Abstract
Introduction
Gold nanoparticles are versatile carriers for delivery of biomacromolecules. Here, we have developed spiky gold nanoparticles (SGNPs) that can efficiently deliver immunostimulatory agents.
Objectives
Our goal was to develop a platform technology for co-delivery of multiple adjuvant molecules for synergistic stimulation and maturation of innate immune cells.
Methods
SGNPs were synthesized by a seed-mediated, surfactant-free synthesis method and incorporated with polyinosinic-polycytidylic acid (pIC) and DNA oligonucleotide containing unmethylated CpG motif (CpG) by an electrostatic layer-by-layer approach. Adjuvant-loaded SGNP nano-complexes were examined for their biophysical and biochemical properties and studied for immune activation using bone marrow-derived dendritic cells (BMDCs).
Results
We have synthesized SGNPs with branched nano-spikes layered with pIC and/or CpG. Adjuvant-loaded SGNP nano-complexes promoted cellular uptake of the adjuvants. Importantly, we achieved spatio-temporal control over co-delivery of pIC and CpG via SGNPs, which produced synergistic enhancement in cytokine release (IL-6, TNF-α) and upregulation of co-stimulatory markers (CD40, CD80, CD86) in BMDCs, compared with pIC, CpG, or their admixtures.
Conclusion
SGNPs serve as a versatile delivery platform that allows flexible and on-demand cargo fabrication for strong activation of innate immune cells.
Keywords: Inorganic nanoparticle, Adjuvant, TLR agonist, Vaccine delivery
Introduction
Induction of robust immune response requires engagement and activation of the innate immune system with danger signals from an exogenous pathogenic source or an endogenous source from tissue damage or cellular stress.35 Danger signals are recognized by pattern-recognition receptors (PRRs), which include Toll-like receptors (TLRs), C-type lectin receptors (CLRs), Retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), and NOD-like receptors (NLRs).58 PRRs can recognize conserved microbial molecular structures, termed pathogen-associated molecular patterns (PAMPs), and the interaction between PRRs and PAMPs triggers inflammatory responses.58 Pathogen recognition during infection involves simultaneous or sequential engagement of PRRs on innate immune cells by multiple PAMPs, which activate distinct or shared signaling pathways and induce immune stimulation in a synergistic manner.30,39,61 Synthetic molecules mimicking the structures and functions of PAMPs can serve as effective adjuvants for stimulation of the innate immune system. In particular, TLR agonists have been widely investigated as vaccine adjuvants.23,26,43,56 However, they typically require high doses for in vivo administration and immune activation, thus raising potential safety concerns, such as reactogenicity at the injection site.15,55
On the other hand, particulate carriers may improve the potency and delivery of adjuvants by enhancing their solubility, stability, tissue and cell targeting.21,37 Thus, particle-based delivery of adjuvants may limit dose-dependent injection site toxicity and allow for dose-sparing of immunostimulatory agents.13 Our goal in this study was to develop a nanoparticle (NP) platform that can induce activation of innate immune cells and to perform initial characterization studies. In particular, gold nanoparticles (GNP) are one of the most extensively investigated inorganic NPs for drug delivery applications because of their intrinsic biocompatibility, well-defined synthetic and surface chemistry for targeted and controlled delivery, and in vivo stability.14,17,21,36,38,60,64 Here, we have designed spiky GNPs (SGNPs) as a versatile platform for intracellular co-delivery of multiple adjuvant molecules. Exploiting the high surface area-to-volume ratio of SGNPs attributed to their unique elongated nano-spikes, we have decorated their surfaces with TLR agonists and endowed them with immunostimulatory properties. We have achieved this by employing the electrostatic layer-by-layer assembly process10–12,51,68,69 with cationic polyelectrolytes that mediate charge interaction between anionic surfaces of SGNP and adjuvants. Specifically, we coated spiky surfaces of SGNPs with polyinosinic-polycytidylic acid (pIC) and oligonucleotide containing unmethylated CpG motif (CpG). pIC is a TLR3 agonist based on a synthetic double-stranded viral RNA analogue that promotes activation of macrophages and dendritic cells, while CpG is a DNA oligonucleotide-based TLR9 agonist that promotes robust innate and adaptive immune responses.2,3,33,54,57,62,63,70 Notably, the combination of pIC and CpG has been demonstrated to induce synergistic immune activation.3,62,70
Our proof-of-concept studies presented here were performed with bone-marrow derived dendritic cells (BMDCs), a widely-used model for innate immune cells. Our results indicate that these adjuvant-SGNP nano-complexes can promote efficient cellular uptake of pIC and CpG by innate immune cells and mediate co-delivery of multiple adjuvant species to endolysosomal compartments, where TLR3 and TLR9 are expressed,30,39,61 in a spatio-temporally controlled manner. In particular, co-localized delivery of dual adjuvants mediated by SGNP nano-complexes induced potent, synergistic immune activation of BMDCs with much lower concentrations of adjuvants than free soluble adjuvants. Our studies described here suggest that the SGNP system offers a simple yet versatile synthetic platform for dose-sparing of adjuvants and co-delivery of multiple immunostimulatory ligands to innate immune cells.
Materials and Methods
Reagents and Instruments
L-ascorbic acid was obtained from Fisher Chemical. Methoxy poly(ethyleneglycol) propionic acid N-hydroxysuccinimide (MW 5000, Methoxy-PEG-NHS) was purchased from Nanocs. pIC (high molecular weight, 1.5–8 kb) was purchased from Invivogen, and CpG (CpG 1826) was obtained from Integrated DNA Technology. Other chemicals were obtained from Sigma-Aldrich, and all reagents were used as received. UV–Vis absorption spectra were obtained using BioTek synergy neo microplate reader. Transmission electron microscope (TEM) images were acquired using JEOL 1400-plus, and confocal microscope images were taken with Nikon A1Rsi Confocal Microscope. Hydrodynamic size and zeta potential were measured using Malvern Zetasizer Nano ZSP. The amount of pIC and CpG loaded on particles was quantified by gel permeation chromatography (GPC, Shimadzu). Flow cytometric analyses were performed using Cyan 5 (Beckman Coulter), and the data were analyzed using FlowJo 10.2 software.
Synthesis of Citrate-Stabilized GNPs
Five mM deionized (DI) water solution of HAuCl4 (300 ml, 1.5 mmol) was boiled to reflux for 30 min, then 1.5 M sodium citrate tribasic dehydrate (3 ml, 4.5 mmol) was quickly added with vigorous stirring. The solution color changed from yellow to red within 5 min as gold ion was reduced to form GNPs. The mixture was boiled for 10 min and then cooled for 30 min at room temperature. The resulting citrate-stabilized GNPs were stored at 4 °C until further use.
Synthesis of SGNPs
SGNPs were prepared as described in the literature with slight modifications.67 Ten ml of the above citrate-stabilized seed GNPs were diluted in 300 ml DI water and sequentially mixed with 3 ml of HAuCl4 (20 mM, 60 μmol), 300 μl of 1 M HCl, and 3 ml of AgNO3 (3 mM, 9 μmol) with vigorous stirring. After stirring for 1 min, 3 ml of L-ascorbic acid (40 mM, 120 μmol) was added, which induced abrupt color change from red to greenish black in 10 s indicating the SGNP formation. Finally, 300 μl of poly(ethylene glycol) methyl ether thiol (MW 6000; 10 mM, 3 μmol) was added to quench the reaction and stabilize the SGNPs. The mixture was stirred for another 2 h at room temperature and centrifuged at 3,000×g for 60 min to remove excess unreacted reagents. The pellet was re-dispersed in 1 ml DI water and passed through illustra NAP-10 column (GE healthcare life sciences) for further purification. The resulting SGNPs were stored at 4 °C until further use. For physicochemical characterization of SGNPs, TEM images, hydrodynamic size, and zeta potentials were obtained after dilution in DI water.
