ABSTRACT
Norovirus (NoV) is the leading cause of gastroenteritis outbreaks linked to oyster consumption. In this study, we investigated the potential of F-specific RNA bacteriophages (FRNAPH) as indicators of viral contamination in oysters by focusing especially on FRNAPH subgroup II (FRNAPH-II). These viral indicators have been neglected because their behavior is sometimes different from that of NoV in shellfish, especially during the depuration processes usually performed before marketing. However, a significant bias needs to be taken into account. This bias is that, in the absence of routine culture methods, NoV is targeted by genome detection, while the presence of FRNAPH is usually investigated by isolation of infectious particles. In this study, by targeting both viruses using genome detection, a significant correlation between the presence of FRNAPH-II and that of NoV in shellfish collected from various European harvesting areas impacted by fecal pollution was observed. Moreover, during their depuration, while the long period of persistence of NoV was confirmed, a similar or even longer period of persistence of the FRNAPH-II genome, which was over 30 days, was observed. Such a striking genome persistence calls into question the relevance of molecular methods for assessing viral hazards. Targeting the same virus (i.e., FRNAPH-II) by culture and genome detection in specimens from harvesting areas as well as during depuration, we concluded that the presence of genomes in shellfish does not provide any information on the presence of the corresponding infectious particles. In view of these results, infectious FRNAPH detection should be reconsidered as a valuable indicator in oysters, and its potential for use in assessing viral hazard needs to be investigated.
IMPORTANCE This work brings new data about the behavior of viruses in shellfish, as well as about the relevance of molecular methods for their detection and evaluation of the viral hazard. First, a strong correlation between the presence of F-specific RNA bacteriophages of subgroup II (FRNAPH-II) and that of norovirus (NoV) in shellfish impacted by fecal contamination has been observed when both viruses are detected using molecular approaches. Second, when reverse transcription-PCR and culture are used to detect FRNAPH-II in shellfish, it appears that the genomes of the viruses present a longer period of persistence than infectious virus, and thus, virus genome detection fails to give information about the concomitant presence of infectious viruses. Finally, this study shows that FRNAPH persist at least as long as NoV does. These data are major arguments to reconsider the potential of FRNAPH as indicators of shellfish viral quality.
KEYWORDS: norovirus, F-specific RNA bacteriophages, shellfish, viral pollution
INTRODUCTION
The link between seafood and foodborne illness has been recognized for over a century (1), and shellfish in particular are well-known vectors of human enteric viruses, such as norovirus (NoV) or hepatitis A virus (2–4). Among shellfish, oysters are particularly involved in virus transmission to humans because they are generally consumed raw.
The contamination of shellfish with enteric viruses occurs mostly when harvesting areas are affected by fecally contaminated waters, due, for example to inefficient wastewater treatments or plant overflow following rainfall events (3). Microorganisms accumulate in shellfish digestive tissues through the filtration of large volumes of seawater (5–7) and probably because of the presence of particular ligands which may favor NoV retention (8–10). The winter season is typically associated with shellfish-borne NoV outbreaks because, during this period, (i) pathogenic viruses circulate in populations and are excreted in massive amounts, (ii) better virus stability is observed in the natural environment due to lower water temperatures and low levels of UV radiation, and (iii) NoV is known to accumulate to high concentrations in shellfish (11).
Currently, shellfish contamination by pathogenic viruses is difficult to evaluate. Indeed, European regulations (12), which are based on a bacterial criterion, Escherichia coli detection in shellfish, have been shown to have many limitations for the identification of viral contamination, and thus, shellfish compliant with the regulations may be the source of NoV outbreaks (13–16). This phenomenon, which is mainly due to different times of virus persistence in water as well as the different accumulation and elimination kinetics between pathogenic viruses and bacterial indicators in shellfish, has led the scientific community to conclude that the detection of E. coli is not a suitable means to track viral pollution in shellfish (16, 17). In such a situation, a new virological indicator is needed for shellfish risk management, and two completely different strategies may be considered.
The first strategy, which seems to be increasingly favored, is the specific detection of pathogenic viral genomes by the use of molecular tools. To this end, a new ISO standard method has recently been proposed (18). This approach is a relevant tool for retrospective studies to demonstrate the link between foodstuffs and outbreaks. This should not hide the major disadvantages of its application to prospective investigations concerning the evaluation of viral hazards in vulnerable foodstuffs, like oysters. Indeed, even though it has never been demonstrated in oysters, it is very well recognized that viral genomes have a better persistence than infectious viruses in the environment, and thus, only a negligible part of the detected genomes in fact corresponds to infectious particles (19–22), especially after inactivation by UV light (23). More alarmingly, while the absence of genomes is necessarily a sign of the absence of the corresponding infectious viruses, the strict application of the ISO 15216-1 standard expresses a theoretical limit of detection (LOD) of greater than the 18 particles, usually defined to be the minimal NoV infective dose (24, 25). It can be 10 to 100 times higher because of the small volume analyzed by reverse transcription (RT)-PCR, the low recovery rate during genome extraction, and the presence of (RT-)PCR inhibitors. Thus, except in some specific cases (i.e., in cases with very high levels of pollution or artificial contamination) (26, 27), the number of genome copies of NoV in oysters is usually close to the limit of detection (16, 28–31), and all these considerations highlight the difficulties in interpreting a positive or a negative result when using such an approach.
The second strategy, which has been proposed for years, is the use of other indicators with the goal of tracking overall viral pollution. Great interest in the use of fecal bacteriophages as fecal indicators in the natural environment has been shown (32, 33), and among them, F-specific RNA bacteriophages (FRNAPH) have numerous advantages. They have been extensively studied because of their structural similarity to many waterborne pathogens. The genome of FRNAPH can be detected by RT-PCR (34, 35), but infectious particles can also be easily quantified (36) or rapidly detected (37). The specific detection of FRNAPH subgroup II (FRNAPH-II) and FRNAPH-III may also be used to track more specifically human fecal contamination (19, 38, 39), which provides essential information in view of the high degree of host specificity of pathogenic viruses. In shellfish, these indicators persist longer than E. coli (40), and as for NoV, better accumulation is observed during the winter period (6, 41). Despite their many advantages, FRNAPH are not, however, commonly used to evaluate the virological quality of oysters. A recent review of the literature shows the advantages and limitations of FRNAPH as viral indicators in shellfish management (42). FRNAPH have been discredited because, even though a number of studies have pointed out a positive correlation between the presence of FRNAPH and that of enteric viruses in shellfish (24, 26, 43–47), others have expressed opposite views (48–50). A closer look reveals that most studies compare the detection of infectious FRNAPH with that of the human NoV genome. This is due to the fact that human NoV cannot be routinely cultivated, although progress with the cultivation of human NoV has recently been made (51). An inconsistent correlation between the presence of NoV and that of infectious FRNAPH is therefore not so surprising when the well-established longer persistence of viral genomes than infectious viruses, as underlined above, is taken into account. It may also be assumed that good correlation cases may be linked to recent fecal pollution, while poor correlation cases may rather be linked to past pollution or the analysis of shellfish subjected to the depuration process (i.e., shellfish in which infectious particles were inactivated but genomes may have been detected). The two cases should not be associated with the same virological health hazard, and such differences are very difficult to discuss because no studies have compared the presence of infectious particles and genomes for a single virus in oysters. In the light of this, a strict comparison should be made only by investigating the genomes of both NoV and FRNAPH. In doing so, a good correlation between the presence of NoV and that of FRNAPH, especially when human FRNAPH-II is considered, has been observed (24, 26). Nevertheless, this observation was made with a limited number of data.
