Abstract
Hereditary pulmonary alveolar proteinosis (hPAP) is a rare disorder of pulmonary surfactant accumulation and hypoxemic respiratory failure caused by mutations in CSF2RA (encoding the granulocyte/macrophage colony-stimulating factor [GM-CSF] receptor α-chain [CD116]), which results in reduced GM-CSF-dependent pulmonary surfactant clearance by alveolar macrophages. While no pharmacologic therapy currently exists for hPAP, it was recently demonstrated that endotracheal instillation of wild-type or gene-corrected mononuclear phagocytes (pulmonary macrophage transplantation [PMT]) results in a significant and durable therapeutic efficacy in a validated murine model of hPAP. To facilitate the translation of PMT therapy to human hPAP patients, a self-inactivating (SIN) lentiviral vector was generated expressing a codon-optimized human CSF2RA-cDNA driven from an EF1α short promoter (Lv.EFS.CSF2RAcoop), and a series of nonclinical efficacy and safety studies were performed in cultured macrophage cell lines and primary human cells. Studies in cytokine-dependent Ba/F3 cells demonstrated efficient transduction, vector-derived CD116 expression proportional to vector copy number, and GM-CSF-dependent cell survival and proliferation. Using a novel cell line constructed to express a normal GM-CSF receptor β subunit and a dysfunctional α subunit (due to a function-altering CSF2RAG196R mutation) that reflects the macrophage disease phenotype of hPAP patients, it was demonstrated that Lv.EFS.CSF2RAcoop transduction restored GM-CSF receptor function. Further, Lv.EFS.CSF2RAcoop transduction of healthy primary CD34+ cells did not adversely affect cell proliferation or affect the cell differentiation program. Results demonstrate Lv.EFS.CSF2RAcoop reconstituted GM-CSF receptor α expression, restoring GM-CSF signaling in hPAP macrophages, and had no adverse effects in the intended target cells, thus supporting testing of PMT therapy of hPAP in humans.
Keywords: : hematopoietic gene therapy, hPAP, lentivirus, CSF2RA, HSCs, macrophages
Introduction
Hereditary pulmonary alveolar proteinosis (hPAP) is a rare, life-threatening lung disease characterized by progressive accumulation of surfactant within alveoli and resulting hypoxemic respiratory failure.1–3 It is caused by recessive homozygous or compound-heterozygous mutations in genes encoding the granulocyte/macrophage colony-stimulating factor receptor (CSF2R), a heterodimeric, high-affinity receptor composed of an α-subunit (CSF2RA, GM-CSFRα, CD116) conferring GM-CSF specificity and a β-subunit (CSF2RB, GM-CSFRβ, CD131) common to the receptors for interleukin (IL)-3 and IL-5 and β subunit-associated Janus kinase 2 (JAK2). Upon binding of GM-CSF to the α-subunit, the intracytoplasmic portion of the β-subunit becomes phosphorylated by JAK2 and initiates signaling through multiple pathways, including phosphorylation of signal transducer and activator of transcription 5 (STAT5).4 Disruption of CSF2R-mediated GM-CSF signaling impairs alveolar macrophage (AM) differentiation and function but has no effect on other baseline hematopoietic parameters.5 Without GM-CSF, AMs have a reduction in their ability to clear surfactant from the alveolar surface, which slowly accumulates and causes the cardinal clinical manifestation of hPAP, progressive dyspnea of insidious onset.
Current therapy of hPAP is whole lung lavage (WLL), a procedure performed under general anesthesia, with endotracheal intubation, and mechanical ventilation of one lung while the other is repeatedly filled with saline and drained to remove the excess surfactant physically. Retroviral vector-mediated transfer of a normal Csf2rb cDNA into Csf2rb–/– mouse hematopoietic stem/progenitor cells (HSPCs) followed by bone-marrow transplantation corrected GM-CSF signaling in AMs and lung disease in Csf2rb–/– mice.6 Notwithstanding the success of this approach in mice and of bone-marrow transplantation in various other diseases,7–9 it was unsuccessful in hPAP and resulted in death from overwhelming infection, presumably due to an inability to protect the lung as a result of the disease-related immune defects and the required preparative myeloablation.2
Recently, transplantation of macrophages or monocytes into the lungs without myeloablation, a new therapeutic approach known as pulmonary macrophage transplantation (PMT), was shown to be highly efficacious using either wild-type or gene-corrected cells in murine hPAP models.10–12 These and other results support the feasibility of translating PMT therapy of hPAP to humans using autologous, patient CD34+ cell-derived, lentiviral vector-mediated gene-corrected macrophages for transplantation.
As part of a PMT clinical development program, a state-of-the-art third-generation self-inactivating (SIN) lentiviral vector construct was generated expressing a codon-optimized CSF2RA cDNA driven by an internal elongation factor 1α (short; EFS) promoter (Lv.EFS.CSF2RAcoop). This article reports the results of nonclinical, in vitro safety and function studies for this vector in cultured macrophages and primary human cells.