Synthesis of Poly(ethyleneglycol)-Grated Polyethyleneimine (PEG-PEI)
Methoxy-PEG-NHS (400 mg, 80 μmol) was dissolved in 4 ml dimethyl sulfoxide (DMSO) by sonication. Polyethyleneimine (PEI, MW 25,000; 100 mg, 4 μmol) dissolved in 1 ml DMSO was added to the above methoxy-PEG-NHS solution in a dropwise manner to achieve the final stoichiometry of 1:20 (PEI:PEG). The mixture was reacted for 24 h at room temperature with vigorous stirring and then dialyzed 6 times against DI water using Amicon ultra 10 kDa MW cutoff centrifugal filters to remove unreacted methoxy-PEG-NHS. The purified PEG-PEI conjugate was stored at −20 °C until further use.
Preparation of Adjuvant-Loaded SGNPs
PEI and PEG-PEI were dissolved in phosphate buffered saline (PBS), and the pH was adjusted to 7.4. DI water suspension of SGNPs (10 nM, 100 μl) was rapidly mixed with 100 μl PBS solution of PEI (2 mg/ml) and incubated for 10 min at room temperature. Excess free PEI was removed by centrifugation at 3000×g for 10 min. The pellet was dispersed in 200 μl PBS and mixed with 200 μl PBS solution of pIC (100 μg) and CpG (100 μg), either separately or together as indicated in the result section. After 10 min, the mixture was added to 400 μl PBS solution of PEG-PEI at a varying weight ratio (0:1–20:1 = PEG-PEI:adjuvants) and further incubated for 10 min. The crude mixture was purified from unloaded free adjuvants and PEIs by 2 rounds of centrifugation at 3,000×g for 10 min, using PBS (0.01% tween 20) and DI water, respectively. The adjuvant-loaded SGNPs were dispersed in DI water for further characterizations using dynamic light scattering and zeta potential. To verify the loading of adjuvant-PEG-PEI complexes, TEM images were acquired after staining SGNP complexes with 2% uranyl acetate (Electron Microscopy Sciences). Loading efficiency was calculated by releasing adjuvant-PEG-PEI complexes from SGNPs with treatment of heparin sulfate (1 mg/ml), followed by GPC analysis. Specifically, SGNP complexes were diluted in PBS (0.01% tween 20, 1 mg/ml heparin) and sonicated for 1 min at 40% amplitude (Qsonica Q125) to facilitate heparin-mediated dissociation of adjuvants from SGNPs. Free adjuvant was separated from SGNPs by centrifugation at 10,000×g for 5 min and quantified by GPC equipped with TSKgel G3000SWxl column (7.8 mm ID × 300 mm, Tosoh Bioscience LLC).
Fluorophore Labeling of pIC and CpG for Confocal Microscopy
For fluorophore labeling, 5′ phosphate group of pIC and CpG was crosslinked with ethylenediamine via the 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) coupling reaction in methyl imidazole (MeIm) buffer, which tethers primary amine to the 5′ phosphate group.52,53 Amine-reactive fluorophore dyes were then conjugated to the resulting ethylenediamine-crosslinked pIC and CpG. For pIC conjugation, 1 ml DI water solution of pIC (2.5 mg) was mixed with 19 μl of EDC (1 mg/ml, 100 nmol) and 13 μl of ethylenediamine (1 μl/ml, 200 nmol), and the volume was brought up to 1.2 ml using DI water and 1 M MeIm buffer to conduct the coupling reaction in 0.1 M MeIm buffer (pH 7.4). After 2 h reaction at room temperature, the mixture was dialyzed three times using Amicon ultra 3 kDa MW cutoff centrifugal filters with buffer exchange to 0.1 M NaHCO3 (pH 8.0). To 1 ml of the above solution, 10 μl of Alexa Fluor® 647 NHS Ester (10 mg/ml, Invitrogen) was added and reacted for 2 h at room temperature. The mixture was dialyzed three times, freeze-dried, and stored at −20 °C. For CpG conjugation, 2 mg of CpG, 30 μmol of EDC, 60 μmol of ethylenediamine were used for amine functionalization. Then 40 μl of Alexa Fluor® 488 NHS Ester (10 mg/ml, Invitrogen) was conjugated to CpG. Concentrations of fluorophore-tagged pIC (pIC-AF647) and CpG (CpG-AF488) were measured using GPC, and they were used at the same amount (100 μg) for loading on SGNPs.
Preparation and Maintenance of Bone Marrow-Derived Dendritic Cells (BMDCs)
BMDCs were prepared according to the literature.41 BMDCs were maintained in RPMI 1640 (Gibco) supplemented 10% fetal bovine serum (Corning), 1% penicillin–streptomycin (Gibco), 20 ng/ml granulocyte–macrophage colony-stimulating factor (GM-CSF, Genscript), and 50 μM beta-mercaptoethanol (β-ME, Gibco). For incubation with samples, BMDCs were treated in the medium without GM-CSF and β-ME to prevent their potential impact on BMDC activation and surface modification of SGNPs, respectively. BMDC isolation was performed in compliance with the animal study protocol reviewed and approved by the Institutional Animal Care and Use Committee at the University of Michigan.
Visualization of Intracellular Distribution of pIC and CpG Using Confocal Microscopy
BMDCs were grown onto 12 mm glass coverslips in 24 well plates at a density of 5 × 105 cells/well and incubated overnight at 37 °C under 5% CO2. BMDCs were then treated with SP-P/C, SP-P + SP-C (formulated using pIC-AF647 and CpG-AF488), pIC-AF647 + CpG-AF488. Equivalent concentrations of adjuvants were used at 1 μg/ml CpG-AF488 and 1.8 μg/ml pIC-AF647. After 24 h, cells were washed three times with serum-free media and further incubated with 2 μg/ml Hoechst 33258 and 100 nM LysoTracker Red DND-99 (Invitrogen) in serum-free media for 30 min to stain nuclei and lysosomes, respectively. Cells were washed three times using PBS and fixed with 4% formaldehyde in PBS. Coverslips were mounted on slide glass using Fluoroshield Mounting Medium with anti-fade agent (Abcam), and the samples were visualized using Nikon A1Rsi Confocal Microscope.
Activation of BMDCs
Immature BMDCs were plated at a density of 1 × 105 cells/well in 96 well plates and incubated overnight at 37 °C under 5% CO2. Cells were then incubated with SGNP complexes or free soluble adjuvants. Specifically, we used SP-P/C, SP-P, SP-C, SP-P + SP-C, pIC, CpG, or pIC + CpG with their concentrations at 1 μg/ml CpG and 1.8 μg/ml pIC or 0.1 μg/ml CpG and 0.18 μg/ml pIC for 2 h and 24 h incubation, respectively. Cell culture media were collected for cytokine analysis with IL-6 and TNF-α ELISA kits (R&D system).
Flow Cytometric Analysis of BMDC Maturation Markers
BMDCs were plated at a density of 5 × 105 cells/well in 24 well plates and incubated overnight at 37 °C under 5% CO2. Cells were treated with free adjuvants or SGNP complexes with the concentrations at 0.1 μg/ml CpG and 0.18 μg/ml pIC. After 24 h, cells were collected and washed with 1% bovine serum albumin (BSA) in PBS, followed by centrifugation at 2,000×g for 5 min. The cell pellet was suspended in 1% BSA in PBS and incubated with CD16/32 FcR blocking antibody (0.5 µg, eBioscience) for 10 min at room temperature. Cells were then stained with fluorophore-conjugated antibodies against cell surface markers including FITC-CD80 (0.2 µg, BD Biosciences), PE-CD86 (0.1 µg, eBioscience), APC-CD40 (0.1 µg, eBioscience), and PECy7-CD11c (0.1 µg, BD Biosciences) for 30 min at room temperature. Cells were washed twice with 1% BSA in PBS and suspended in 1% BSA in PBS containing 2 µM DAPI. Cells were then analyzed by flow cytometric analysis for quantification of upregulation of surface maturation markers.
Statistical Analyses
All values are reported as mean ± SD For statistical comparison of multiple samples, data were analyzed by one-way ANOVA with post hoc Turkey HSD test using Graphpad Prism 6.07 software. p values less than 0.05 were considered statistically significant. The level of statistical significance was defined as *p < 0.05, **p < 0.01, ***p < 0.001.