Because oysters are usually consumed raw, the only treatment which may be used for their decontamination is their storage in unpolluted water (i.e., the depuration process). In such a context, the elimination of FRNAPH and NoV by shellfish has also been investigated. Again, while NoV elimination kinetics is currently assessed using molecular methods (13, 52–55), results concerning FRNAPH are usually obtained by infectious virus detection (40, 56–58), and comparison of the results of most studies may show a significant bias according to the differences in stability between the genome and the infectious virus. Thus, while the persistence of infectious FRNAPH seems to be shorter than that of the NoV genome in shellfish, no information, to our knowledge, about the concomitant elimination of FRNAPH and NoV when these two viruses are targeted using molecular approaches is currently available. In addition, a large number of studies have been conducted using shellfish artificially contaminated with viruses, leading to concentrations very different from those observed in the environment.
In this context, the aim of this study was to explore if FRNAPH-II (infectious FRNAPH-II and/or the FRNAPH-II genome) may be good indicators of the virological quality of oysters, especially during the depuration processes. We first verified the correlation between the presence of the FRNAPH-II genome and that of the NoV genome in a significant number of samples (n = 111) that were collected from different class B harvesting areas over a 1-year period and in which E. coli and infectious FRNAPH were also monitored to evaluate the overall level of fecal pollution on the basis of viable microbiological criteria. Second, we compared the elimination of infectious FRNAPH to that of the corresponding genome, as well as the elimination of the genomes of both FRNAPH and NoV, in oysters. To be as close as possible to environmental conditions, the study was performed with three different oyster batches naturally impacted by fecal contamination and subjected to two different depuration processes.
RESULTS
Relation between the presence of the FRNAPH genome and that of the NoV genome in shellfish collected from harvesting areas.
The collected oysters (n = 111) were found to be significantly impacted by fecal pollution, given the prevalence of E. coli and infectious FRNAPH (i.e., 31.5% and 93.7%, respectively). Hence, the detection of infectious FRNAPH is proof of fecal contamination, even in the absence of E. coli. Among the analyzed batches, infectious FRNAPH-I and -II were detected in 84.7% and 64.9% of cases, respectively (Table 1), while only two samples (i.e., 1.8%) contained infectious FRNAPH-III.
TABLE 1.
Prevalence of NoV, FRNAPH, and E. coli in oysters
| Virus or bacterium | Winter (Oct. to Apr.) (n = 98) |
Summer (May to Sept.) (n = 13) |
Total % of samples positive (n = 111) | ||
|---|---|---|---|---|---|
| % of samples positive | No. of samples positive | % of samples positive | No. of samples positive | ||
| Genome | |||||
| NoV GI | 15.3 | 15 | 0 | 13.5 | |
| NoV GII | 46.9 | 46 | 0 | 41.4 | |
| FRNAPH-I | 9.2 | 9 | 7.7 | 1 | 9.0 |
| FRNAPH-II | 52.0 | 51 | 7.7 | 1 | 46.8 |
| FRNAPH-III | 2.0 | 2 | 7.7 | 1 | 2.7 |
| Infectious particles | |||||
| FRNAPH-I | 87.6 | 86 | 61.5 | 8 | 84.7 |
| FRNAPH-II | 72.5 | 71 | 7.7 | 1 | 64.9 |
| FRNAPH-III | 1.0 | 1 | 7.7 | 1 | 1.8 |
| E. coli | 32.7 | 32 | 23.1 | 3 | 31.5 |
The monitoring of NoV and FRNAPH genomes was conducted according to the ISO 15216-1 standard (18). A high degree of heterogeneity both in the recovery rates for genome extraction (average, 29.6% ± 27.0%) and in the RT-PCR inhibition rates (23.3% ± 17.9%) was observed, but the values for all samples fell within the performance criteria defined by the standard (i.e., >1% for the extraction rate and <75% for the RT-PCR inhibition rate). For the mean recovery and inhibition rates, the LOD was estimated to be about 600 genome copies (gc)/g of hepatopancreas (HP), nearly 15 times higher than the theoretical value. In the worst case, the recovery rate (1.2%) and inhibition rate (25%) for a single sample led to an LOD of greater than 104 gc/g of HP. However, as suggested in the ISO procedure, these two rates were not used to correct the measured genome concentrations.
Among the analyzed batches, 13.5% were positive for NoV genogroup I (GI) genomes (Table 1), and the concentrations were estimated to be between 42 and 437 gc/g of HP. Thus, genome concentrations appeared to be relatively low, not exceeding 10 times the LOD. Similarly, NoV genogroup II (GII) genomes were detected in 41.4% of the samples, with the concentrations ranging from 40 to 1,229 gc/g of HP. A higher prevalence of the NoV GII genome was observed in shellfish during the winter period (October to April) than during the summer period (May to September) (Fisher's exact test, P = 0.0006), while the NoV GI genome prevalence was not significantly different between the two seasons (P = 0.2089). Concerning FRNAPH genomes, 52.3% of the analyzed batches were positive, and, more precisely, the FRNAPH-I, FRNAPH-II, and FRNAPH-III genomes were detected in 9.0%, 46.8%, and 2.7% of samples, respectively (Table 1). As with NoV GII, a significant seasonal difference was observed for FRNAPH-II, with a higher prevalence being detected during the winter period for both the genome (Fisher's exact test, P = 0.0025) and the infectious particles (P < 10−4), and an association between these two parameters was observed (Table 2) (P = 0.0004). Concerning FRNAPH-I, while a slight seasonal difference in infectivity was detected (P = 0.0277), no difference in the prevalence of its genome was observed between these two periods (P = 1) (Table 1).
TABLE 2.
Contingency table for detection of infectious FRNAPH-II particles and the FRNAPH-II genome in oyster batches
| FRNAPH-II genome detection | No. of samples with the following result for infectious FRNAPH-II detection: |
||
|---|---|---|---|
| − | + | ++ | |
| − | 28 | 17 | 14 |
| + | 10 | 6 | 15 |
| ++ | 1 | 5 | 15 |
Finally, when the three-level classification used to rank the microorganism concentrations in shellfish was considered, a significant association between the presence of the NoV GII genome and that of the FRNAPH-II genome in shellfish (P < 10−4) was observed (Table 3). Conversely, no association between the presence of the FRNAPH-I genome and that of the NoV genome was observed (P = 0.2797). The same conclusions might be drawn when a two-level classification (i.e., a positive or negative result for microorganism detection) was used (P < 10−4 and P = 0.1910, respectively).
TABLE 3.
