Abstract
Metabolites are the small biological molecules involved in energy conversion and biosynthesis. Studying metabolism is inherently challenging due to metabolites’ reactivity, structural diversity, and broad concentration range. Herein, we review the common pitfalls encountered in metabolomics and provide concrete guidelines for obtaining accurate metabolite measurements, focusing on water-soluble primary metabolites. We show how seemingly straightforward sample preparation methods can introduce systematic errors (e.g., owing to interconversion among metabolites) and how proper selection of quenching solvent (e.g., acidic acetonitrile:methanol:water) can mitigate such problems. We discuss the specific strengths, pitfalls, and best practices for each common analytical platform: liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), nuclear magnetic resonance (NMR), and enzyme assays. Together this information provides a pragmatic knowledge base for carrying out biologically informative metabolite measurements.
Keywords: metabolomics, metabonomics, metabolite extraction, stability, accuracy, mass spectroscopy
1. SCOPE OF THE CHALLENGE
In contrast to DNA, RNA, and proteins, which are made by genetically encoded polymerization of a small number of building blocks, metabolites do not follow a fixed structural template and accordingly have diverse physical properties. As a result, no single analytical tool can measure all metabolites. In addition, the complete scope of metabolic networks remains unknown, and this uncertainty complicates metabolite analysis because a definitive list of relevant analytes (analogous to genes in the genome) is lacking. The Human Metabolome Database (HMDB) (1, 2) is a good illustration of this uncertainty: The HMDB has identified more than 40,000 putative human metabolites, but only 680 of the HMDB’s water-soluble compounds have been mapped to standard metabolic pathways in humans (3). The biological significance of many unmapped compounds remains unknown.
Metabolites can be classified in both biological and chemical dimensions. Biologically, primary metabolites are required for growth, development, and reproduction, whereas secondary metabolites are involved in communication, defense, or other life cycle-independent functions. Chemically, water-soluble versus insoluble compounds must be extracted in different solvents, and therefore, solubility plays a major role in real-world metabolomics studies. Primary water-soluble metabolites are key players in the conversion of nutrients into usable energy, provide the building blocks for biomass, turn over rapidly, and collectively carry the vast majority of metabolic flux. They are among the most evolutionarily conserved biomolecules.
This review focuses on the practical challenges that are encountered in the analysis of water-soluble primary metabolites with particular attention to issues arising from their reactivity and fast cellular turnover. These issues are substantially different from those in analyzing lipids, whose measurements were discussed in a previous article (4). We begin by discussing quenching and extraction, steps that are nearly universal in metabolite measurement and critical to measurement accuracy. This is followed by in-depth discussion of liquid chromatography-mass spectrometry (LC-MS), an important and versatile metabolite measurement technology. Several general issues in metabolite quantitation are also illustrated within the context of LC-MS. We then discuss important alternative measurement approaches, including gas chromatography-mass spectrometry (GC-MS), nuclear magnetic resonance (NMR), and enzyme assays. Throughout, we focus on both the strengths and weaknesses of each technology, with an eye on potential pitfalls and how they can be avoided.
2. ABSOLUTE VERSUS RELATIVE QUANTITATION
In most cases, the absolute concentration of a metabolite cannot be determined based solely on the intensity of the associated instrument signal. For example, in LC-MS, different metabolites ionize with dramatically different efficiencies. Thus, depending on the metabolite, the same number of ion counts can reflect a millimolar or nanomolar concentration in the sample. A standard curve shows the relationship between signal intensity and analyte concentration. When a signal falls in the linear range of the standard curve, a relative signal change equals a fold concentration change. When the signals from different biological samples fall out of the linear range, fold changes can be either under- or overestimated.
Absolute metabolite concentrations provide additional information. They determine enzyme binding-site occupancies, the thermodynamics of metabolic reactions, and the relevant concentration range for biochemical experiments. There are two ways to determine absolute concentrations: by comparison to internal standards (for MS, this is accomplished by measuring intensity difference between 13C or 15N labeled standards and unlabeled metabolites; for NMR, a single reference metabolite can frequently be used) or by external comparison to metabolite standards prepared at a range of concentrations. To account for matrix effects, external calibration curves are preferably made by adding standards into samples. To equate losses during extraction and handling between standards and endogenous analyte, standards should be added in the original extraction solvent, not to the final samples.
Although isotopic internal standards are both convenient and reliable, they are not commercially available for many metabolites. Accordingly, it is sometimes more pragmatic to feed cells with a labeled nutrient (e.g., 13C6-glucose) and compare the levels of labeled intracellular metabolites with unlabeled standards. In such experiments, correction for incomplete cellular metabolite labeling is important. Detailed methods for this purpose are provided in Reference 5. Using this approach, we have measured absolute concentrations for ~100 of the most important metabolites in Escherichia coli, yeast, and mammalian cells (6, 7). Once such absolute concentration measurements have been made, absolute concentration measurements in related specimens can be made by relative quantitation, multiplying the known absolute concentration in the previously measured sample by the experimentally observed fold change in the new condition.
3. QUENCHING
The goal of quenching and extraction is to produce a stable extract that quantitatively reflects the metabolites present in the biological specimen of interest. Quenching is particularly important for highly metabolically active specimens like cells and tissues and is of less concern for serum, plasma, or urine specimens. Two main problems can arise during quenching: (a) perturbation of metabolite levels in the harvesting steps or (b) incomplete or insufficiently fast termination of enzyme activity during quenching. For analysis of primary metabolites from cellular and tissue samples, rapid quenching is necessary as the turnover rate can be on the order of 1 s for compounds like ATP and glucose 6-phosphate (8–12).
For cellular studies, the first step in quenching is isolation of the cells from media. Pelleting is slow and perturbs the nutrient environment. Accordingly, for harvesting from suspension cultures, we recommend fast filtration followed by immediately placing the filter in quenching solvent (5). For adherent cultures, we prefer to simply aspirate the media and directly add the quenching solvent (13).
Washing with phosphate-buffered saline (PBS), although typically considered innocuous, is a metabolic perturbation as it removes all media nutrients. Cold PBS is not necessarily better because cold shock can lead to leakage of intracellular metabolites (8, 14). Thus, unless there are compelling reasons for washing, we avoid it. This makes the downstream analytical chemistry somewhat more challenging, but that is a small price to pay compared to the risk of systematic error. Nevertheless, sometimes there are compelling reasons for washing. For example, in standard mammalian tissue culture, it is infeasible to measure intracellular amino acids without washing due to the high concentrations of media amino acids. In such cases, we typically wash quickly (<10 s) with warm PBS. Important findings are then validated, whenever possible, using an orthogonal approach.
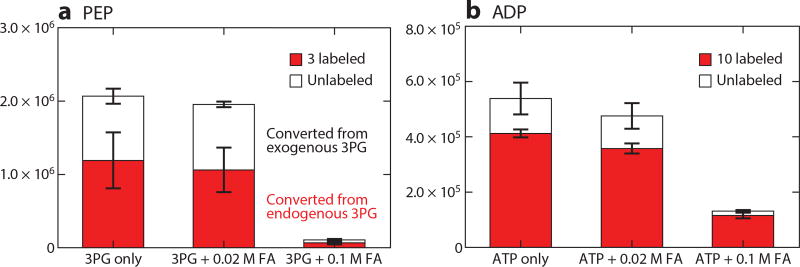
In terms of quenching metabolism, the basic requirement is to stop enzymatic activity. This is achieved with some combination of organic solvent, cold, heat, acid, or base. For tissue specimens, the typical first step is deep freezing at liquid nitrogen temperature. Owing to slow heat transfer from liquid nitrogen into warm biological specimens, smashing the tissue against precooled metal plates (Wohlenberger clamp) is faster (15). For cultured cells, it is more common to directly add hot or cold organic solvent. A classical approach, which works well for many analytes, is boiling ethanol (16–18). Although the boiling solvent raises concerns about thermal degradation, it reliably denatures enzymes. In contrast, cold organic solvent may not fully denature enzymes or may do so too slowly such that some metabolic reactions continue, interconverting metabolites during the quenching process. Incomplete quenching can be experimentally tracked by looking for transformations of isotope-labeled standards spiked into the quenching solvent (19). Figure 1 shows two examples: (a) transformation of 3-phosphoglycerate into phosphoenolpyruvate and (b) ATP into ADP. Phosphoenolpyruvate has a higher standard free energy then 3-phosphoglycerate; however, phosphoenolpyruvate’s concentration in most biological settings is very low, and, accordingly, residual enolase activity leads to its production. We observe that interconversion during quenching is prevented by use of acidic solvent: addition of 0.1 M (mol/L) formic acid (~0.5% v/v) (0.02 M is insufficient) (Figure 1). After metabolism is quenched, we neutralize with ammonium bicarbonate (NH4HCO3) to avoid acid-catalyzed degradation in the resulting extract. Although we find that acid generally accelerates quenching, even brief exposure to acid may be deleterious for some analytes. Also, enzymes in certain organisms (such as acid-tolerant microbes) may be acid resistant. Thus, when working on new analytes or organisms, spiking experiments should be performed to check for effective quenching and analyte stability. Nevertheless, for common organisms (including mammals) and metabolites, quenching in cold acidic organic solvent is effective.
Figure 1.
Metabolite interconversion owing to incomplete quenching of enzymatic activity is prevented by 0.1 M formic acid (FA). (a) HEK293 T cells were grown in 13C6-glucose media to completely label glycolytic intermediates. Unlabeled 3-phosphoglycerate (3PG) was added to the extraction solvent of 80:20 methanol:H2O (−70°C), which contained no FA, or 0.02 M FA, or 0.1 M FA. Phosphoenolpyruvate (PEP) can be made from 3PG through enolase activity. Extraction with 0.1 M FA eliminates unlabeled PEP made from the added 3PG standard. The same reaction occurs with endogenous (in this case, labeled) 3PG. Therefore, quenching without FA would greatly overestimate cellular PEP. (b) Yeasts were grown in 13C6-glucose media to completely label cellular metabolites. Unlabeled ATP was added to the extraction solvent of 80:20 methanol:H2O (−70°C), which contained no FA, or 0.02 M FA, or 0.1 M FA. Extraction with 0.1 M FA eliminated unlabeled ADP made from the added ATP standard. Therefore, quenching without FA would greatly overestimate ADP and underestimate the energy charge. Note that the FA should be neutralized after quenching to prevent acid-catalyzed degradation in the extract.
4. EXTRACTION
The goal of extraction is to obtain quantitative yields of metabolites in the specimen. Because certain metabolites may be formed during quenching or extraction, more is not always better. For example, some low-energy metabolites are also low-concentration ones (e.g., [ATP] > [ADP] > [AMP] > [adenosine]). High yields of such metabolites do not indicate successful extraction, but rather artifactual production from more abundant higher-energy species. Another consideration is that some metabolites may be protein bound. The total metabolome concentration is approximately 300 mM, whereas the protein concentration is approximately 7 mM (6), which implies that most cellular metabolites are in free form. However, certain metabolites, such as NADP+, both bind to many proteins and are present in cells at low concentration; such metabolites may be largely protein bound. Depending on the application, users might ideally want to extract only free metabolites, but effective methods for selective extraction have yet to be developed.
4.1. Extraction Methods
Cells can be extracted by adding organic solvent. Tissue samples need to first be pulverized into fine powders (11, 20–23). This can be achieved by manual grinding using a mortar and pestle at a cold temperature. Alternatively, a cryomill machine (such as the one manufactured by Retsch GmbH, Germany) operated at a liquid nitrogen temperature is effective. Extraction can be performed by mixing the specimen and solvent (e.g., on a shaker) at a cold temperature for approximately 15 min. Studies involving serial extraction show substantial metabolite yields (typically 20–40% of the total) in a second round of extraction and diminishing returns thereafter (24). Therefore, we usually perform two rounds of extraction.