Results and Discussion
The Overall Schematic and Synthesis of SGNPs
Overall, we have developed SGNPs as a delivery platform for adjuvants by utilizing an electrostatic layer-by-layer assembly approach (Fig. 1a). pIC and CpG are oligonucleotide-based adjuvants with multiple negative charges in their repeating phosphate backbone units. Our approach exploits the anionic charges of naïve pIC and CpG for self-assembly, thereby avoiding other complex chemical and structural modifications that may harm their immunological activities.9,24,25,27 We employed cationic polyelectrolytes to mediate the electrostatic assembly between SGNPs and nucleic acid-based adjuvants. Branched PEI (MW 25,000) was used as a model polyelectrolyte since it can readily form strong electrostatic complexation with oligonucleotides due to its high cationic charge density.5,40 SGNPs were coated with a PEI layer through electrostatic attraction (SGNP@PEI), onto which pIC and/or CpG were further layered and complexed. We prepared the SGNP complexes with either single (SP-P and SP-C) or dual (SP-P/C) adjuvants, and the outer pIC and/or CpG layers were coated with PEG-grafted PEI (PEG-PEI) to protect the adjuvants from degradation as well as to enhance colloidal stability of the complexes.
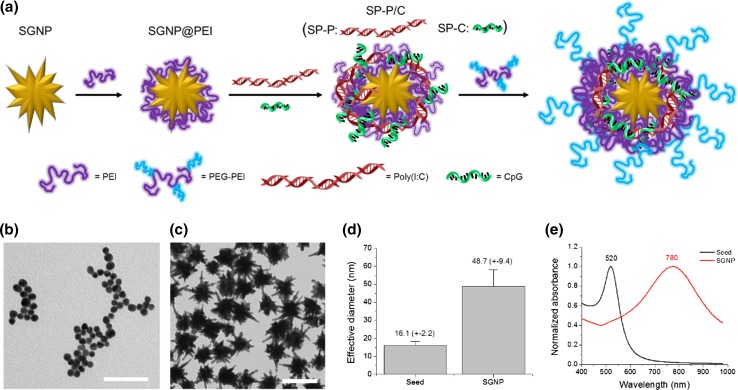
Figure 1.
Synthesis of SNGPs and the schematic of their modification with adjuvants by a layer-by-layer assembly approach (a). SGNPs were layered consecutively with PEI, pIC and/or CpG, followed by coating with PEG-PEI via utilization of multiple electrostatic interactions. TEM images of the seed GNPs (b) and SGNPs (c), and their effective diameter (d) and UV–Vis absorption spectra (e). Scale bars = 100 nm.
For the synthesis of core SGNPs, we have adopted a surfactant-free, seed-mediated growth method20 in order to alleviate potential toxicity associated with surfactants typically employed during the conventional synthesis of GNPs. Specifically, citrate-stabilized GNPs were prepared following the citrate reduction method16 and utilized as the seed for synthesis of SGNPs (Fig. 1b). The branched structures were grown from the seed GNPs using AgNO3 and ascorbic acid as the structure-directing agent and reducing agent, respectively. Our “bare” SGNPs, which were likely adsorbed with small molecules, such as citrate and ascorbate, easily aggregated during the purification processes. Hence, we have improved their colloidal stability by partially passivating their surfaces with PEG methyl ether thiol. The reaction was carefully controlled using the minimum concentration of PEG needed to preserve the initial anionic surfaces for the subsequent electrostatic assembly with cationic polyelectrolytes while at the same time maintaining colloidal stability during purification.
The synthesis of SGNPs was monitored using TEM and UV–Vis absorption measurements. The seed GNPs initially had spherical shape and small size, which turned into larger anisotropic SGNPs with multiple branched nano-spike structures after the growth reaction (Fig. 1c). The effective diameter, deduced from the surface area of each particle with the assumption of their spherical morphology, increased from 16.1 (± 2.2) nm for GNPs to 48.7 (± 9.4) nm for SGNPs (Fig. 1d). The size distribution of SGNPs was reasonably narrow without any apparent particles as small as the seed GNPs. This indicates that the growth was very efficient with uniform reaction on most seed GNPs. The growth of branched structures was accompanied by the appearance of new surface plasmon resonance modes. SGNPs showed an intense UV–Vis absorption peak at 780 nm, which was redshifted by ~260 nm from that of the seed GNPs (Fig. 1e). The redshifted absorption band is ascribed to the unique surface plasmon resonance (SPR) mode that is governed by the aspect ratio of branches, whereas the small absorbance band that remained at 520 nm is due to the spherical cores.20
Synthesis and Characterization of Adjuvant-Loaded SGNP Nano-Complexes
SGNP complexes were prepared by coating “bare” SGNPs with PEI (termed SGNPs@PEI), followed by loading of pIC and CpG either separately or together and the final PEG-PEI treatment, resulting in nano-complexes referred to as SP-P for pIC, SP-C for CpG, SP-P/C for pIC/CpG. The amounts of SGNP (1 pmol), PEI (200 µg), pIC and CpG (100 µg each) were fixed, while the amount of PEG-PEI was adjusted in a range of 0:1–20:1 weight ratio of PEG-PEI:adjuvants for all complexations. The number after the abbreviation of adjuvants denotes the weight ratio of PEG-PEI relative to adjuvants.
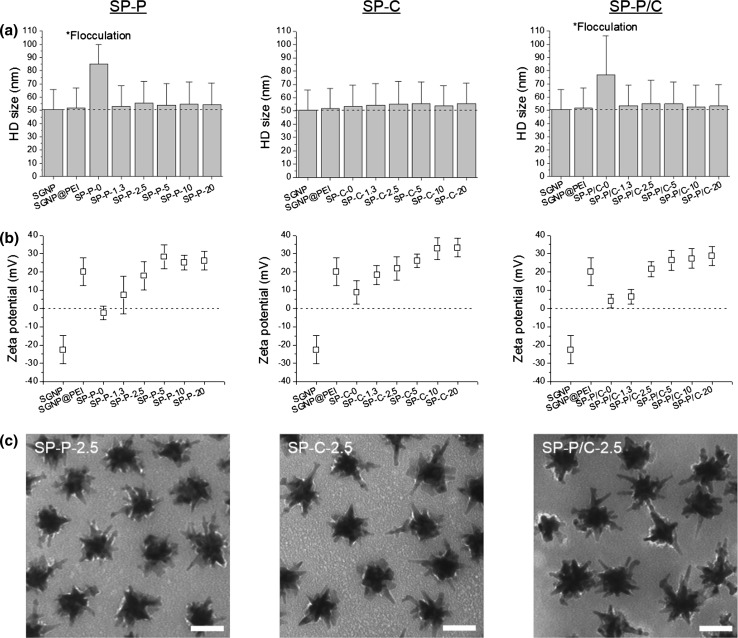
The complexation process was monitored by measuring their hydrodynamic (HD) size and zeta potential throughout the assembly processes (Figs. 2a, 2b). The HD size and zeta potential of the “bare” SGNPs were 50.6 (± 15.2) nm and −22.6 (± 7.7) mV, respectively. The HD size was well correlated to the actual size measured by TEM in Fig. 1d and indicated singly dispersed particles. The anionic zeta potential of SGNPs allowed for electrostatic assembly with cationic PEI. After coating SGNPs with PEI, the anionic surface charge was converted to 20.2 (± 7.5) mV, which facilitated electrostatic complexation with pIC and CpG. Complexation with pIC caused a large increase in HD size (>30 nm) for both SP-P and SP-P/C, suggesting flocculation due to strong inter-particle charge interaction between pIC and PEI (Fig. 2a). However, the final PEG-PEI treatment reversed the flocculation, reducing the HD size to a level similar to that of well-dispersed SGNPs, suggesting that the PEG layer reduced inter-particle interaction by providing steric barrier. In contrast, CpG complexation did not cause flocculation, and SP-C remained stable in size regardless of the PEG-PEI treatment. The HD size increased slightly with the addition of adjuvant(s) and the PEG-PEI layer (Fig. 2a). Upon complexation with pIC and/or CpG, the positive charge of SGNP@PEI was dropped to the range of ± 10 mV (Fig. 2b). The subsequent PEG-PEI coating restored the cationic surface charge, which stabilized SP-P and SP-P/C from flocculation. Their zeta potential gradually increased with higher weight ratio of PEG-PEI and reached a plateau at 5 for all complexes, indicating that complete PEG-PEI coating was achieved at the weight ratio of 2.5–5. To further verify successful complexation, SGNP complexes formulated with the PEG-PEI weight ratio 2.5 were negatively stained with 2% uranyl acetate and examined under TEM (Fig. 2c). The TEM images clearly showed a thin adjuvant-PEG-PEI complex layer surrounding SGNPs as a white shade. It is also noted that SGNP complexes were well separated individually. This confirms substantial loading and stable complexation of adjuvants using our layer-by-layer approach.