Material and Methods
Cell culture
Murine Ba/F3 cells were cultured in RPMI1640 (GIBCO; Life Technologies, Paisley, United Kingdom) supplemented with 10% fetal calf serum (FCS), 100 IU/mL of penicillin/streptomycin (PAA, Pasching, Austria), and 2 ng/mL of mIL-3 (Peprotech, Hamburg, Germany) on suspension culture plates. Murine alveolar macrophage (mAM) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS, 10 IU/mL of penicillin/streptomycin, 2 mM HEPES (PAA) on adherent culture plates. All cell lines were cultured in standard conditions at 37°C and 5% CO2. Human CD34+ cells were isolated from umbilical cord blood (purchased from Hannover Medical School). All donors have given informed consent. After gradient centrifugation of peripheral blood mononuclear cells (PBMCs), CD34+ cells were enriched from PBMCs by magnetic separation using CD34+ MicroBead kit (Miltenyi Biotec, Bergisch-Gladbach, Germany). Cells were cultured in StemSpan (STEMCELL Technologies, Vancouver, Canada) containing 100 IU/mL of penicillin/streptomycin, 2 mM of L-glutamine (Thermo Fisher Scientific, Waltham, MA), 100 ng/mL of hSCF, 100 ng/mL of hFlt3l, and 50 ng/mL of hTPO (Peprotech) at 37°C and 5% CO2. For differentiation toward macrophages, CD34+ cells were transferred to RPMI1640 containing 10% FCS, 100 IU/mL of penicillin/streptomycin, 100 ng/mL of hM-CSF, 100 ng/mL of hGM-CSF, 100 ng/mL of hFlt3l, 20 ng/mL of hIL-3, and 20 ng/mL of hIL-6 (Peprotech) for at least 10 days.
Lentiviral vector construction and production
Codon optimization of CSF2RA was performed based on PUBMED NM_006140. Codon-optimized CSF2RA cDNA was flanked by AgeI and SalI restriction sites and synthesized by GeneScript (Piscataway, NJ). Using restriction digestion (AgeI, SalI), CSF2RA cDNA was inserted into a third-generation self-inactivating lentiviral backbone (pRRL.cPPT.EFS.GFP), which was used as a control vector throughout the experiments. The final vector pRRL.cPPT.EFS.CSF2RAcoop (Lv.EFS.CSF2RAcoop) was sequence verified by DNA sequencing (GATC, Konstanz, Germany). For production of viral particles, a transient four-vector transfection of HEK293T cells was used, as previously described.13 HEK293T cells were cultured in DMEM (PAA) containing 10% FCS, 100 IU/mL penicillin/streptomycin, 20 mM of HEPES (PAA), and 25 μM of chloroquine (Sigma–Aldrich, Steinheim, Germany). Cells were transfected using calcium phosphate precipitation in the presence of 8 μg/mL of gag/pol, 5 μg/mL of pRSV-Rev, 5 μg/mL of lentiviral vector plasmid, and 1.5 μg/mL of vesicular stomatitis virus glycoprotein (VSVg). Viral supernatants were harvested 36 and 48 h post transfection, filtered, and concentrated by ultracentrifugation (Becton Dickinson, Krefeld, Germany) for 16 h at 14,000 g and 4°C. Viral titers were determined by several dilutions on SC-1 fibroblasts and flow cytometry analysis.
Lentiviral transduction
For lentiviral transduction, 100,000 mAM or Ba/F3 cells were transferred to respective culture medium containing 4 μg/mL of protamine sulfate. Viral transduction was performed for 24 h. Thereafter, cells were washed and transferred back to standard culture medium. Transduction efficiency was analyzed 72 h after transduction using flow cytometry.
CD34+ cells were transduced using RetroNectin® (Takara Bio, Inc., Shiga, Japan), with a multiplicity of infection (MOI) of 20, according to the manufacturer's instructions.
Generation of mAM cell lines
The mAM cell line is a murine AM cell line previously derived from GM-CSF-deficient mice.14 The mAM-hGM-R cell line is a murine AM cell line expressing functional human GM-CSF receptors previously derived from mAM cells by retroviral-mediated expression of normal human GM-CSF α- and β-subunits (MIEG3-CSF2RA, MSCF2.1-CSF2RB vectors, respectively).1 The mAM-hPAP cell line is a murine AM cell line expressing non-functional human GM-CSF receptors previously derived from mAM cells by retroviral-mediated expression of mutant human GM-CSF α and normal β-subunits (MIEG3-CSF2RAG196R, MSCF2.1-CSF2RB vectors, respectively).1
Cell sorting
Before sorting, Ba/F3 cells were stained with CD116 PE antibody for 45 min at 4°C and separated on a XDP flow cytometer (Beckman Coulter, Krefeld, Germany). Single cells from high, medium, and low expressing fractions were sorted and cultured, as previously described.
Cell cytology
Approximately 50,000 cells were re-suspended in 200 μL of phosphate-buffered saline and centrifuged onto glass slides using a medite Cytofuge® (medite, Burgdorf, Germany) at 600 g for 7 min. Glass slides were subsequently stained using Pappenheim staining.
hGM-CSF-dependent survival assay
After Ba/F3 cells had been cultured in X-VIVO 15 (Lonza, Basel, Switzerland) for 24 h without cytokines, 100,000 cells per condition were transferred either to X-VIVO 15 only as a negative control or X-VIVO 15 (Lonza) supplemented with 2 ng/mL of mIL-3 (Peprotech) as a positive control. In addition, transduced cells were also cultured in the presence of 5, 10, 20, 50, or 100 ng/mL of hGM-CSF (Peprotech) and incubated for 72 h. The percentage of surviving cells was analyzed using FSC/SSC gating in flow cytometry analysis.