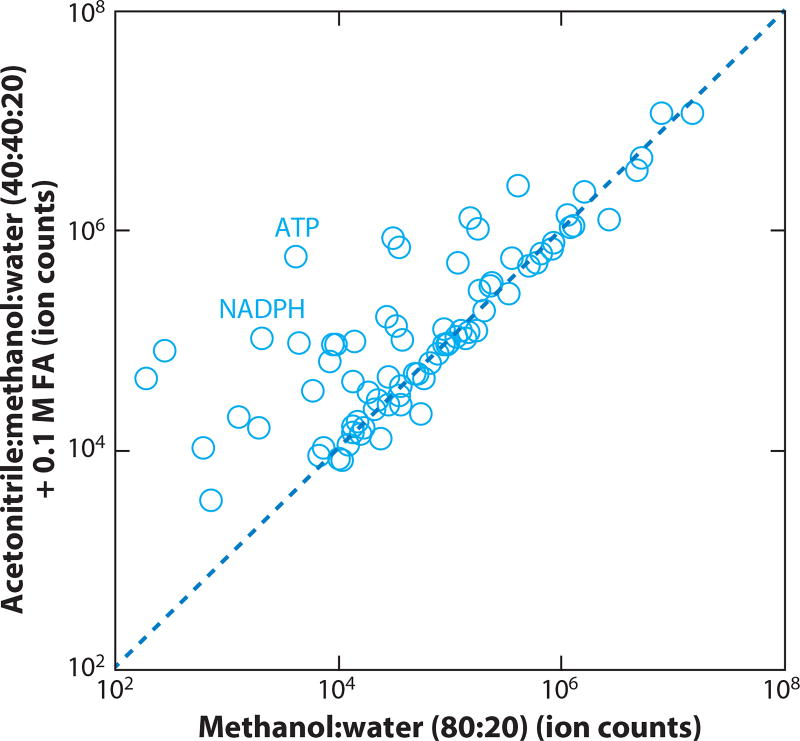
Existing literature is ambiguous about the best solvent systems for metabolite extraction (12, 25–31). This reflects that many metabolites, including amino acids, Krebs cycle intermediates, and glycolytic intermediates, are adequately extracted using a variety of solvents. It further reflects the failure of many studies to monitor harder to extract compounds, such as ATP and NADPH. The reported concentrations of cellular ATP are notoriously inconsistent because of incomplete quenching and incomplete extraction. In our experience, for both cell and tissue specimens, 40:40:20 acetonitrile:methanol:water with 0.1M formic acid (and subsequent neutralization with ammonium bicarbonate) is generally an effective solvent system for both quenching and extraction, including for ATP and other high-energy phosphorylated compounds (Figure 2) (24). We typically use approximately 1 mL of solvent mix to extract 25 mg of biological specimen. For serum and plasma, simpler approaches like mixing with methanol may be sufficient to precipitate protein and thereby yield an analysis-ready sample. Identification of the best extraction procedures, as a function of sample type and compounds of interest, is an important ongoing area of investigation.
Figure 2.
Comparison of two different extraction methods for polar metabolites from mouse liver samples. Mouse liver was ground using a cryomill, and 25 mg of ground tissue was then extracted in a 2 mL Eppendorf tube using the following methods: The first protocol (80% methanol) is to add 870 µL 80:20 methanol:water (v/v) at −70° C, vortex for 10 s, and place on dry ice for 20 min. The second protocol (addition of acetonitrile and formic acid) is to add 800 µL 40:40:20 acetonitrile:methanol:water + 0.1 M formic acid (solvent A) at −20°C, vortex for 10 s, place on ice for 2 min, add 70 µL 15% (w/v) NH4HCO3 in H2O (solvent B), place in −20°C freezer for 20 min. For both protocols, samples were spun for 10 min at 16,000 × gravity, and the supernatant was analyzed by liquid chromatography-mass spectrometry (LC-MS). Control experiments (not shown) demonstrated that the difference in yields was due primarily to the solvent and not the temperature.
4.2. Sample Degradation and Metabolite Interconversion After Extraction
Metabolite extracts contain a large number of chemical species, which can potentially degrade or interconvert (32, 33). Reducing the time interval between sample preparation and analysis helps mitigate such problems. In some cases, however, preservatives are needed. For example, for folate species including ascorbic acid in the extraction solvent mitigates folate degradation and interconversion (34, 35).
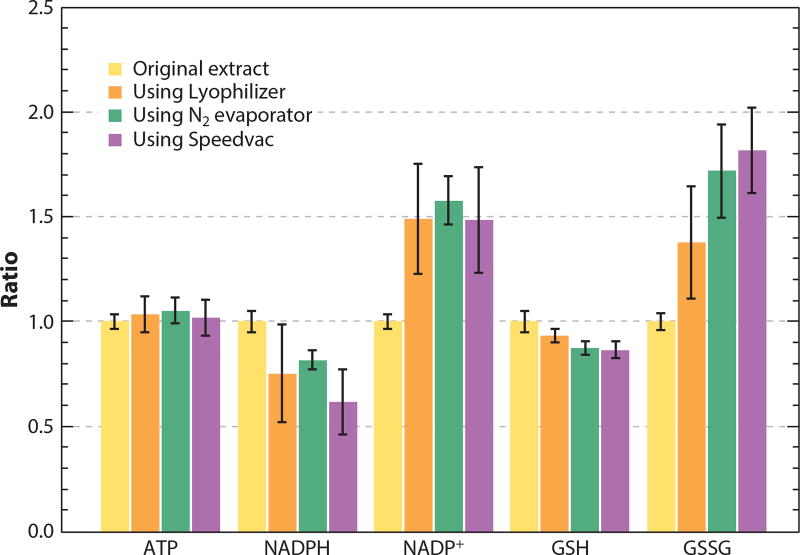
A challenge in building a well-functioning metabolite analysis pipeline is that solvents used for quenching and extraction may interfere with downstream analysis. For example, most organic solvents interfere with retention on reversed-phase LC. Solvent removal addresses this problem, as well as concentrating metabolites and facilitating long-term storage (11). During solvent removal, however, the sample becomes more concentrated, and reactions between metabolites accelerate. We studied the recovery of selected metabolites from a tissue extract upon drying using three different approaches: a lyophilizer, a N2 evaporator, or a room temperature Speedvac. The dried extracts were each redissolved into an equal volume of water with 10 mM ammonium bicarbonate. Most metabolites, including ATP, were recovered nearly quantitatively (Figure 3). By contrast, NADPH and reduced glutathione (GSH) were depleted under all drying conditions, in part through oxidation to NADP+ and glutathione disulfide (GSSG). Because the reduced forms (NADPH and GSH) are substantially more abundant in cells, modest oxidation during drying strongly perturbs the concentrations of the oxidized species and leads to substantial underestimation of the cellular NADPH:NADP+ and GSH:GSSG ratios. Thus, although drying is acceptable for most metabolites, care must be taken with redox-active species.
Figure 3.
Effects of solvent evaporation on selected metabolites. Approximately 30 mg of mouse liver tissue was extracted using 1.2 mL 40:40:20 acetonitrile:methanol:water with 0.1 M formic acid, followed by neutralization by NH4HCO3. The extract was divided into multiple 200 µL portions, dried using indicated methods, and redissolved in 200 µL of 10 mM NH4HCO3 in H2O. Samples were analyzed by liquid chromatography-mass spectrometry (LC-MS), and the peak area of each compound was compared to those in the original extract. It was found that the majority of metabolites, including ATP, were not affected by the solvent evaporation process. By contrast, there was a significant decrease in NADPH and reduced glutathione (GSH) and an increase in NADP+ and oxidized glutathione disulfide (GSSG).
4.3. Take Home Message
The above examples illustrate the potential for the loss of abundant high-energy compounds and the gain of less-abundant, lower-energy derivatives at multiple steps in the quenching and extraction process. Modest degradation of the abundant compounds can lead to large fold changes in the less-abundant degradation products. Each of these errors tends to decrease the ATP:ADP, NADPH:NADP+, and GSH:GSSG ratios.
An equally important consideration is avoiding metabolite perturbations during the handling steps leading up to quenching. Such biological perturbations have unpredictable directional effects but nevertheless can be reproducible for a given system, leading to false discoveries. For this reason, it is critical to minimize prequenching handling of cells, tissues, and animals (36).
5. LIQUID CHROMATOGRAPHY-MASS SPECTROMETRY
Diverse analytical methods are available for quantitating extracted metabolites. The most sensitive detection, and thus the broadest metabolome coverage, is achieved by MS-based methods (37–41). Among MS techniques, LC-MS is the most versatile. Accordingly, we begin by discussing LC-MS in detail. In so doing, we also cover several general issues in MS and metabolite measurement.
In LC-MS, analytes are separated on column, ionized at an ion source, separated by a mass analyzer, and detected. Specificity is achieved through the combination of retention time from the column and the MS signature. Specific MS signatures can be achieved through tandem MS (MS/MS) and/or high-resolution MS.
5.1. Mass Spectrometry
The first requirement for MS detection is ionization. Most LC-MS methods for metabolomics rely on electrospray ionization, which works effectively for charged metabolites. LC-MS is blind to metabolites that do not ionize. For example, cholesterol is invisible on LC-MS with electrospray ionization. For metabolites that do not ionize via electrospray, alternative LC ionization methods, such as atmospheric pressure chemical ionization, are available.
Once ions are obtained, the major choice is whether to use a targeted measurement approach, which looks only for a predetermined set of ions, or a full-scan approach that measures all ions. The main instruments used for targeted analysis are triple quadrupoles. These are low-resolution mass spectrometers, which use MS/MS to achieve specificity: monitoring the fragmentation of a parent ion into a characteristic daughter ion. The first and third quadruples act as mass filters, selecting the parent ion and daughter ion mass, respectively. The intervening quadrupole is used to carry out the fragmentation reaction. The approach of monitoring for a specific parent/product pair is sometimes called selected reaction monitoring (SRM). When multiple SRMs are performed in series (typically for ~0.05 s each), the approach is known as multiple reaction monitoring. The primary advantage of SRM is sensitivity. The main downside is its targeted nature: The instruments must be preprogrammed with the parent/product scan events and have finite scan capacity, limiting the total number of metabolites (or, in tracer experiments, labeled forms) that can be measured.
To avoid the limitations of targeted analysis, especially for studies with isotope labeling, we prefer full-scan approaches. Full-scan LC-MS can be performed on a low-resolution mass analyzer like a single quadrupole, but with limited sensitivity and specificity. Accordingly, high-resolution mass analyzers are preferred: time-of-flight (TOF) or Orbitrap mass analyzers. Specificity is achieved by resolving ions of almost identical mass. Resolution is defined as M/ΔM, where M is the ion mass and ΔM is the minimum mass difference that allows two ions to be distinguished. A TOF mass analyzer has a typical resolution of 10,000–40,000, whereas an Orbitrap mass analyzer can reach 400,000 (42). The TOF mass analyzer is a long-standing technology based on the principle that lower m/z ions accelerate faster in an electric field. Recent years have seen steady improvement in TOF performance. Orbitraps are a relatively new type of mass analyzer, with the first commercial instrument introduced in 2005 (43, 44). They are based on the principle of trapping ions in an electrostatic field, with ions simultaneously circling around an inner spindle electrode and oscillating along the electrode’s Z-axis, with the Z-axis oscillation frequency inversely related to ion mass (45).
Both TOF and Orbitrap mass analyzers are well suited to measuring metabolites, whether targeted or untargeted. Typical metabolomics runs detect on the order of 10,000 ion peaks, but some of these are adducts (i.e., pairs of ions) or fragments, not molecular ions: [M+H]+ in positive mode or [M-H]− in negative mode. Buried among these are molecular ion peaks for a few dozen to a few hundred primary water-soluble metabolites. With knowledge of the retention time of targeted metabolites, it is possible to read their peaks directly off the high-resolution MS chromatograms. Moreover, it is possible to compare all peaks across biological conditions to find ones that change significantly in response to a biological perturbation and thus merit identification (46–48).
Quadrupoles can also be coupled to high-resolution detection in a hybrid Q-TOF or Q-Orbitrap instrument. Even with access to such a high-resolution MS/MS instrument, we find that high-resolution full-scan MS is the most straightforward approach for measuring large numbers of metabolites (and their labeled forms) in parallel. The MS/MS capabilities are, however, useful for fragmentation-based identification of unknowns, distinguishing certain isomers and maximizing detection sensitivity for selected metabolites.
