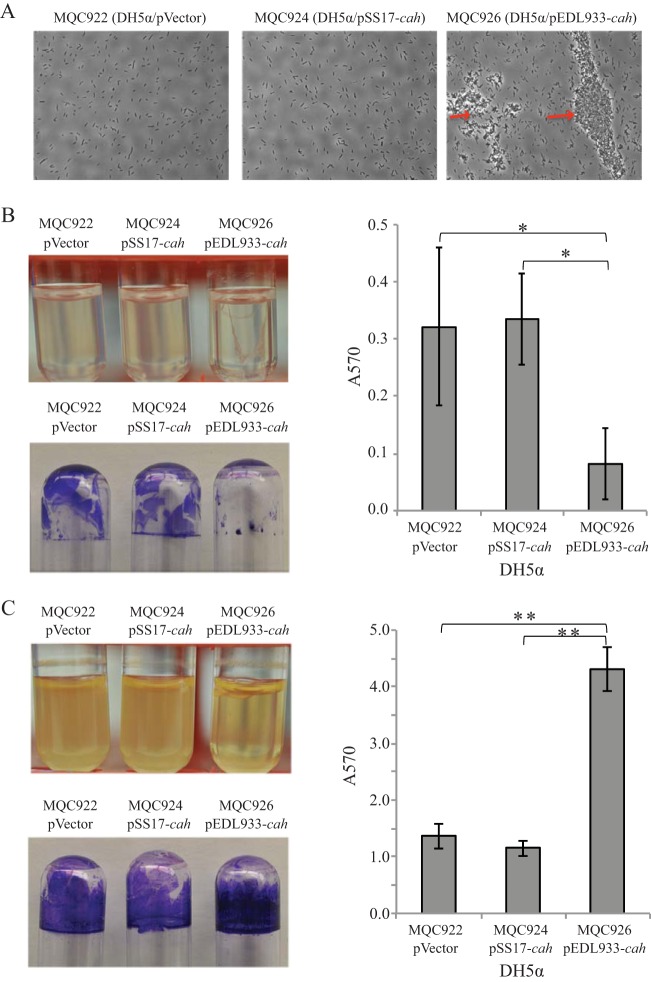
To confirm that the mutations in STEC cah are loss-of-function mutations, we chose to perform comparative functional analyses of EDL933 cah with SS17 cah since SS17 cah retained the largest coding sequence of cah among all the mutations detected in this study (Fig. 2B). The EDL933 cah and SS17 cah genes were cloned and expressed in nonpathogenic E. coli strain DH5α (Table 2). We first examined the growth of E. coli DH5α carrying the empty expression vector (MQC922), cloned SS17 cah (MQC924), and cloned EDL933 cah (MQC926) in LB broth under static and agitated growth conditions. For each of the three strains, growth under the agitated condition was better than that under the static condition. However, under either of the two growth conditions, there was no difference in growth among the three strains (data not shown). We next examined biofilm formation by strains transformed with plasmid carrying EDL933 cah or SS17 cah under both static and agitated growth conditions. When DH5α cells were grown statically, a strong autoaggregative phenotype was observed only for DH5α cells transformed with cloned EDL933 cah (Fig. 3A, MQC926). No visible aggregates in static cultures were observed for either the control strain (DH5α cells transformed with empty expression vector) (Fig. 3A, MQC922) or DH5α cells transformed with cloned SS17 cah (Fig. 3A, MQC924). Biofilm formation on glass surfaces appeared to be inversely correlated with the formation of cell aggregates since strain MQC926 produced noticeably less biofilm on glass surfaces (borosilicate culture tubes) than did either strain MQC922 or strain MQC924 under the conditions examined (Fig. 3B). Further quantitative assays revealed a significant reduction in biofilm on glass surfaces in DH5α cells expressing EDL933 cah (MQC926) but not in DH5α cells expressing SS17 cah (MQC924), compared with the control strain (MQC922) (Fig. 3B).
TABLE 2.
E. coli strains and plasmids used in this study
| Strain or plasmid | Antibiotic resistancea | Descriptionb | Source or reference |
|---|---|---|---|
| Strains | |||
| DH5α | Cms | Escherichia coli strain K-12 derivative for general laboratory use | S. Lory |
| SS17 | Cms | Escherichia coli O157:H7 strain isolated from supershedder cattle | 19 |
| MB912 | Kmr | A cah deletion mutant of strain SS17 (Δcah::Km) | This study |
| MQC922 | Cmr | pBBR1MCS-transformed DH5α | This study |
| MQC924 | Cmr | pXQ32-transformed DH5α | This study |
| MQC926 | Cmr | pXQ31-transformed DH5α | This study |
| MQC928 | Cmr | pBBR1MCS-transformed SS17 | This study |
| MQC930 | Cmr | pXQ31-transformed SS17 | This study |
| MQC932 | Cmr | pXQ32-transformed SS17 | This study |
| MQC940 | Kmr Cmr | pBBR1MCS-transformed MB912 | This study |
| MQC942 | Kmr Cmr | pXQ31-transformed MB912 | This study |
| MQC944 | Kmr Cmr | pXQ32-transformed MB912 | This study |
| MQC966 | Cmr | pBBR1MCS-transformed EDL933 | This study |
| MQC1046 | Kmr | A cah deletion mutant of strain EDL933 (Δcah::Km) | This study |
| MQC1048 | Kmr Cmr | pBBR1MCS-transformed MQC1046 | This study |
| MQC1050 | Kmr Cmr | pXQ31-transformed MQC1046 | This study |
| Plasmids | |||
| pACYC177 | Ampr | Lambda Red recombinase expression plasmid | New England BioLabs |
| pKD119 | Kmr | Template plasmid for amplification of Km resistance cassette | New England BioLabs |
| pBBR1MCS | Cmr | Expression vector for complementation analysis | 54 |
| pXQ31 | Cmr | The cah gene and its native promoter in E. coli O157:H7 strain EDL933 were cloned into vector pBBR1MCS | This study |
| pXQ32 | Cmr | The cah gene and its native promoter in E. coli O157:H7 strain SS17 were cloned into vector pBBR1MCS | This study |
Ampr, Kmr, and Cmr, ampicillin, kanamycin, and chloramphenicol resistance, respectively; Cms, chloramphenicol susceptibility.