Contingency table for detection of FRNAPH-II and NoV GII genomes in oyster batches
| FRNAPH-II genome detection | No. of samples with the following result for NoV GII genome detection: |
||
|---|---|---|---|
| − | + | ++ | |
| − | 49 | 9 | 1 |
| + | 11 | 13 | 7 |
| ++ | 5 | 7 | 9 |
Elimination of FRNAPH and NoV GII during the depuration process of oysters.
In view of the association between the presence of the NoV GII genome and that of the FRNAPH-II genome in the samples, further investigations were performed to study the behavior of these microorganisms during shellfish depuration. Shellfish batches (n = 3) were subjected to two different storage conditions, namely, in a tank with UV-treated seawater and in natural seawater. During their storage, both FRNAPH-II and NoV GII were monitored by targeting of their genomes, while infectious FRNAPH were also quantified by plaque assay, and the presence of the FRNAPH subgroup was investigated by a sensitive qualitative approach (37).
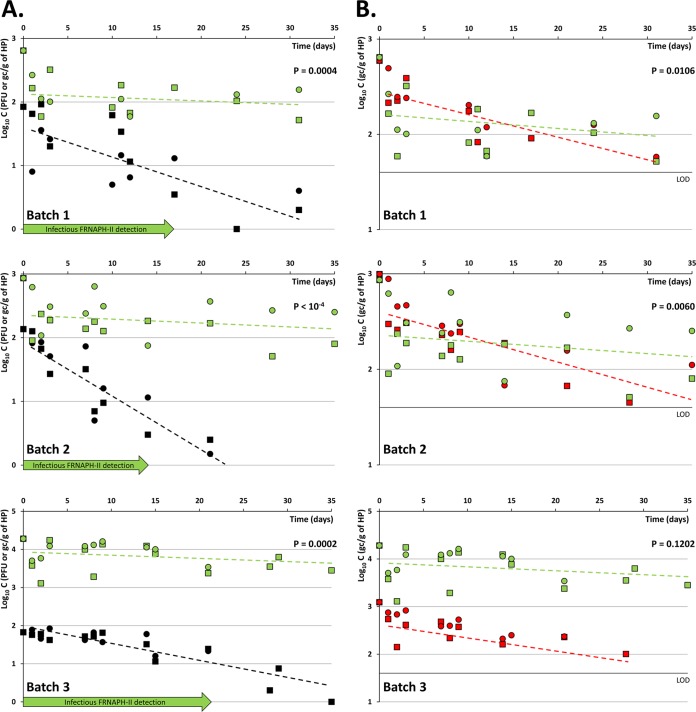
Despite the absence of E. coli in the 3 batches, the analyzed samples were found to be significantly impacted by fecal pollution, with an initial concentration of total infectious FRNAPH of 84, 136, and 67 PFU/g of HP in batches 1, 2, and 3, respectively. Initial NoV GII genome concentrations were 593, 992, and 1,230 gc/g of HP, respectively. FRNAPH-II genome concentrations were of the same order of magnitude in batches 1 and 2 (i.e., 644 and 860 gc/g of HP, respectively), while in the third batch, the concentration reached 1.9 × 104 gc/g of HP. From a qualitative point of view, infectious FRNAPH-I was detected throughout the study period (i.e., over 30 days), whereas infectious FRNAPH-II were last detected in batches 1, 2, and 3 at days 17, 14, and 21, respectively (Fig. 1A). Concerning the genomes, both the NoV GII and FRNAPH-II genomes were detected throughout the study period, except in batch 3, in which the NoV genome was last detected at day 28 (Fig. 1B).
FIG 1.
Behavior of FRNAPH and NoV GII during oyster depuration processes. (A) Comparison of the decrease of FRNAPH infectious particles with that of the FRNAPH-II genome. (B) Comparison of the decrease of the NoV GII genome with that of the FRNAPH-II genome. The limit of detection was 40 gc/g of shellfish hepatopancreas (HP). Squares, storage in UV-treated seawater tanks; circles, storage in natural seawater; black symbols, FRNAPH infectious particles; green symbols, FRNAPH-II genome; red symbols, NoV GII genome; C, concentration. Dotted lines correspond to the log-linear decays observed after day 1. The P value corresponding to the t test for comparison of the two slopes is indicated in each graph.
A statistically significant difference in the concentrations between the two depuration conditions was observed for the NoV GII genome (Wilcoxon signed test, P = 0.0027), with the geometric mean value measured during storage in natural seawater being 21% higher than that measured during depuration in the tank (Fig. 1B). On the other hand, no significant differences between the two conditions were shown for the FRNAPH-II genome (P = 0.0543) and FRNAPH infectious particles (P = 0.7156). This result led us to consider the use of the paired values obtained under the two conditions at a given time as replicates for use in subsequent statistical analyses. However, it must be emphasized that the variability corresponding to each of such formed pairs includes both the lab variability and the variability induced by the possible differences between the depuration processes.
Comparison of the variability of the experimental results within the pairs obtained for each of the three parameters during depuration showed that the variances observed within and/or among batches could be considered homogeneous (Cochran's test, P > 0.05). This outcome prompted the calculation of a unique coefficient (i.e., the coefficient of variation [CV]), which may characterize the variability associated with each parameter. Thus, CV was estimated to be 54.01% (95% confidence interval [CI0.95] = 38.79%, 67.27%) for NoV GII genomes, 75.62% (CI0.95 = 52.25%, 97.00%) for FRNAPH-II genomes, and 70.84% (CI0.95 = 37.89%, 98.61%) for FRNAPH infectious particles. These values point out the high level of variability in the experimental results which were obtained with each of the considered microorganisms.
For each of the parameters, decay coefficients were calculated and are reported in Table 4. Interestingly, a two-phase decrease may be described for each of them, with a breaking point being observed after the first day of storage. This phenomenon seemed to be particularly marked for the FRNAPH-II genome, for which a sharp slowdown in depletion was observed after the first day (i.e., the decay coefficient was <0.28 for the first day and >0.98 after day 1).
TABLE 4.
Estimated decay coefficients for each of the parameters during the depuration of oysters
| Batch | Timea (day) | Estimated decay coefficient (CI0.95) |
||
|---|---|---|---|---|
| NoV GII genome | FRNAPH-II genome | Infectious FRNAPH | ||
| 1 | ≤1 | 0.55 | 0.32 | 0.27 |
| >1 | 0.92 (0.89, 0.95) | 0.94 (0.91, 0.97) | 0.94 (0.90, 0.97) | |
| 2 | ≤1 | 0.52 | 0.28 | 0.75 |
| >1 | 0.91 (0.89, 0.94) | 0.98 (0.95, 1.01) | 0.86 (0.83, 0.89) | |
| 3 | ≤1 | 0.52 | 0.23 | 0.99 |
| >1 | 0.93 (0.89, 0.96) | 1.04 (1.03, 1.08) | 0.93 (0.90, 0.96) | |
| All | ≤1 | 0.53 | 0.27 | 0.59 |
| >1 | 0.90 (0.88, 0.92) | 0.99 (0.97, 1.01) | 0.90 (0.89, 0.92) | |
For times of ≤1 day, estimation of the decay coefficients was computed from a limited number of observations (i.e., n ≤ 6), hence the absence of CI0.95.