Figure 2.
Characterization of adjuvant-loaded SGNPs. HD size (a) and zeta potential (b) of SGNP complexes incorporated with pIC (SP-P), CpG (SP-C), or pIC/CpG (SP-P/C). The number after the abbreviation of adjuvants denotes the weight ratio of PEG-PEI relative to adjuvants. The dashed line signifies the HD size of “bare” SGNPs (a) and zeta potential value of zero (b). Representative TEM images of SGNP complexes formulated at the PEG-PEI weight ratio of 2.5 (c). TEM images were taken after 2% uranyl acetate staining. Scale bars = 50 nm.
Quantification Method for pIC and CpG
We have developed a method for quantifying the amount and loading efficiencies of pIC and CpG on SGNPs. When measured by UV–Vis absorption, both pIC and CpG in free forms exhibited a strong concentration-dependent absorption peak at ~260 nm (Figs. 3a, 3c). In particular, pIC had two distinctive peaks at ~245 and ~265 nm, corresponding to inosinic and cytidylic strands, respectively.7,48 We further developed a method to identify pIC and CpG simultaneously in a mixture solution, which could not be achieved by the UV–Vis absorption measurement alone because of their overlapping spectra. For this, we exploited the large differences between the molecular weights of pIC and CpG; pIC used in our studies has 1500–8000 base pairs whereas CpG has only 20 single bases (5′-tccatgacgttcctgacgtt-3′), resulting in >100-fold higher molecular weight for pIC, compared with CpG. Gel permeation chromatography (GPC) was performed for size-based separation of pIC and CpG, while the concentration was determined by measuring the absorbance at 260 nm (Figs. 3b, 3d). Differences in the elution time were large enough to identify pIC at ~7.7 min and CpG ~13.8 min, respectively, without any cross talk. pIC exhibited additional small peaks at ~15.7 min, probably due to residual amount of impurities. However, their peak intensity was negligible, accounting for just ~3% of all peaks, and the peak position did not overlap with that of CpG. The integrated peak areas were linearly proportional to the concentrations of pIC and CpG with the coefficient of determination higher than 0.9999. Together with the UV–Vis absorbance-based concentration measurement, our GPC-based method allows for accurate identification and quantification of pIC and CpG simultaneously from their mixture solution.
Figure 3.
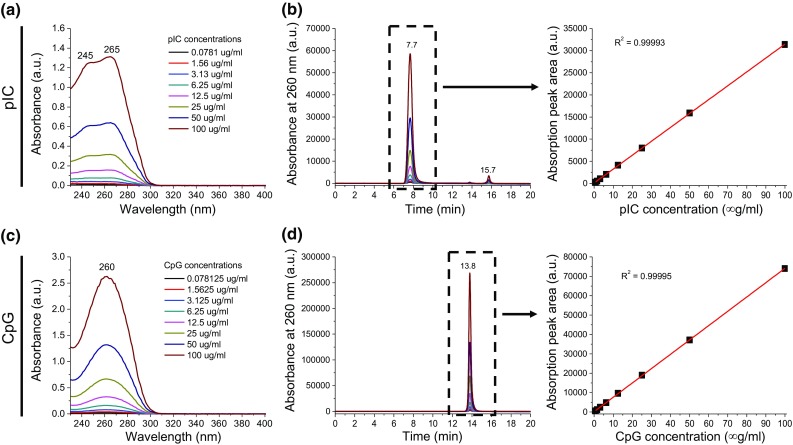
Quantification of dual pIC and CpG adjuvants. pIC and CpG were identified and quantified accurately by absorption measurement and GPC. UV–Vis absorption spectra (a, c) and GPC spectra acquired based on the absorbance at 260 nm (b, d) for pIC (a, b) and CpG (c, d). The linear fitting of absorption peak area vs. concentration was obtained from selected peaks in GPC.
Determination of Adjuvant Loading on SGNP Nano-Complexes
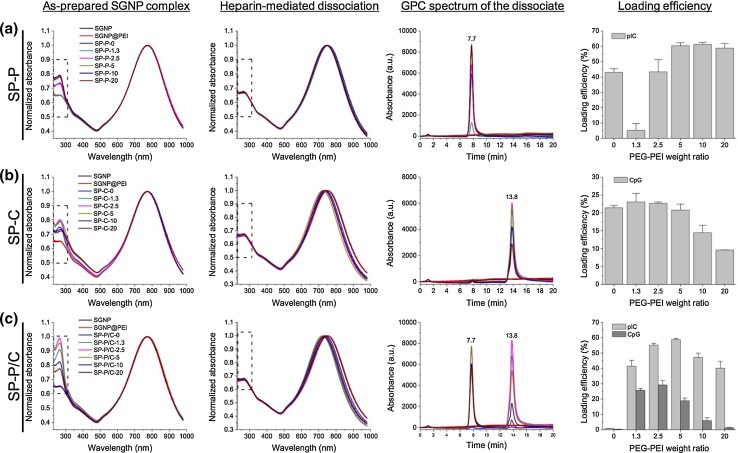
Adjuvant loading was confirmed using the UV–Vis absorbance and GPC methods as described above. As-prepared adjuvant-SGNP complexes showed an increased absorption peak at ~260 nm, compared with “bare” SGNPs and SGNP@PEI due to the presence of adjuvants (Fig. 4, the first column, boxed area). We note that uncomplexed free adjuvants were removed from SGNP complexes by centrifugation (3000×g) and washing. SP-P/C showed a larger increase in their absorption peak at ~260 nm, compared with SP-P and SP-C, due to the loading of the dual adjuvants. The adjuvant loading efficiency was also dependent on the weight ratio of PEG-PEI, indicating that PEG-PEI contributed to the complexation. On the other hand, there were no noticeable changes in the absorption peak of the core SGNPs at ~780 nm, suggesting that the adjuvant complexation process did not directly affect the physicochemical properties of the base SGNPs.
Figure 4.
Characterization of pIC and CpG loading on SGNPs with UV–Vis absorbance and GPC. UV–Vis absorption spectra of SGNP complexes were measured before (first column) and after (second column) heparin-mediated release of the adjuvants from SGNPs. Adjuvants released from SGNP complexes were analyzed using GPC (third column), and the concentrations of pIC and CpG and their loading efficiencies were calculated from the standard curve of GPC absorption peak area vs. concentration (forth column). The values are reported as mean ± SD with n = 2–4. SP-P (a), SP-C (b), or SP-P/C (c) were analyzed separately. The values on the x-axes in the bar graphs (fourth column) indicate the weight ratio of PEG-PEI relative to the adjuvants.
For more quantitative assessment of adjuvant loading efficiency, we released surface-adsorbed pIC and CpG from SGNPs by heparin sulfate treatment and sonication. The increased absorption peaks that resulted from adjuvant loading completely disappeared after this heparin treatment, confirming complete release of adjuvants from SGNPs (Fig. 4, the second column, boxed area). The sonication step facilitated heparin-mediated adjuvant dissociation from SGNPs not only by physically disturbing the complexation but also by mildly heating the solution, thereby deforming the unstable branched structures and causing slight blueshift of the SGNP absorption peaks.8 The released adjuvants were separated from SGNPs and quantified using the GPC-based quantification developed above (Fig. 4, the third column). SP-P and SC-C showed intense peaks corresponding to pIC and CpG, respectively (Figs. 4a, 4b, the third column), while SP-P/C showed two clearly separated peaks, each corresponding to pIC and CpG (Fig. 4c, third column). The peak positions of pIC and CpG released from SGNP complexes were identical to those of free adjuvants in Fig. 3, indicating that the complexation and heparin-mediated dissociation did not compromise the structural integrity of the adjuvants. “Bare” SGNPs and SGNP@PEI did not exhibit noticeable peaks in the elution time range, confirming the indicated peaks are wholly from adjuvants released from SGNP complexes.
