ABSTRACT
Bacteriocins from lactic acid bacteria (LAB) are of increasing interest in recent years due to their potential as natural preservatives against food and beverage spoilage microorganisms. In a screening study for LAB, we isolated from olives a strain, Lactobacillus plantarum NI326, with activity against the beverage-spoilage bacterium Alicyclobacillus acidoterrestris. Genome sequencing of NI326 enabled the identification of a gene cluster (designated plc) encoding a putative circular bacteriocin and proteins involved in its modification, transport, and immunity. This novel bacteriocin, named plantaricyclin A (PlcA), was grouped into the circular bacteriocin subgroup II due to its high degree of similarity with other gassericin A-like bacteriocins. Purification of PlcA from the supernatant of Lb. plantarum NI326 resulted in an active peptide with a molecular mass of 5,570 Da, corresponding to that predicted from the (processed) PlcA amino acid sequence. The plc gene cluster was cloned and expressed in Lactococcus lactis NZ9000, resulting in the production of an active 5,570-Da bacteriocin in the supernatant. PlcA is believed to be produced as a 91-amino-acid precursor with a 33-amino-acid leader peptide, which is predicted to be removed, followed by joining of the N and C termini via a covalent linkage to form the mature 58-amino-acid circular bacteriocin PlcA. We report the characterization of a circular bacteriocin produced by Lb. plantarum. The inhibition displayed against A. acidoterrestris highlights its potential use as a preservative in food and beverages.
IMPORTANCE In this work, we describe the purification and characterization of an antimicrobial peptide, termed plantaricyclin A (PlcA), produced by a Lactobacillus plantarum strain isolated from olives. This peptide has a circular structure, and all genes involved in its production, circularization, and secretion were identified. PlcA shows antimicrobial activity against different strains, including Alicyclobacillus acidoterrestris, a common spoilage bacterium, which causes substantial economic losses in the beverage industry every year. In this study, we describe a circular antimicrobial peptide, PlcA, for a Lactobacillus plantarum strain.
KEYWORDS: circular bacteriocin, Alicyclobacillus acidoterrestris, Lactobacillus plantarum, immunity
INTRODUCTION
Bacteriocins are ribosomally synthesized antimicrobial peptides produced by bacteria to inhibit the growth of other, often closely related, strains (1). Bacteriocin production is a common feature among food-grade lactic acid bacteria (LAB), and bacteriocins have, for this reason, attracted considerable interest for their potential use as natural and nontoxic food preservatives (2, 3). Some of these peptides have demonstrated greater efficacy than conventional antibiotics against numerous pathogenic and drug-resistant bacteria, while not displaying any toxicity toward eukaryotic cells (4). For this reason, bacteriocins may also be useful in human and veterinary applications as a powerful weapon in the ongoing battle against antibiotic resistance, including for the treatment of local and systemic bacterial infections (4–6).
Within the different families of bacteriocins, circular bacteriocins constitute a unique group of active proteins in which the N- and C-terminal ends are covalently linked to form a circular backbone (7). This additional bond is thought to enhance the thermodynamic stability and structural integrity of the peptide and consequently improve its biological activity (8–10). To date, only a small number of circular bacteriocins have been described. These can be subdivided in two major groups according to their physicochemical characteristics and level of sequence identity (10). Subgroup I encompasses circular bacteriocins with a high content of positively charged amino acids and a high isoelectric point (pI, ∼10). This includes the best-studied circular bacteriocin, enterocin AS-48 (11), together with other bacteriocins, such as carnocyclin A (12), circularin A (13), lactocyclin Q (14), and garvicin ML (15). Subgroup II circular bacteriocins include bacteriocins with a smaller number of positively charged amino acid residues and a medium to low isoelectric point (pI, between ∼4 and 7). Currently this group comprises just three members, gassericin A (16), butyrivibriocin AR10 (17), and acidocin B (18).
In this study, we screened 50 colonies, isolated from olives, for their potential to inhibit growth of the beverage-spoilage strain Alicyclobacillus acidoterrestris sp1. We report the purification and genetic characterization of a circular gassericin A-like bacteriocin, termed plantaricyclin A, produced by Lactobacillus plantarum NI326, with antimicrobial activity against various microorganisms, including A. acidoterrestris sp1.
RESULTS AND DISCUSSION
Alicyclobacillus acidoterrestris is considered to be one of the species with the highest food spoilage impact worldwide (19). A. acidoterrestris is a thermoacidophilic spore-forming bacterium with a strong spoiling potential, especially in low-pH juices. The presence of A. acidoterrestris in juices is difficult to detect visually, but its presence is associated with an unpleasant odor caused by the production of guaiacol and other halophenols by the strain. Bacteriocins, such as the lantibiotic nisin A or the circular bacteriocin enterocin AS-48, have shown some promising results when used as strategies to inhibit growth of A. acidoterrestris in juices (20, 21).
Isolation and identification of Lactobacillus plantarum NI326.
In this study, we screened 50 presumed LAB isolates from olives with the aim of finding an isolate exhibiting antimicrobial activity against A. acidoterrestris sp1. Only one out of the 50 obtained isolates was shown to exhibit a zone of inhibition against the indicator strain. This colony was identified as Lb. plantarum by 16S rRNA sequencing and designated Lb. plantarum NI326. No zone of inhibition was apparent when the cell-free culture supernatant (CFS) was first treated with proteinase K, confirming the proteinaceous nature of the antimicrobial compound (data not shown).