Taking into account the two-phase decrease, regression analyses were performed after the first day of depuration, and their outcomes are summarized in Table 5. From these results, it can be pointed out that the slopes of all the fitted lines represented in Fig. 1A and B corresponding to both the NoV GII genome and FRNAPH-II infectious particles are significantly different from zero (t test, P < 10−3), in contrast to the findings for the FRNAPH-II genome (P > 0.25). With regard to the F tests (data not shown) which were performed to compare the variances of both the departures from the linear regression and the replications, 8 of the 9 tests carried out yielded a result that was not statistically significant (P > 0.20). Despite the high level of variability in the experimental results obtained in this study, all these results favor the suitability of the log-linear model. In other words, this means an exponential decay for each of the three parameters as a function of time after day 1. Note that the decay coefficients estimated from the slopes of the regression lines (Table 5) are very similar to those reported in Table 4, which were calculated from the geometric weighted means of the empirical decay rates.
TABLE 5.
Regression analyses for each of the parameters during depuration of oysters after the first daya
| Virus | Batch 1 |
Batch 2 |
Batch 3 |
All batches |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | P value | dc | r2 | a | P value | dc | r2 | a | P value | dc | r2 | a | P value | dc | r2 | |
| NoV GII genome | −0.0283 | <10−4 | 0.9370 | 0.6747 | −0.0284 | <10−4 | 0.9366 | 0.6804 | −0.0234 | 0.0012 | 0.9476 | 0.4901 | −0.0275 | <10−4 | 0.9387 | 0.6321 |
| FRNAPH-II genome | −0.0061 | 0.3596 | 0.9860 | 0.0527 | −0.0064 | 0.2941 | 0.9855 | 0.0609 | −0.0082 | 0.2884 | 0.9814 | 0.0591 | −0.0071 | 0.0870 | 0.9838 | 0.0505 |
| Infectious FRNAPH | −0.0466 | 0.0002 | 0.8982 | 0.5969 | −0.0797 | <10−4 | 0.8324 | 0.7641 | −0.0461 | <10−4 | 0.8993 | 0.7894 | −0.0468 | <10−4 | 0.8978 | 0.4934 |
a, slope; dc, decay coefficient.
Furthermore, the estimated decay coefficient for the FRNAPH-II genome was significantly higher than the corresponding coefficient for both FRNAPH infectious particles and the NoV GII genome (t test, P < 10−4). When seen in another way, this means that the decay of the FRNAPH-II genome is significantly slower than that of FRNAPH infectious particles and the NoV GII genome. More precisely, the times required for a 90% reduction in concentration (T90 values), which were estimated before and after the day 1 breaking point, were 3.7 day−1 and 36.4 day−1, respectively, for the NoV GII genome, 1.8 day−1 and 140.7 day−1, respectively, for the FRNAPH-II genome and 4.3 day−1 and 21.3 day−1, respectively, for FRNAPH infectious particles.
DISCUSSION
The first objective of this study was to evaluate the interest in FRNAPH as indicators of NoV pollution in oysters by using comparable molecular approaches (i.e., RT-PCR) for the detection of both viruses. The second objective was then to elucidate the difference between infectious viral particles and the viral genome in terms of their prevalence and behavior in oysters naturally affected by fecal contamination, as well as during depuration processes, focusing on a single virus (i.e., FRNAPH).
Shellfish were collected over a 1-year period from several class B harvesting areas (n = 111 samples), which explain the significant number of samples positive for E. coli (i.e., 31.5%). The use of a qualitative approach (37) underlined that more than 90% of the specimens were positive for FRNAPH infectious particles (i.e., 84.7% were positive for infectious FRNAPH-I and 64.9% were positive for infectious FRNAPH-II). We confirmed, in accordance with other works, the lack of a relation between the regulatory indicator E. coli and infectious viral indicators, such as FRNAPH (Fisher's exact test, P = 0.8374) (24, 41, 44). Compared to the results obtained during a 1-year period of surveillance of marketed oysters, for which only 34% and 7% of the samples were positive for infectious FRNAPH-I and -II, respectively, the oysters sampled in the present study seemed to be more impacted by fecal pollution when the same qualitative method was used (37).
Under such conditions, a significant relationship between the presence of the NoV GII genome and that of the FRNAPH-II genome in oysters was observed (P < 10−4), with a higher prevalence being found in winter. On the basis of the analysis of a large number of batches from several areas in Europe, this means that both FRNAPH-II and NoV GII infectious particles and FRNAPH-II and NoV GII genomes have similar behaviors, including occurrence in water, adhesion to oyster tissues, as well as persistence and release from shellfish.
When the focus was only on the depuration conditions applied to oysters coming from polluted areas, many studies have shown not only the impact of the physicochemical conditions (i.e., water temperature, salinity, dissolved oxygen) and shellfish physiology but also the impact of the initial microorganism load (i.e., natural or artificial contamination) (59–62). For these reasons, we decided to work with oysters that were naturally contaminated with NoV and that were also positive for FRNAPH because of the high correlation of the presence of the two viruses discussed above. The initial viral loads were therefore moderate, but conditions were much closer to those found in the environment. Interestingly, we demonstrated no differences in FRNAPH decay between storage in natural seawater and storage in UV-treated seawater tanks and minor differences in NoV decay in favor of depuration in the tank.
We confirmed, as has also been reported in numerous studies (13, 52, 53, 55), the very long period of persistence of the NoV genome during shellfish storage. Such a long period of persistence (i.e., >30 days in our study) has, however, compromised the use of any other proxy indicators, including the presence of FRNAPH. Moreover, the suspected involvement of specific ligands explaining the persistence of NoV in shellfish has been an additional argument for rejecting FRNAPH as an indicator (8, 63). However, it is important to note that in all aforementioned studies, a comparison of NoV persistence with FRNAPH persistence was rarely made, and if so, only infectious FRNAPH were followed.
We fully confirmed the results described above in terms of the persistence of the NoV genome compared to that of infectious FRNAPH during the depuration of oysters. However, when investigating the persistence of FRNAPH by genome targeting, we clearly demonstrated the similar behaviors of the NoV and FRNAPH-II genomes or even the longer period of persistence of the FRNAPH-II genome. These observations may reopen the debate on the use of FRNAPH as an indicator of the presence of NoV in shellfish.
Under these circumstances, the second point that needs to be discussed here is the relevance of the detection of viral genomes to assessment of the presence of infectious particles in shellfish. To that end, conclusions should be drawn on the basis of data collected during the monitoring of the same virus by following both infectious particles and the associated genome by culture and molecular detection, respectively. Thus, while the FRNAPH-II genome was present in shellfish throughout their storage, no corresponding infectious particles were detected in the last days when the sensitive qualitative approach proposed by Hartard et al. (37) was used. The long period of persistence of the FRNAPH-II genome and the inability to eliminate it even after an extended depuration process illustrate the fact that linking the presence of genomes with that of the corresponding infectious microorganisms appears to be an unconvincing strategy, including for NoV risk management. In that sense, it should also be underlined that, despite moderate initial concentrations, NoV GII genomes were also detected in shellfish during the entire study period (i.e., over 30 days).