Although each of the above approaches enables specific, sensitive, and quantitative metabolite measurement, they also share certain pitfalls, mainly related to limitations of the upfront LC and electrospray ionization steps.
5.2. Liquid Chromatography
A major challenge in metabolomics is the lack of a single LC method that can analyze all metabolites. Instead, to obtain optimal coverage, one must employ multiple LC-MS approaches (49). Reversed-phase chromatography using a C18 column, while providing good separation of fatty acids and lipids, is not ideal for the analysis of polar metabolites due to poor retention on column. Retention of negatively charged metabolites can be improved by including a cationic ion-pairing agent in the running buffer (50, 51). Several different ion-pairing agents have been used, including hexylamine and tributylamine. These also improve the peak shape for phosphate-containing metabolites (52–54). The drawback is that the ion-pairing agent, which takes days to wash out of an LC system, suppresses ionization of positively charged metabolites. Thus, metabolites that only ionize in the positive ion mode, including carnitines and S-adenosylmethionine, cannot be measured.
An alternative chromatographic approach is hydrophilic interaction chromatography (HILIC), a variant of normal phase chromatography, which uses only water-miscible solvents. In HILIC, metabolites bind to a polar stationary phase and are eluted with a gradient of increasing water content (55, 56). Several different stationary phases are available, for example, silica hydride, aminopropyl, amide, cyano, or zwitterionic (57). In our experience, performance of HILIC is quite sensitive to the choice of column, solvent, and gradient, with optimized methods enabling effective measurement of a wide diversity of metabolites, as well as good separation of many isomers (52, 58). For example, Pesek et al. (59) used a stepped gradient with acidic mobile phases on a silica hydride stationary phase to separate leucine and isoleucine. Zhang et al. (60) separated various isomers using a zwitterionic column. We used a slow gradient with acidic mobile phases on an aminopropyl column to separate the isomers leucine and isoleucine and 2-aminobutyrate and 4-aminobutyrate (61). Overall, although no single method currently provides complete isomer resolution or primary metabolite coverage, HILIC methods continue to improve and are probably the most versatile current choice for metabolomics aimed at primary water-soluble metabolites.
5.3. Peak Misidentification
To convert LC-MS raw data into metabolite abundances, it is necessary to match peaks with metabolites. To this end, we rely on the peak’s retention time and mass. In some cases, the MS/MS fragmentation pattern can also be useful. Although the combination of retention time and mass signature provides great specificity, peak misannotation remains a major problem. In our experience, there are three major causes: isomers, similar molecular-weight interferences, and in-source degradation products.
5.3.1. Isomers
Isomers are compounds with identical molecular formulas but different structures. Important examples in metabolism include hexose phosphates/inositol phosphates, pentose phosphates, citrate/isocitrate, leucine/isoleucine, AMP/dGMP, ATP/dGTP, and alanine/sarcosine. Measurement of isomers is a long-standing challenge in analytical chemistry. Isomers have identical mass, so high-resolution MS alone is not sufficient for their separation. Possible ways to differentiate them include chromatography separation (59), ion mobility MS (62, 63), or MS/MS fragmentation (64). In some cases, when isomers cannot be readily differentiated directly, they can be separated after chemical derivatization.
Both reversed-phase ion-pairing chromatography and HILIC can separate isomeric metabolites. Additional separation power can be achieved by MS/MS. It is well known that the acyl tail components of glycerophospholipids can be identified by MS/MS (64). By combining LC with MS/MS, Buescher et al. (54) measured a large number of isomers of hexose phosphate. Some of these species were only partially separated on column, but they generated different fragmentation product masses (including 79, 97, 169, 241). We find that, although 3-phosphoglycerate and 2-phosphoglycerate are difficult to chromatographically resolve, they generate different ratios of product ions of 79 (PO3−) and 97 (H2PO4−), enabling deconvolution of their relative concentrations by linear algebra (65).
When LC and MS/MS are insufficient for separating isomers, another option is chemical derivatization (66). For example, R-2-hydroxyglutarate and S-2-hydroxyglutarate can be separated by LC after derivatization with diacetyl-L-tartaric anhydride (67). GC separation using the corresponding O-acetyl-di-(D)-2-butyl esters provides similar results (68). Some other useful derivatization reagents include dansyl chloride and N,N’-dicyclohexylcarbodiimide. Dansyl chloride reacts with amines to form stable sulfonamides, enabling separation of leucine and isoleucine, as well as sarcosine, α-alanine, and β-alanine, by standard reversed-phase LC (69, 70). Similar results for the alanine isomers have also been obtained using N,N’-dicyclohexylcarbodiimide as the derivatization reagent, with the added benefit of increased detection sensitivity in positive ion mode because the reaction products have two additional nitrogen atoms (71).
In summary, separation of isomers is important to enable proper biological interpretation of metabolite measurements. For example, it is important to know whether an apparent decrease in alanine could actually be the result of decreased sarcosine. Although there is still no generic solution for separation of important metabolic isomers, GC-MS or LC-MS/MS are often sufficient.
5.3.2. Interfering compounds
The complexity of metabolite extracts frequently results in interferences between compounds of similar molecular weight (e.g., ± 20 ppm). A key benefit of high-resolution MS is the ability to minimize such interferences. In this regard, it is important to recognize the difference between mass accuracy (which in a well-calibrated TOF or Orbitrap mass analyzer is <2 ppm) and resolving power. Accuracy reflects the ability to properly assign mass to a single isolated ion, whereas resolving power reflects the ability to separate ions with similar mass. A TOF instrument with < 2 ppm mass accuracy and 40,000 resolving power cannot separate ions differing in mass by <25 ppm. Thus, because of both the continued use of low-resolution instruments and the finite resolving power of TOF instruments, interfering compounds remain a persistent issue. A simple example involves lysine and glutamine, two amino acids with accurate masses in positive mode of 147.1128 and 147.0764. The resolution required to distinguish these is 4,000, which is readily achieved on a TOF or Orbitrap mass analyzer. On a triple quadruple mass spectrometer, however, both molecules have same SRM transition of m/z 147→84. Thus, low-resolution MS/MS alone is not sufficient to differentiate these two compounds. Isotopic tracers usually amplify the interfering compound issue by bringing in additional labeled features. For example, the mass of 13C3 pyroglutamate is only 6 ppm smaller than that of 13C0 asparagine (131.04538 versus 131.04622).
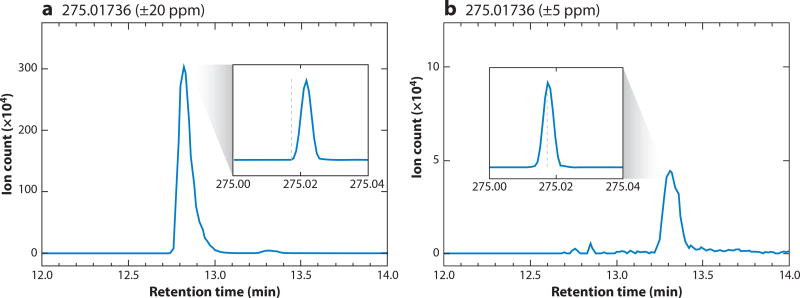
When an interfering compound elutes far away from a targeted analyte, chromatography provides a straightforward fix. Trouble arises, however, with interfering compounds that elute near to the expected retention time of a metabolite of interest. In Figure 4, we illustrate such a case: detection of 6-phosphogluconate with a theoretical mass of 275.0174 in negative ion mode by reversed-phase ion-pairing chromatography coupled to a stand-alone Orbitrap mass spectrometer operated at 100,000 resolution. Using an m/z window of 275.0174 ± 20 ppm, we detected two peaks near the expected retention time, a prominent peak at 12.8 min and a weaker peak at 13.3 min. When using a narrower mass window of 275.0174 ± 5 ppm, only the weaker peak at 13.3 minute remains. The feature at 13.3 min was confirmed to be the 6-phosphogluconate based on coelution with the authenticated standard. We have yet to identify the feature at 12.8 min.
Figure 4.
Measurement of the 6-phosphogluconate (M–H)− ion by liquid chromatography-mass spectrometry (LC-MS) showing the effect of interfering compounds with similar mass from a mammalian cell extract. (a) Within a mass range of 275.01736 ± 20 ppm, two features were detected, one at ~12.8 min and one at ~13.3 min. (b) With a narrower range of 275.01736 ± 5 ppm, only the feature at ~13.3 min is observed. The feature at ~13.3 min is 6-phosphogluconate, with a measured mass of 275.0176 (+1.0 ppm error) and a retention time match to the authenticated standard. The other feature has a measured mass of 275.02167 (+16 ppm difference from 275.0174) and comes from an unknown interfering compound. Main panels a and b show mass-specific chromatograms. Insets show mass spectra with X-axis in units of m/z and the dashed line indicates the 6-phosphogluconate mass of 275.01736.
5.3.3. In-source degradation products
Electrospray ionization is a complex process and may produce many types of by-product ions: fragments owing to the simple loss of H2O, CO2, or H3PO4; products of more complicated rearrangement reactions; multiple-charged ions such as [M+2H]2+; dimers or trimers or heterodimers; and adducts from attachment of Na+, NH4+, K+, Fe2+, etc. In-source fragmentation both reduces the signal for the metabolite ion and produces lower molecular-weight fragments that can mimic other metabolites. In many cases, the fragment ion has the same molecular formula (and even structure)as the molecular ion of anothermetabolite. Accordingly, the fragment ion cannot be differentiated by mass resolution.
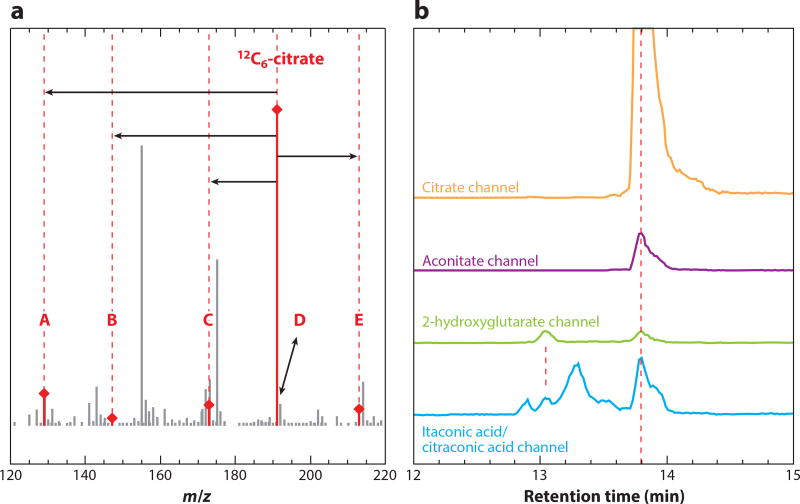
In one example, Purwaha et al. (72) reported the in-source cyclization of glutamine and glutamate to pyroglutamate. The three compounds are separated on a column, and the m/z channel of pyroglutamate shows peaks at each of their respective retention times. It is important to recognize that only one of these reflects biological pyroglutamate. We observe similar phenomena for many metabolites (61). For example, using our reversed-phase ion-pairing method (53), citrate elutes at 13.8 min, and the corresponding mass spectrum shows five other major ions associated with it, including three fragmentation products (Figure 5a). These can mimic other metabolites, as seen in Figure 5b, which shows mass-specific chromatograms at the m/z of aconitate, 2-hydroxyglutarate, and itaconic acid. The only peak at the aconitate mass is from citrate H2O loss. There is no detectable aconitate in this sample. There are two peaks at the 2-hydroglutarate mass, one from 2-hydroxyglutarate and one from CO2 loss from citrate. At the mass of itaconic acid, there are four peaks: itaconic acid itself, H2O loss from 2-hydroglutarate, citraconic acid (an isomer of itaconic acid typically formed by heating of citrate), and loss of CO2 + H2O from citrate. Other examples abound, with one of the most important of involving nucleotides, with higher-energy species yielding lower-energy fragments (e.g., AMP can be formed from in-source fragmentation of ATP or ADP) (61); due to such fragmentation, energy charge determinations require chromatographic separation.