Primers used for gene deletion and cloning are described in Table 4.
FIG 3.
Autoaggregation and biofilm formation by E. coli DH5α when cells were grown in LB broth. (A) DH5α cells visualized using a phase-contrast microscope at a magnification of 40×. The red arrows indicate the cell aggregates. (B) Results under static conditions: cultures of DH5α strains (upper left), crystal violet staining of the biofilm biomass on the glass culture tubes (bottom left), and quantitative assay of biofilms (right). (C) Results under agitated conditions: cultures of DH5α strains (upper left), crystal violet staining the biofilm biomass on the glass culture tubes (bottom left), and quantitative assay of the biofilms (right). Strain MQC922 is strain DH5α transformed with the expression vector pBBR1MCS and served as the control strain. Strain MQC926 is strain DH5α transformed with the cloned EDL933 cah, and strain MQC924 is the DH5α transformed with cloned SS17 cah (Table 2). Each data set represents the mean absorbance and standard errors of the means (SEM) from at least five biological replicates. Significant differences in means between a given strain and each of the two other strains by the t test are indicated with asterisks: *, P < 0.05; **, P < 0.001.
In contrast to the static cultures, when DH5α cells were grown under the agitated condition, a strong biofilm of bacteria was observed on glass surfaces for all three strains (Fig. 3C). Quantitative comparison of biofilms among the three strains revealed that expressing EDL933 cah in DH5α (MQC926) significantly enhanced biofilm on the glass surfaces compared with the control strain (MQC922) (Fig. 3C). In contrast, no significant difference was observed between the control strain (MQC922) and the strain transformed with cloned SS17 cah (MQC924), indicating that the deletion in SS17 cah indeed was a loss-of-function mutation.
Strain-dependent contribution of Cah to E. coli biofilm formation.
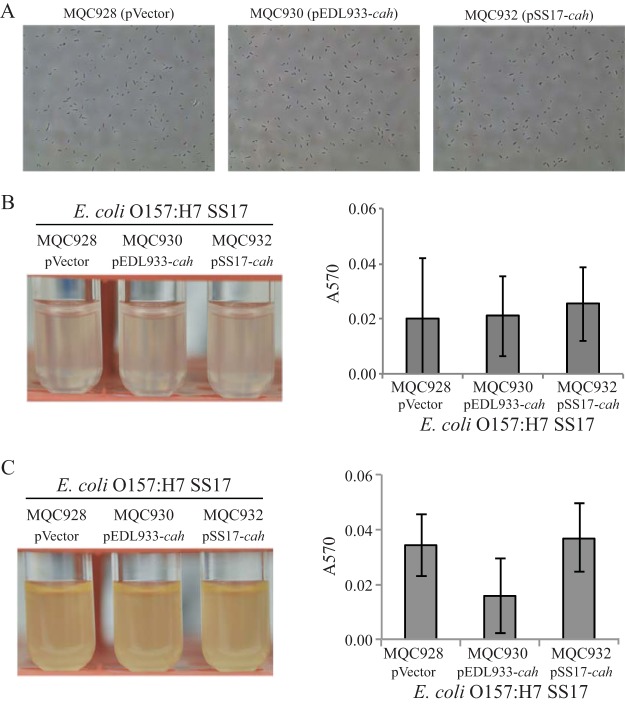
We next examined the contribution of EDL933-Cah to biofilm formation of STEC strain SS17 in vitro. Similar to what was seen for E. coli DH5α, there was no growth difference among the SS17 strains carrying an empty expression vector (MQC928), cloned EDL933 cah (MQC930), and cloned SS17 cah (MQC932) under either static or agitated condition. Expression of EDL933 cah in trans in STEC strain SS17 failed to confer on SS17 cells the autoaggregative phenotype that was observed in DH5α cells under static conditions (Fig. 4A, MQC930). Consistently, the quantitative biofilm assay revealed that there was no significant difference in biofilm formation between the control strain (MQC928) and the strain transformed with EDL933 cah (MQC930) or the strain transformed with SS17 cah (MQC932) under static growth conditions (Fig. 4B). Furthermore, unlike that of DH5α cells, the expression of EDL933 cah in SS17 (MQC930) did not increase biofilm on glass surfaces under agitated conditions compared with either the control strain (MQC928) or SS17 transformed with the cloned SS17 cah (MQC932) (Fig. 4C). A similar result was observed for the SS17Δcah mutant (MB912). There was no difference in biofilm formation under either static or agitated conditions among strains carrying the empty expression vector (MQC940), cloned EDL933 cah (MQC942), and cloned SS17 cah (MQC944) (data not shown).
FIG 4.
Biofilm formation by E. coli O157:H7 strain SS17 when cells were grown in LB broth. (A) SS17 cells visualized using a phase-contrast microscope at a magnification of 40×. (B) Cultures of SS17 strains (left) and quantitative assay of biofilms (right) under static conditions. (C) Cultures of SS17 strains (left) and quantitative assay of the biofilms (right) under agitated conditions. Strain MQC928 is strain SS17 transformed with the expression vector pBBR1MCS and served as the control strain. Strain MQC930 is strain SS17 transformed with the cloned EDL933 cah, and strain MQC932 is strain SS17 transformed with the cloned SS17 cah (Table 2). Each data set represents the mean absorbance and SEM from at least five biological replicates.
Cah is not required for biofilm formation in spinach lysates.
E. coli O157:H7 has the ability to form thick biofilms in lysates derived from homogenized spinach leaves, which have served as a model system to investigate the behavior of this enteric pathogen in injured leaf tissue (22, 23). Different adhesins are involved in the formation of E. coli O157:H7 biofilms in spinach lysates compared with LB broth (11). We thus assessed if Cah plays a role in biofilm production by STEC strains in spinach lysates. Deletion of cah in strain EDL933 (MQC1048) did not alter its biofilm formation on glass surfaces compared with the wild-type strain (MQC966) when cells were grown in spinach lysates at 28°C for 24 h (Fig. 5A). Furthermore, expression of EDL933 cah in trans in the EDL933Δcah strain (MQC1050) did not impact the biofilm formation of the mutant strain (MQC1048) under the conditions examined (Fig. 5A). A similar result was observed in STEC strain SS17. There was no significant difference in biofilm on glass surfaces between any two pairs of the three SS17 strains examined: MQC928 (SS17 transformed with empty expression vector), MQC930 (SS17 transformed with cloned EDL933 cah), and MQC932 (SS17 transformed with cloned SS17 cah) (Fig. 5B). Similar observations were made for E. coli DH5α and SS17Δcah mutant (data not shown), suggesting that Cah plays a minimal role in biofilm formation in spinach lysates.