As previously observed using murine NoV (64), a two-phase elimination could thus be considered during genome decay, with a pronounced decrease occurring in the first days of storage, followed by subsequent stabilization with a lower elimination rate, leading to a residual basal concentration of viruses in shellfish. Hence, the first phase could be associated with the elimination of viral particles, while the second phase could just be the result of the degradation of residual genomes present in shellfish, leading to concentrations close to the LOD, as determined using the ISO 15216-1 standard (18) (i.e., 40 gc/g of HP in the best case), and, thus, amounts hardly quantifiable in such samples. Taking into account the striking persistence of genomes compared to that of viral particles, the 28-day minimum closure period for shellfish production areas, established following a significant pollution incident or a known NoV outbreak, may need to be discussed (30, 65).
Finally, similar conclusions concerning the presence of infectious particles and viral genomes may be drawn from the analyses of the 111 oyster samples impacted by fecal contamination in the several class B harvesting areas studied. Although an association between the presence of the FRNAPH-II genome and that of FRNAPH-II infectious particles (P < 10−3) was observed, the presence of the genome was predictive of infectivity in only 62% of cases. Furthermore, among the samples negative for FRNAPH-II infectious particles, 28% were positive for the corresponding genome (i.e., false-positive results); this finding was mainly linked to the better persistence of the genome than infectious particles (19–22). In opposition to this finding, among the samples positive for infectious particles, 43% were negative for the genome (i.e., false-negative results), which is probably explained by the difference between the LODs of the methods used (i.e., 1 PFU/20 g of whole shellfish by culture and up to 104 gc/g of HP in the worst-case scenario by use of the ISO 15216-1 standard, taking into account the recovery and inhibition rates).
To conclude, this study reveals that genome detection provides limited information about the presence of associated infectious viral particles in oysters on the basis of targeting of FRNAPH. This fact was observed both in harvesting areas and when oyster depuration processes were monitored. Indeed, a significant difference between infectious particle and genome decay was observed, and viral genomes may still be present in shellfish and, more broadly, in food commodities in the absence of infectious particles. Conversely, the presence of infectious viral particles is undoubtedly associated with the presence of the corresponding genomes, but the latter is commonly unquantifiable or even undetectable using currently available standardized methods.
Concerning the use of a proxy indicator which may thus be useful to assess viral pollution more effectively, similar trends and a relationship between the behavior of NoV and that of FRNAPH-II have been observed, with the same limitations in the detection of their genomes being present. However, contrary to the infectivity of NoV, the infectivity of FRNAPH-II is easily assessable, and their presence should therefore be reconsidered as potential indicators of a viral hazard in shellfish. Further investigations are now needed to gain a better understanding of the behavior of infectious FRNAPH and that of infectious NoV in shellfish and in the associated environment. This will be made possible when the cell culture approach is more routinely handled for NoV.
MATERIALS AND METHODS
Oyster samples.
In order to study the relationship between the presence of NoV and that of FRNAPH-II in shellfish, 111 batches of oysters (Crassostrea gigas) were collected from different harvesting areas in France (n = 1) and Ireland (n = 8) between January 2016 and January 2017. All sampled harvesting areas were classified as class B harvesting areas (i.e., they had E. coli concentrations of between 230 and 4,600 most probable number [MPN]/100 g of flesh and intravalvular liquid [FIL]), according to European regulations (12).
Concerning the behavior of microorganisms during depuration processes, three oyster batches that were collected from class B harvesting areas and that were positive for NoV genome detection were investigated. After collection, the specimens were split into two different subbatches and subjected to different depuration conditions over 31- to 35-day periods. After sampling, all specimens were kept at 4°C, and analyses were performed within 7 days.
Depuration processes.
The first half of the batches was stored in a pilot tank containing seawater disinfected by continued UV treatment (80 to 90 mJ/cm2). The water temperature was 12.1°C ± 0.7°C.
The second half was placed in an area with natural class A seawater. The water temperature of the area was 8.7°C ± 2.7°C during the study period.
NoV and FRNAPH genome detection.
Human NoV genogroup I (GI) and NoV genogroup II (GII) genome detection from oysters was performed by strictly following the NF EN ISO 15216-1 standard (18). Briefly, after dissection of 10 live specimens, 2 g of hepatopancreas (HP) was finely chopped, supplemented with 10 μl of a suspension of 104 infectious particles/ml bovine enterovirus type 1 (ATCC VR-248) as a process extraction control, and then incubated with 2 ml of proteinase K (3 U/ml) at 37°C for 1 h with stirring, followed by a second incubation at 60°C for 15 min. After centrifugation (3,000 × g for 5 min), the supernatant was collected and RNA extraction from 500 μl was performed using NucliSens magnetic extraction reagents (bioMérieux, Marcy l'Etoile, France) in 100 μl of elution buffer, according to the manufacturer's recommendations. NoV GI and GII genomes were detected using an RNA UltraSense one-step quantitative RT-PCR system (Life Technologies, Carlsbad, CA, USA) according to the recommendations of the ISO 15216-1 standard, and quantification was carried out using a standard curve with a concentration range of 101 to 105 genome copies (gc)/reaction mixture.
For each subgroup, FRNAPH genome detection was performed from the same RNA extracts produced for NoV detection. Quantification was performed using an RNA UltraSense one-step quantitative RT-PCR system (Life Technologies, Carlsbad, CA, USA) from 5 μl of RNA in a 20-μl reaction volume using the primers (1,000 nM) and probes (300 nM) designed by Wolf et al. (35). The choice of these RT-PCR systems was motivated here by their high sensitivity, even if other systems seem more appropriate for specifically detecting human FRNAPH (19). Quantification was carried out using a standard curve with a concentration range of from 2.5 to 2.5 × 104 gc/reaction mixture, and the reaction was carried out at 50°C for 30 min (for reverse transcription) and 5 min at 95°C, followed by 45 cycles of 15 s at 95°C and 40 s at 58°C.
According to the sample preparation procedure and the instructions given in the ISO 15216-1 standard, the theoretical LOD for NoV and FRNAPH, corresponding to the presence of 1 gc in a PCR well, was close to 40 gc/g of HP.
Infectious FRNAPH detection and genotyping.
Infectious FRNAPH were detected by two different methods using Salmonella enterica serovar Typhimurium WG49 (NCTC 12484) as the host strain (66). The first method was derived from the ISO 10705-1 standard (36). For each oyster sample, HP tissues from 5 specimens were dissected and mixed with 2 volumes of phosphate-buffered saline–0.3% peptone for 3 min in a DT-20 tube with an Ultra-Turrax tube drive (IKA-Werke GmbH & Co. KG, Staufen, Germany). After centrifugation (2,000 × g for 5 min), the supernatant was collected. The culture of 1.5 ml of supernatant was performed four times in 150-mm-diameter petri dishes, allowing the analysis of 6 ml of supernatant (corresponding to 2 g of HP). The infectious FRNAPH concentration was expressed as the number of PFU per gram of HP after 18 h of incubation.