STAT5 phosphorylation assay
After cultivation of Ba/F3 cells in X-VIVO 15 without cytokines for 24 h, cells were stimulated for 15 min in X-VIVO 15 supplemented with either 2 ng/mL of mIL-3 or 10 ng/mL of hGM-CSF (Peprotech). Cells were harvested and lysed using RIPA buffer with proteinase inhibitor cocktail. Cell lysates were analyzed by Western blot using antibodies of STAT5 (Santa Cruz, Dallas, TX), phospho-STAT5 (Millipore, Billerica, MA), and actin (Santa Cruz) as a loading control.1
Quantitation of vector copy number
Genomic DNA was isolated using GeneElute Mammalian Genomic DNA Miniprep Kit (Sigma–Aldrich). Quantitative real-time polymerase chain reaction was performed on a StepOnePlus light cycler (Applied Biosystems, Foster City, CA) using Fast SybrGreen reagent (Qiagen, Hilden, Germany) and primers detecting either wPRE (Fwd: GAGGAGTTGTGGCCCGTTGT; Rev: TGACAGGTGGTGG-CAATGCC) or the polypyrimidine tract binding protein 2 (PTBP2; Fwd: TCTCCATTCCC-TATGTTCATGC; Rev: GTTCCCGCAGAATGGTGAGGTG) sequence as the internal control, respectively. Normalization was performed using a plasmid standard harboring the relevant PTBP2 sequences. Copy number calculations were performed by the Pfaffl method.15
GM-CSF clearance assay
For GM-CSF clearance, 100,000 mAM cells were seeded onto a 24-well plate in standard culture medium. After 24 h, the medium was replaced by X-VIVO 15 containing 1 ng/mL of hGM-CSF. Medium samples were taken after 0, 1, 3, 6, 12, and 24 h of stimulation, and hGM-CSF concentration was determined using a Human GM-CSF ELISA Ready-SET-Go! Kit (Thermo Fisher Scientific).
Southern blotting analysis
Briefly, 10 μg of genomic DNA was digested using restriction enzyme AflII (Thermo Fisher Scientific). Plasmid vector DNA (10 and 100 pg) Lv.EFS.CSF2RAcoop served as positive controls. Digested DNA was separated on a TAE agarose gel and transferred to a Biodyne B nylon membrane (Pall Life Sciences, Portsmouth, United Kingdom). Blotted DNA was analyzed with a P32-labeled EcoRI fragment (∼600 bp) of the woodchuck hepatitis element (wpre) using the DecaLabel™ DNA labeling kit (Fermentas/Thermo Fisher Scientific). After alkaline stripping, the membrane was rehybridized with a radioactively labeled BamHI fragment (∼900 bp) of the codon-optimized CSFR2RA cDNA. Both probes were isolated from the pEFS.hCSF2RAcoop plasmid vector. HindIII digested P32-labeled bacteriophage lambda DNA served as a size standard.
Colony-forming assay
In order to evaluate clonogenic potential of CD34+ cells, 1,500 cells/mL were seeded in methylcellulose (R&D Systems, Minneapolis, MN) HSC003 containing Epo, GM-CSF, IL-3, and SCF in 8.8 cm2 cell culture dishes with a 2 mm grid (Thermo Scientific Nunc™ cell culture) and cultured for 7 days in the incubator at standard culture conditions in a separately closed humid chamber. The overall number of colonies was counted after 7–10 days.
Statistical analysis
Statistical analysis was performed using Prism 6 software (GraphPad, La Jolla, CA). Unless otherwise noted, analysis of variance (ANOVA) with respective post hoc testing (see figure legends) was used. All data were reproducible and were performed as a minimum of three independent experiments.
Results
Lentiviral vector design and restoration of GM-CSF receptor function in murine Ba/F3 cells
To lay the foundation for a human gene therapy approach using HSPC-derived macrophages, a SIN-LV backbone equipped with an EFS promoter was utilized to constitutively express a codon-optimized cDNA of CSF2RA (Lv.EFS.CSF2RAcoop; Fig. 1a). As a control vector, the same vector architecture was used to express a green fluorescent protein instead of CSF2RA (Lv.EFS.GFP; Fig. 1a). To enable gene transfer into both human and murine hematopoietic cells, the lentiviral vectors were pseudotyped with VSVg.