Figure 5.
In-source fragments from electrospray ionization of citrate mimic other metabolites. (a) Mass spectrum taken at the retention time of citrate shows isotopic forms, adducts, and fragments: 13C1-natural abundance peak (D), Na+ adduct (E), H2O loss (C), CO2 loss (B), and the loss of CO2 + H2O (A). (b) Mass-specific chromatograms show that citrate fragment ions mimic other metabolites. Channels refer to signals at the m/z of the stated metabolite molecular anion ±5 ppm. All of the peaks at 13.8 min are from citrate. The apparent itaconic acid peak at 13.1 min is from 2-hydroxyglutarate fragmentation.
These examples demonstrate the need for careful selection of the correct peak in each mass-specific chromatogram. At present, we are unaware of automated algorithms that do this effectively. Development of ion sources that reduce in-source fragmentation could mitigate this issue. Alternatively, more standardized chromatographic approaches would facilitate automation of peak picking and thus accurate data interpretation.
5.4. Quantitative Error in Liquid Chromatography-Mass Spectrometry
Metabolite measurements by LC-MS are subject to many of the same quantitative considerations as other analytical methods. These include precision (reproducibility), sensitivity (limit of detection), and linearity (dynamic range). Each of these can be assessed by running the authentic standards at different concentrations. Absolute signal intensity in LC-MS can drift from day to day (and sometimes even run to run) due to various factors, including changing ionization efficiency. For this reason, when comparing metabolite levels across groups of samples, it is important to run the samples in a randomized or interwoven order, rather than all samples from one group followed by all samples from another. Such randomization of sample order is in general good practice for metabolite measurement.
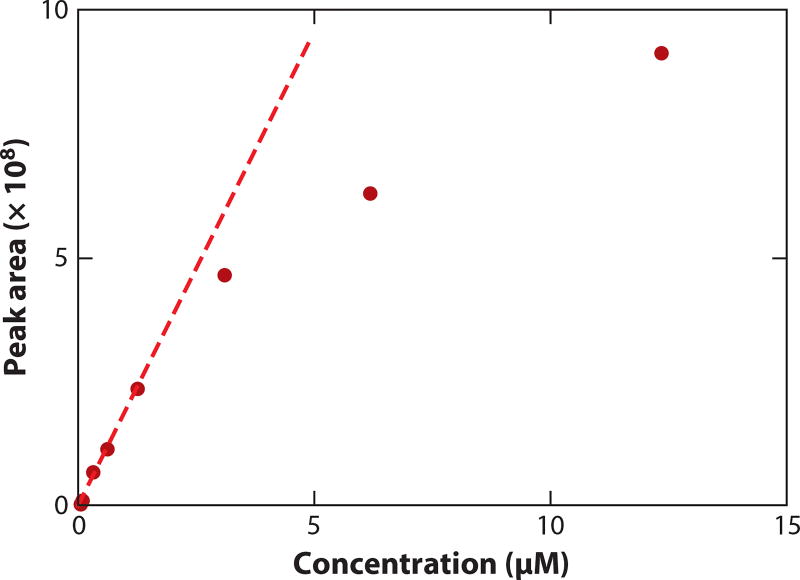
LC-MS is not particularly precise. The typical relative standard deviation (RSD) for repeat analysis of the same metabolomics sample varies by compound, with the median RSD for well-defined peaks usually ~10% (and worse for peaks near the limit of detection). LC-MS is, however, sensitive and has a broad linear range, typically two to four orders of magnitude. As shown for carnitine in Figure 6, signal response often becomes sublinear at high concentrations. The underlying reason depends on the instrument. TOF instruments are subject to detector saturation, which blunts the signal for abundant compounds and thereby underestimates their concentrations. By contrast, ion trap instruments, including Orbitraps, are more often subject to trap filling that leads to across-the-board signal decreases due to a space-charge effect. In an effort to see low-abundance peaks, it is common to overload LC-MS systems, compromising quantitative performance.
Figure 6.
Assessment of linearity. Plot of the liquid chromatography-mass spectrometry (LC-MS) peak areas (dark red dots) versus the concentration of the carnitine standard. The LC-MS method is reversed-phase chromatography on a C18 column coupled to a Q-Exactive plus mass spectrometer operating in positive ion mode. The signal is linear from 0.0005 to 1.2 µM and sublinear at higher concentrations.
A particularly important issue in LC-MS is ion suppression, wherein a high-abundance ion suppresses the electrospray ionization and thus the signal of coeluting ions (73). Electrospray ionization involves the formation of small charged droplets at the spray tip. Evaporation of solvent from these droplets leads to increasing charge density on the droplets and eventually emission of gas-phase ions. Because emission of any appropriately charged ion alleviates charge accumulation, ions effectively compete to enter the gas phase. In practice, for metabolomics, ion suppression can be caused by ion-pairing agents, salts, and metabolites. An extreme example involves tributylamine, which is a useful ion-pairing reagent for negative mode analysis but in positive ion mode produces a peak at m/z 186.2, which is so strong as to suppress virtually all other signals. Biological samples contain many different metabolites as well as salts, such as NaCl and KH2PO4, which are abundant in cell culture medium and tissues. When these coelute with metabolites, signals are suppressed.
A related pitfall in LC-MS is differential adduct formation, wherein coelution of metabolites and salts increases formation of adduct peaks at the expense of the metabolite molecular ion peak. A problematic scenario is when an abundant compound or salt varies in concentration across samples, as this can induce apparent signal changes in coeluting metabolites.
Ways to minimize ion suppression and differential adduct formation include upfront sample cleanup (e.g., by solid-phase extraction), sample dilution, better chromatography separation, and the use of isotope-labeled internal standards to correct for any changes in ionization efficiency across samples (73).In practice, we find that a combination of awareness and good chromatography, along with restraint in not overloading the LC-MS system, usually suffices. Because in complex samples it is difficult to rule out ion suppression, important results should be validated with isotopic internal standards, sample dilution, or an orthogonal measurement approach.
6. GAS CHROMATOGRAPHY-MASS SPECTROMETRY
GC-MS is a versatile technique that can measure a broad spectrum of primary water-soluble metabolites (74). Its strength is measurement of low-molecular-weight and volatile analytes, including small species that typically do not retain well on LC and uncharged species that ionize poorly by electrospray. For some compound classes, especially for essential oils and volatiles, GC and GC-MS are the only universally applicable analytical methods (75).
To obtain broad metabolome coverage, GC-MS must be coupled with up-front chemical derivatization to increase metabolite stability and volatility. This is typically achieved by the trimethylsilylation derivatization reaction on samples that have been completely dried. The reaction works at room temperature with pyridine as the catalyst (76) and can be performed manually in tubes or vials, or automatically using robotic equipment, which can be part of GC autosamplers. Derivatization success is impacted by solvent and sample cleanliness, dryness, and only to a lesser extent by reaction time or temperature (77).
With derivatization, GC-MS is the preferred measurement option for many metabolites that are hard to measure by LC-electrospray ionization-MS, including very-short-chain fatty acids and alcohols, hydroxy acids, sugars and monophosphorylated sugars (78), and sterols. It is also well suited to measurements of amino acids, fatty acids, polyamines, aromatics, and catecholamines. For general metabolomics, the strengths of GC-MS include outstanding chromatographic peak sharpness and extensive mass spectral libraries for peak identification (79), such as the NIST14 Mass Spectral Library (80). The effectiveness of GC as a separation method takes the pressure off MS analysis, with single quadruple detection cost effective and robust. TOF mass spectrometers yield yet better coverage and sensitivity. Similar to LC-MS, quantitative precision for technical replicates depends on the specific compound and matrix. Precision may range from 2% RSD, e.g., for sugars and acids, to more than 20% RSD for low-abundance compounds and metabolites that are affected by matrix compositions (81).
GC requires, however, that molecules evaporate during the injection procedure. Hot injections preclude undamaged quantification of thermolabile compounds, such as di- and triphosphates (including ATP or NADPH). Even after trimethylsilylation derivatization, molecules can decompose during the injection, for example, the guanidinium group of arginine decomposes to yield ornithine (82). This major caveat has been recognized for decades. Yet, in metabolomics, all signals are recorded, including decomposition products, which can confound biochemical interpretations or lead to structures that are absent from chemical libraries such as PubChem. For example, UDP-N-acetylglucosamine and UDP-glucuronide lead to decomposition products that cleave off the UDP moiety, forming a double bond in a residual molecule that is not known as a naturally occurring metabolite (83).
Unknown compound identification in GC-MS is complicated by the use of hard electron ionization. This type of ionization yields many highly reproducible fragments and rearrangement ions, making identification of known compounds straightforward, based on comparison to mass spectra and retention indices of authenticated standards that have been deposited in databases. However, the intact molecular ions are often absent from the mass spectra (84). An alternative ionization approach for GC-MS is chemical ionization, which is softer and thus more commonly yields a molecular ion peak (74). With chemical ionization, molecular ions are observable even for high molecular-weight trimethylsilylated metabolites, but fragmentation still occurs, and different chemical adducts abound, depending on the type of chemical reactant gas used. This results in many of the same challenges as discussed above in Section 5.3.3.
Another problem in GC-MS is the robustness of quantification, at least for some metabolite classes. On the one hand, unlike electrospray ionization in LC-MS, GC-MS does not suffer from ion suppression or differential adduct formation. On the other hand, compared to LC-MS, the injection step is more challenging in GC-MS: The sample is introduced into the inlet liner and volatilized, and the resulting vapor is mixed with carrier gas. During this process, thermal degradation of analytes can occur and can be accelerated by degradation products arising from an unclean injector or the sample itself. Nonvolatile compounds, such as phosphatidylcholines, can degrade in the injector, cross contaminate samples, and lead to undesired catalytic degradation of analytes of interest. One concern is the loss of trimethylsilyl groups introduced during derivatization. O-trimethylsilyl groups are stable, rendering GC-MS robust for measurement of sugars, phosphates, and hydroxy acids. N-trimethylsilyl groups are less stable, however, and may be lost from trimethylsilylated amines and amino acids. During analysis, such compounds can show different ratios of fully and partially derivatized compounds (lacking the N-trimethylsilyl group) across samples, owing to sensitivity of the N-trimethylsilyl linkage to matrix loads in the sample as well as to the cleanliness of the injector system.
In general, cleanliness of both the sample and the injector system (from the injection needle to involatile depositions in the liner and the beginning of the column) are critical to GC-MS performance (77). Detailed protocols have been published to maintain analysis quality (85), but staff needs to be trained in detail. First, liners (used to hold the injection gas cloud) need to be kept meticulously clean and changed regularly. Automatic liner exchangers are available, and their use helps maintain analysis quality. Second, users should employ empty guard columns to protect the analytical GC column. Involatile matrix components deposit on the guard column’s start site. The guard column may then be cut in quality maintenance procedures without compromising GC separations (85). Alternatively, users can resort to using more than one derivatization reaction (and more than one analytical run) to enhance precision and accuracy, e.g., trimethylsilylation for general metabolome profiling (for sugars, hydroxy acids, and similar compounds) and tertiary-butyldimethylsilylation for amines and amino acids (86, 87).
Overall, LC-MS and GC-MS are complementary techniques, which work best for different analytes. Although some of the pitfalls in LC-MS and GC-MS are conceptually similar, they are unlikely to incorrectly measure the same analyte. Thus, obtaining the same quantitative results in both methods is reassuring. The difficulty is that both methods require not only advanced instrumentation but also substantial practical expertise, and few labs do both well. More optimized—and thus easier—workflows will hopefully change this moving forward.
7. HIGH-THROUGHPUT MASS SPECTROMETRY
An important constraint in LC-MS and GC-MS is the time required for each chromatography run. This is a particularly important issue given the expense of the back-end mass spectrometer. Accordingly, there is substantial interest in direct MS to increase sample throughput. This includes MS without chromatography separation, desorption ionization MS, and microfluidics coupled to MS (88). One commercial example of a chromatography-free system is the RapidFire instrument from Agilent (89–91). The benefit is the analysis time of < 1 min per sample, versus approximately 30 min per sample with typical metabolomics LC-MS methods.