The loading efficiency was obtained for pIC and CpG by calculating their concentrations based on the standard curve of peak area vs. concentration shown in Fig. 3. For SP-P, the loading efficiency of pIC was generally increased with the weight ratio of PEG-PEI, reaching a plateau at 5. The only exception was found at the ratio of 1.3, at which PEG-PEI seemed to disturb the complexation (Fig. 4a, the forth column). For SP-C, the loading efficiency of CpG was unaffected or only marginally increased with PEG-PEI at the weight ratio of 1.3–5 and gradually decreased thereafter at higher ratio (Fig. 4b, forth column). In contrast, efficient co-loading of pIC and CpG on SP-P/C required the PEG-PEI treatment; there was almost no adjuvant loading without the PEG-PEI (Fig. 4c, forth column). With the PEG-PEI treatment at various ratios, the pIC loading efficiency was generally maintained at a substantial level, and its maximum loading efficiency was ~60% at the weight ratio of 5. On the other hand, the CpG loading efficiency was heavily governed by the PEG-PEI ratio, reaching the highest level at ~30% with the PEG-PEI weight ratio of 2.5. Similar to the cases of single adjuvant loading (SP-P and SP-C), high concentration of PEG-PEI disturbed complexation of CpG whereas pIC complexation remains robust. This difference is thought to be from different degrees of charge interaction for each adjuvant. pIC possesses thousands of anionic charges per molecule, thus mediating strong complexation with PEG-PEI. In contrast, CpG has only ~20 charges per molecule capable of weaker charge interactions, leading to disruption and loss of CpG from SGNPs at high concentration of PEG-PEI. The structure of adjuvants may also affect the complexation strength as PEG-PEI is known to preferentially complex with supercoiled DNA rather than linearized DNA.6 The outer PEG-PEI layer may also improve the in vivo stability of adjuvant-SGNP complexes by forming compact oligonucleotide condensate,49,50 while posing negligible cytotoxicity both in vitro and in vivo.31,47,59 Based on these results, the PEG-PEI weight ratio of 2.5 was used to prepare SP-P, SP-C, and SP-P/C complexes for subsequent studies since it achieved complete passivation with reasonable loading of both pIC and CpG.
Intracellular Distribution of Adjuvants
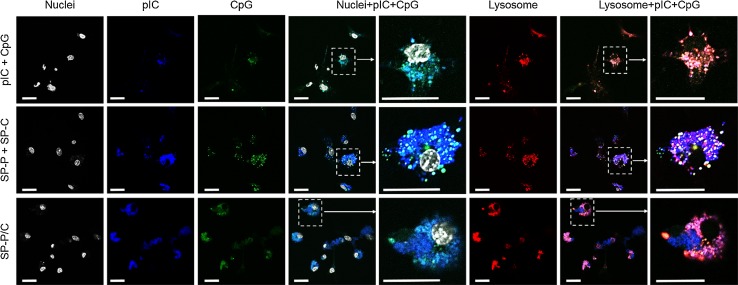
We monitored intracellular distribution of adjuvants either as a free form or after complexation with SGNPs by utilizing pIC and CpG labeled with Alexa Fluor® 647 and Alexa Fluor® 488, respectively (Fig. 5, as detailed in the Materials and Method section). The fluorophore-conjugated adjuvants were complexed with SGNPs as described above. The cell experiments were conducted using bone marrow-derived dendritic cells (BMDCs) to study the effects of adjuvants and their SGNP complexes on innate immune responses. BMDCs were incubated with pIC + CpG (free adjuvant combination), SP-P + SP-C (admixture of single SGNP complex), or SP-P/C (combinational SGNP complex) with equivalent concentrations of adjuvants at 1 μg/ml CpG and 1.8 μg/ml pIC. After 24 h, cells were stained for nuclei and lysosomes, fixed with 4% formaldehyde, and visualized with confocal microscopy. Free pIC + CpG exhibited only dim intracellular fluorescence (Fig. 5, first row), whereas SGNP complexes (SP-P + SP-C and SP-P/C) showed much brighter fluorescence, indicating that the complexation promoted cellular uptake of the adjuvants (Fig. 5, second and third row). This may be attributed to the positive surface charge of SGNP complexes, which allows favorable adsorption on negatively charged cellular membrane and facilitates subsequent cellular uptake.18,42,44,45 The nano-spike structures of SGNPs may also play a role by increasing the contact area with cellular membrane and promoting particle uptake via phagocytosis.22
Figure 5.
Intracellular distribution of adjuvants as visualized by confocal microscopy. BMDCs were incubated with pIC + CpG, SP-P + SP-C, or SP-P/C, all prepared using fluorophore-conjugated pIC and CpG. Nuclei and lysosomes were stained using Hoechst and lysotracker, respectively. Selected regions as indicated by the dashed boxes were magnified to more clearly visualize the distribution and co-localization of pIC and CpG components. Scale bars = 20 µm.
Although the extent of cellular uptake was similar for BMDCs treated with either SP-P + SP-C or SP-P/C, they showed a stark difference in intracellular distribution of adjuvants. Following the SP-P + SP-C co-treatment, pIC and CpG were distributed separately with limited co-localization, whereas significant overlap was observed in BMDCs treated with SP-P/C (Fig. 5, forth and fifth columns). We also examined localization of adjuvants in endolysosomes (Fig. 5, last two columns). Free pIC + CpG were found in endolysosomes, whereas SGNP complexes showed endolysosomal co-localization of the adjuvants as well as a portion of adjuvants localized in the cytosol with substantial fluorescence signal from pIC found in the cytosol for SP-P + SP-C and SP-P/C. The cytosolic localization may have resulted from the buffering capacity of PEI, which is known to mediate endosomal escape and cytosolic drug delivery by inducing swelling and rupturing of endolysosomes.1,65 Overall, these results demonstrate that adjuvant-loaded SGNP complexes can achieve spatio-temporal control over the combinational adjuvant delivery while enabling cytosolic delivery of adjuvants.
Activation and Maturation of BMDCs
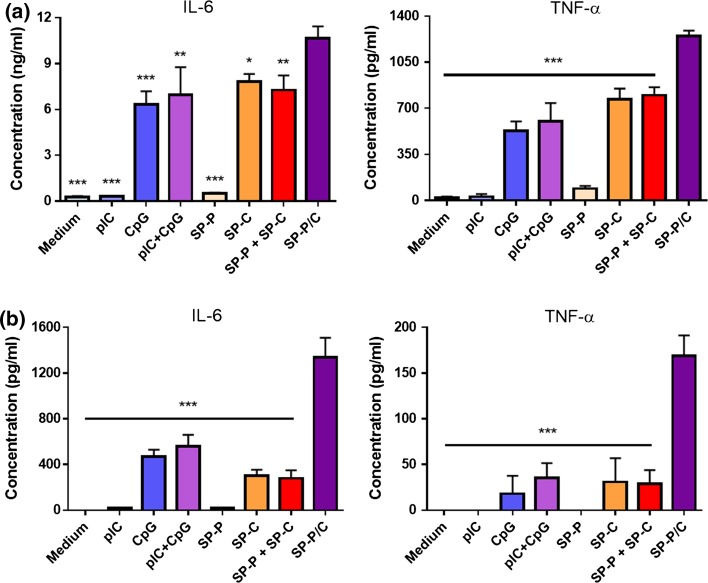
We studied activation and maturation of BMDCs after incubation of BMDCs with free adjuvants or SGNP complexes, followed by analyses of cytokine release and surface marker expression. SGNP complexes containing dual adjuvants (SP-P/C) were compared with single adjuvant complexes treated either separately (SP-P or SP-C) or as an admixture (SP-P + SP-C). Free adjuvants (pIC, CpG, and pIC + CpG) were also used as control samples for the SGNP complexes. To examine BMDC activation, the samples were incubated with BMDCs for 2 h at concentrations of 1 μg/ml CpG and 1.8 μg/ml pIC, and the release of pro-inflammatory cytokines (IL-6 and TNF-α) was measured after 24 h (Fig. 6a). We found that CpG triggered substantial cytokine release, either as a free form or after complexation with SGNPs. In contrast, cytokine release was hardly detected with either form of pIC, indicating sub-optimal concentration of pIC for immune activation.34,46,62 BMDCs incubated with free pIC + CpG showed slightly higher cytokine levels than those exposed to free CpG, but the increase was only marginal. SP-P + SP-C did not further increase cytokine release over SP-C. In contrast, SP-P/C significantly increased release of IL-6 and TNF-α, compared with any of the treatment groups. We did not detect any cytokine release from BMDCs incubated with plain SGNPs without any adjuvants (data not shown). Overall, these results suggest synergistic BMDC activation by SP-P/C-mediated co-delivery and potentiation of pIC and CpG. It is also noted that SP-C or SP-P + SP-C induced slightly higher levels of cytokine release than the respective free adjuvant samples, probably by enhancing cellular uptake as demonstrated in Fig. 5.