Genome sequence analysis of Lb. plantarum NI326.
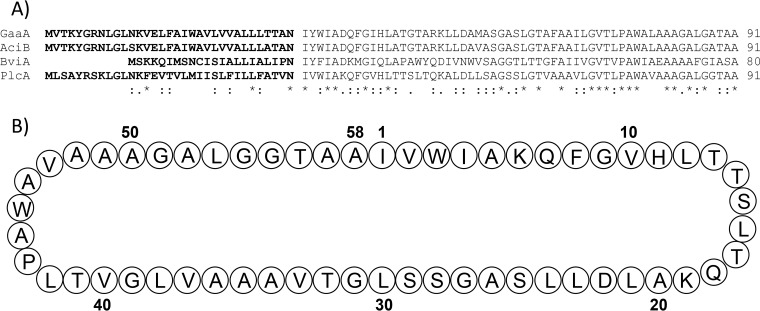
To find potential bacteriocin-encoding gene clusters, the genome of Lb. plantarum NI326 was sequenced, generating 84 contigs following sequence assembly. In silico analysis of the 84 contigs with BAGEL3 detected a potential bacteriocin gene cluster (designated here as plc) predicted to encode a peptide with a 43-amino-acid (aa) putative conserved domain corresponding to the subgroup II gassericin A-like circular bacteriocins. This putative peptide, designated plantaricyclin A (PlcA), exhibits 67% similarity to the circular bacteriocin gassericin A. An alignment of this peptide with all other members of the gassericin A-like circular bacteriocin group, gassericin A (GaaA), acidocin B (AciB), and butyrivibriocin AR10 (BviA), revealed a high degree of similarity. This alignment facilitated the prediction of the cleavage site of the signal peptide from the mature peptide to be between amino acids N33 and I34 (Fig. 1). Both GaaA and AciB are synthesized as 91-aa prepeptides with 33-aa leader peptides that are cleaved off, followed by a covalent linkage between the N and C terminus, to form the mature 58-aa circular bacteriocin. In previous studies, sequence alignments between characterized and hypothetical subgroup II circular bacteriocins did reveal the presence of a fully conserved asparaginyl cleavage site (18), which is also present in PlcA.
FIG 1.
(A) Sequence alignment of all the members of subgroup II circular bacteriocins with plantaricyclin A, using MUSCLE (41). Conserved, conservative, and semiconservative substitutions are indicated by asterisks, colons, and semicolons, respectively. Bold letters depict the leader sequence. (B) Schematic plantaricyclin A mature peptide.
The function of these leader peptides and mechanism through which peptide circularization occurs is still unclear. One of the biggest challenges in the field of circular proteins is finding out how their ends are stitched together from their linear precursors (22). Identification of this mechanism has the potential to facilitate the creation of new, highly stable antimicrobial agents for use in food, veterinary, and medical applications (12). PlcA has a predicted mass of 5,588 Da and represents a new bacteriocin within the subgroup II and the first (predicted) circular bacteriocin isolated from Lb. plantarum.
Analysis of the plc gene cluster revealed the presence of seven open reading frames (ORFs) downstream of the PlcA-encoding gene (plcA), with sequence and organizational similarity to those found in the gene clusters responsible for GaaA and AciB production (Table 1 and Fig. 2). Accordingly, plcA is followed by plcD, which encodes a putative 157-aa membrane-associated protein with a DUF95 conserved domain. Recent research suggests that DUF95 proteins play a dual role in the biosynthesis of circular peptides, as an immunity-associated transporter protein and as a secretion-aiding agent (23). The ORF, which is designated plcI, is located immediately downstream of plcD, and encodes a 54-aa protein with a hypothetical function as an immunity protein. Kawai et al. (24) showed that heterologous expression of GaaI, which is similar to the plcI protein product, in Lactococcus lactis confers a 7-fold-higher resistance to gassericin A compared to that of a control strain.
TABLE 1.
Putative proteins derived from the plc operon
| ORF | Product length (aa) | Amino acid identity (%) relative to gassericin A gene cluster homologs | Hypothetical function |
|---|---|---|---|
| plcA | 90 | 56 | Plantaricyclin A precursor |
| plcD | 157 | 33 | Unknown, DUF95 family |
| plcI | 54 | 33 | Immunity |
| plcT | 227 | 45 | ATP-binding protein |
| plcE | 214 | 37 | Membrane transporter |
| plcB | 173 | 30 | Unknown |
| plcC | 56 | 35 | Unknown |
FIG 2.
Schematic representation of the gene clusters involved in the production of the circular bacteriocins gassericin A (24), acidocin B (18) and plantaricyclin A. The known or putative biochemical function or properties are denoted by color, as indicated in the key.
The next two genes of the cluster (plcT and plcE) encode proteins of 227 aa and 214 aa, respectively (Table 1). Both have conserved ATP-binding domains linked to proteins of the ABC transporter family, and based on homology to their equivalents from the gassericin A and acidocin B associated gene clusters, they are most likely involved in the secretion of PlcA. The downstream plcB and plcC genes are in positions that are different from their homologs in the clusters for GaaA and AciB production (Fig. 2). The function of the proteins coded by these two genes is still unknown, but their presence in all of the clusters from circular bacteriocins clearly indicates that they play an important role (9).