FIG 5.
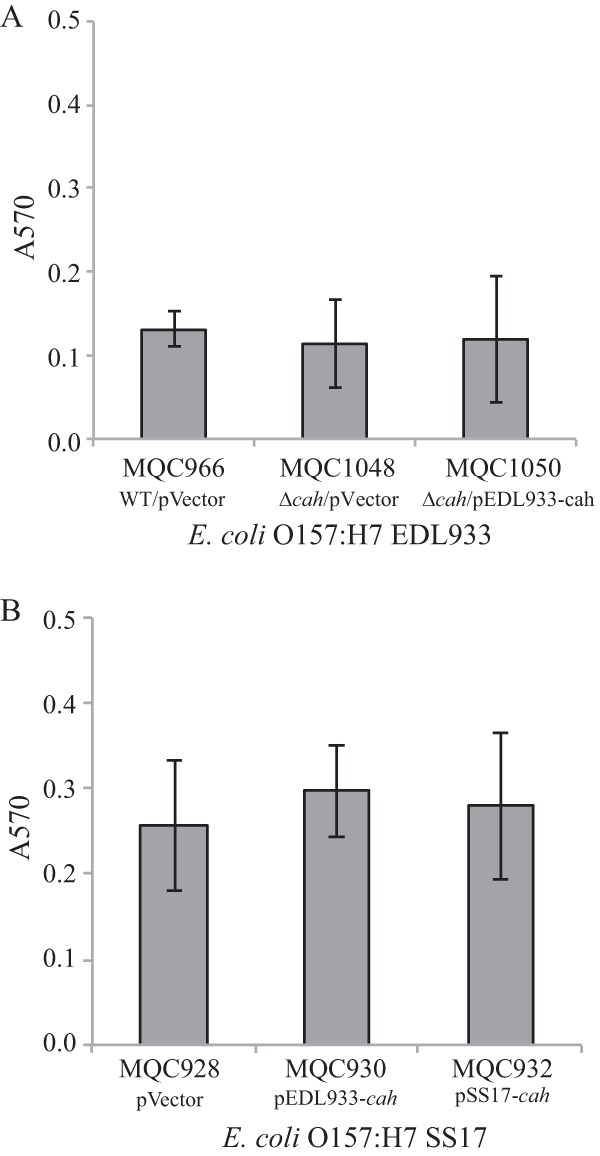
Biofilm formation by E. coli O157:H7 strains when cells were grown in spinach lysates. (A) Quantitative assays of biofilm formation by strain EDL933 and its derivatives. Strain MQC966 is strain EDL933 transformed with the expression vector pBBR1MCS and served as the control strain. Strain MQC1048 is strain EDL933Δcah transformed with the expression vector pBBR1MCS. Strain MQC1050 is EDL933Δcah-transformed cloned strain EDL933 cah. (B) Quantitative assays of biofilm formation by strain SS17 and its derivatives. Strain MQC928 is strain SS17 transformed with the expression vector pBBR1MCS and served as the control strain. Strain MQC930 is strain SS17 transformed with the cloned EDL933 cah, and strain MQC932 is strain SS17 transformed with the cloned SS17 cah. Each data set represents the mean absorbance and SEM from at least five biological replicates.
Strain-dependent role of Cah in attachment of STEC cells to spinach leaves.
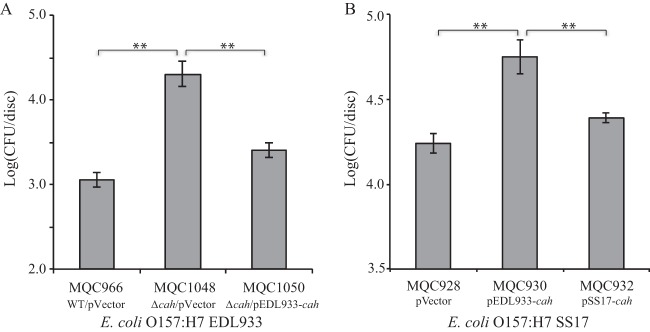
The function of cah was further assessed for its role in attachment to plant tissue. In strain EDL933, deletion of cah (strain MQC1048) led to a 15.3-fold increase in of the number of EDL933 cells that attached to spinach leaves over 2 h of incubation compared with the wild-type strain (MQC966) (Fig. 6A). This function of Cah was further confirmed by complementation analysis. When EDL933 cah was expressed in trans in the EDL933Δcah strain (MQC1050), the population of bacterial cells attached to spinach leaves was restored to a level similar to that of wild-type EDL933 strain MQC966. In contrast, the transformed SS17 that expressed EDL933 cah (MQC930) attached to the leaf surfaces at densities significantly greater (3.2-fold) than those of the SS17 transformed with the empty vector (MQC928) and at densities 2.3-fold greater than the strain transformed with cloned SS17 cah (MQC932) (Fig. 6B). There was no significant difference in the population of SS17 attached to leaf surfaces between strains MQC928 and MQC932 (Fig. 6B). A similar result was observed for the SS17Δcah mutant. Complementation of the SS17Δcah mutant with EDL933 cah (MQC942) resulted in a 2.6-fold increase in pathogen population attached to leaf surfaces compared with the strain transformed with an empty expression vector (MQC940), while complementation of SS17Δcah with its own cah gene (MQC944) did not alter the pathogen population attached to leaf surfaces significantly (see Fig. S2 in the supplemental material).
FIG 6.