The second method used for infectious FRNAPH detection was a qualitative integrated cell culture (ICC)-RT-quantitative PCR (qPCR) approach performed on the whole shellfish without any dissection step, as described by Hartard et al. (37), with slight modifications. Briefly, 10 oyster specimens were mixed for 3 min with a neck blender. Culture of infectious FRNAPH was then performed in 250-ml Erlenmeyer flasks by adding 20 ml of the oyster mixture, 25 ml of 2× tryptone-yeast extract-glucose broth (TYGB), 500 μl of a calcium-glucose solution, 200 μl of a 25-mg/ml kanamycin and nalidixic acid solution, and 30 PFU of FRNAPH-IV, used as a culture positive control. Finally, 5 ml of a Salmonella enterica serovar Typhimurium WG49 suspension prepared as described in the ISO 10705-01 standard (36) was added. Biological amplification was performed at 37°C for 4 h under agitation (110 rpm). Genome extraction from FRNAPH infectious particles was then performed with 1 ml of the total suspension. After centrifugation (18,000 × g, 3 min), 500 μl of the supernatant was collected and extraction was performed using a NucliSens EasyMag system (bioMérieux, Marcy l'Etoile, France) in 50 μl of elution buffer. The genomes of each FRNAPH subgroup were detected using the primers and probes developed by Wolf et al. (35) under the conditions described in a previous study (19).
The detection limits of the ISO 10705-01 standard and the ICC-RT-qPCR method were 1 PFU/2 g of HP and 1 PFU/20 g of whole shellfish flesh, respectively. Considering that FRNAPH are mainly found in shellfish HP and that HP corresponds to 5% to 10% of the total shellfish mass, the detection limits of these two methods were similar.
E. coli detection.
E. coli in shellfish FIL was detected by direct impedance measurement, according to NF V08-106 (67). The results were expressed as the most probable number (MPN), and the LOD was 66 MPN of E. coli/100 g of FIL.
Statistical analyses.
In order to rank the abundance of each microorganism in shellfish, a three-level classification was applied. Thus, concerning the FRNAPH and NoV genomes, specimens were considered negative (−) if no genomes were detected (<40 gc/g of HP), positive (+) if the concentrations were between 40 and 400 gc/g of HP (i.e., 10 times the LOD), and quantifiable (++) if the concentrations were over 400 gc/g of HP. Concerning infectious FRNAPH, samples were considered negative (−) when the ICC-RT-qPCR approach gave a quantification cycle value greater than 37. Because FRNAPH-I have a better growth rate, the threshold used to differentiate positive (+) from strongly positive (++) samples was 20 for this subgroup, while it was 32 for FRNAPH-II and -III (37). Finally, concerning the presence of E. coli, samples were considered negative (−) when the concentrations were below 66 MPN/100 g of FIL, and the threshold used to differentiate positive (+) from strongly positive (++) specimens was 4,600 MPN/100 g of FIL (i.e., the threshold used in European regulations). The relationships between these parameters were investigated using Fisher's exact test.
Concerning microorganism behavior during the depuration processes of the oysters, the raw data were subjected to logarithmic transformation before the statistical analysis was carried out. The nonparametric Wilcoxon signed-rank test was used to compare the medians of the concentrations under the two conditions. Cochran's test for the homogeneity of variances was used to compare the variances associated with the measurements made at a given time for each of the microorganisms considered. Regression analysis was performed to describe the variation in concentrations which was observed for each microorganism under the given experimental conditions as a linear function of time. The linearity of the depletion models thus obtained was tested by using the F test. In practice, the F test compares the variance of the departures from the fitted linear regression line with the experimental variance. Note that if the linear model is adequate, the previous two variances are equal. All statistical analyses were generated using R statistical software (v.3.4.0).
Finally, the decay coefficients corresponding to the given experimental conditions were estimated in two different ways, that is, (i) from the slope of the fitted regression decay line and (ii) as the weighted mean of the log empirical decay rate calculated from each observation weighted by the associated duration.
ACKNOWLEDGMENTS
The results of this study were obtained within the scope of OxyVir, a project funded by the Fonds Européen pour les Affaires Maritimes et la Pêche (FEAMP). This study was supported by the Joint Technological Unit ACTIA VIROcontrol, the Institut Carnot Energie et Environnement en Lorraine (ICEEL), and the Conseil Régional de Normandie.
REFERENCES
- 1.Richards GP. 1988. Microbial purification of shellfish: a review of depuration and relaying. J Food Prot 51:218–251. doi: 10.4315/0362-028X-51.3.218. [DOI] [PubMed] [Google Scholar]
- 2.Koopmans M, von Bonsdorff CH, Vinjé J, de Medici D, Monroe S. 2002. Foodborne viruses. FEMS Microbiol Rev 26:187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos CJA, Lees DN. 2014. Environmental transmission of human noroviruses in shellfish waters. Appl Environ Microbiol 80:3552–3561. doi: 10.1128/AEM.04188-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellou M, Kokkinos P, Vantarakis A. 2013. Shellfish-borne viral outbreaks: a systematic review. Food Environ Virol 5:13–23. doi: 10.1007/s12560-012-9097-6. [DOI] [PubMed] [Google Scholar]
- 5.Schwab KJ, Neill FH, Estes MK, Metcalf TG, Atmar RL. 1998. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J Food Prot 61:1674–1680. doi: 10.4315/0362-028X-61.12.1674. [DOI] [PubMed] [Google Scholar]
- 6.Burkhardt W, Calci KR. 2000. Selective accumulation may account for shellfish-associated viral illness. Appl Environ Microbiol 66:1375–1378. doi: 10.1128/AEM.66.4.1375-1378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atmar RL, Neill FH, Woodley CM, Manger R, Fout GS, Burkhardt W, Leja L, McGovern ER, Le Guyader F, Metcalf TG, Estes MK. 1996. Collaborative evaluation of a method for the detection of Norwalk virus in shellfish tissues by PCR. Appl Environ Microbiol 62:254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian P, Bates AH, Jensen HM, Mandrell RE. 2006. Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Lett Appl Microbiol 43:645–651. doi: 10.1111/j.1472-765X.2006.02010.x. [DOI] [PubMed] [Google Scholar]
- 9.Le Guyader FS, Loisy F, Atmar RL, Hutson AM, Estes MK, Ruvoën-Clouet N, Pommepuy M, Le Pendu J. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg Infect Dis 12:931–936. doi: 10.3201/eid1206.051519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian P, Engelbrektson AL, Jiang X, Zhong W, Mandrell RE. 2007. Norovirus recognizes histo-blood group antigens on gastrointestinal cells of clams, mussels, and oysters: a possible mechanism of bioaccumulation. J Food Prot 70:2140–2147. doi: 10.4315/0362-028X-70.9.2140. [DOI] [PubMed] [Google Scholar]