Figure 6.
Cytokine release from BMDCs treated with free adjuvants or adjuvant-SGNP complexes. Release of IL-6 and TNF-α from BMDCS treated with the indicated samples at concentrations of 1 μg/ml CpG and 1.8 μg/ml pIC (a), or 0.1 μg/ml CpG and 0.18 μg/ml pIC (b). The data show mean ± SD of representative results (n = 3) from 2 to 4 independent experiments. Statistical significance was analyzed by one-way ANOVA with post hoc Tukey’s HSD test and reported with respect to the SP-P/C group (*p < 0.05, **p < 0.01, ***p < 0.001).
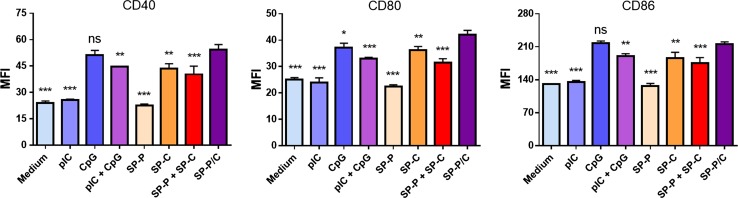
Next BMDC activation was further tested at tenfold lower sample concentrations (0.1 μg/ml CpG and 0.18 μg/ml pIC) (Fig. 6b). Our aim in this study was to validate the synergistic effect of SP-P/C at this low adjuvant concentration by minimizing the concentration-dependent diffusional overlap of separate adjuvants. Indeed, release of IL-6 and TNF-α were generally reduced by ~ tenfold due to the low adjuvant concentrations. However, SP-P/C treatment triggered robust release of IL-6 and TNF-α, with even greater relative differences from other control groups than those observed at higher adjuvant concentration (Fig. 6a). BMDC activation was accompanied by upregulation of co-stimulatory markers, including CD40, CD80, and CD86 (Fig. 7). Consistent with what we observed in the cytokine release assay, SP-P/C increased expression of co-stimulatory markers, compared with other control samples (with the exception of free CpG).
Figure 7.
Maturation of BMDCs treated with free adjuvants or adjuvant-SGNP complexes. Up-regulation of DC maturation markers was measured for CD40, CD80, and CD86 among BMDCs treated with the indicated samples at concentration of 0.1 μg/ml CpG and 0.18 μg/ml pIC. The data show mean ± SD of representative results (n = 3) from 2 to 4 independent experiments. Statistical significance was analyzed by one-way ANOVA with post hoc Tukey’s HSD test and reported with respect to the SP-P/C group (ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001).
Overall, these results demonstrate that SP-P/C can mediate spatio-temporally concerted delivery of combinational adjuvants, thereby promoting synergistic stimulation and maturation of innate immune cells. Our results showed that CpG induced immune stimulation of BMDCs whereas only negligible stimulation was found for pIC at the concentration range tested (0.1–1 μg/ml for CpG and 0.18–1.8 μg/ml for pIC), indicating sub-optimal concentration of pIC for immune activation by itself (Fig. 6).34,46,62 However, when they were co-formulated on SGNPs, pIC greatly enhanced the immunostimulatory effect of CpG (Figs. 6, 7). Previous reports have shown synergistic immune stimulation by the combination of free pIC and CpG, but high concentration of pIC (>10 μg/ml) was required.3,4,34,62,70 Compared with previous studies, our SP-P/C achieved increased immunostimulatory efficacy at much lower concentration of pIC. We speculate that such synergistic immune activation is attributed to SGNP-mediated co-delivery of pIC and CpG to the same cellular compartments (Fig. 5). In contrast, when the adjuvants were co-treated as an admixture either in free pIC + CpG form or SP-C + SP-P nano-complexes, the adjuvants were distributed separately with limited cellular co-localization due to poor spatial and temporal coordination of their uptake, leading to decreased cytokine release and maturation of BMDCs (Figs. 5, 6, 7). In addition to endosomal TLR3 and TLR9 (i.e. the canonical receptors for pIC and CpG, respectively), pIC is known to be detected by cytosolic dsRNA pattern recognition receptors, such as retinoic-inducible gene-1 (RIG-1) and melanoma differentiation-associated gene 5 (MDA5), which are RNA helicases that detect viral RNA species in the cytoplasm and induce type I interferon for viral clearance.28,29,66 Thus, SGNP-mediated adjuvant delivery may trigger activation of multiple receptors, potentially broadening the breadth and strength of innate immune responses. We are currently working to delineate the mechanism underlying the adjuvant-receptor interactions and how sub-cellular localization of adjuvant(s) mediated by SGNPs vs. plain spherical gold nanoparticles affects innate and adaptive immune responses. In addition, SGNPs with strong near-infrared (NIR) SPR characteristics may be employed in NIR-based imaging and therapy,19,32 potentially providing a versatile platform for theranostic applications in vaccines and immunotherapies.
Conclusions
In this study, we have developed SGNPs as a platform for efficient intracellular delivery of immunostimulatory agents. We prepared SGNPs by a seed-mediated, surfactant-free synthesis method, followed by surface-decoration with pIC/CpG molecules assembled via a layer-by-layer approach. As pIC and CpG are synthetic analogues of PAMPs, SGNPs complexed with pIC and/or CpG mimic certain aspects of immune activation by microbes. We have demonstrated that the combinational SP-P/C can co-deliver both adjuvants into BMDCs in a spatio-temporally concerted manner, leading to synergistic enhancement in immune stimulation and allowing for dose-sparing of adjuvants, whereas free soluble adjuvants or admixture of individually loaded adjuvant-SGNPs failed to achieve simultaneous, concerted delivery of adjuvants to the same sub-cellular compartments. In summary, SGNPs serve as a versatile delivery platform that allows flexible and on-demand cargo fabrication for strong activation of innate immune cells.
Acknowledgments
This work was supported in part by NIH R01-AI127070, NIH R01-EB022563, NIH R01-CA210273, MTRAC for Life Sciences Hub, Emerald Foundation, and University of Michigan Comprehensive Cancer Center’s Forbes Institute for Cancer Discovery. J.J.M. is a Young Investigator supported by the Melanoma Research Alliance (348774), NSF CAREER Award (1553831), DoD/CDMRP Peer Reviewed Cancer Research Program (W81XWH-16-1-0369), and Emerald Foundation. Opinions interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.
Conflict of interest
Jutaek Nam, Sejin Son, and James J. Moon declare no conflicts of interest.
Ethical Standards
No animal or human studies were performed in this work.