Heterologous production of PlcA in L. lactis NZ9000.
To further confirm that PlcA is responsible for the activity shown by Lb. plantarum NI326, the entire plc cluster was cloned into the nisin-inducible plasmid pNZ8048 (pNZPlc) and transformed into L. lactis NZ9000, a naturally non-bacteriocin-producing strain. The CFS from L. lactis pNZPlc was shown to exhibit antimicrobial activity against A. acidoterrestris sp1 similar to that from the wild-type Lb. plantarum NI326 (Fig. 3A). The production of PlcA by L. lactis confirms that the cluster contains all necessary information for the correct production, modification, and secretion of PlcA. Based on these results and the similarity of the plc cluster to those from GaaA and AciB, we hypothesize that the biosynthetic machinery for all members of this bacteriocin subgroup is similar.
FIG 3.
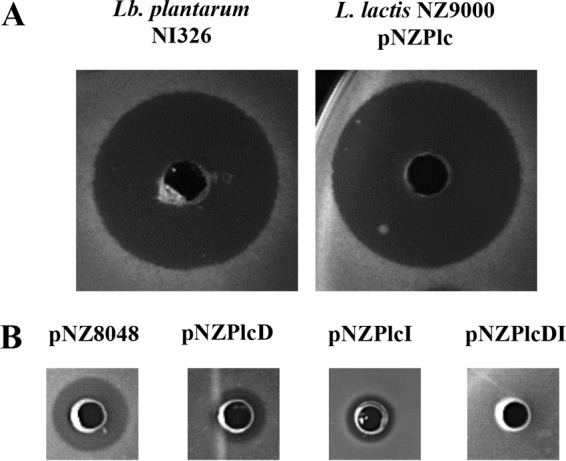
(A) Antimicrobial activity of the CFS of L. plantarum NI326 and nisin A-induced L. lactis NZ9000(pNZPlc) against A. acidoterrestris sp1. (B) Antimicrobial activity of the CFS of Lb. plantarum NI326 against cultures of L. lactis NZ9000(pNZ8048), L. lactis NZ9000(pNZPlcD), L. lactis NZ9000(pNZPlcI), and L. lactis NZ9000(pNZPlcDI) uninduced (−) or induced (+) with nisin A.
Analysis of immunity to PlcA.
In order to determine if plcD and/or plcI encode immunity proteins for PlcA, the genes were cloned individually or together in the NisA-inducible vector pNZ8048 and transformed into L. lactis NZ9000. The recombinant strain L. lactis NZ9000(pNZPlcDI) induced with nisin A displayed full resistance to PlcA, while strains L. lactis NZ9000(pNZPlcD) and L. lactis NZ9000(pNZPlcI) induced with NisA still exhibited sensitivity to PlcA, but at a visibly lower level compared to the control strain L. lactis NZ9000(pNZ8048) (Fig. 3B). Therefore, although both proteins individually appear to confer partial immunity to L. lactis NZ9000 against the antimicrobial activity of PlcA, the recombinant strain was fully protected against the action of PlcA when both proteins were being produced concomitantly. Similar results have been observed with other circular bacteriocins, such as carnocyclin A, where the production of the immunity protein (CclI) was not enough to confer full protection to the producer, and only when CclD and CclI were coproduced did the strain show full immunity (25).
Purification and MALDI-TOF analyses of the antimicrobial activity of Lb. plantarum NI326.
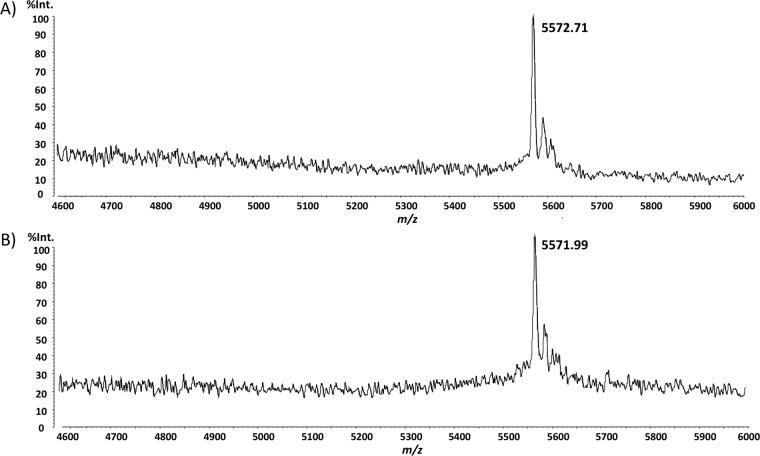
The antimicrobial peptide produced in the CFS by Lb. plantarum NI326 and L. lactis pNZPlc was purified by using reversed phase high-performance liquid chromatography (RP-HPLC), and the molecular mass was analyzed by using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). In both cases, a single mass of 5,572 Da was detected in the active fractions (Fig. 4). The 18-Da difference between the molecular mass of PlcA and its theoretical mass calculated from the AA sequence corresponds to the loss of a molecule of water that occurs during circularization of the peptide, as reported for other circular bacteriocins (18, 26).
FIG 4.
MALDI-TOF mass spectrometry analysis of the purified plantaricyclin A produced by L. lactis pNZPlca (A) and L. plantarum NI326 (B). %Int., percentage of intensity.