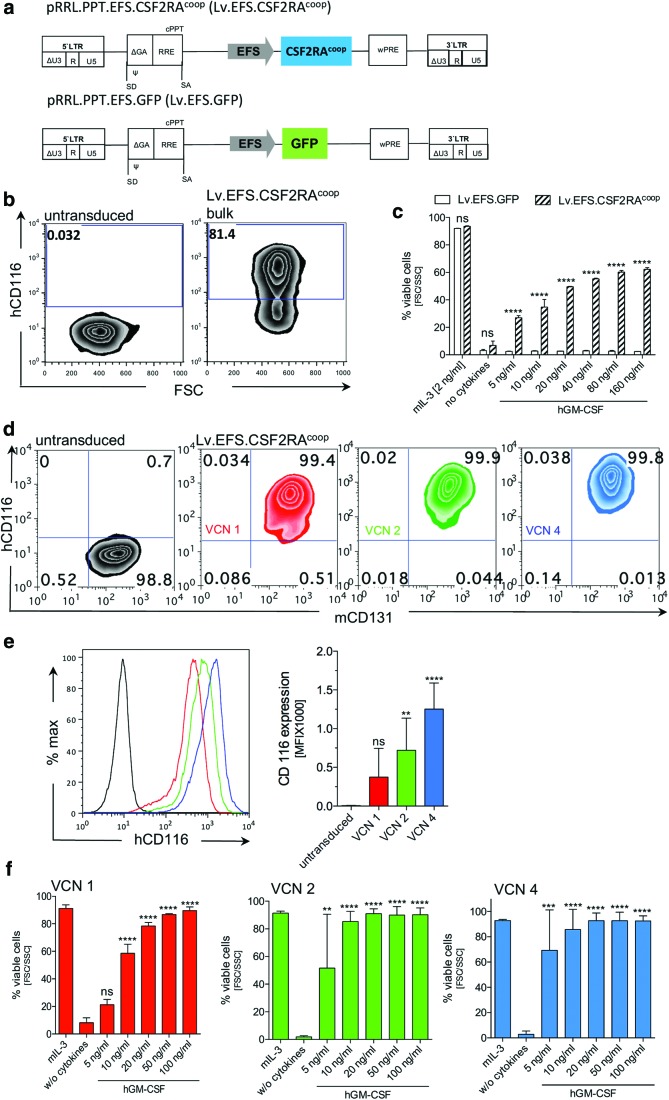
Figure 1.
Vector design and functionality in the Ba/F3 cell line. (a) Schematic picture of the third-generation self-inactivating (SIN) lentiviral constructs Lv.EFS.CSF2RAcoop expressing the human codon-optimized cDNA of CSF2RA (upper picture) and Lv.EFS.GFP control vector (lower picture). (b) Human CD116 expression in untransduced (left) and Lv.EFS.CSF2RAcoop non-sorted transduced (right) Ba/F3 cells. (c) Human granulocyte/macrophage colony-stimulating factor (hGM-CSF)-dependent survival of Lv.EFS.GFP and Lv.EFS.CSF2RAcoop transduced and non-sorted Ba/F3 cells with mIL-3 as positive control, without cytokines as a negative control and increasing hGM-CSF concentrations (5–160 ng/mL; n = 3; two-way analysis of variance [ANOVA] using Sidak's post hoc testing). (d) Representative plots analyzing mCD131 and hCD116 expression in untransduced and Lv.EFS.CSF2RAcoop transduced Ba/F3 cells with defined vector copy numbers (VCNs) of 1, 2, and 4. (e) Mean fluorescence intensity (MFI) analysis of hCD116 expression in untransduced and Lv.EFS.CSF2RAcoop transduced Ba/F3 cells with defined VCNs of 1, 2, and 4 (left), and a bar graph depicting the CD116 MFI (right; n = 3; one-way ANOVA using Dunnett's post hoc testing). (f) hGM-CSF-dependent survival of Lv.EFS.CSF2RAcoop transduced Ba/F3 cells with defined VCNs of 1, 2, and 4 (n = 3; one-way ANOVA using Dunnett's post hoc testing). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. Color images available online at www.liebertpub.com/hgtb
To evaluate the functionality of transgenic human CSF2RA expression, the murine mIL3/mGM-CSF-dependent pro B cell line Ba/F3 was used. Physiologically, GM-CSFRα initiates GM-CSF-dependent signaling by extracellular binding to GM-CSF. Upon pairing with GM-CSFRβ, high-affinity binding of GM-CSF is induced, which in turn is necessary for intracellular GM-CSF-dependent signal transduction.16 The same process has been described for the interaction of murine GM-CSFRβ, and human GM-CSFRα in the presence of hGM-CSF,16 highlighting their suitability to study proliferation and cell survival directly after lentiviral transduction. To test the functionality of the preclinical vector, Ba/F3 cells were transduced with an MOI of 1, and GM-CSFRα expression (CD116) was analyzed by flow cytometry. In contrast to non-transduced control cells, CD116 was detectable on >80% of Lv.EFS.CSF2RAcoop-transduced cells (Fig. 1b). Next, the functionality of CSF2RA in Lv.EFS.CSF2RAcoop and Lv.EFS.GFP transduced cells was assessed by culturing them in the presence or absence of hGM-CSF. Maintenance culture of transduced Ba/F3 cells in the presence of mIL3 had no effect on cell viability, irrespective of the construct (Fig. 1c). In contrast, withdrawal of mIL3 resulted in <10% viable cells after 3 days in culture. Similar observations were made when Lv.EFS.GFP transduced Ba/F3 cells were cultured in the presence of accelerating concentrations of hGM-CSF. Similar to the cultures without cytokines, administration of hGM-CSF could not rescue cell viability (Fig. 1c). The opposite effect was observed when Ba/F3 cells were transduced with Lv.EFS.CSF2RAcoop. Expression of GM-CSFRα was able to rescue hGM-CSF-dependent growth in Ba/F3 cells in a concentration-dependent manner. While only 30% of cells could be rescued by 5 ng/mL of hGM-CSF, administration of 160 ng/mL was able to provide cell survival in >60% of the cells (Fig. 1c). Moreover, constitutive overexpression of CSF2RA in Ba/F3 cells did not lead to any differences in cell proliferation (Supplementary Fig. S1a and b; Supplementary Data are available online at www.liebertpub.com/hgtb) or apoptosis (Supplementary Fig. S1c and d). After evaluating CSF2RA functionality