The direct-injection approach is ideally suited to certain applications, such as measuring metabolite levels in enzyme assays consisting of purified protein and a few metabolites. In such a context, direct measurement of all metabolites by MS has important advantages over monitoring the fluorescence of a single indicator compound, which was the main option for high-throughput analysis until recently.
Direct-injection MS has also shown substantial potential for metabolomics analysis of cellular extracts (92–94). Even over the brief injection window, a large number of ions are detected, corresponding to a similar number of primary metabolite masses as in a standard LC-MS run. Thus, ion suppression is not so severe as to eliminate signals for most metabolites. The improved throughput has enabled some important applications, including screening of whole-genome knockout collections and fine-grained kinetic analysis of the metabolome of bacteria undergoing dynamic perturbations (94).
That said, direct-injection MS is not a suitable substitute for LC-MS for quantitative metabolite analysis in complex mixtures because without any chromatographic separation the potential problems with LC-MS are greatly magnified: (a) Isomers cannot be distinguished; (b) in-source fragments cannot be distinguished from true molecular ions, precluding measurement of many central carbon metabolites and of energy charge; (c) matrix effects and/or ion suppression may alter quantitative measurements in unpredictable ways that result in both random and systematic errors. Thus, in the arena of metabolomics, direct-injection MS is a promising discovery tool but, absent confirmatory analysis by LC-MS or other methods, is likely to generate many artifactual observations in addition to true discoveries.
8. NUCLEAR MAGNETIC RESONANCE
NMR spectroscopy played a foundational role in the development of metabolomics (95, 96) and has the important advantage (compared to MS-based methods) in that it sees all organic molecules with signal intensities proportional to metabolite concentrations (97). NMR also provides more extensive structural information than MS, which is valuable for unknown identification or(in labeling studies) position-specific isotope enrichment measurements. The most widely acknowledged downside of NMR relative to MS is lower sensitivity [~10 µM is the practical limit for one-dimensional (1D) 1H NMR experiments] (98–100). In practice, a yet more important challenge may be limited resolution of individual metabolites from complex samples.
In 1D 1H NMR, the most sensitive and widely used NMR experiment, metabolite signals populate a narrow frequency range from ~1–10 ppm. Consequently, almost all aliphatic signals (−CH, −CH2, −CH3 groups; 1–5 ppm) overlap with peaks from multiple metabolites. This overlap makes peak integration, multiplicity analysis, measurement of J-couplings, and other basic structural determination procedures infeasible. Although the aromatic signals (5–10 ppm) are generally well resolved, most of these peaks are singlets and provide limited structural information. Moreover, traditional structural assignment requires definitive molecule-specific peak lists, which cannot be generated directly from complex 1D spectra. These challenges have driven the development of alternative methods; the most successful of these are database searches (1, 101, 102), peak fitting (103), statistical deconvolution (104), and multidimensional NMR (105–107).
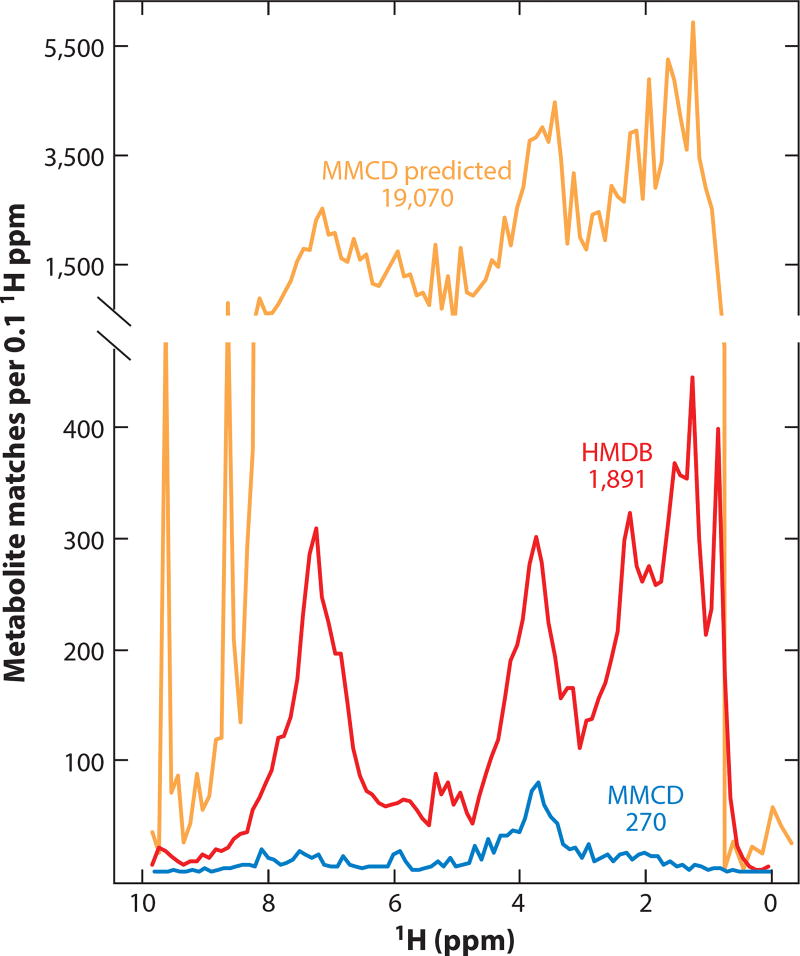
Database searching is the most popular approach for assigning metabolomics spectra. Several databases (e.g., the Human Metabolome Database, http://www.hmdb.ca) allow users to upload NMR spectra to generate putative metabolite assignments. These automated assignments are complicated by the fact that 1H NMR shifts are sensitive to pH, osmolality, and concentrations of certain ions (e.g., Ca2+). As a result, database searches must allow for considerable uncertainty in peak position. This tolerance, along with the many similarities in reference spectra, leads to a high false discovery rate in database-driven assignments. More than 137 metabolites, for example, can be mapped to every 0.1 ppm segment of a 1H NMR spectrum (Figure 7). This issue can be mitigated by combining database searches with peak fitting (103, 108), leveraging a priori knowledge of the sample composition (109), or statistical deconvolution of the data (104, 110). Although each of these approaches significantly improves assignments, results derived from overlapped spectra should be treated with skepticism until the assignments are cross validated. Multidimensional NMR offers a validation mechanism. Specifically, 1H-13C heteronuclear single quantum coherence (HSQC), 1H-13C heteronuclear multiple bond correlation (HMBC), and 1H-1H correlation spectroscopy (COSY) allow molecular connectivity to be established, and 1H-1H total correlation spectroscopy (TOCSY) allows signals from individual metabolites to be differentiated in complex spectra (105).
Figure 7.
Signal specificity and false discovery in one-dimensional (1D) 1H-NMR-based metabolomics. Database searches using a hand-curated standard set (N = 270 common metabolites) from the Madison Metabolomics Consortium Database (MMCD; http://mmcd.nmrfam.wisc.edu) returns an average of 12 matches per 0.1 ppm region in a 1D 1H NMR spectrum. This increases to an average of 137 metabolites per 0.1 ppm using the online tools available from the Human Metabolome Database (HMDB; http://www.hmdb.ca/) and 1,710 per 0.1 ppm for MMCD’s predicted chemical shift dataset (N = 19,070).
Difficulties in resolving individual peaks also increase quantitative error. Even under idealized conditions, error rates in overlapped 1D NMR spectra exceed 16% (111); real-world error reaches 20% (112) and is likely higher for low-abundance compounds. This quantitative performance is disappointing given that NMR can achieve error rates under 0.1% (113). Peak fitting and multidimensional NMR provide potential routes to improved measurement precision. Peak fitting is unfortunately a skill-dependent manual procedure where incorrect assignments can lead to systematic error. Multidimensional NMR analyses are not subject to these user-dependent problems, but the complicated multidimensional pulse sequences introduce numerous quantitative variables. Sophisticated new NMR approaches are being developed that allow two-dimensional (2D) signals to be quantified directly (114, 115); however, the most practical approach for non-NMR experts is to calibrate observed 2D signal intensities using mixtures of standards prepared at known concentrations (99, 111, 116, 117). This strategy serves two purposes: It enforces robust peak assignments and allows real-world metabolomics studies to achieve error rates in the range of 5% or less (111).
8.1. Practical Recommendations for Identifying and Quantifying Metabolites by NMR
To facilitate metabolite identification, a representative sample should be carefully prepared to match the solution conditions used by the large reference databases (e.g., pH 7.4, 100% D2O, and 0.5 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid for matching data to the Biological Magnetic Resonance Bank). Then high-resolution 2D NMR spectra (e.g.,1H-13C HSQC, 1,024 increments, 8,192 points in the direct dimension) should be recorded for metabolite identification. These spectra can be assigned via various software and databases (1, 101, 102, 118, 119). Metabolite assignments should be validated using mixtures of standards. With the metabolites of interest thus identified, the relative abundances of metabolites can be determined using 2D NMR (generally HSQC, TOCSY, or COSY), selecting well-resolved signals in the 1D spectra, or carefully fitting known metabolite signals from the overlapped spectra. For absolute quantification, reference mixtures of standards prepared at known concentrations should be used to calibrate the signals observed in the experimental data.
8.2. NMR-Based Measurement of Isotope Labeling
NMR is a powerful tool for measuring position-specific isotope labeling in metabolites (120–122), which makes NMR valuable for quantifying flux through metabolic networks. Differential isotopic enrichment in the 2-C versus 3-C positions of lactate in cells incubated with 2-13C glucose, for example, allows one to probe flux through the pentose phosphate pathway versus glycolysis (123–125). Though powerful, these isotope-based analyses introduce quantitative pitfalls. Most significantly, signals from 1H attached to 13C relax more quickly than those attached to 12C, which artificially deflates the 1H-13C signal (120). Although this quantitative defect can be corrected using standards, the isotopomers needed for this calibration are frequently expensive or unavailable. A practical solution is to include a paramagnetic relaxation agent in each sample (e.g., 1 mM Fe-EDTA solution) (114) to normalize T1 relaxation rates across molecules. The main disadvantage of this approach is that the paramagnetically broadened lines further reduce NMR’s sensitivity.
8.3. In Vivo NMR
NMR’s ability to interrogate the metabolism of intact cells and organisms (126, 127) is one of its most exciting attributes. Because metabolically active cells that are present at sufficient density for NMR analyses quickly deplete their nutrients and oxygen over the course of an NMR experiment, careful attention must be paid to the solution conditions to ensure that the data remain physiologically relevant. In addition, the magnetic permittivity of cells differs from the surrounding solution, which diminishes NMR sensitivity and quantitative reliability. Although a variety of hyperpolarization techniques are being developed that may transform this field by boosting NMR’s sensitivity > 10,000 fold (128), these strategies are currently inaccessible for routine metabolomics. Nevertheless, moving toward cellular and in vivo measurements is of great importance, especially given the myriad of ways that metabolite concentrations can be altered during quenching and extraction.
9. ENZYME ASSAYS
Although MS and NMR have grabbed most of the recent headlines in terms of metabolite measurement, enzyme assays remain among the most commonly used approaches (129). Enzymatic reaction followed by colorimetric detection uses only standard lab equipment. Here, we sought to assess whether LC-MS and kit-based measurements of ATP, NADPH/NADP+, and GSH/GSSG agree, and, if not, where the discrepancies arise. We find that the commonly used chemiluminescence assay for ATP provides a reliable ATP measurement. In contrast, the kits for redox cofactors are subject to systematic measurement error owing to metabolite interconversion during the aqueous extraction steps.