Contribution of Cah to attachment of E. coli O157:H7 to spinach leaves. Bacterial adhesion to baby spinach leaves was assessed in STEC strains EDL933 (A) and SS17 (B). The population of attached STEC cells on spinach leaves was expressed in CFU per disc. Strain MQC966 is strain EDL933 transformed with the expression vector pBBR1MCS and served as the control strain. Strain MQC1048 is strain EDL933Δcah transformed with the expression vector pBBR1MCS. Strain MQC1050 is the EDL933Δcah-transformed cloned strain EDL933 cah. Strain MQC928 is strain SS17 transformed with the expression vector pBBR1MCS and served as the control strain. Strain MQC930 is strain SS17 transformed with the cloned EDL933 cah, and strain MQC932 is strain SS17 transformed with cloned SS17 cah (Table 2). Each bar represents the average number of attached cells and SEM from two discs of tissue per leaf and from four leaves. **, P < 0.001 (indicating significant differences in the means between the two strains by Tukey's multiple-comparison test).
Cah contributes to bovine RSE cell adherence in a strain-dependent manner.
EDL933-Cah enhanced the quantitative adherence of DH5α (MQC926 [DH5α transformed with cloned EDL933 cah]) to RSE cells compared with the control strain MQC922 (DH5α transformed with empty expression vector) and strain MQC924 (DH5α transformed with cloned SS17 cah) (Table 3; see also Fig. S3 in the supplemental material). In contrast, expression of SS17 cah in trans in DH5α (MQC924) did not impact either the adherence pattern or the number of SS17 cells that adhered to RSE cells compared with the control strain (MQC922). In STEC strain EDL933, inactivation of Cah altered the adherence pattern of EDL933 to RSE cell from an aggregative, moderate pattern to a diffuse, moderate pattern (Table 3; Fig. S3). Similarly, only EDL933 cah (MQC1050) could restore the wild-type adherence phenotype (aggregative, moderate) in the EDL933Δcah strain (MQC1048), implying a potential role of Cah in colonization of animal hosts.
TABLE 3.
Quantification of RSE cells with adherent DH5α, EDL933, and SS17 strains
| Strainsa | Bacterial adherence patternb | % of eukaryotic cells with adherent bacteria in given rangesc |
|
|---|---|---|---|
| >10 | 1–10 | ||
| E. coli DH5α | |||
| MQC922 (wt/pVector) | Diffuse, moderate | 0.0 | 100.0 |
| MQC926 (wt/pEDL933-cah) | Diffuse, strong | 77.0 ± 8.0 | 17.0 ± 2.0 |
| MQC924 (wt/pSS17-cah) | Diffuse, moderate | 0.0 | 100.0 |
| E. coli O157:H7 EDL933 | |||
| MQC966 (wt/pVector) | Aggregative, moderate | 41 ± 8 | 60 ± 9 |
| MQC1048 (wtΔcah/pVector) | Diffuse, moderate | 40 ± 6 | 60 ± 6 |
| MQC1050 (wtΔcah/pEDL933-cah) | Aggregative, moderate | 23.5 ± 9.5 | 76 ± 8 |
| E. coli O157:H7 SS17 | |||
| MQC928 (wt/pVector) | Aggregative, moderate | 9.5 ± 9.5 | 79.0 ± 2.0 |
| MQC930 (wt/pEDL933-cah) | Aggregative, moderate | 46.5 ± 7.5 | 53.5 ± 7.5 |
| MQC932 (wt/pSS17-cah) | Aggregative, moderate | 30.0 ± 11.0 | 62.5 ± 3.5 |
wt, wild type.
The bacterial adherence pattern was classified as described previously (19), and the immunofluorescence images are supplied in Fig. S2 in the supplemental material.
Data represent mean percentages of RSE cells of a total of 80 with the number of attached E. coli cells in two ranges (>10 and 1–10) from two replicate experiments. Each trial included one slide per bacterial strain, and each slide had 4 technical replicate spots; 20 well-dispersed RSE cells were evaluated per spot for a total of 80. The mean percentage for each range was used to rate adherence as strong, moderate, or nonadherent.
In contrast, transformation with EDL933 cah or SS17 cah did not alter the adherence patterns of SS17 (Table 3, MQC928, MQC930, and MQC932). Although a greater number of SS17 isolates expressing EDL933 cah (MQC930; 46.5%; P = 0.0001) and SS17 cah isolates (MQC932; 30%; P = 0.0041) adhered to RSE cells in the “>10” range than the control strain (MQC928; 9.5%), it was not sufficient to alter the adherence pattern compared with MQC928. Deletion of cah in SS17 and transformation of this strain with either EDL933 cah or SS17 cah did not alter the bacterial adherence pattern of STEC strain SS17 (data not shown).
DISCUSSION
While cah is widespread in the STEC population, strains of STEC differ in the number of genes encoding Cah and Cah homologs, including Ag43. Strain EDL933 carries two copies of cah, which are both functional, whereas strain SS17 carries a deletion mutation in cah that naturally inactivates Cah in this strain. Unlike STEC, E. coli K-12 strain DH5α lacks cah but carries a gene encoding Ag43 (also known as Flu). Our observation that the complementation of DH5α with EDL933-Cah increased its ability to form biofilms on glass in agitated culture but inhibited biofilm production under static conditions was likely caused by the strong cell-to-cell autoaggregation in DH5α conferred by EDL933-Cah. This inhibition of biofilm production correlated with the presence of cell aggregates at the bottom of culture tubes in the autoaggregation test performed with static suspensions of this complemented strain, which was observed previously (14). On the other hand, culture agitation may have prevented the formation of such large aggregates, making planktonic cells of DH5α expressing EDL933 cah available for attachment to the glass surface and initiation of a biofilm.