Sensitivity of plantaricyclin A to heat, pH, and proteolytic enzymes.
The antimicrobial activity of partially purified PlcA was the same as the initial antagonistic activity following exposure to temperatures ranging from 30°C to 100°C for 10 min, suggesting the relative stability of the bacteriocin. No antimicrobial activity was lost when PlcA was adjusted to pH values 2 to 10. The antimicrobial activity of PlcA was completely lost when treated with proteinase K and pronase, whereas pepsin and α-chymotrypsin treatments resulted in the retention of 100% and 78% of the initial antagonistic antimicrobial activity, respectively (results not shown).
The resistance of circular bacteriocins to temperature, pH variations, and proteolytic enzymes is due mainly to their three-dimensional conformation. The solution structure of acidocin B has recently been solved. Accordingly, AciB is composed of four α-helices of similar length folded to form a compact, globular bundle that allows the formation of a central pore, resembling the structure of the saposins. The surfaces of acidocin B and gassericin A are dominated by hydrophobic and uncharged residues and, therefore, it is believed that the initial contact between these circular peptides and the target strains is mediated by hydrophobic interactions (18).
Antimicrobial spectrum of plantaricyclin A.
Aliquots of HPLC purified fractions of PlcA were evaluated for their antimicrobial activities and inhibitory spectra against different indicator microorganisms. The strains A. acidoterrestris sp1, Lb. bulgaricus UCC, Pediococcus inopinatus 1011, and all tested lactococcal strains were inhibited by the bacteriocin produced by Lb. plantarum NI326 (Table 2).
TABLE 2.
Strains used in this study, sources, and activity of PlcA
| Strain | Sourcea | Activityb |
|---|---|---|
| Alicyclobacillus acidoterrestris sp1 | Coca Cola Co. | + |
| Lactococcus lactis HP | UCC | + |
| L. lactis KH | UCC | + |
| L. lactis MG1363 | UCC | + |
| L. lactis RT28 | UCC | + |
| L. lactis NZ9000 | UCC | + |
| Lactobacillus bulgaricus UCC | UCC | + |
| Lactobacillus plantarum PARA | UCC | − |
| L. plantarum WCFSI | UCC | − |
| Lactobacillus brevis MB124 | UCC | − |
| L. brevis SAC12 | UCC | − |
| L. brevis L102 | UCC | − |
| L. brevis L94 | UCC | − |
| Pediococcus claussenii H5 | UCC | − |
| Pediococcus inopinatus 1011 | UCC | + |
| Enterococcus faecium DPC1146 | UCC | − |
| Listeria innocua UCC | UCC | − |
| Listeria monocytogenes EGD-e | UCC | − |
| L. monocytogenes 33077 | UCC | − |
| Escherichia coli EC10B | UCC | − |
| Staphylococcus aureus DPC5243 | UCC | − |
| Streptococcus uberis ATCC 700407 | UCC | − |
| Streptococcus dysgalactiae GrpC | UCC | − |
| Salmonella enterica serovar Typhimurium UTC1lux | UCC | − |
| Klebsiella pneumoniae UCC | UCC | − |
| Bacillus cereus DPC6087 | UCC | − |
UCC, University College Cork.
+, zone of inhibition observed; −, no zone of inhibition observed.
In addition to the spectra of inhibition, we observed some other differences between PlcA and the other members of subgroup II, such as a higher isoelectric point (8.6) and a net charge of +1 (27). In fact, some authors use the pI values and net charges to differentiate between circular bacteriocins of subgroup I (pI, ∼10 and positively charged) from circular bacteriocins of subgroup II (pI, 4 to 7 and uncharged or slightly negative) (10). According to this classification system, PlcA should be placed in an intermediate position between subgroups I and II. However, we strongly believe that this peptide should be classified within subgroup II and propose to modify the classification criteria and broaden the pI range for this subgroup to be between 4 and ∼9.
Plantaricyclin A represents the first circular bacteriocin isolated and characterized from an Lb. plantarum strain. The antimicrobial activity observed against the food and beverage spoilage microorganism Alicyclobacillus acidoterrestris should be further studied, as this strain represents a significant problem for the food industry. The use of bacteriocins, such as nisin A and enterocin AS-48, as preservatives in low-pH beverages and juices has shown some promising results to control the growth of A. acidoterrestris (28). The circular nature of PlcA makes it especially interesting for industrial applications, as this peptide could survive and retain most of the activity under changing conditions (temperature and pH, for example) during food/beverage manufacture. Moreover, the narrow spectrum of activity from PlcA can be considered an advantage, especially in fermented beverages. In comparison to other broad-spectrum bacteriocins, such as nisin A or enterocin AS-48, PlcA could be used to specifically target A. acidoterrestris spp., while having little or no effect against other desirable microorganisms present in the beverage. However, a more detailed investigation, including that of more indicator strains and other A. acidoterrestris, is needed in order to further assess the full potential and applicability of this biofunctional peptide.
MATERIALS AND METHODS
Cultures and growth conditions.