- 11.Maalouf H, Zakhour M, Le Pendu J, Le Saux J-C, Atmar RL, Le Guyader FS. 2010. Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Appl Environ Microbiol 76:5621–5630. doi: 10.1128/AEM.00148-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Parliament. 2004. Regulation (EC) no. 854/2004 of the European Parliament and of the Council of 29 April 2004 laying down specific rules for the organisation of official controls on products of animal origin intended for human consumption. Off J Eur Union L 226:83–127. [Google Scholar]
- 13.Doré B, Keaveney S, Flannery J, Rajko-Nenow P. 2010. Management of health risks associated with oysters harvested from a norovirus contaminated area, Ireland, February-March 2010. Euro Surveill 15(19):pii=19567 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19567. [PubMed] [Google Scholar]
- 14.Le Guyader FS, Bon F, DeMedici D, Parnaudeau S, Bertone A, Crudeli S, Doyle A, Zidane M, Suffredini E, Kohli E, Maddalo F, Monini M, Gallay A, Pommepuy M, Pothier P, Ruggeri FM. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J Clin Microbiol 44:3878–3882. doi: 10.1128/JCM.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalmers JWT, McMillan JH. 1995. An outbreak of viral gastroenteritis associated with adequately prepared oysters. Epidemiol Infect 115:163–167. doi: 10.1017/S0950268800058222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowther JA, Avant JM, Gizynski K, Rangdale RE, Lees DN. 2010. Comparison between quantitative real-time reverse transcription PCR results for norovirus in oysters and self-reported gastroenteric illness in restaurant customers. J Food Prot 73:305–311. doi: 10.4315/0362-028X-73.2.305. [DOI] [PubMed] [Google Scholar]
- 17.Lees D. 2010. International standardisation of a method for detection of human pathogenic viruses in molluscan shellfish. Food Environ Virol 2:146–155. doi: 10.1007/s12560-010-9042-5. [DOI] [Google Scholar]
- 18.International Organization for Standardization. 2017. NF EN ISO 15216-1. Microbiology of the food chain—horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR. Part 1. Method for quantification. International Organization for Standardization Geneva, Switzerland. [Google Scholar]
- 19.Hartard C, Rivet R, Banas S, Gantzer C. 2015. Occurrence of and sequence variation among F-specific RNA bacteriophage subgroups in feces and wastewater of urban and animal origins. Appl Environ Microbiol 81:6505–6515. doi: 10.1128/AEM.01905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogorzaly L, Bertrand I, Paris M, Maul A, Gantzer C. 2010. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl Environ Microbiol 76:8019–8025. doi: 10.1128/AEM.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gassilloud B, Schwartzbrod L, Gantzer C. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl Environ Microbiol 69:3965–3969. doi: 10.1128/AEM.69.7.3965-3969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S, Jiang SC. 2005. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl Environ Microbiol 71:7426–7433. doi: 10.1128/AEM.71.11.7426-7433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonet J, Gantzer C. 2006. Inactivation of poliovirus 1 and F-specific RNA phages and degradation of their genomes by UV irradiation at 254 nanometers. Appl Environ Microbiol 72:7671–7677. doi: 10.1128/AEM.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartard C, Banas S, Loutreul J, Rincé A, Benoit F, Boudaud N, Gantzer C. 2016. Relevance of F-specific RNA bacteriophages in assessing human norovirus risk in shellfish and environmental waters. Appl Environ Microbiol 82:5709–5719. doi: 10.1128/AEM.01528-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 26.Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W. 2013. Norovirus and FRNA bacteriophage determined by RT-qPCR and infectious FRNA bacteriophage in wastewater and oysters. Water Res 47:5222–5231. doi: 10.1016/j.watres.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W. 2012. Concentration of norovirus during wastewater treatment and its impact on oyster contamination. Appl Environ Microbiol 78:3400–3406. doi: 10.1128/AEM.07569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowther JA, Gustar NE, Hartnell RE, Lees DN. 2012. Comparison of norovirus RNA levels in outbreak-related oysters with background environmental levels. J Food Prot 75:389–393. doi: 10.4315/0362-028X.JFP-11-360. [DOI] [PubMed] [Google Scholar]
- 29.Lowther JA, Gustar NE, Powell AL, Hartnell RE, Lees DN. 2012. Two-year systematic study to assess norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Appl Environ Microbiol 78:5812–5817. doi: 10.1128/AEM.01046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassard F, Sharp JH, Taft H, LeVay L, Harris JP, McDonald JE, Tuson K, Wilson J, Jones DL, Malham SK. 2017. Critical review on the public health impact of norovirus contamination in shellfish and the environment: a UK perspective. Food Environ Virol 9:123–141. doi: 10.1007/s12560-017-9279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Mennec C, Parnaudeau S, Rumebe M, Le Saux J-C, Piquet J-C, Le Guyader SF. 2017. Follow-up of norovirus contamination in an oyster production area linked to repeated outbreaks. Food Environ Virol 9:54–61. doi: 10.1007/s12560-016-9260-6. [DOI] [PubMed] [Google Scholar]
- 32.Jofre J, Lucena F, Blanch A, Muniesa M. 2016. Coliphages as model organisms in the characterization and management of water resources. Water 8:199. doi: 10.3390/w8050199. [DOI] [Google Scholar]
- 33.Skraber S, Gassilloud B, Gantzer C. 2004. Comparison of coliforms and coliphages as tools for assessment of viral contamination in river water. Appl Environ Microbiol 70:3644–3649. doi: 10.1128/AEM.70.6.3644-3649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogorzaly L, Gantzer C. 2006. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J Virol Methods 138:131–139. doi: 10.1016/j.jviromet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Wolf S, Hewitt J, Greening GE. 2010. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl Environ Microbiol 76:1388–1394. doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Organization for Standardization. 2001. ISO 10705-1. Water quality: detection and enumeration of bacteriophages. Part 1. Enumeration of F-specific RNA bacteriophages. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 37.Hartard C, Banas S, Rivet R, Boudaud N, Gantzer C. 2017. Rapid and sensitive method to assess human viral pollution in shellfish using infectious F-specific RNA bacteriophages: application to marketed products. Food Microbiol 63:248–254. doi: 10.1016/j.fm.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Vergara GGRV, Goh SG, Rezaeinejad S, Chang SY, Sobsey MD, Gin KYH. 2015. Evaluation of FRNA coliphages as indicators of human enteric viruses in a tropical urban freshwater catchment. Water Res 79:39–47. doi: 10.1016/j.watres.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 39.Arredondo-Hernandez LJR, Diaz-Avalos C, Lopez-Vidal Y, Castillo-Rojas G, Mazari-Hiriart M. 2017. FRNA bacteriophages as viral indicators of faecal contamination in Mexican tropical aquatic systems. PLoS One 12:e0170399. doi: 10.1371/journal.pone.0170399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doré WJ, Lees DN. 1995. Behavior of Escherichia coli and male-specific bacteriophage in environmentally contaminated bivalve molluscs before and after depuration. Appl Environ Microbiol 61:2830–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doré WJ, Mackie M, Lees DN. 2003. Levels of male-specific RNA bacteriophage and Escherichia coli in molluscan bivalve shellfish from commercial harvesting areas. Lett Appl Microbiol 36:92–96. doi: 10.1046/j.1472-765X.2003.01268.x. [DOI] [PubMed] [Google Scholar]