Abbreviations
- BMDC
Bone-marrow derived dendritic cell
- CLR
C-type lectin receptor
- CpG
Oligonucleotide containing unmethylated CpG motif
- DMSO
Dimethyl sulfoxide
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- GNP
Gold nanoparticle
- GPC
Gel permeation chromatography
- MDA5
Melanoma differentiation-associated gene 5
- MeIm
Methyl imidazole
- Methoxy-PEG-NHS
Methoxy poly(ethyleneglycol) propionic acid N-hydroxysuccinimide
- NIR
Near-infrared
- NLR
NOD-like receptor
- NP
Nanoparticle
- PAMP
Pathogen-associated molecular pattern
- PEG
Poly(ethyleneglycol)
- PEG-PEI
PEG-grafted PEI
- PEI
Poly(ethyleneimine)
- pIC
Polyinosinic-polycytidylic acid
- PRR
Pattern-recognition receptor
- RIG-1
Retinoic-inducible gene-1
- RLR
Retinoic acid-inducible gene (RIG)-I-like receptor
- SGNP
Spiky gold nanoparticle
- SGNP@PEI
SGNP coated with PEI
- SP-C
SGNP nano-complex layered with CpG
- SP-P
SGNP nano-complex layered with pIC
- SP-P/C
SGNP nano-complex layered with pIC and CpG
- SPR
Surface plasmon resonance
- TEM
Transmission electron microscope
- TLR
Toll-like receptor
Footnotes
Dr. James Moon
is John Gideon Searle Assistant Professor in the Department of Pharmaceutical Sciences and Biomedical Engineering at the University of Michigan, Ann Arbor. His translational research program aims to develop novel engineering tools for improving vaccines and immunotherapies. His work has been published in Nature Materials, Nature Medicine, PNAS, ACS Nano, and Sci Transl Med, and led to two new biotech companies, Vedantra Pharmaceuticals (Cambridge, MA) and EVOQ Therapeutics (Ann Arbor, MI) that focus on clinical translation of new nano-vaccine technologies. Dr. Moon has received numerous awards, including 2017 Emerald Foundation Distinguished Investigator Award, 2016 National Science Foundation CAREER Award, 2016 DOD-CDMRP Career Development Award, and 2015 Melanoma Research Alliance Young Investigator Award. Dr. Moon received his B.S. in Bioengineering from University of California, Berkeley, obtained his Ph.D. in Bioengineering from Rice University with Prof. Jennifer West, and completed his postdoctoral training with Prof. Darrell Irvine at MIT.
This article is part of the 2017 CMBE Young Innovators special issue.
References
- 1.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 3.Arsenault RJ, Kogut MH, He H. Combined CpG and poly I: C stimulation of monocytes results in unique signaling activation not observed with the individual ligands. Cell. Signal. 2013;25:2246–2254. doi: 10.1016/j.cellsig.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin H-S, et al. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J. Immunol. 2007;178:1164–1171. doi: 10.4049/jimmunol.178.2.1164. [DOI] [PubMed] [Google Scholar]
- 5.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronich T, Kabanov AV, Marky LA. A thermodynamic characterization of the interaction of a cationic copolymer with DNA. J. Phys. Chem. B. 2001;105:6042–6050. doi: 10.1021/jp004395k. [DOI] [Google Scholar]
- 7.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13–e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng L-C, Huang J-H, Chen HM, Lai T-C, Yang K-Y, et al. Seedless, silver-induced synthesis of star-shaped gold/silver bimetallic nanoparticles as high efficiency photothermal therapy reagent. J. Mater. Chem. 2012;22:2244–2253. doi: 10.1039/C1JM13937A. [DOI] [Google Scholar]
- 9.Cong Y-P, Song SS, Bhagat L, Pandey RK, Yu D, et al. Self-stabilized CpG DNAs optimally activate human B cells and plasmacytoid dendritic cells. Biochem. Biophys. Res. Commun. 2003;310:1133–1139. doi: 10.1016/j.bbrc.2003.09.134. [DOI] [PubMed] [Google Scholar]
- 10.DeMuth PC, Min Y, Huang B, Kramer JA, Miller AD, et al. Polymer multilayer tattooing for enhanced DNA vaccination. Nat. Mater. 2013;12:367–376. doi: 10.1038/nmat3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMuth PC, Min Y, Irvine DJ, Hammond PT. Implantable silk composite microneedles for programmable vaccine release kinetics and enhanced immunogenicity in transcutaneous immunization. Adv. Healthcare Mater. 2014;3:47–58. doi: 10.1002/adhm.201300139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Geest BG, Willart MA, Hammad H, Lambrecht BN, Pollard C, et al. Polymeric multilayer capsule-mediated vaccination induces protective immunity against cancer and viral infection. ACS Nano. 2012;6:2136–2149. doi: 10.1021/nn205099c. [DOI] [PubMed] [Google Scholar]
- 13.de Titta A, Ballester M, Julier Z, Nembrini C, Jeanbart L, et al. Nanoparticle conjugation of CpG enhances adjuvancy for cellular immunity and memory recall at low dose. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19902–19907. doi: 10.1073/pnas.1313152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. The golden age: gold nanoparticles for biomedicine. Chem. Soc. Rev. 2012;41:2740–2779. doi: 10.1039/C1CS15237H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel AL, Holt GE, Lu H. The pharmacokinetics of Toll-like receptor agonists and the impact on the immune system. Expert Rev. Clin. Pharmacol. 2011;4:275–289. doi: 10.1586/ecp.11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat. Phys. Sci. 1973;241:20–22. doi: 10.1038/physci241020a0. [DOI] [Google Scholar]
- 17.Ghosh P, Han G, De M, Kim CK, Rotello VM. Gold nanoparticles in delivery applications. Adv. Drug Deliv. Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl. Acad. Sci. USA. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen S, Huang R, Wall MA, Karabeber H, Samii JM, et al. Surface-enhanced resonance Raman scattering nanostars for high-precision cancer imaging. Sci. Transl. Med. 2015;7:271ra277–271ra277. doi: 10.1126/scitranslmed.3010633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiangkuo Y, Christopher GK, Hanjun H, Christy MW, Gerald AG, et al. Gold nanostars: surfactant-free synthesis, 3D modelling, and two-photon photoluminescence imaging. Nanotechnology. 2012;23:075102. doi: 10.1088/0957-4484/23/7/075102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbell JA, Thomas SN, Swartz MA. Materials engineering for immunomodulation. Nature. 2009;462:449–460. doi: 10.1038/nature08604. [DOI] [PubMed] [Google Scholar]
- 22.Hutter E, Boridy S, Labrecque S, Lalancette-Hébert M, Kriz J, et al. Microglial response to gold nanoparticles. ACS Nano. 2010;4:2595–2606. doi: 10.1021/nn901869f. [DOI] [PubMed] [Google Scholar]
- 23.Kaczanowska S, Joseph AM, Davila E. TLR agonists: our best frenemy in cancer immunotherapy. J. Leukoc. Biol. 2013;93:847–863. doi: 10.1189/jlb.1012501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandimalla ER, Bhagat L, Cong Y-P, Pandey RK, Yu D, et al. Secondary structures in CpG oligonucleotides affect immunostimulatory activity. Biochem. Biophys. Res. Commun. 2003;306:948–953. doi: 10.1016/S0006-291X(03)01080-5. [DOI] [PubMed] [Google Scholar]
- 25.Kandimalla ER, Bhagat L, Yu D, Cong Y, Tang J, et al. Conjugation of ligands at the 5′-End of CpG DNA affects immunostimulatory activity. Bioconjugate Chem. 2002;13:966–974. doi: 10.1021/bc0200374. [DOI] [PubMed] [Google Scholar]
- 26.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat. Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 27.Kariko K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: Implication for therapeutic RNA development. Curr. Opin. Drug. Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 28.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 30.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Kichler A, Chillon M, Leborgne C, Danos O, Frisch B. Intranasal gene delivery with a polyethylenimine–PEG conjugate. J. Control. Release. 2002;81:379–388. doi: 10.1016/S0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 32.Kim C, Song H-M, Cai X, Yao J, Wei A, et al. In vivo photoacoustic mapping of lymphatic systems with plasmon-resonant nanostars. J. Mater. Chem. 2011;21:2841–2844. doi: 10.1039/c0jm04194g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 34.Krummen M, Balkow S, Shen L, Heinz S, Loquai C, et al. Release of IL-12 by dendritic cells activated by TLR ligation is dependent on MyD88 signaling, whereas TRIF signaling is indispensable for TLR synergy. J. Leukoc. Biol. 2010;88:189–199. doi: 10.1189/jlb.0408228. [DOI] [PubMed] [Google Scholar]