in non-sorted bulk cell densities, single cell sorting of Lv.EFS.CSF2RAcoop cells was performed to establish individual cell clones harboring one, two, or four vector copies (vector copy number [VCN] 1, 2, or 4). Evidence of stable CSF2RA expression was present in >99% of cells in all established lentiviral vector-transduced cell clones (Fig. 1d). Further, the level of transgene expression increased in proportion to VCN, as shown by the mean fluorescence intensity (MFI) after CD116 immunostaining: VCN = 1 → MFI = 374; VCN = 2 → MFI = 720; VCN = 4 → MFI = 1,252 (Fig. 1e). Of note, expression of murine Csf2rb was not affected by overexpression of CSF2RA (Fig. 1d). In order to discriminate VCN-dependent functionality in cells transduced with Lv.EFS.CSF2RAcoop, the individual clones were cultured in the presence or absence of hGM-CSF. As expected, maintenance culture in the presence of mIL-3 had no effect on cell survival, irrespective of the VCN. In contrast, a concentration of 5 ng/mL of hGM-CSF was sufficient to rescue >50% of the transduced cells harboring a VCN of >2, which was similar to the observation in bulk cells. However, a more pronounced GM-CSF dose dependency of cell growth was observed for cells harboring a VCN of 1. Although 60% of the cells could be rescued by 10 ng/mL of of hGM-CSF, only 20% of viable cells could be detected in 5 ng/mL of hGM-CSF cultures (Fig. 1f). Similar results occurred when the Lv.EFS.CSF2RAcoop vector was compared to SIN-lentiviral vectors expressing wild-type CSF2RA from either the EFS (Lv.EFS.CSF2RA) or phosphoglycerate kinase (PGK; Lv.PGK.CSF2RA) promoters (Supplementary Fig. 2a). Individual clones of Ba/F3 cells transduced with either Lv.EFS.CSF2RA or Lv.PGK.CSF2RA exhibited VCN-dependent expression of CSF2RA (Supplementary Fig. 2b). Comparison of all three vectors revealed the highest expression of CSF2RA for Ba/F3 cells transduced with Lv.EFS.CSF2RAcoop (Supplementary Fig. 2c). The increased expression of the CSF2RAcoop cDNA was reflected in GM-CSF-dependent Ba/F3 cell survival. Lv.EFS.CSF2RAcoop-transduced cells accumulated to slightly higher numbers in response to GM-CSF compared to the other vector constructs (Supplementary Fig. 2d). These data indicate that the Lv.EFS.CSF2RAcoop lentiviral vector restores GM-CSF receptor signaling function in Ba/F3 cells.
Evaluation of vector function in murine AM cell lines expressing normal and abnormal human GM-CSF receptors
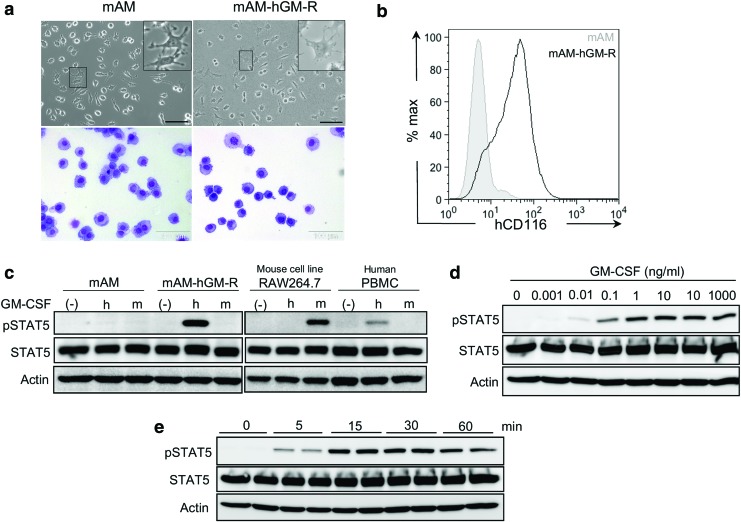
In order to study potential effects of transgene-mediated CSF2RA overexpression, an AM cell line was utilized previously created from GM-CSF–/– AMs (mAM) that lacks expression of functional GM-CSF receptors and does not respond to murine or human GM-CSF14 and a derivative cell line (mAM-hGM-R) that was transduced to express normal human GM-CSF receptors conferring responsiveness to human GM-CSF.17 Both mAM and mAM-hGM-R cells had similar macrophage morphology (Fig. 2a) and comparable growth characteristics (not shown). Human CD116 expression was detected on mAM-hGM-R but not mAM cells (Fig. 2b). Stimulation by human but not murine GM-CSF resulted in STAT5 phosphorylation in mAM-hGM-R cells, while mAM cells did not respond to either (Fig 2c). As controls, murine RAW264.7 cells responded to murine but not human GM-CSF, while human monocytes responded to human but not mouse GM-CSF, as measured by STAT5 phosphorylation (Fig. 2c). The GM-CSF receptor signaling response of mAM-hGM-R cells, as measured by STAT5 phosphorylation, was dose dependent (Fig. 2d), detectable at 5 min, and peaked 15 min after exposure to human GM-CSF (10 ng/mL; Fig. 2e). These results support the utility of mAM cells in studies evaluating the function of human GM-CSF receptor subunits.