The contribution of Cah to the initial attachment of bacteria to plant tissue appeared to be strain specific. Expression of EDL933 cah in trans in SS17 enhanced attachment of SS17 cells to spinach leaves compared with that of its parental strain. This observation supports a previous report that introduction of cah into E. coli DH5α increased its attachment to alfalfa sprouts (18). This suggests that like curli and the self-associating autotransporter AIDA-1, which both are secreted mediators of adherence to alfalfa sprouts (18), Cah is likely part of the multifactorial interaction of STEC with plant tissue and may contribute to the colonization of produce by enteric pathogens. Transformation of SS17 with its own cah did not increase attachment of the pathogen to spinach leaves, providing further evidence that SS17 cah carries a loss-of-function mutation. On the other hand, Cah appeared to inhibit the initial interaction of EDL933 cells with the spinach leaf surface since its inactivation resulted in an increased attachment of this pathogen to spinach leaves. The lower attachment of Cah-expressing EDL933 cells to spinach leaves was unlikely to have been caused by autoaggregation that would hamper single-cell attachment to the leaf surfaces, since observation under the microscope failed to reveal aggregate formation during the attachment assay (our unpublished data). It is probable that this distinct role of Cah in an SS17 and EDL933 background results from differences in the combined role of various adhesins and colonization factors that are specific to each strain, such that absence of Cah in strain EDL933 allows for enhanced attachment to leaves via another adhesion factor, whereas its presence in SS17 promotes the bacterial cell-to-plant tissue interaction either directly or indirectly. The interdependence of attachment phenotype in the light of a large number of adhesins present in E. coli is discussed below.
Cah-mediated adherence of bacteria to eukaryotic cells also appeared to be conditional. A Cah-mediated “aggregative, moderate” adherence pattern was observed only in STEC strain EDL933. In trans expression of EDL933 cah in STEC strain SS17 had no effect on the adherence of SS17 to RSE cells. We observed that the presence of a functional Cah strongly increased the adherence of E. coli DH5α to RSE cells, similarly to the autoaggregative protein AIDA-1 shown to facilitate binding of E. coli to HeLa and HT29 human cells (17). However, this result is in contrast to a previous report that the Cah-positive DH5α showed reduced adherence to HeLa cells compared with the control strain due to the formation of cell aggregates by the Cah-expressing DH5α strain (14). This cell line-dependent adherence of E. coli cells to eukaryotic cells was reported also in our previous study, in which both RSE cells and HEp-2 cells were used to assess the adherence of supershedder strain SS17 (19) and for the role of curli fimbriae in adherence of STEC O157 to RSE cells (24) and to HEp-2 cells (25).
Our study demonstrates that the role of Cah in adhesion and autoaggregation is largely dependent on the bacterial genetic background. The strong autoaggregation conferred by EDL933-Cah was observed only in DH5α and not in SS17 or EDL933. Similarly, while expression of EDL933-Cah in trans restored the wild-type adherence phenotype to EDL933Δcah mutant, it increased adherence of only DH5α but not EDL933 or SS17 to bovine RSE cells. As mentioned above, this distinct strain-specific function of Cah could be in part attributed to the presence of different fimbrial and nonfimbrial adhesins in E. coli DH5α and STEC strains; STEC strains are also known to carry more adhesin genes than E. coli K-12 (15, 26). Furthermore, STEC strains vary greatly in both abundance and regulation of genes encoding adhesins and colonizing factors (19, 27). Attachment and biofilm formation in E. coli are multifactorial and may result from the combined effects of several adhesins, e.g., the competitive relationship between type I fimbriae and Ag43 in K-12 biofilm formation (16), and the interdependent regulation of long polar fimbriae and curli expression in STEC O157:H7 that affects its adherence to intestinal epithelial cells (28). Like many other STEC strains, EDL933 and SS17 carry genes encoding adhesins known to play a role in biofilm formation and attachment to plant tissue, such as curli, cellulose, poly-N-acetylglucosamine, and colanic acid, as well as adhesins known to mediate adherence of E. coli O157:H7 to epithelial cells, such as long polar fimbriae, curli, pili, and other autotransporter proteins (11, 13, 18). In particular, curli fimbriae, which promote adhesion to abiotic and biotic surfaces such as human cells and plant surfaces (18), are known to trigger strong cell-cell interactions that result in autoaggregation (29), similarly to intercellular interactions via the self-recognizing adhesins Cah (14) and Ag43 (16). Some of these adhesins may interfere with Cah function or may strengthen the role of Cah in adhesion when they are expressed in a particular niche in E. coli. Similar to Cah, the aggregative and strong biofilm phenotypes conferred by Ag43 were observed only in K-12 and not in uropathogenic E. coli (UPEC) isolate CFT073, although agn43 alleles were associated with the persistence of CFT073 in the urinary tract (30). These findings suggest that regulatory controls in different niches or factors lacking in K-12 strains are likely involved in modulating Cah and Ag43 levels or function in pathogenic E. coli. This may also explain the distinct role of Cah among SS17, EDL933, and DH5α in adherence to RSE cells and in attachment of SS17 and EDL933 to spinach leaves.
Among the 48 STEC strains examined, 83% carry at least one allele of cah or cah homolog, indicating the biological importance of the autotransporter protein encoded by this gene. However, loss-of-function mutations in cah appeared to be common in STEC since 31.3% of the cah-positive strains that we examined in this study carry a cah mutation. These common loss-of-function mutations in cah might be selected in distinct ecological niches, a phenomenon known as adaptive mutation (31). Adaptive mutations often enhance the fitness of bacterial cells under a particular selective pressure, resulting, for example, in improved nutrient scavenging or increased resistance to stresses or antibiotics in bacterial subpopulations (32–36). Several genes encoding fimbrial adhesins such as fimH, involved in the biogenesis of type I fimbriae, and curli loci, encoding the curli fimbriae, are subject to adaptive mutation (37–41). Variation of FimH in uropathogenic E. coli strains was linked to increased bacterial uroepithelial adhesion and bladder colonization (42), whereas loss of curli was suggested to improve pathogen survival in the host following infections (41, 43–45). Adaptive mutations also were observed in STEC O157:H7 genes encoding the response regulator of the two-component signal transduction cassette RcsB, as well as the alternative sigma factor RpoS (46, 47). Inactivation of RcsB or RpoS in E. coli O157:H7 was suggested to be beneficial to the bacterial population as a whole, since two subpopulations with an array of distinct phenotypes emerged. Remarkably, the polymorphic nature of autoaggregation observed in a collection of STEC O111 strains was attributed to loss-of-function mutations in RpoS (29, 48), suggesting that adaptive mutation is a common mechanism in STEC to fine-tune its colonization and survival under changing habitat conditions.