- 35.Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- 36.Lee I-H, Kwon H-K, An S, Kim D, Kim S, et al. Imageable antigen-presenting gold nanoparticle vaccines for effective cancer immunotherapy in vivo. Angew. Chem. Int. Ed. 2012;51:8800–8805. doi: 10.1002/anie.201203193. [DOI] [PubMed] [Google Scholar]
- 37.Leleux J, Roy K. Micro and nanoparticle-based delivery systems for vaccine immunotherapy: an immunological and materials perspective. Adv. Healthcare Mater. 2013;2:72–94. doi: 10.1002/adhm.201200268. [DOI] [PubMed] [Google Scholar]
- 38.Lin AY, Mattos Almeida JP, Bear A, Liu N, Luo L, et al. Gold nanoparticle delivery of modified CpG stimulates macrophages and inhibits tumor growth for enhanced immunotherapy. PLoS ONE. 2013;8:e63550. doi: 10.1371/journal.pone.0063550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Ding JL. The molecular mechanisms of TLR-signaling cooperation in cytokine regulation. Immunol. Cell Biol. 2016;94:538–542. doi: 10.1038/icb.2016.18. [DOI] [PubMed] [Google Scholar]
- 40.Lungwitz U, Breunig M, Blunk T, Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Lutz MB, Kukutsch N, Ogilvie ALJ, Rößner S, Koch F, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods. 1999;223:77–92. doi: 10.1016/S0022-1759(98)00204-X. [DOI] [PubMed] [Google Scholar]
- 42.Makino K, Yamamoto N, Higuchi K, Harada N, Ohshima H, et al. Phagocytic uptake of polystyrene microspheres by alveolar macrophages: effects of the size and surface properties of the microspheres. Colloids Surf. 2003;27:33–39. doi: 10.1016/S0927-7765(02)00042-5. [DOI] [Google Scholar]
- 43.McGettrick AF, O’Neill LAJ. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr. Opin. Immunol. 2010;22:20–27. doi: 10.1016/j.coi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc. Natl. Acad. Sci. USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mutsaers SE, Papadimitriou JM. Surface charge of macrophages and their interaction with charged particles. J. Leukoc. Biol. 1988;44:17–26. doi: 10.1002/jlb.44.1.17. [DOI] [PubMed] [Google Scholar]
- 46.Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 48.Parker-Esquivel B, Flores KJ, Louiselle D, Craig M, Dong L, et al. Association of poly I: C RNA and plasmid DNA onto MnO nanorods mediated by PAMAM. Langmuir. 2012;28:3860–3870. doi: 10.1021/la203998r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen H, Fechner PM, Fischer D, Kissel T. Synthesis, characterization, and biocompatibility of polyethylenimine-graft-poly(ethylene glycol) block copolymers. Macromolecules. 2002;35:6867–6874. doi: 10.1021/ma012060a. [DOI] [Google Scholar]
- 50.Petersen H, Fechner PM, Martin AL, Kunath K, Stolnik S, et al. Polyethylenimine-graft-poly(ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjugate Chem. 2002;13:845–854. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- 51.Sexton A, Whitney PG, Chong S-F, Zelikin AN, Johnston APR, et al. A protective vaccine delivery system for in vivo T cell stimulation using nanoengineered polymer hydrogel capsules. ACS Nano. 2009;3:3391–3400. doi: 10.1021/nn900715g. [DOI] [PubMed] [Google Scholar]
- 52.Shukoor MI, Natalio F, Ksenofontov V, Tahir MN, Eberhardt M, et al. Double-stranded RNA polyinosinic-polycytidylic acid immobilized onto γ-Fe2O3 nanoparticles by using a multifunctional polymeric linker. Small. 2007;3:1374–1378. doi: 10.1002/smll.200600664. [DOI] [PubMed] [Google Scholar]
- 53.Shukoor MI, Natalio F, Metz N, Glube N, Tahir MN, et al. dsRNA-functionalized multifunctional γ-Fe2O3 nanocrystals: a tool for targeting cell surface receptors. Angew. Chem. Int. Ed. 2008;47:4748–4752. doi: 10.1002/anie.200704735. [DOI] [PubMed] [Google Scholar]
- 54.Sloat BR, Cui Z. Nasal immunization with anthrax protective antigen protein adjuvanted with polyriboinosinic-polyribocytidylic acid induced strong mucosal and systemic immunities. Pharm. Res. 2006;23:1217–1226. doi: 10.1007/s11095-006-0206-9. [DOI] [PubMed] [Google Scholar]
- 55.Sparwasser T, Miethke T, Lipford G, Borschert K, Hacker H, et al. Bacterial DNA causes septic shock. Nature. 1997;386:336–337. doi: 10.1038/386336a0. [DOI] [PubMed] [Google Scholar]
- 56.Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-based immune adjuvants. Vaccine. 2011;29:3341–3355. doi: 10.1016/j.vaccine.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeshita F, Gursel I, Ishii KJ, Suzuki K, Gursel M, et al. Signal transduction pathways mediated by the interaction of CpG DNA with Toll-like receptor 9. Semin. Immunol. 2004;16:17–22. doi: 10.1016/j.smim.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 59.Tang GP, Zeng JM, Gao SJ, Ma YX, Shi L, et al. Polyethylene glycol modified polyethylenimine for improved CNS gene transfer: effects of PEGylation extent. Biomaterials. 2003;24:2351–2362. doi: 10.1016/S0142-9612(03)00029-2. [DOI] [PubMed] [Google Scholar]
- 60.Tao Y, Ju E, Li Z, Ren J, Qu X. Engineered CpG-antigen conjugates protected gold nanoclusters as smart self-vaccines for enhanced immune response and cell imaging. Adv. Funct. Mater. 2014;24:1004–1010. doi: 10.1002/adfm.201302347. [DOI] [Google Scholar]
- 61.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Drug Discov. 2007;7:179–190. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 62.Tross D, Petrenko L, Klaschik S, Zhu Q, Klinman DM. Global changes in gene expression and synergistic interactions induced by TLR9 and TLR3. Mol. Immunol. 2009;46:2557–2564. doi: 10.1016/j.molimm.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trumpfheller C, Caskey M, Nchinda G, Longhi MP, Mizenina O, et al. The microbial mimic poly IC induces durable and protective CD4 + T cell immunity together with a dendritic cell targeted vaccine. Proc. Natl. Acad. Sci. USA. 2008;105:2574–2579. doi: 10.1073/pnas.0711976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei M, Chen N, Li J, Yin M, Liang L, et al. Polyvalent immunostimulatory nanoagents with self-assembled CpG oligonucleotide-conjugated gold nanoparticles. Angew. Chem. Int. Ed. 2012;51:1202–1206. doi: 10.1002/anie.201105187. [DOI] [PubMed] [Google Scholar]
- 65.Yang S, May S. Release of cationic polymer-DNA complexes from the endosome: a theoretical investigation of the proton sponge hypothesis. J. Chem. Phys. 2008;129:185105. doi: 10.1063/1.3009263. [DOI] [PubMed] [Google Scholar]
- 66.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 67.Yuan H, Khoury CG, Hwang H, Wilson CM, Grant GA, et al. Gold nanostars: surfactant-free synthesis, 3D modelling, and two-photon photoluminescence imaging. Nanotechnology. 2012;23:075102. doi: 10.1088/0957-4484/23/7/075102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng Q, Gammon JM, Tostanoski LH, Chiu Y-C, Jewell CM. In vivo expansion of melanoma-specific T cells using microneedle arrays coated with immune-polyelectrolyte multilayers. ACS Biomaterials Science & Engineering. 2017;3:195–205. doi: 10.1021/acsbiomaterials.6b00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang P, Chiu Y-C, Tostanoski LH, Jewell CM. Polyelectrolyte multilayers assembled entirely from immune signals on gold nanoparticle templates promote antigen-specific T cell response. ACS Nano. 2015;9:6465–6477. doi: 10.1021/acsnano.5b02153. [DOI] [PubMed] [Google Scholar]
- 70.Zhu Q, Egelston C, Vivekanandhan A, Uematsu S, Akira S, et al. Toll-like receptor ligands synergize through distinct dendritic cell pathways to induce T cell responses: Implications for vaccines. Proc. Natl. Acad. Sci. USA. 2008;105:16260–16265. doi: 10.1073/pnas.0805325105. [DOI] [PMC free article] [PubMed] [Google Scholar]