9.1. ATP
The luminescent ATP assay kit is based on the production of light from the luciferase-catalyzed reaction of ATP with D-luciferin (129, 130). Either 6 M guanidine-HCl or 5% trichloroacetic acid can be used to inactivate ATP consuming enzymes. In our hands, we obtained equal ATP yields (within ~20% error) for quenching and extraction with 6 M guanidine-HCl in Tris-EDTA (100 mM Tris, 4 mM EDTA, pH 7.8) and with 40:40:20 acetonitrile:methanol:water + 0.1 M formic acid. The luciferase ATP assay can measure less than 1 picomole of ATP with high specificity and no inhibition from ADP or AMP at up to equimolar concentrations. The signal decays quickly (t1/2 ~10 min); therefore, the timing between reagent addition and measurement needs to be consistent. Importantly, whereas the insensitivity of luciferase to ADP and AMP enables accurate ATP measurement, ATP concentration is more reflective of overall purine abundance than cellular energetics. The ATP:ADP and ATP:AMP ratios, which can be quantified by LC-MS but not directly by luciferase, are more biologically informative measures of energy status.
9.2. NADPH/NADP+
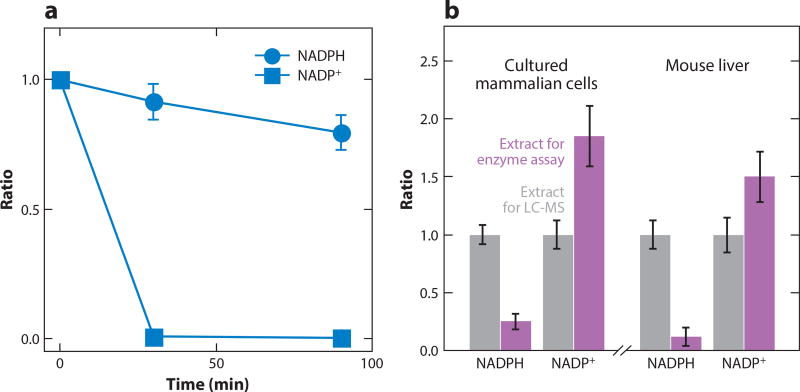
Measurement of NADPH/NADP+ is based on conversion of NADP+ to NADPH with glucose-6-phosphate dehydrogenase, with the resulting NADPH used to reduce thiazolyl blue to formazan (570 nm absorbance) (131–133). Selective measurement of NADPH is based on its superior stability to NADP+ under basic conditions: Incubation for 30 min at 60°C and pH 12 destroys NADP+ (134, 135). Determination of NADP+ is by subtraction. We independently validated that the heating stepatbasicpH destroys > 99% of NADP+ by using LC-MS (Figure 8a). Although most of the NADPH is retained, given the much greater concentration of NADPH than NADP+ in cells, even minor degradation of NADPH results in substantial mismeasurement of NADP+ by subtraction.
Figure 8.
Evaluation of enzymatic assay for quantitation of NADPH/NADP+. (a) Consistent with the principle of the enzymatic assay, basic pH selectively degrades NADP+. 10 µM NADPH and 10 µM NADP in aqueous buffer containing 20mM nicotinamide, 20mM NaHCO3, and 100mM Na2CO3 were heated at 60°C for the indicated times, and their concentrations were compared to the unheated samples by liquid chromatography-mass spectrometry (LC-MS). Note that modest degradation of NADPH is sufficient to alter NADP+ measurements, which are determined by subtraction. (b) Aqueous extraction without detergent using typical enzymatic assay conditions results in a dramatic loss of NADPH and a gain in NADP+, undermining utility of the enzyme assay. NADPH and NADP+ were measured from cultured HEK293T cells and mouse liver using two extraction methods: the suggested conditions of the enzymatic assay kit (20mM nicotinamide, 20mM NaHCO3, 100 mM Na2CO3 in water) or our suggested extraction for LC-MS analysis (40:40:20 acetontrile:methanol:water + 0.1 M formic acid, followed by neutralization with NH4HCO3). Results were normalized to the LC-MS extraction buffer.
The greater problem, however, involves the extraction step. We compared extraction using the kit buffer (20mMnicotinamide,20 mMNaHCO3, 100mMNa2CO3 in water)with acidic40:40:20 acetonitrile:methanol:water. The aqueous extract is dramatically lower in NADPH and higher in NADP+, consistent with residual enzyme activity that oxidizes NADPH (Figure 8b). Extraction buffer containing detergent [e.g., 0.05% Triton X-100 + 1% (v/v) Dodecyltrimethylammonium bromide] mitigates NADPH loss but does not fully prevent interconversion. Thus, although acidic organic extraction followed by LC-MS can give more accurate NADP(H) measurement, when applying enzyme assays, it is critical to include detergent in the extraction buffer to limit interconversion.
9.3. Glutathione
The reduced GSH cofactor is measured through its reaction with 5,5-dithio-bis (2-nitrobenzoic acid) to produce 5’-thio-2-nitrobenzoic acid (412 nm absorbance). This step both measures and consumes the GSH. To measure GSSG, it is converted to GSH by glutathione reductase in the presence of NADPH, and the colorimetric assay is repeated. Although the principles of the assay appear to be solid, GSH may oxidize to GSSG during sample handling (136, 137). At neutral pH in water, substantial GSH is oxidized to GSSG within a few minutes (136). We observe ~20% loss of 10 µM GSH standard in 40:40:20 methanol:acetonitrile:water solution after 16 h at 4°C; losses in neutral pH water have been reported to be faster (136). Giustarini et al. (138) recently reported a method that derivatizes GSH with the alkylating agent N-ethylmaleimide (NEM) to form GS-NEM (137). The NEM is added immediately upon sample collection. This prevents the oxidation of GSH. GS-NEM can be measured by LC, MS, or spectrophotometry. This NEM-based method yields a dramatically higher GSH:GSSG ratio (> 100 across rat tissues) than that typically measured by enzyme assay. We currently consider NEM derivatization the most reliable way of measuring GSH/GSSG.
10. OUTLOOK
Over the past decade, advanced analytical technologies, especially MS, have revolutionized metabolite measurement. Incremental improvements in protocols and automation are poised to enable the measurement of a majority of primary metabolites via straightforward LC-MS methods. Although technology-focused labs will surely want to continuously test new measurement procedures, there would be great value in the field in standardizing as much as possible a few measurement pipelines, enabling their facile adaptation across labs and automation of data analysis. In so doing, it is important to give equal weight to sample preparation as to extract measurement.
An important benefit of measuring large numbers of metabolites of each sample is that their respective concentrations must fit together. Metabolism is subject to thermodynamic constraints, with downstream metabolites in a pathway lower in free energy (chemical potential) than their upstream precursors. In practice, systems-level thermodynamic analysis can be used to identify mismeasured metabolites, to quantitate unmeasured metabolites, and to more precisely measure many species (7, 139, 140). Such analyses are most powerful when combined with flux measurements that further constrain metabolite concentrations. Greater utilization of such information is a significant opportunity.
Looking forward, a critical frontier in metabolomics is spatial resolution. Metabolites are compartmentalized at both the cellular and tissue level. Isolation of subcellular organelles can unfortunately perturb metabolite concentrations: There is no validated method to quench metabolic activity without causing leakage between compartments. Faster organelle isolation can help mitigate such problems, but the current standard for fast isolation of ~10 min remains slow relative to turnover of many metabolites (141–143). Nevertheless, some biological insights may be obtained, although it is important to validate their accuracy. Metabolite-sensitive fluorescent reporter proteins enable compartmentalized measurement, but current versions have modest signal-to-noise ratios and remain challenging to use (144–146). Spatially resolved MS holds the potential to measure many more metabolites, but issues remain with quenching, signal specificity (e.g., due to metabolite mimics made during ionization), and spatial resolution. Hyperpolarized NMR holds great potential for measurements in live mammals. Ultimately, progress along several of these dimensions is likely to provide complementary information to standard extract analysis and, with proper care to avoid measurement pitfalls, is poised to take metabolomics to new heights over the coming decade.
Acknowledgments
We thank members of Rabinowitz lab for discussion and support. This work is supported by National Institutes of Health grants CA163591 and DK113643 to J.D.R., CA211437 to W.L., DK097154 to O.F., and P30DK019525, which supports a joint Princeton-University of Pennsylvania Metabolomics Core for Diabetes Research. It was also supported by Alberta Innovates-Health Solutions, Canada Foundation for Innovation Grant CFI-JELF 34986, and the Natural Sciences and Engineering Research Council of Canada Discovery Grant 04547 to I.A.L.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wishart DS, Mandal R, Stanislaus A, Ramirez-Gaona M. Cancer metabolomics and the Human Metabolome Database. Metabolites. 2016;6:10. doi: 10.3390/metabo6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31:419–25. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011;80:301–25. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008;3:1299–311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–99. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JO, Rubin SA, Xu YF, Amador-Noguez D, Fan J, et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 2016;12:482–89. doi: 10.1038/nchembio.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mashego MR, Rumbold K, De Mey M, Vandamme E, Soetaert W, Heijnen JJ. Microbial metabolomics: past, present and future methodologies. Biotechnol. Lett. 2007;29:1–16. doi: 10.1007/s10529-006-9218-0. [DOI] [PubMed] [Google Scholar]
- 9.van Gulik WM. Fast sampling for quantitative microbial metabolomics. Curr. Opin. Biotechnol. 2010;21:27–34. doi: 10.1016/j.copbio.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Van Gulik WM, Canelas AB, Taymaz-Nikerel H, Douma RD, de Jonge LP, Heijnen JJ. Fast sampling of the cellular metabolome. Methods Mol. Biol. 2012;881:279–306. doi: 10.1007/978-1-61779-827-6_10. [DOI] [PubMed] [Google Scholar]
- 11.Vuckovic D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2012;403:1523–48. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- 12.Martano G, Delmotte N, Kiefer P, Christen P, Kentner D, et al. Fast sampling method for mammalian cell metabolic analyses using liquid chromatography-mass spectrometry. Nat. Protoc. 2015;10:1–11. doi: 10.1038/nprot.2014.198. [DOI] [PubMed] [Google Scholar]
- 13.Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 2008;26:1179–86. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittmann C, Kromer JO, Kiefer P, Binz T, Heinzle E. Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Anal. Biochem. 2004;327:135–39. doi: 10.1016/j.ab.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Palladino GW, Wood JJ, Proctor HJ. Modified freeze clamp technique for tissue assay. J. Surg. Res. 1980;28:188–90. doi: 10.1016/0022-4804(80)90161-4. [DOI] [PubMed] [Google Scholar]
- 16.Winder CL, Dunn WB, Schuler S, Broadhurst D, Jarvis R, et al. Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal. Chem. 2008;80:2939–48. doi: 10.1021/ac7023409. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez B, Francois J, Renaud M. A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast. 1997;13:1347–55. doi: 10.1002/(SICI)1097-0061(199711)13:14<1347::AID-YEA176>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.Maharjan RP, Ferenci T. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 2003;313:145–54. doi: 10.1016/s0003-2697(02)00536-5. [DOI] [PubMed] [Google Scholar]
- 19.Kimball E, Rabinowitz JD. Identifying decomposition products in extracts of cellular metabolites. Anal. Biochem. 2006;358:273–80. doi: 10.1016/j.ab.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Rammouz R, Letisse F, Durand S, Portais JC, Moussa ZW, Fernandez X. Analysis of skeletal muscle metabolome: evaluation of extraction methods for targeted metabolite quantification using liquid chromatography tandem mass spectrometry. Anal. Biochem. 2010;398:169–77. doi: 10.1016/j.ab.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Geier FM, Want EJ, Leroi AM, Bundy JG. Cross-platform comparison of Caenorhabditis elegans tissue extraction strategies for comprehensive metabolome coverage. Anal. Chem. 2011;83:3730–36. doi: 10.1021/ac2001109. [DOI] [PubMed] [Google Scholar]
- 22.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7:872–81. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 24.Rabinowitz JD, Kimball E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 2007;79:6167–73. doi: 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- 25.Shryock JC, Rubio R, Berne RM. Extraction of adenine nucleotides from cultured endothelial cells. Anal. Biochem. 1986;159:73–81. doi: 10.1016/0003-2697(86)90309-x. [DOI] [PubMed] [Google Scholar]