In this study, we observed a mutation rate in cah similar to that reported in rpoS (22.4%) (49) and in fimH (0.8 to 26.2%) (38) and further demonstrated that the Cah autotransporter protein confers on E. coli DH5α cells a strong autoaggregative phenotype that is inversely correlated with its ability to form biofilms and plays a strain-specific role in plant and animal colonization by STEC. Formation of cell aggregates due to increased bacterium-to-bacterium interactions may be disadvantageous to bacteria under certain conditions, such as during the initiation of a new biofilm or disassembly of a mature biofilm or the need to seek nutrient sources. Therefore, the loss-of-function mutations in cah may be a selective trait under conditions that favor a planktonic state in STEC, thus contributing to the adaptability of this common pathovar to fluctuating environments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth medium.
The E. coli strains used in this study are listed in Table 2. Strains were grown routinely in Luria-Bertani half-salt medium (LB) (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl). Chloramphenicol (Cm) was supplemented at a final concentration of 15 μg/ml for strains carrying the expression plasmids.
DNA analysis.
The complete genomes of STEC that are available in GenBank as of September 2016 were used to create a database (Table 1) for search of cah or cah homologs using Geneious8.1.8 (BioNumeric). BLAST was performed using EDL933 cah as a query. The retrieved cah and cah homologs were translated using the bacterial translation codon table. Each Cah and Cah homolog was aligned using ClustalW with BLOSUM matrix to score the alignment. The gap open penalty was set at 10, and the gap extension penalty was set at 0.1. A neighbor-joining tree was constructed using the Jukes-Cantor genetic distance mode with 10,000 bootstrap replicates.
Gene deletion and cloning.
cah was deleted using lambda Red-mediated gene replacement (50). Briefly, the kanamycin resistance cassette was amplified from pACYC177 with primers cah-UFP1 and cah-DRP2 (Table 4) and electroporated into E. coli strains carrying pKD119 (Table 2). The replacement of cah with the kanamycin resistance gene was verified by PCR using primers flanking cah (Table 4) and confirmed by DNA sequencing.
TABLE 4.
Primers used in this study
| Name | Oligonucleotide sequence (5′ to 3′)a | Purpose |
|---|---|---|
| cah_UFP1 | tctcttgcgtgactgctctactgttaatagaataaaacgatcgataaaacCAACAAAGCCACGTTGTGTC | Deletion of cah |
| cah_DRP2 | accgtcatcatccttaacatcaacggaagaatggcctgcagcaccatacaTCCCGTCAAGTCAGCGTAAT | Deletion of cah |
| cah-UFHindIII | gatcaagcttatcactgacctgcccg | Cloning of cah |
| cah-DRBamHI | gatcggatccatccccggtttgctgc | Cloning of cah |
| pBBR1MCS-F | gttttcccagtcacgacgtt | Specific to plasmid pBBR1MCS, upstream of the MCSb site |
| pBBR1MCS-R | ggctcgtatgttgtgtggaa | Specific to plasmid pBBR1MCS, downstream of the MCS site |
| cah-F1 | atgcccctcctgtttctctt | Sequencing of cloned cah |
| cah-F2 | accacaaatggtcgtcaggt | Sequencing of cloned cah |
| cah-F3 | ggtaaggctgacggtgttgt | Sequencing of cloned cah |
| cah-F4 | gttgtggatgcacagaatgg | Sequencing of cloned cah |
| cah-F5 | ggagccacaactgcagtaca | Sequencing of cloned cah |
| cah-R1 | ttcagaactgcggtgaacag | Sequencing of cloned cah |
| cah-R2 | gacataccggcaacctctgt | Sequencing of cloned cah |
| cah-R3 | gatgtccgacagtggtgttg | Sequencing of cloned cah |
| cah-R4 | ccactgctcgcctctgttat | Sequencing of cloned cah |
| cah-R5 | cgcaggccattaaccactat | Sequencing of cloned cah |
Nucleotides in uppercase font are specific to gene replacement vector pACYC177, and nucleotides in bold lowercase font represent the endonuclease restriction sites incorporated in the primers to facilitate cloning.
MCS, multiple-cloning site.
DNA fragments containing the cah gene were PCR amplified, KpnI and HindIII digested, and cloned into pBBR1MCS (Table 2). The constructs were verified by DNA sequencing using primers specific to pBBR1MCS and to the cah gene (Table 4). The sequence-confirmed constructs were transformed into the target strains by electroporation. The promoter and coding sequences of cah in strains SS17 and EDL933 are provided in Fig. S1 in the supplemental material.
Microscopy.
A 10-μl aliquot of each bacterial culture grown statically in LB was placed on a glass slide with a coverslip. E. coli cells were visualized under a Leica DMRB microscope (Leica Microsystems, Wetzlar, Germany), and images were captured with a Hamamatsu Orca C4742-95 camera (Bridgewater, NJ).
Biofilms.
Biofilm assays in LB or spinach lysates were carried out as described previously (11, 51). Briefly, 1 ml of LB or spinach lysate inoculated with 5 × 103 cells/ml was aliquoted into borosilicate glass tubes and incubated for 24 h at 37°C or 28°C, respectively. After incubation, the biofilms were rinsed twice with sterile distilled water. The crystal violet bound to the biofilm biomass on the glass tube was solubilized in 1 ml of 33% acetic acid and quantified using a microplate reader (SpectraMax 340; Molecular Devices, Sunnyvale, CA). Each data set was the average of results from at least five biological replicates.
Attachment to spinach leaves.
Cells from overnight cultures at 28°C in LB (with appropriate antibiotics) were washed twice in potassium phosphate buffer (KPB; 10 mM; pH 7.0) and resuspended in diluted KPB (1 mM) at 2 × 107 cells/ml. Four young leaves sampled randomly from greenhouse-grown spinach plants (Spinacia oleracea L. cv. Avenger) were taped by their petiole to the inner side of the ridge of a beaker and immersed into the suspension. The leaves were incubated in the static suspension for 2 h and rinsed by gently moving the leaves up and down in distilled deionized (DDI) water three times in each of three beakers. Two discs on each side of the main vein were then cut out of each replicate leaf with a cork borer #5 (diameter, 9 mm) and were homogenized individually in 2 ml KPB with a mortar and pestle. The homogenate was dilution plated onto LB agar for bacterial counts.