Figure 2.
Characteristics of the new murine alveolar macrophage cell line (mAM). (a) Morphology of the mAM parental line mAM (left) and mAM-hGM-R line expressing the human GM-CSFR (right); scale bar: ∼100 μm. (b) Representative histogram depicting hCD116 expression of mAM and mAM-hGM-R cells. (c) Western blot analysis of overall STAT5 expression and phosphorylated STAT5 in response to human and murine GM-CSF in mAM and mAM-hGMR cells to confirm species-specific response. No addition of cytokines (–) served as a negative control, human PBMCs as a positive control for hGM-CSF, the mouse cell line RAW264.7 as a positive control for mGM-CSF, and actin expression as loading control. (d) Western blot depicting dose-dependent phosphorylation of STAT5 in response to hGM-CSF (0–1,000 ng/mL) with actin expression as loading control. (e) Western blot depicting time-dependent phosphorylation of STAT5 after stimulation with hGM-CSF (0, 5, 15, 30, and 60 min of stimulation) with actin expression as loading control. PBMC, peripheral blood mononuclear cells; STAT5, signal transducer and activator of transcription 5; pSTAT5, phosphorylated STAT5. Color images available online at www.liebertpub.com/hgtb
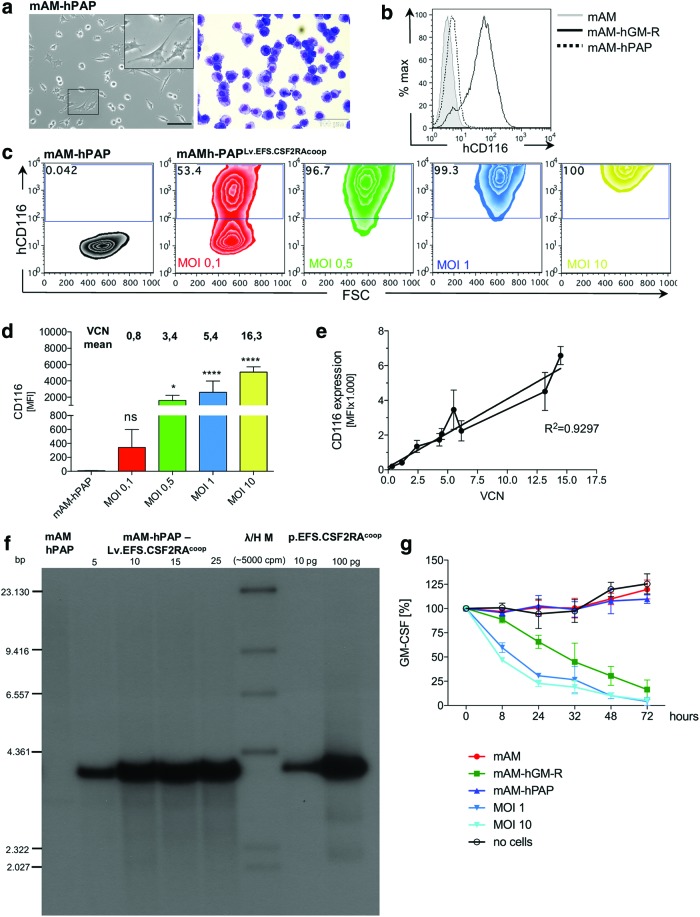
To begin to study the effects of transgene overexpression, another mAM cell-derived cell line (mAM-hPAP) was used, which was previously created (by T.S. and B.C.T., unpublished results) to express a dysfunctional α-subunit and a normal β-subunit mimicking the precise GM-CSF receptor alleles in an index hPAP patient (CSF2RAG196R, CSF2RBNormal).1 These cells also had typical macrophage morphology (Fig. 3a) and no detectable cell-surface GM-CSFRα (Fig. 3b). First, mAM-hPAP cells were transduced with Lv.EFS.CSF2RAcoop while varying the MOI between 0.1 and 10 infectious particles per cell shown to exhibit GM-CSF receptor α-subunit expression that increased with MOI (MOIs of 0.1, 0.5, 1, or 10 resulted in VCNs of 0.8, 3.4, 5.4, or 16.3, respectively; Fig. 3c and d). Further, transgene product expression was highly correlated in direct linear fashion with VCN (Fig. 3e). Southern blotting with CSF2RA- (Fig. 3f) or vector-specific (Supplementary Fig. 3a and b) probes demonstrated integration of the vector into mAP-hPAP cell chromosomal DNA in a MOI-dependent fashion across a broad MOI, ranging from 5 to 25. Vector-mediated restoration of GM-CSF receptor function was evaluated by measuring the capacity of cells to clear human GM-CSF from the culture media, as previously reported.1 Lv.EFS.CSF2RAcoop-transduced mAM-hPAP cells cleared human GM-CSF similar to mAM-hGM-R cells (which have functioning GM-CSF receptors), while no GM-CSF clearance was observed in non-transduced cells (which do not have functioning GM-CSF receptors; Fig. 3g). These results show that Lv.EFS.CSF2RAcoop transduction of an AM cell line mimicking the genetic defect of hPAP patient cells results in vector dose-dependent transduction, transgene expression, and restoration of GM-CSF receptor signaling.