- 26.Grob MK, O’Brien K, Chu JJ, Chen DD. Optimization of cellular nucleotide extraction and sample preparation for nucleotide pool analyses using capillary electrophoresis. J. Chromatogr. B. 2003;788:103–11. doi: 10.1016/s1570-0232(02)01033-4. [DOI] [PubMed] [Google Scholar]
- 27.Ritter JB, Genzel Y, Reichl U. Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal. Biochem. 2008;373:349–69. doi: 10.1016/j.ab.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 28.Dietmair S, Timmins NE, Gray PP, Nielsen LK, Kromer JO. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal. Biochem. 2010;404:155–64. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 2011;83:3406–14. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ser Z, Liu X, Tang NN, Locasale JW. Extraction parameters for metabolomics from cultured cells. Anal. Biochem. 2015;475:22–28. doi: 10.1016/j.ab.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay GM, Zheng L, van den Broek NJF, Gottlieb E. Analysis of cell metabolism using LC-MS and isotope tracers. Methods Enzymol. 2015;561:171–96. doi: 10.1016/bs.mie.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Siegel D, Permentier H, Reijngoud DJ, Bischoff R. Chemical and technical challenges in the analysis of central carbon metabolites by liquid-chromatography mass spectrometry. J. Chromatogr. B. 2014;966:21–33. doi: 10.1016/j.jchromb.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 33.Gil A, Siegel D, Permentier H, Reijngoud DJ, Dekker F, Bischoff R. Stability of energy metabolites—an often overlooked issue in metabolomics studies: a review. Electrophoresis. 2015;36:2156–69. doi: 10.1002/elps.201500031. [DOI] [PubMed] [Google Scholar]
- 34.Lu W, Kwon YK, Rabinowitz JD. Isotope ratio-based profiling of microbial folates. J. Am. Soc. Mass Spectrom. 2007;18:898–909. doi: 10.1016/j.jasms.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon YK, Lu W, Melamud E, Khanam N, Bognar A, Rabinowitz JD. A domino effect in antifolate drug action in Escherichia coli. Nat. Chem. Biol. 2008;4:602–8. doi: 10.1038/nchembio.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overmyer KA, Thonusin C, Qi NR, Burant CF, Evans CR. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLOS ONE. 2015;10:e0117232. doi: 10.1371/journal.pone.0117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theodoridis G, Gika HG, Wilson ID. Mass spectrometry-based holistic analysis approaches for metabolite profiling in systems biology studies. Mass. Spectrom. Rev. 2011;30:884–906. doi: 10.1002/mas.20306. [DOI] [PubMed] [Google Scholar]
- 38.Junot C, Fenaille F, Colsch B, Becher F. High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass. Spectrom. Rev. 2014;33:471–500. doi: 10.1002/mas.21401. [DOI] [PubMed] [Google Scholar]
- 39.Kuehnbaum NL, Britz-McKibbin P. New advances in separation science for metabolomics: resolving chemical diversity in a post-genomic era. Chem. Rev. 2013;113:2437–68. doi: 10.1021/cr300484s. [DOI] [PubMed] [Google Scholar]
- 40.Milne SB, Mathews TP, Myers DS, Ivanova PT, Brown HA. Sum of the parts: mass spectrometry- based metabolomics. Biochemistry. 2013;52:3829–40. doi: 10.1021/bi400060e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–69. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crutchfield CA, Lu WY, Melamud E, Rabinowitz JD. Mass spectrometry-based metabolomics of yeast. Methods Enzymol. 2010;470:393–426. doi: 10.1016/S0076-6879(10)70016-1. [DOI] [PubMed] [Google Scholar]
- 43.Makarov A, Denisov E, Kholomeev A, Baischun W, Lange O, et al. Performance evaluation of a hybrid linear ion trap/Orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–20. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 44.Eliuk S, Makarov A. Evolution of Orbitrap mass spectrometry instrumentation. Annu. Rev. Anal. Chem. 2015;8:61–80. doi: 10.1146/annurev-anchem-071114-040325. [DOI] [PubMed] [Google Scholar]
- 45.Makarov A. Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 2000;72:1156–62. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 46.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 47.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–39. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahieu NG, Genenbacher JL, Patti GJ. A roadmap for the XCMS family of software solutions in metabolomics. Curr. Opin. Chem. Biol. 2016;30:87–93. doi: 10.1016/j.cbpa.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patti GJ. Separation strategies for untargeted metabolomics. J. Sep. Sci. 2011;34:3460–69. doi: 10.1002/jssc.201100532. [DOI] [PubMed] [Google Scholar]
- 50.Coulier L, Bas R, Jespersen S, Verheij E, van der Werf MJ, Hankemeier T. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 2006;78:6573–82. doi: 10.1021/ac0607616. [DOI] [PubMed] [Google Scholar]
- 51.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J. Chromatogr. A. 2007;1147:153–64. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 52.Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B. 2008;871:236–42. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone Orbitrap mass spectrometer. Anal. Chem. 2010;82:3212–21. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buescher JM, Moco S, Sauer U, Zamboni N. Ultrahigh performance liquid chromatography- tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 2010;82:4403–12. doi: 10.1021/ac100101d. [DOI] [PubMed] [Google Scholar]
- 55.Cubbon S, Bradbury T, Wilson J, Thomas-Oates J. Hydrophilic interaction chromatography for mass spectrometric metabonomic studies of urine. Anal. Chem. 2007;79:8911–18. doi: 10.1021/ac071008v. [DOI] [PubMed] [Google Scholar]
- 56.Spagou K, Tsoukali H, Raikos N, Gika H, Wilson ID, Theodoridis G. Hydrophilic interaction chromatography coupled to MS for metabonomic/metabolomic studies. J. Sep. Sci. 2010;33:716–27. doi: 10.1002/jssc.200900803. [DOI] [PubMed] [Google Scholar]
- 57.Buszewski B, Noga S. Hydrophilic interaction liquid chromatography (HILIC)—a powerful separation technique. Anal. Bioanal. Chem. 2012;402:231–47. doi: 10.1007/s00216-011-5308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bajad SU, Lu WY, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 59.Pesek JJ, Matyska MT, Fischer SM, Sana TR. Analysis of hydrophilic metabolites by high-performance liquid chromatography-mass spectrometry using a silica hydride-based stationary phase. J. Chromatogr. A. 2008;1204:48–55. doi: 10.1016/j.chroma.2008.07.077. [DOI] [PubMed] [Google Scholar]
- 60.Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal. Chem. 2012;84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- 61.Xu YF, Lu W, Rabinowitz JD. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal. Chem. 2015;87:2273–81. doi: 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J. Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 63.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014;6:281–94. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 64.Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, et al. Focused lipidomics by tandem mass spectrometry. J. Chromatogr. B. 2005;823:26–36. doi: 10.1016/j.jchromb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Ross KL, Dalluge JJ. Liquid chromatography/tandem mass spectrometry of glycolytic intermediates: deconvolution of coeluting structural isomers based on unique product ion ratios. Anal. Chem. 2009;81:4021–26. doi: 10.1021/ac9004698. [DOI] [PubMed] [Google Scholar]
- 66.Xu FG, Zou L, Liu Y, Zhang ZJ, Ong CN. Enhancement of the capabilities of liquid chromatography-mass spectrometry with derivatization: general principles and applications. Mass. Spectrom. Rev. 2011;30:1143–72. doi: 10.1002/mas.20316. [DOI] [PubMed] [Google Scholar]
- 67.Struys EA, Jansen EEW, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride. Clin. Chem. 2004;50:1391–95. doi: 10.1373/clinchem.2004.033399. [DOI] [PubMed] [Google Scholar]
- 68.Gibson KM, ten Brink HJ, Schor DS, Kok RM, Bootsma AH, et al. Stable-isotope dilution analysis of D- and L-2-hydroxyglutaric acid: application to the detection and prenatal diagnosis of D - and L-2-hydroxyglutaric acidemias. Pediatr. Res. 1993;34:277–80. doi: 10.1203/00006450-199309000-00007. [DOI] [PubMed] [Google Scholar]
- 69.Guo K, Li L. Differential C-/ C-isotope dansylation labeling and fast liquid chromatography/ mass spectrometry for absolute and relative quantification of the metabolome. Anal. Chem. 2009;81:3919–32. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 70.Meyer TE, Fox SD, Issaq HJ, Xu X, Chu LW, et al. A reproducible and high-throughput HPLC/MS method to separate sarcosine from α- and β-alanine and to quantify sarcosine in human serum and urine. Anal. Chem. 2011;83:5735–40. doi: 10.1021/ac201003r. [DOI] [PubMed] [Google Scholar]
- 71.Chen J, Zhang J, Zhang WP, Chen ZL. Sensitive determination of the potential biomarker sarcosine for prostate cancer by LC-MS with N,N’-dicyclohexylcarbodiimide derivatization. J. Sep. Sci. 2014;37:14–19. doi: 10.1002/jssc.201301043. [DOI] [PubMed] [Google Scholar]
- 72.Purwaha P, Silva LP, Hawke DH, Weinstein JN, Lorenzi PL. An artifact in LC-MS/MS measurement of glutamine and glutamic acid: in-source cyclization to pyroglutamic acid. Anal. Chem. 2014;86:5633–37. doi: 10.1021/ac501451v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Annesley TM. Ion suppression in mass spectrometry. Clin. Chem. 2003;49:1041–44. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 74.Halket JM, Waterman D, Przyborowska AM, Patel RKP, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005;56:219–43. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 75.Zimmermann D, Hartmann M, Moyer MP, Nolte J, Baumbach JI. Determination of volatile products of human colon cell line metabolism by GC/MS analysis. Metabolomics. 2007;3:13–17. [Google Scholar]
- 76.Grundy SM, Ahrens EH, Miettinen TA. Quantitative isolation and gas-liquid chromatographic analysis of total fecal bile acids. J. Lipid Res. 1965;6:397–410. [PubMed] [Google Scholar]
- 77.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, et al. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 78.Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–42. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 79.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009;81:10038–48. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stein SE. The NIST14 mass spectral library. Natl. Inst. Stand. Technol.; Gaithersburg, MD: 2014. updated Jan. 13, 2017. https://www.nist.gov/srd/nist-standard-reference-database-1a-v14. [Google Scholar]
- 81.Koek MM, Jellema RH, van der Greef J, Tas AC, Hankemeier T. Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics. 2011;7:307–28. doi: 10.1007/s11306-010-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaspar H, Dettmer K, Gronwald W, Oefner PJ. Automated GC-MS analysis of free amino acids in biological fluids. J. Chromatogr. B. 2008;870:222–32. doi: 10.1016/j.jchromb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 83.Kumari S, Stevens D, Kind T, Denkert C, Fiehn O. Applying in-silico retention index and mass spectra matching for identification of unknown metabolites in accurate mass GC-TOF mass spectrometry. Anal. Chem. 2011;83:5895–902. doi: 10.1021/ac2006137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abate S, Ahn YG, Kind T, Cataldi TRI, Fiehn O. Determination of elemental compositions by gas chromatography/time-of-flight mass spectrometry using chemical and electron ionization. Rapid Commun Mass Spectrom. 2010;24:1172–80. doi: 10.1002/rcm.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and un-targeted profiling. Curr. Protoc. Mol. Biol. 2016;114:30.4.1–32. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Niehaus TD, Nguyen TND, Gidda SK, ElBadawi-Sidhu M, Lambrecht JA, et al. Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell. 2014;26:3010–22. doi: 10.1105/tpc.114.126854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Raad M, Fischer CR, Northen TR. High-throughput platforms for metabolomics. Curr. Opin. Chem. Biol. 2016;30:7–13. doi: 10.1016/j.cbpa.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 89.Jonas M, LaMarr WA, Ozbal C. Mass spectrometry in high throughput screening: a case study on acetyl-coenzyme a carboxylase using RapidFire®-mass spectrometry (RF-MS) Comb. Chem. High Throughput Screen. 2009;12:752–59. doi: 10.2174/138620709789104924. [DOI] [PubMed] [Google Scholar]