Adherence to bovine RSE cells.
The bacterial adherence assay with RSE eukaryotic cells was performed as described previously (52) with a multiplicity of infection of 106 bacteria for 105 RSE cells. Bacterial adherence patterns were qualitatively recorded as diffuse or aggregative (clumps). Well-dispersed RSE cells were quantitatively analyzed for the number of adhering bacteria by immunofluorescence microscopy as previously described (19, 52, 53).
Statistical analysis.
For both biofilm and leaf attachment assays, an unpaired t test was performed for two-group comparisons, and analysis of variance (ANOVA) followed by Tukey's multiple-comparison test was performed for multiple comparisons with GraphPad Prism 7 (GraphPad Software, Inc.). For the t test, if the equal variance test failed, the Mann-Whitney rank sum test was performed. For the ANOVA, if the normality test or the equal variance test failed, the Kruskal-Wallis one-way ANOVA on ranks was performed followed by Dunn's test for multiple comparisons. Data obtained about adherence to RSE cells were statistically analyzed using the two-tailed Fisher's exact test (with P values of <0.05 considered significant) with GraphPad Prism 6.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jacqueline Louie for assistance with the biofilm assay and Yaguang Zhou for assistance with the spinach leaf attachment assay. Technical assistance provided by Bryan Wheeler at the National Animal Disease Center (NADC), Ames, IA, with the RSE cell adherence assays is acknowledged.
This work was supported by USDA-ARS CRIS projects 2030-42000-050-00D and 5030-32000-100-00D.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01739-17.
REFERENCES
- 1.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandl MT. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol 44:367–392. doi: 10.1146/annurev.phyto.44.070505.143359. [DOI] [PubMed] [Google Scholar]
- 3.Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P, Frankel G. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol 12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 4.Cooley MB, Quinones B, Oryang D, Mandrell RE, Gorski L. 2014. Prevalence of shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front Cell Infect Microbiol 4:30. doi: 10.3389/fcimb.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strawn LK, Grohn YT, Warchocki S, Worobo RW, Bihn EA, Wiedmann M. 2013. Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl Environ Microbiol 79:7618–7627. doi: 10.1128/AEM.02831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Lopez VM, Marin A, Allende A, Beuchat LR, Gil MI. 2013. Postharvest handling conditions affect internalization of Salmonella in baby spinach during washing. J Food Prot 76:1145–1151. doi: 10.4315/0362-028X.JFP-12-539. [DOI] [PubMed] [Google Scholar]
- 7.Beuchat LR, Ryu JH. 1997. Produce handling and processing practices. Emerg Infect Dis 3:459–465. doi: 10.3201/eid0304.970407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch MF, Tauxe RV, Hedberg CW. 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol Infect 137:307–315. doi: 10.1017/S0950268808001969. [DOI] [PubMed] [Google Scholar]
- 9.Frank JF, Chmielewski R. 2001. Influence of surface finish on the cleanability of stainless steel. J Food Prot 64:1178–1182. doi: 10.4315/0362-028X-64.8.1178. [DOI] [PubMed] [Google Scholar]
- 10.Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, Surette M, Kay WW. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol 188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter MQ, Louie JW, Feng D, Zhong W, Brandl MT. 2016. Curli fimbriae are conditionally required in Escherichia coli O157:H7 for initial attachment and biofilm formation. Food Microbiol 57:81–89. doi: 10.1016/j.fm.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Carter MQ, Brandl MT. 2015. Biofilms in fresh vegetables and fruits, p 176–204. In Pometto AL III, Demirci A (ed), Biofilms in the food environment, 2nd ed Wiley Blackwell, Selangor, Malaysia. [Google Scholar]
- 13.Matthysse AG, Deora R, Mishra M, Torres AG. 2008. Polysaccharides cellulose, poly-beta-1,6-N-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol 74:2384–2390. doi: 10.1128/AEM.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres AG, Perna NT, Burland V, Ruknudin A, Blattner FR, Kaper JB. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol Microbiol 45:951–966. doi: 10.1046/j.1365-2958.2002.03094.x. [DOI] [PubMed] [Google Scholar]
- 15.Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 16.Hasman H, Chakraborty T, Klemm P. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181:4834–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherlock O, Schembri MA, Reisner A, Klemm P. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J Bacteriol 186:8058–8065. doi: 10.1128/JB.186.23.8058-8065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres AG, Jeter C, Langley W, Matthysse AG. 2005. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl Environ Microbiol 71:8008–8015. doi: 10.1128/AEM.71.12.8008-8015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cote R, Katani R, Moreau MR, Kudva IT, Arthur TM, DebRoy C, Mwangi MM, Albert I, Raygoza Garay JA, Li L, Brandl MT, Carter MQ, Kapur V. 2015. Comparative analysis of super-shedder strains of Escherichia coli O157:H7 reveals distinctive genomic features and a strongly aggregative adherent phenotype on bovine rectoanal junction squamous epithelial cells. PLoS One 10:e0116743. doi: 10.1371/journal.pone.0116743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper KK, Mandrell RE, Louie JW, Korlach J, Clark TA, Parker CT, Huynh S, Chain PS, Ahmed S, Carter MQ. 2014. Complete genome sequences of two Escherichia coli O145:H28 outbreak strains of food origin. Genome Announc 2(3):e00482-. doi: 10.1128/genomeA.00482-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper KK, Mandrell RE, Louie JW, Korlach J, Clark TA, Parker CT, Huynh S, Chain PS, Ahmed S, Carter MQ. 2014. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genomics 15:17. doi: 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyle JL, Parker CT, Goudeau D, Brandl MT. 2010. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl Environ Microbiol 76:1375–1387. doi: 10.1128/AEM.02461-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter MQ, Xue K, Brandl MT, Liu F, Wu L, Louie JW, Mandrell RE, Zhou J. 2012. Functional metagenomics of Escherichia coli O157:H7 interactions with spinach indigenous microorganisms during biofilm formation. PLoS One 7:e44186. doi: 10.1371/journal.pone.0044186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudva IT, Carter MQ, Sharma VK, Stasko JA, Giron JA. 2017. Curli temper adherence of Escherichia coli O157:H7 to squamous epithelial cells from the bovine recto-anal junction in a strain-dependent manner. Appl Environ Microbiol 83:e02594-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SH, Kim YH. 2004. Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J Vet Sci 5:119–124. [PubMed] [Google Scholar]