The strains used in this study are summarized in Table 2. All Lactobacillus, Pediococcus and Leuconostoc strains were grown in de Man-Rogosa-Sharpe (MRS) medium (Oxoid, Hampshire, United Kingdom) at 30°C, A. acidoterrestris sp1 was grown in Bacillus acidoterrestris (BAT) broth (Pronadisa, Spain) at 45°C, while some of the other indicator strains were grown in LB broth (1% peptone, 1% NaCl, 0.5% yeast extract) at 37°C (Escherichia coli, Salmonella enterica serovar Typhimurium, and Klebsiella pneumoniae), brain heart infusion (BHI) broth (Oxoid) at 37°C (Staphylococcus aureus, Listeria monocytogenes, Listeria innocua, and Bacillus cereus), TSB broth (Oxoid) at 37°C (Streptococcus uberis and Streptococcus dysgalactiae) and M17 broth (Oxoid) supplemented with 0.5% glucose (Sigma-Aldrich, USA) at 30°C (Lactococcus lactis) or at 37°C (Enterococcus faecium). Chloramphenicol (Sigma-Aldrich) was added at 5 μg/ml where required. All of these microorganisms were grown under aerobic conditions. All strains were stored at −80°C in their respective media with 20% glycerol until required for use.
Isolation of LAB strains from olives.
Over 50 isolates were isolated from olives as previously described (29). Briefly, 5 g of olives (obtained from the English Market, Cork, Ireland) was homogenized with 45 ml of Ringer's solution using a stomacher at 300 bpm for 1 min (stomacher circular 400; Seward, UK). The resulting homogenate was serially diluted in Ringer's solution, and 100 μl of each dilution plated on MRS agar (Oxoid) plates supplemented with 100 μg/ml cycloheximide (Sigma) to suppress fungal growth. Plates were then incubated at 30°C anaerobically for 2 days. Colonies obtained were handpicked and inoculated into 250-μl aliquots of MRS broth in 96-well plates. Cultures were grown anaerobically overnight at 30°C and stored at −80°C with 20% glycerol for further analysis.
Isolation of anti-A. acidoterrestris sp1 bacteriocin-producing LAB.
Presumed LAB isolates exerting antimicrobial activity were identified using the spot-on-lawn method (29). Briefly, 5-μl aliquots of LAB cultures were spotted onto MRS agar plates and grown at 30°C anaerobically for 48 h. Plates were then overlaid with 5 ml of MRS soft agar (MRS broth supplemented with 0.8% bacteriological agar) seeded with 105 to 106 CFU/ml of an overnight culture of L. lactis HP. Plates were incubated at 30°C for 48 h, after which zones of inhibition surrounding the LAB colony were measured.
The LAB isolate showing inhibition against L. lactis HP was further cultured in 10 ml MRS broth and grown at 30°C overnight. Cell-free culture supernatant (CFS) was obtained by centrifugation of the culture at 12,000 × g, 4°C for 10 min and filtered through 0.2-μm-pore-size filters (Whatman International Ltd., Maidstone, UK). The activity of the CFS against A. acidoterrestris sp1 was analyzed using an agar diffusion test (ADT) (30). Briefly, 100-μl aliquots of CFS were placed in wells (6-mm diameter) bored in cooled Alicyclobacillus agar (Pronadisa) plates (30 ml) previously seeded (105 CFU/ml) with A. acidoterrestris sp1. Plates were incubated at 50°C to allow growth of the target organism and checked for zones of inhibition after 24 to 48 h.
Identification of LAB isolates.
Individual colonies were used as the templates for PCR. The primers Luc-F (5′ CTT GTT ACG ACT TCA CCC 3′) and Luc-R (5′ TGC CTA ATA CAT GCA AGT 3′) (Eurofins MWG, Ebersberg, Germany) were used to amplify a variable region of the 16S rRNA gene (31). The following conditions were used for the PCRs: 95°C for 60 s, 53°C for 60 s, and 72°C for 95 s for 30 cycles. The DNA from individually purified amplicons was subjected to Sanger sequencing (Eurofins MWG), and the corresponding species identity was obtained by comparative sequence analysis (BLASTN) against available sequence data in the National Center for Biotechnology Information (NCBI) database (32).
Lactobacillus plantarum NI326 genome sequencing, genome annotation, and bacteriocin screening.
The genome of Lb. plantarum NI326 was sequenced using a combined Roche GS-FLX Titanium and Illumina HiSeq 2000 approach (GATC Biotech, Constance, Germany) to a final coverage of ∼490-fold. Sequences obtained were first quality checked using IlluQC.pl from the NGS QC Toolkit v2.3 (http://www.nipgr.res.in/ngsqctoolkit.html) (33) and assembled with AbySS v1.9.0 (34).
Following sequence assembly, the generated contigs were employed to perform open reading frame (ORF) prediction with Prodigal v2.5 prediction software (http://gensoft.pasteur.fr/docs/prodigal/2.50/_README), supported by BLASTX v2.2.26 alignments (35). ORFs were automatically annotated using BLASTP v2.2.26 (35) analysis against the nonredundant protein database curated by the NCBI database. Following automatic annotation, ORFs were manually curated using the Artemis v16 genome browser and annotation tool (http://www.sanger.ac.uk/science/tools/artemis). The software tool was used to inspect and validate ORF results, to adjust start codons where necessary, and to aid in the identification of pseudogenes. The resulting ORF annotations were further refined, where required, using the alternative databases Pfam (36) and Uniprot/EMBL (http://www.uniprot.org/). Transfer tRNA was predicted using tRNA-scan-SE v1.4 (37). The whole genome was analyzed with the Web-based bacteriocin genome mining tool BAGEL3 (http://bagel.molgenrug.nl/) (38) to search for known and/or potential novel bacteriocins.