Figure 3.
Vector analysis in the mAM cell line. (a) Morphology of the mAM-hPAP line expressing the mutated GM-CSFRα (mAM-hPAP); scale bar: ∼100 μm. (b) Representative histogram depicting hCD116 expression of mAM, mAM-hGM-R, and mAM-hPAP cells. (c) mAM-hPAP cells were transduced with Lv.EFS.CSF2RAcoop using multiplicities of infection (MOIs) of 0.1, 0.5, 1, and 10. Representative plots depicting hCD116 expression of mAM-hPAP and mAM-hPAP transduced with Lv.EFS.CSF2RAcoop. (d) MFI bargraph of hCD116 expression summarizing three independent transductions (n = 3; one-way ANOVA using Dunnett's post hoc testing). (e) Correlation between hCD116 MFI and VCN indicating a linear relationship between VCN and CD116 expression. (f) Southern blot analysis using a CSF2RAcoop probe to trace the transgene integrated to the genome of mAM-hPAP cells transduced with Lv.EFS.CSF2RAcoop. No transgene expression is detectable in mAM-hPAP cells, whereas the signal is increasing with higher MOIs (5, 10, 15, and 25) used to transduce the cells. Lentiviral transfer plasmid in different concentrations was used as a positive control. λ/H M = HindIII digested P32-labeled bacteriophage lambda DNA marker. (g) hGM-CSF uptake from cell culture medium. mAM and mAM-hPAP cells were not able to clear hGM-CSF from the cell culture supernatant, whereas mAM and mAM-hPAPLv.EFS.CSF2RAcoop cells efficiently cleared hGM-CSF over a period of 72 h. A well with no cells was used as a negative control. bp, base pairs. *p < 0.05; ****p < 0.0001. Color images available online at www.liebertpub.com/hgtb
Evaluation of vector safety
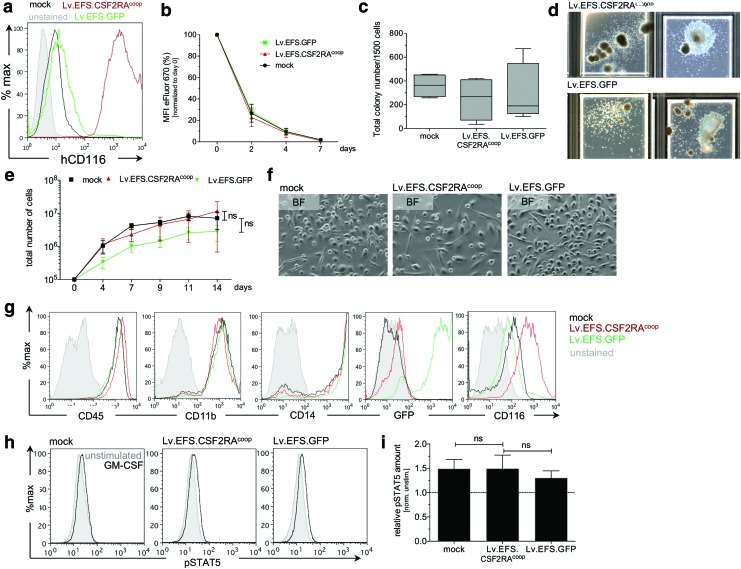
Primary human CD34+ cells (the cell type that will be transduced in a future clinical trial) were used to address potential side effects of transgene-mediated CSF2RA expression. Cells transduced with Lv.EFS.CSF2RAcoop exhibited readily detectable expression of the GM-CSF receptor α-subunit in contrast to Lv.EFS.GFP-transduced cells (Fig 4a). The potential effects of overexpression were evaluated in transduced CD34+ cells and derived by evaluating proliferation using the eFluor670 method.13,18 Lv.EFS.CSF2RAcoop-, Lv.EFS.GFP-, or mock-transduced cells had a similar cell proliferation-related dye-dilution profile, indicating a lack of effect of transgene expression on CD34+ cell proliferation (Fig. 4b). Using a standard methylcellulose assay,19,20 these three cell populations gave rise to similar total colony counts (Fig. 4c), and both Lv.EFS.CSF2RAcoop- and Lv.EFS.GFP-transduced cells gave rise to CFU-GM/GEMM or BFU-E colonies (Fig. 4d), indicating transduction did not affect cell proliferation or multi-lineage differentiation potential. These three cell populations also had similar cellular proliferation kinetics during cytokine-mediated macrophage differentiation in vitro over 14 days (Fig. 4e), a similar macrophage appearance (Fig. 4f), and similar pattern of macrophage cell-surface markers (Fig. 4g). Importantly, Lv.EFS.CSF2RAcoop transduction did not affect the level of GM-CSF-stimulated STAT5 phosphorylation, indicating that CSF2RA overexpression did not increase the magnitude of GM-CSF signaling significantly (Fig. 4h and i) in CD34+ cell-derived macrophages. These studies did not identify adverse effects of Lv.EFS.CSF2RAcoop transduction on CD34+ cells or their ability to proliferate and differentiate into mature, functioning macrophages.