- 26.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Ali GS, Ouellette LM, Henderson ST, Lacher DW, Riordan JT, Whittam TS, Manning SD. 2010. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One 5:e10167. doi: 10.1371/journal.pone.0010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd SJ, Ritchie JM, Torres AG. 2012. Fimbriation and curliation in Escherichia coli O157:H7: a paradigm of intestinal and environmental colonization. Gut Microbes 3:272–276. doi: 10.4161/gmic.20661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diodati ME, Bates AH, Miller WG, Carter MQ, Zhou Y, Brandl MT. 2016. The polymorphic aggregative phenotype of Shiga toxin-producing Escherichia coli O111 depends on RpoS and curli. Appl Environ Microbiol 82:1475–1485. doi: 10.1128/AEM.03935-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster PL. 2000. Adaptive mutation: implications for evolution. Bioessays 22:1067–1074. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Spira B, Zhou Z, Feng L, Maharjan RP, Li X, Li F, McKenzie C, Reeves PR, Ferenci T. 2010. Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol Evol 2:478–487. doi: 10.1093/gbe/evq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillemet ML, Moreau PL. 2012. Activation of the cryptic PhnE permease promotes rapid adaptive evolution in a population of Escherichia coli K-12 starved for phosphate. J Bacteriol 194:253–260. doi: 10.1128/JB.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall BG. 1989. Selection, adaptation, and bacterial operons. Genome 31:265–271. doi: 10.1139/g89-044. [DOI] [PubMed] [Google Scholar]
- 35.Sung HM, Yasbin RE. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J Bacteriol 184:5641–5653. doi: 10.1128/JB.184.20.5641-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogstam A, Larsson JT, Kjelgaard P, von Wachenfeldt C. 2007. Mechanisms of adaptation to nitrosative stress in Bacillus subtilis. J Bacteriol 189:3063–3071. doi: 10.1128/JB.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trinchina EV. 2003. Structural and functional study of the receptor binding site for FimH adhesin in uropathogenic strains of Escherichia coli. Bull Exp Biol Med 136:380–384. doi: 10.1023/B:BEBM.0000010958.02737.e7. [DOI] [PubMed] [Google Scholar]
- 38.Hommais F, Gouriou S, Amorin C, Bui H, Rahimy MC, Picard B, Denamur E. 2003. The FimH A27V mutation is pathoadaptive for urovirulence in Escherichia coli B2 phylogenetic group isolates. Infect Immun 71:3619–3622. doi: 10.1128/IAI.71.6.3619-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronald LS, Yakovenko O, Yazvenko N, Chattopadhyay S, Aprikian P, Thomas WE, Sokurenko EV. 2008. Adaptive mutations in the signal peptide of the type 1 fimbrial adhesin of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 105:10937–10942. doi: 10.1073/pnas.0803158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kisiela DI, Chattopadhyay S, Libby SJ, Karlinsey JE, Fang FC, Tchesnokova V, Kramer JJ, Beskhlebnaya V, Samadpour M, Grzymajlo K, Ugorski M, Lankau EW, Mackie RI, Clegg S, Sokurenko EV. 2012. Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin. PLoS Pathog 8:e1002733. doi: 10.1371/journal.ppat.1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakellaris H, Hannink NK, Rajakumar K, Bulach D, Hunt M, Sasakawa C, Adler B. 2000. Curli loci of Shigella spp. Infect Immun 68:3780–3783. doi: 10.1128/IAI.68.6.3780-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissman SJ, Beskhlebnaya V, Chesnokova V, Chattopadhyay S, Stamm WE, Hooton TM, Sokurenko EV. 2007. Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect Immun 75:3548–3555. doi: 10.1128/IAI.01963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bian Z, Brauner A, Li Y, Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis 181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 44.Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Baumler AJ. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 45.Kai-Larsen Y, Luthje P, Chromek M, Peters V, Wang X, Holm A, Kadas L, Hedlund KO, Johansson J, Chapman MR, Jacobson SH, Romling U, Agerberth B, Brauner A. 2010. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog 6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter MQ, Parker CT, Louie JW, Huynh S, Fagerquist CK, Mandrell RE. 2012. RcsB contributes to the distinct stress fitness among Escherichia coli O157:H7 curli variants of the 1993 hamburger-associated outbreak strains. Appl Environ Microbiol 78:7706–7719. doi: 10.1128/AEM.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carter MQ, Louie JW, Huynh S, Parker CT. 2014. Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157:H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol 44:108–118. doi: 10.1016/j.fm.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 48.Diodati ME, Bates AH, Cooley MB, Walker S, Mandrell RE, Brandl MT. 2015. High genotypic and phenotypic similarity among Shiga toxin-producing Escherichia coli O111 environmental and outbreak strains. Foodborne Pathog Dis 12:235–243. doi: 10.1089/fpd.2014.1887. [DOI] [PubMed] [Google Scholar]
- 49.Waterman SR, Small PL. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect Immun 64:2808–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carter MQ, Brandl MT, Louie JW, Kyle JL, Carychao DK, Cooley MB, Parker CT, Bates AH, Mandrell RE. 2011. Distinct acid resistance and survival fitness displayed by curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl Environ Microbiol 77:3685–3695. doi: 10.1128/AEM.02315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kudva IT, Dean-Nystrom EA. 2011. Bovine recto-anal junction squamous epithelial (RSE) cell adhesion assay for studying Escherichia coli O157 adherence. J Appl Microbiol 111:1283–1294. doi: 10.1111/j.1365-2672.2011.05139.x. [DOI] [PubMed] [Google Scholar]
- 53.Kudva IT, Krastins B, Torres AG, Griffin RW, Sheng H, Sarracino DA, Hovde CJ, Calderwood SB, John M. 2015. The Escherichia coli O157:H7 cattle immunoproteome includes outer membrane protein A (OmpA), a modulator of adherence to bovine rectoanal junction squamous epithelial (RSE) cells. Proteomics 15:1829–1842. doi: 10.1002/pmic.201400432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.