Cloning of the plc gene cluster in L. lactis NZ9000.
Primers, PCR fragments, and plasmids used in this study are listed in Table 3. All primers were ordered from Eurofins. Plasmid derivatives were constructed as follows: primers Plc-F/Plc-R were used for PCR amplification of a 3,172-bp fragment from total genomic DNA of Lb. plantarum NI326, which encompassed the entire plc gene cluster, including its promoter(s). Using this plc gene cluster fragment as the template and the primer pairs NcoI-Plc/XbaI-Plc, NcoI-PlcD/XbaI-PlcD, NcoI-PlcI/XbaI-PlcI, and NcoI-PlcD/XbaI-PlcI, fragments encompassing plcADITEB, plcD, plcI, and plcDI, respectively, were amplified (Table 3). Such fragments were digested with NcoI and XbaI and ligated into pNZ8048, digested with the same enzymes. The ligation mixtures were used to transform L. lactis NZ9000 competent cells as previously described (39). The plasmid derivatives pNZPlc, pNZPlcD, pNZPlcI, and pNZPlcDI, were checked by colony-PCR and sequencing of the inserts using primers PNZ-F/PNZ-R (Table 3).
TABLE 3.
Primers, PCR products, and plasmids used in this study
| Primer, PCR fragment, or plasmid | Nucleotide sequence (5′ to 3′) or descriptiona |
|---|---|
| Primers (PCR fragment) | |
| Plc-F (Plc-Clust) | AACGCAAATGTTCCACACGG |
| Plc-R (Plc-Clust) | GGATTGGACTAGTAGCTCTAGGGT |
| NcoI-Plc (PlcADITEB) | CACTCACCATGGGTTAATGCTTTCAGCATATCGTAGTAAAT |
| XbaI-Plc (PlcADITEB) | ATCTATCTAGACTATAAAAAAATCAAGCTATATATAGG |
| NcoI-PlcD (PlcD/PlcDI) | CACTCACCATGGTGAATAAACCGCGGAGTAATATC |
| XbaI-PlcD (PlcD) | ATCTATCTAGATTAATCTCCTAACAACCATAAGGC |
| NcoI-PlcI (PlcI) | CACTCACCATGGTTGTTAGGAGATTAATTATGAAGAATTTAG |
| XbaI-PlcI (PlcI/PlcDI) | ATCTATCTAGATTAATCTGTATGCCGTTTAATTAGCTGA |
| pNZ-F | TGTCGATAACGCGAGCATAA |
| pNZ-R | CAAAGCAACACGTGCTGTAA |
| PCR fragments | |
| Plc-Clust | 3,172-bp fragment external to Plc cluster |
| PlcADITEB | 2,908-bp NcoI/XbaI fragment containing genes plcA, plcD, plcI, plcT, plcE, and plcB |
| PlcD | 495-bp NcoI/XbaI fragment containing gene plcD |
| PlcI | 204-bp NcoI/XbaI fragment containing gene plcI |
| PlcDI | 662-bp NcoI/XbaI fragment containing genes plcD and plcI |
| Plasmids | |
| pNZ8048 | Cmr; inducible expression vector carrying the nisA promoter (42) |
| pNZPlc | pNZ8048 derivative containing PlcADITEB |
| pNZPlcD | pNZ8048 derivative containing PlcD |
| pNZPlcI | pNZ8048 derivative containing PlcI |
| pNZPlcDI | pNZ8048 derivative containing PlcDI |
Cleavage site for restriction enzymes is underlined. Cmr, chloramphenicol resistance.
Purification and MALDI-TOF mass spectrometry analysis of PlcA.
PlcA was purified from Lb. plantarum NI326 and L. lactis NZ9000 was transformed with pNZPlc, as described previously (40), with modifications. Briefly, a 1-liter CFS of Lb. plantarum NI326 was obtained as previously described. Recombinant L. lactis NZ9000(pNZPlc) was induced for the production of PlcA at an optical density at 600 nm (OD600) of 0.5, using nisin A (Nisaplin; Dupont, USA) at a final concentration of 10 ng/ml. The induced culture was grown at 32°C for 3 h. CFS was obtained by centrifugation of the culture at 12,000 × g at 4°C for 10 min. Activity of the CFS from either strain against A. acidoterrestris sp1 was confirmed on an ADT, as previously described. CFS was applied to a 10-g (60-ml) Varian C18 bond elution column (Varian, Harbor City, CA) preequilibrated with methanol and water. The column was washed with 20% ethanol, and the inhibitory peptide was eluted in 100 ml of 70% 2-propanol 0.1% trifluoroacetic acid (TFA). Aliquots (15 ml) were concentrated to 2 ml through the removal of 2-propanol by rotary evaporation (Buchi). Samples were then applied to a semipreparative Vydac C4 mass spectrometry (10 by 250 mm, 300 Å, 5 μm) RP-HPLC column (Grace, Columbia, USA) running an acetonitrile and propan-2-ol gradient described as follows: 5% to 55% buffer B and 0% to 5% buffer C over 25 min, followed by and 55% to 19% buffer B and 5% to 65% buffer C over 60 min, 19% to 5% buffer B and 65% to 95% buffer C over 5 min, where buffer A is Milli-Q water containing 0.1% TFA, buffer B is 90% acetonitrile 0.1% TFA, and buffer C is 90% propan-2-ol–0.1% TFA. The eluent was monitored at 214 nm, and fractions were collected at 1-min intervals. Fractions were assayed on Lactobacillus bulgaricus (a highly sensitive strain) indicator plates and active fractions assayed for the antimicrobial mass of interest using MALDI-TOF mass spectrometry (MALDI-TOF MS). MALDI-TOF MS was performed with an Axima TOF2 MALDI-TOF mass spectrometer (Shimadzu Biotech, Manchester, UK), as described by Field et al. (40).