- 42.Hodgson KR, Torok VA, Turnbull AR. 2017. Bacteriophages as enteric viral indicators in bivalve mollusc management. Food Microbiol 65:284–293. doi: 10.1016/j.fm.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Doré WJ, Henshilwood K, Lees DN. 2000. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl Environ Microbiol 66:1280–1285. doi: 10.1128/AEM.66.4.1280-1285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flannery J, Keaveney S, Doré W. 2009. Use of FRNA bacteriophages to indicate the risk of norovirus contamination in Irish oyster. J Food Prot 72:2358–2362. doi: 10.4315/0362-028X-72.11.2358. [DOI] [PubMed] [Google Scholar]
- 45.Lowther JA, Henshilwood K, Lees DN. 2008. Determination of norovirus contamination in oysters from two commercial harvesting areas over an extended period, using semiquantitative real-time reverse transcription PCR. J Food Prot 71:1427–1433. doi: 10.4315/0362-028X-71.7.1427. [DOI] [PubMed] [Google Scholar]
- 46.FDA. 2015. National Shellfish Sanitation Program (NSSP)—guide for the control of molluscan shellfish 2015. FDA, Rockville, MD. [Google Scholar]
- 47.Goblick GN, Anbarchian JM, Woods J, Burkhardt W, Calci K. 2011. Evaluating the dilution of wastewater treatment plant effluent and viral impacts on shellfish growing areas in Mobile Bay, Alabama. J Shellfish Res 30:979–987. doi: 10.2983/035.030.0341. [DOI] [Google Scholar]
- 48.Formiga-Cruz M, Allard AK, Conden-Hansson A-C, Henshilwood K, Hernroth BE, Jofre J, Lees DN, Lucena F, Papapetropoulou M, Rangdale RE, Tsibouxi A, Vantarakis A, Girones R. 2003. Evaluation of potential indicators of viral contamination in shellfish and their applicability to diverse geographical areas. Appl Environ Microbiol 69:1556–1563. doi: 10.1128/AEM.69.3.1556-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muniain-Mujika I, Calvo M, Lucena F, Girones R. 2003. Comparative analysis of viral pathogens and potential indicators in shellfish. Int J Food Microbiol 83:75–85. doi: 10.1016/S0168-1605(02)00324-0. [DOI] [PubMed] [Google Scholar]
- 50.Croci DL, De Medici D, Scalfaro C, Fiore A, Divizia M, Donia D, Cosentino AM, Moretti P, Costantini G. 2000. Determination of enteroviruses, hepatitis A virus, bacteriophages and Escherichia coli in Adriatic Sea mussels. J Appl Microbiol 88:293–298. doi: 10.1046/j.1365-2672.2000.00966.x. [DOI] [PubMed] [Google Scholar]
- 51.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi C, Kingsley DH. 2016. Temperature-dependent persistence of human norovirus within oysters (Crassostrea virginica). Food Environ Virol 8:141–147. doi: 10.1007/s12560-016-9234-8. [DOI] [PubMed] [Google Scholar]
- 53.Ueki Y, Shoji M, Suto A, Tanabe T, Okimura Y, Kikuchi Y, Saito N, Sano D, Omura T. 2007. Persistence of caliciviruses in artificially contaminated oysters during depuration. Appl Environ Microbiol 73:5698–5701. doi: 10.1128/AEM.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nappier SP, Graczyk TK, Schwab KJ. 2008. Bioaccumulation, retention, and depuration of enteric viruses by Crassostrea virginica and Crassostrea ariakensis oysters. Appl Environ Microbiol 74:6825–6831. doi: 10.1128/AEM.01000-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drouaz N, Schaeffer J, Farkas T, Le Pendu J, Le Guyader FS. 2015. Tulane virus as a potential surrogate to mimic norovirus behavior in oysters. Appl Environ Microbiol 81:5249–5256. doi: 10.1128/AEM.01067-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love DC, Lovelace GL, Sobsey MD. 2010. Removal of Escherichia coli, Enterococcus fecalis, coliphage MS2, poliovirus, and hepatitis A virus from oysters (Crassostrea virginica) and hard shell clams (Mercinaria mercinaria) by depuration. Int J Food Microbiol 143:211–217. doi: 10.1016/j.ijfoodmicro.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 57.Olalemi A, Purnell S, Caplin J, Ebdon J, Taylor H. 2016. The application of phage-based faecal pollution markers to predict the concentration of adenoviruses in mussels (Mytilus edulis) and their overlying waters. J Appl Microbiol 121:1152–1162. doi: 10.1111/jam.13222. [DOI] [PubMed] [Google Scholar]
- 58.Muniain-Mujika I, Girones R, Tofio-Quesada G, Calvo M, Lucena F. 2002. Depuration dynamics of viruses in shellfish. Int J Food Microbiol 77:125–133. doi: 10.1016/S0168-1605(02)00052-1. [DOI] [PubMed] [Google Scholar]
- 59.Campos CJA, Kershaw S, Morgan OC, Lees DN. 2017. Risk factors for norovirus contamination of shellfish water catchments in England and Wales. Int J Food Microbiol 241:318–324. doi: 10.1016/j.ijfoodmicro.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 60.Lees D. 2000. Viruses and bivalve shellfish. Int J Food Microbiol 59:81–116. doi: 10.1016/S0168-1605(00)00248-8. [DOI] [PubMed] [Google Scholar]
- 61.Pommepuy M, Caprais M-P, Le Saux J-C, Le Mennec C, Parnaudeau S, Madec Y, Monier M, Brest G, Le Guyader F. 2003. Evaluation of viral shellfish depuration in a semi-professional tank, p 485–499. In Villalba A, Reguera B, Romalde JL, Beiras R (ed), Molluscan shellfish safety. Conselleria de Pesca e Assuntos Maritimos da Xunta de Galicia and Intergovernmental Oceanographic Commission of UNESCO, Santiago de Compostela, Spain. [Google Scholar]
- 62.Olalemi A, Baker-Austin C, Ebdon J, Taylor H. 2016. Bioaccumulation and persistence of faecal bacterial and viral indicators in Mytilus edulis and Crassostrea gigas. Int J Hyg Environ Health 219:592–598. doi: 10.1016/j.ijheh.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Le Guyader FS, Atmar RL, Le Pendu J. 2012. Transmission of viruses through shellfish: when specific ligands come into play. Curr Opin Virol 2:103–110. doi: 10.1016/j.coviro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polo D, Feal X, Varela MF, Monteagudo A, Romalde JL. 2014. Depuration kinetics of murine norovirus in shellfish. Food Res Int 64:182–187. doi: 10.1016/j.foodres.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 65.Direction Générale de l'Alimentation. 2017. Gestion du risque norovirus en lien avec la consommation de coquillages—protocole cadre de gestion DGAL/SDSSA/2017-326 du 11 avril 2017. Direction Générale de l'Alimentation, Paris, France. [Google Scholar]
- 66.Havelaar AH, Hogeboom WM. 1984. A method for the enumeration of male-specific bacteriophages in sewage. J Appl Bacteriol 56:439–447. doi: 10.1111/j.1365-2672.1984.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 67.Association Française de Normalisation. 2010. NF V08-106. Enumeration of Escherichia coli in live shellfish—indirect technique using direct impedance measurement. Association Française de Normalisation, La Plaine Saint-Denis, France. [Google Scholar]