- 90.Holt TG, Choi BK, Geoghagen NS, Jensen KK, Luo Q, et al. Label-free high-throughput screening via mass spectrometry: a single cystathionine quantitative method for multiple applications. Assay DrugDev. Technol. 2009;7:495–506. doi: 10.1089/adt.2009.0200. [DOI] [PubMed] [Google Scholar]
- 91.Lange HC, Eman M, van Zuijlen G, Visser D, van Dam JC, et al. Improved rapid sampling for in vivo kinetics of intracellular metabolites in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001;75:406–15. doi: 10.1002/bit.10048. [DOI] [PubMed] [Google Scholar]
- 92.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011;83:7074–80. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- 93.Fuhrer T, Zamboni N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015;31:73–78. doi: 10.1016/j.copbio.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 94.Link H, Fuhrer T, Gerosa L, Zamboni N, Sauer U. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat. Methods. 2015;12:1091–97. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 95.Holmes E, Foxall PJ, Spraul M, Farrant RD, Nicholson JK, Lindon JC. 750 MHz H NMR spectroscopy characterisation of the complex metabolic pattern of urine from patients with inborn errors of metabolism: 2-hydroxyglutaric aciduria and maple syrup urine disease. J. Pharm. Biomed. Anal. 1997;15:1647–59. doi: 10.1016/s0731-7085(97)00066-6. [DOI] [PubMed] [Google Scholar]
- 96.Holmes E, Foxall PJ, Nicholson JK, Neild GH, Brown SM, et al. Automatic data reduction and pattern recognition methods for analysis of 1Hnuclear magnetic resonance spectra of human urine from normal and pathological states. Anal. Biochem. 1994;220:284–96. doi: 10.1006/abio.1994.1339. [DOI] [PubMed] [Google Scholar]
- 97.Emwas AH. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015;1277:161–93. doi: 10.1007/978-1-4939-2377-9_13. [DOI] [PubMed] [Google Scholar]
- 98.Serkova NJ, Rose JC, Epperson LE, Carey HV, Martin SL. Quantitative analysis of liver metabolites in three stages of the circannual hibernation cycle in 13-lined ground squirrels by NMR. Physiol. Genom. 2007;31:15–24. doi: 10.1152/physiolgenomics.00028.2007. [DOI] [PubMed] [Google Scholar]
- 99.Gronwald W, Klein MS, Kaspar H, Fagerer SR, Nurnberger N, et al. Urinary metabolite quantification employing 2D NMR spectroscopy. Anal. Chem. 2008;80:9288–97. doi: 10.1021/ac801627c. [DOI] [PubMed] [Google Scholar]
- 100.Klein MS, Almstetter MF, Schlamberger G, Nurnberger N, Dettmer K, et al. Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J. Dairy Sci. 2010;93:1539–50. doi: 10.3168/jds.2009-2563. [DOI] [PubMed] [Google Scholar]
- 101.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–8. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44:D463–70. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of H NMR metabolomics data. Anal. Chem. 2006;78:4430–42. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 104.Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, et al. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic H NMR data sets. Anal. Chem. 2005;77:1282–89. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- 105.Bingol K, Bruschweiler R. Multidimensional approaches to NMR-based metabolomics. Anal. Chem. 2014;86:47–57. doi: 10.1021/ac403520j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qiu F, McAlpine JB, Lankin DC, Burton I, Karakach T, et al. 2D NMR barcoding and differential analysis of complex mixtures for chemical identification: the Actaea triterpenes. Anal. Chem. 2014;86:3964–72. doi: 10.1021/ac500188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Komatsu T, Ohishi R, Shino A, Kikuchi J. Structure and metabolic-flow analysis of molecular complexity in a C-labeled tree by 2D and 3D NMR. Angew. Chem. Int. Ed. Engl. 2016;55:6000–3. doi: 10.1002/anie.201600334. [DOI] [PubMed] [Google Scholar]
- 108.Schoenberger T, Menges S, Bernstein MA, Perez M, Seoane F, et al. Improving the performance of high-precision qNMR measurements by a double integration procedure in practical cases. Anal. Chem. 2016;88:3836–43. doi: 10.1021/acs.analchem.5b04911. [DOI] [PubMed] [Google Scholar]
- 109.Ravanbakhsh S, Liu P, Bjorndahl TC, Mandal R, Grant JR, et al. Accurate, fully-automated NMR spectral profiling for metabolomics. PLOS ONE. 2015;10:e0124219. doi: 10.1371/journal.pone.0124219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bruschweiler R, Zhang F. Covariance nuclear magnetic resonance spectroscopy. J. Chem. Phys. 2004;120:5253–60. doi: 10.1063/1.1647054. [DOI] [PubMed] [Google Scholar]
- 111.Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, et al. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional H-1 C NMR spectra. Anal. Chem. 2007;79:9385–90. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Giraudeau P. Challenges and perspectives in quantitative NMR. Magn. Reson. Chem. 2017;55:61–69. doi: 10.1002/mrc.4475. [DOI] [PubMed] [Google Scholar]
- 113.Weber M, Hellriegel C, Ruck A, Sauermoser R, Wuthrich J. Using high-performance quantitative NMR (HP-qNMR®) for certifying traceable and highly accurate purity values of organic reference materials with uncertainties < 0.1% Accredit. Qual. Assur. 2013;18:91–98. [Google Scholar]
- 114.Hu K, Westler WM, Markley JL. Simultaneous quantification and identification of individual chemicals in metabolite mixtures by two-dimensional extrapolated time-zero H-1 C HSQC (HSQC0) J.Am. Chem. Soc. 2011;133:1662–65. doi: 10.1021/ja1095304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mauve C, Khlifi S, Gilard F, Mouille G, Farjon J. Sensitive, highly resolved, and quantitative H-1 C NMR data in one go for tracking metabolites in vegetal extracts. Chem. Commun. 2016;52:6142–45. doi: 10.1039/c6cc01783e. [DOI] [PubMed] [Google Scholar]
- 116.Klein MS, Oefner PJ, Gronwald W. MetaboQuant: a tool combining individual peak calibration and outlier detection for accurate metabolite quantification in 1D H and H-1 C HS QC NMR spectra. BioTechniques. 2013;54:251–56. doi: 10.2144/000114026. [DOI] [PubMed] [Google Scholar]
- 117.Hu F, Furihata K, Kato Y, Tanokura M. Nondestructive quantification of organic compounds in whole milk without pretreatment by two-dimensional NMR spectroscopy. J. Agric. Food Chem. 2007;55:4307–11. doi: 10.1021/jf062803x. [DOI] [PubMed] [Google Scholar]
- 118.Lewis IA, Schommer SC, Markley JL. rNMR: open source software for identifying and quantifying metabolites in NMR spectra. Magn. Reson. Chem. 2009;47(01):S123–26. doi: 10.1002/mrc.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, et al. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008;26:162–64. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 120.Lewis IA, Karsten RH, Norton ME, Tonelli M, Westler WM, Markley JL. NMR method for measuring carbon-13 isotopic enrichment of metabolites in complex solutions. Anal. Chem. 2010;82:4558–63. doi: 10.1021/ac100565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fan TW, Lane AN. NMR-based stable isotope resolved metabolomics in systems biochemistry. J. Biomol. NMR. 2011;49:267–80. doi: 10.1007/s10858-011-9484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lane AN, Fan TW, Higashi RM. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol. 2008;84:541–88. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- 123.Schrader MC, Eskey CJ, Simplaceanu V, Ho C. A carbon-13 nuclear magnetic resonance investigation of the metabolic fluxes associated with glucose metabolism in human erythrocytes. Biochim. Biophys. Acta. 1993;1182:162–78. doi: 10.1016/0925-4439(93)90138-q. [DOI] [PubMed] [Google Scholar]
- 124.Delgado TC, Castro MM, Geraldes CF, Jones JG. Quantitation of erythrocyte pentose pathway flux with [2- C]glucoseand H NMR analysis of the lactate methyl signal. Magn. Reson. Med. 2004;51:1283–86. doi: 10.1002/mrm.20096. [DOI] [PubMed] [Google Scholar]
- 125.Lewis IA, Campanella ME, Markley JL, Low PS. Role of band 3 in regulating metabolic flux of red blood cells. PNAS. 2009;106:18515–20. doi: 10.1073/pnas.0905999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tzika AA, Cheng LL, Goumnerova L, Madsen JR, Zurakowski D, et al. Biochemical characterization of pediatric brain tumors by using in vivo and ex vivo magnetic resonance spectroscopy. J. Neurosurg. 2002;96:1023–31. doi: 10.3171/jns.2002.96.6.1023. [DOI] [PubMed] [Google Scholar]
- 127.Rothman DL, Sibson NR, Hyder F, Shen J, Behar KL, Shulman RG. In vivo nuclear magnetic resonance spectroscopy studies of the relationship between the glutamate-glutamine neurotransmitter cycle and functional neuroenergetics. Philos. Trans. R. Soc. B. 1999;354:1165–77. doi: 10.1098/rstb.1999.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. PNAS. 2003;100:10158–63. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68:105–15. doi: 10.1016/j.ymeth.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Park J, Lee SB, Lee S, Kim Y, Song S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 131.Zerez CR, Lee SJ, Tanaka KR. Spectrophotometric determination ofoxidized and reduced pyridine nucleotides in erythrocytes using a single extraction procedure. Anal. Biochem. 1987;164:367–73. doi: 10.1016/0003-2697(87)90506-9. [DOI] [PubMed] [Google Scholar]
- 132.Wagner TC, Scott MD. Single extraction method for the spectrophotometric quantification of oxidized and reduced pyridine nucleotides in erythrocytes. Anal. Biochem. 1994;222:417–26. doi: 10.1006/abio.1994.1511. [DOI] [PubMed] [Google Scholar]
- 133.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–65. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu JT, Wu LH, Knight JA. Stability of NADPH effect of various factors on the kinetics of degradation. Clin. Chem. 1986;32:314–19. [PubMed] [Google Scholar]
- 135.Hofmann D, Wirtz A, Santiago-Schubel B, Disko U, Pohl M. Structure elucidation of the thermal degradation products of the nucleotide cofactors NADH and NADPH by nano-ESI-FTICR-MS and HPLC-MS. Anal. Bioanal. Chem. 2010;398:2803–11. doi: 10.1007/s00216-010-4111-z. [DOI] [PubMed] [Google Scholar]
- 136.Rossi R, Milzani A, Dalle-Donne I, Giustarini D, Lusini L, et al. Blood glutathione disulfide: in vivo factor or in vitro artifact? Clin. Chem. 2002;48:742–53. [PubMed] [Google Scholar]
- 137.Giustarini D, Dalle-Donne I, Milzani A, Fanti P, Rossi R. Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat. Protoc. 2013;8:1660–69. doi: 10.1038/nprot.2013.095. [DOI] [PubMed] [Google Scholar]
- 138.Giustarini D, Dalle-Donne I, Milzani A, Rossi R. Low molecular mass thiols, disulfides and protein mixed disulfides in rat tissues: influence of sample manipulation, oxidative stress and ageing. Mech. Ageing Dev. 2011;132:141–48. doi: 10.1016/j.mad.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 139.Kummel A, Panke S, Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2006;2:2006.0034. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys. J. 2007;92:1792–805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fly R, Lloyd J, Krueger S, Fernie A, van der Merwe MJ. Improvements to define mitochondrial metabolomics using nonaqueous fractionation. Methods Mol. Biol. 2015;1305:197–210. doi: 10.1007/978-1-4939-2639-8_14. [DOI] [PubMed] [Google Scholar]
- 142.Matuszczyk JC, Teleki A, Pfizenmaier J, Takors R. Compartment-specific metabolomics for CHO reveals that ATP pools in mitochondria are much lower than in cytosol. Biotechnol. J. 2015;10:1639–50. doi: 10.1002/biot.201500060. [DOI] [PubMed] [Google Scholar]
- 143.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166:1324–37. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, et al. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 2011;14:555–66. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao Y, Wang A, Zou Y, Su N, Loscalzo J, Yang Y. In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD+/NADH redox state. Nat. Protoc. 2016;11:1345–59. doi: 10.1038/nprot.2016.074. [DOI] [PubMed] [Google Scholar]
- 146.Hou BH, Takanaga H, Grossmann G, Chen LQ, Qu XQ, et al. Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat. Protoc. 2011;6:1818–33. doi: 10.1038/nprot.2011.392. [DOI] [PubMed] [Google Scholar]