Analysis of immunity against PlcA.
The immunity of wild-type L. lactis NZ9000 and recombinant strains L. lactis NZ9000(pNZPlcD), L. lactis NZ9000(pNZPlcI), and L. lactis NZ9000(pNZPlcDI) was tested against the CFS from Lb. plantarum NI326 on an ADT assay as described above. The indicator strains were seeded in GM17–0.8% agar with and without 10 ng/ml nisin A. The areas of zones of inhibition were measured after 24 h growth at 30°C. The absence of a zone indicates that the strain is immune to PlcA. Experiments were performed in triplicate to confirm the results.
Sensitivity of PlcA to heat, pH, and proteolytic enzymes.
Aliquots of a PlcA-containing fraction obtained following reversed-phase HPLC were subjected to the following treatments: (i) 20-fold (vol/vol) dilution with 30% 2-propanol containing 0.1% TFA and heating at 80°C and 100°C for 30 min and at 121°C for 15 min to determine the stability of PlcA to heat; (ii) 20-fold (vol/vol) dilution in 10 mM Tris buffer followed by a pH adjustment at 2, 3, 4, 5, 6, 7, 8, 9, and 10 with 1 M HCl or 1 M NaOH to evaluate the effect of pH on bacteriocin activity; and (iii) dilution as described in step ii, followed by the addition of α-chymotrypsin (Sigma), pepsin (Sigma), pronase (Sigma), and proteinase K (Sigma) at pH 7.0. Each enzyme was added to a final concentration of 1 mg/ml to determine PlcA sensitivity to proteolytic enzymes. After each treatment, the residual antimicrobial activity of PlcA was determined by the agar diffusion test (ADT) with A. acidoterrestris sp1 as the indicator microorganism. Experiments were performed in triplicate.
Antimicrobial spectrum of PlcA.
Aliquots of 100 μl purified PlcA from Lb. plantarum NI326 were used to test its antimicrobial activity against various indicators (Table 2) using an ADT assay as described above. Experiments were performed in triplicate.
Accession number(s).
The genome assembly of Lb. plantarum NI326 is deposited in GenBank under accession number GCA_002407395.1. The nucleotide sequences reported in this study are deposited in GenBank under accession numbers NDXC01000001.1 to NDXC01000084.1.
ACKNOWLEDGMENTS
This work was supported by a grant from Enterprise Ireland–Innovation Partnership Programme IP/2013/0254. D.V.S. is a member of the APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (grant SFI/12/RC/2273). J.M. is the recipient of a Starting Investigator Research Grant funded by SFI (reference no. 15/SIRG/3430).
REFERENCES
- 1.Nes IF, Brede DA, Diep DB. 2013. Chapter 16. Class II non-lantibiotic bacteriocins A2, p 85–92. In Kastin AJ. (ed), Handbook of biologically active peptides, 2nd ed Academic Press, Boston, MA. doi: 10.1016/B978-0-12-385095-9.00016-6. [DOI] [Google Scholar]
- 2.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–88. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 3.Gálvez A, Abriouel H, Lopez RL, Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int J Food Microbiol 120:51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins–a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. [DOI] [PubMed] [Google Scholar]
- 5.Sit CS, Vederas JC. 2008. Approaches to the discovery of new antibacterial agents based on bacteriocins. Biochem Cell Biol 86:116–23. doi: 10.1139/O07-153. [DOI] [PubMed] [Google Scholar]
- 6.Sang Y, Blecha F. 2008. Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim Health Res Rev 9:227–235. doi: 10.1017/S1466252308001497. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Sieiro P, Montalban-Lopez M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong X, Martin-Visscher LA, Nahirney D, Vederas JC, Duszyk M. 2009. The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim Biophys Acta 1788:1797–1803. doi: 10.1016/j.bbamem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 9.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2011. Structure and genetics of circular bacteriocins. Trends Microbiol 19:411–418. doi: 10.1016/j.tim.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Gabrielsen C, Brede DA, Nes IF, Diep DB. 2014. Circular bacteriocins: biosynthesis and mode of action. Appl Environ Microbiol 80:6854–6862. doi: 10.1128/AEM.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maqueda M, Galvez A, Bueno MM, Sanchez-Barrena MJ, Gonzalez C, Albert A, Rico M, Valdivia E. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr Protein Pept Sci 5:399–416. doi: 10.2174/1389203043379567. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC. 2008. Isolation and characterization of carnocyclin a, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl Environ Microbiol 74:4756–4763. doi: 10.1128/AEM.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemperman R, Jonker M, Nauta A, Kuipers OP, Kok J. 2003. Functional analysis of the gene cluster involved in production of the bacteriocin circularin A by Clostridium beijerinckii ATCC 25752. Appl Environ Microbiol 69:5839–5848. doi: 10.1128/AEM.69.10.5839-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawa N, Zendo T, Kiyofuji J, Fujita K, Himeno K, Nakayama J, Sonomoto K. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl Environ Microbiol 75:1552–1558. doi: 10.1128/AEM.02299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrero J, Brede DA, Skaugen M, Diep DB, Herranz C, Nes IF, Cintas LM, Hernandez PE. 2011. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl Environ Microbiol 77:369–373. doi: 10.1128/AEM.01173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai Y, Kemperman R, Kok J, Saito T. 2004. The circular bacteriocins gassericin A and circularin A Curr Protein Pept Sci 5:393–398. [DOI] [PubMed] [Google Scholar]
- 17.Kalmokoff ML, Cyr TD, Hefford MA, Whitford MF, Teather RM. 2003. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: characterization of the gene and peptide. Can J Microbiol 49:763–773. doi: 10.1139/w03-101. [DOI] [PubMed] [Google Scholar]
- 18.Acedo JZ, van Belkum MJ, Lohans CT, McKay RT, Miskolzie M, Vederas JC. 2015. Solution structure of acidocin B, a circular bacteriocin produced by Lactobacillus acidophilus M46. Appl Environ Microbiol 81:2910–2918. doi: 10.1128/AEM.04265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevilacqua A, Sinigaglia M, Corbo MR. 2008. Alicyclobacillus acidoterrestris: new methods for inhibiting spore germination. Int J Food Microbiol 125:103–110. doi: 10.1016/j.ijfoodmicro.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki K, Murakami M, Kawai Y, Inoue N, Matsuda T. 2000. Use of nisin for inhibition of Alicyclobacillus acidoterrestris in acidic drinks. Food Microbiol 17:315–320. doi: 10.1006/fmic.1999.0309. [DOI] [Google Scholar]
- 21.Grande MJ, Lucas R, Abriouel H, Omar NB, Maqueda M, Martínez-Bueno M, Martínez-Cañamero M, Valdivia E, Gálvez A. 2005. Control of Alicyclobacillus acidoterrestris in fruit juices by enterocin AS-48. Int J Food Microbiol 104:289–297. doi: 10.1016/j.ijfoodmicro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Craik DJ. 2006. Chemistry. Seamless proteins tie up their loose ends. Science 311:1563–1564. [DOI] [PubMed] [Google Scholar]
- 23.Mu F, Masuda Y, Zendo T, Ono H, Kitagawa H, Ito H, Nakayama J, Sonomoto K. 2014. Biological function of a DUF95 superfamily protein involved in the biosynthesis of a circular bacteriocin, leucocyclicin Q. J Biosci Bioeng 117:158–164. [DOI] [PubMed] [Google Scholar]
- 24.Kawai Y, Kusnadi J, Kemperman R, Kok J, Ito Y, Endo M, Arakawa K, Uchida H, Nishimura J, Kitazawa H, Saito T. 2009. DNA sequencing and homologous expression of a small peptide conferring immunity to gassericin A, a circular bacteriocin produced by Lactobacillus gasseri LA39. Appl Environ Microbiol 75:1324–1330. doi: 10.1128/AEM.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2010. Cloning and characterization of the gene cluster involved in the production of the circular bacteriocin carnocyclin A probiotics and antimicrobial proteins. 2:218–225. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Bueno M, Maqueda M, Galvez A, Samyn B, Van Beeumen J, Coyette J, Valdivia E. 1994. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J Bacteriol 176:6334–6339. doi: 10.1128/jb.176.20.6334-6339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, New York City, NY. [Google Scholar]
- 28.Tianli Y, Jiangbo Z, Yahong Y. 2014. Spoilage by Alicyclobacillus bacteria in juice and beverage products: chemical, physical, and combined control methods. Comp Rev Food Sci Food Safety 13:771–797. doi: 10.1111/1541-4337.12093. [DOI] [Google Scholar]
- 29.Crowley S, Mahony J, van Sinderen D. 2013. Broad-spectrum antifungal-producing lactic acid bacteria and their application in fruit models. Folia Microbiol (Praha) 58:291–299. doi: 10.1007/s12223-012-0209-3. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez J, Larsen R, Cintas LM, Kok J, Hernandez PE. 2006. High-level heterologous production and functional expression of the sec-dependent enterocin P from Enterococcus faecium P13 in Lactococcus lactis. Appl Microbiol Biotechnol 72:41–51. doi: 10.1007/s00253-005-0233-1. [DOI] [PubMed] [Google Scholar]
- 31.Corsetti A, Settanni L, Van Sinderen D. 2004. Characterization of bacteriocin-like inhibitory substances (BLIS) from sourdough lactic acid bacteria and evaluation of their in vitro and in situ activity. J Appl Microbiol 96:521–534. doi: 10.1111/j.1365-2672.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 32.Coordinators NR. 2017. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 45:D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1723. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. 2013. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Field D, Begley M, O'Connor PM, Daly KM, Hugenholtz F, Cotter PD, Hill C, Ross RP. 2012. Bioengineered nisin A derivatives with enhanced activity against both Gram-positive and Gram-negative pathogens. PLoS One 7:e46884. doi: 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]