Figure 4.
Transduction of primary CD34+ cells and macrophage differentiation. (a) Analysis of hCD116 expression in primary CD34+ cells, transduced with Lv.EFS.CSF2RAcoop (red), Lv.EFS.GFP (green), or mock-treated cells (black). Non-transduced and unstained CD34+ cells have been used as negative controls (gray). (b) Proliferation of transduced CD34+ cells, as determined by dilution of eFluor670. Mean fluorescent intensity values of e670 have been normalized to day 0 time point. (c) Total colony numbers per 1,500 input cells cultivated in methylcellulose containing human SCF, GM-CSF, IL-3, and Epo for 7–10 days. (d) Representative pictures of clonogenic cells observed after 7–10 days in methylcellulose-based assays. (e) Increase in overall cell number during differentiation of Lv.EFS.CSF2RAcoop (red), Lv.EFS.GFP (green), or mock treated cells (black) toward macrophages for 14 days in the presence of IL-3, IL-6, FLT3, M-CSF, and GM-CSF. (f) Representative bright-field images of macrophages on day 14 of differentiation. (g) Phenotypic analysis of macrophages on day 14 of differentiation by flow cytometry. Histograms are shown as overlays of mock-treated (blue), Lv.EFS.CSF2RAcoop (black), and Lv.EFS.GFP (green) cells. (h) Functional analysis of transduced macrophages in the presence of hGM-CSF. Histograms show phosphorylation of STAT5 in the absence (gray) or presence (black) of hGM-CSF. (i) Relative amount of pSTAT5 after stimulation of transduced macrophages with hGM-CSF. Values are calculated and normalized to non-stimulated conditions. Color images available online at www.liebertpub.com/hgtb
Discussion
This article reports on the creation of a third-generation SIN lentiviral vector expressing a codon-optimized CSF2RA cDNA (Lv.EFS.CSF2RAcoop) and its evaluation in multiple cell lines and primary human CD34+ cells. Transduction of murine Ba/F3 cells resulted in VCN-dependent human CSF2RA expression and GM-CSF-dependent proliferation without adverse effects on cell survival or proliferation. Transduction of a murine AM cell line (expressing abnormal α and normal β human GM-CSF receptor subunits) restored GM-CSF receptor function, as measured by STAT5 phosphorylation and receptor-mediated GM-CSF clearance. Transduction of primary healthy human CD34+ cells did not result in any adverse effects on cell survival, cell proliferation, or hematopoietic differentiation potential. These non-clinical results support the feasibility of evaluating Lv.EFS.CSF2RAcoop in human clinical trial of autologous gene-correction/PMT therapy of hPAP.
With respect to clinical translation, a third-generation SIN-lentiviral vector architecture was used, since inclusion of a self-inactivating mutation into the gamma-retroviral or lentiviral vector (SIN-LV) backbones greatly reduce the risk of insertional mutagenesis,21,22 which has been observed in some hematopoietic stem cell gene therapy (HSCGT) clinical trials employing gamma-retroviral vectors.23,24 This choice is of clinical importance, since lentiviral vectors do not show preferential integration in or near enhancer and promoter elements as do gamma-retroviral vectors. To this end, HSCGT using SIN-LVs for the treatment of Wiskott–Aldrich syndrome (WAS), metachromatic leukodystrophy (MLD), X-linked severe combined immunodeficiency (SCID-X1), or most recently beta-thalassemia major could restore functionality of a patient's cells in vivo without any sign of adverse events.22,25–27
Similarly, different promoter enhancer elements have been optimized in order to avoid transgene expression from the long terminal repeat (LTR) region by inserting endogenous promoter elements. The human EFS promoter was used to express the CSF2RA transgene constitutively. The use of lentiviral vectors equipped with an EFS promoter could demonstrate sustained transgene expression in murine and human hematopoietic cells, and no signs of genotoxicity could be observed when this vector combination was analyzed in an in vitro immortalization assay.22,28,29 Similar to the aforementioned data, constitutive overexpression of CSF2RA by the EFS promoter showed no adverse effects on CD34+ cells with regard to their proliferation and differentiation potential toward macrophages. Moreover, CSF2RA-transgenic macrophages showed similar functionality as their mock-treated or control-transduced counterparts. While these data are reassuring, far more detailed studies of the functional and safety characteristics of this vector construct will be mandatory prior to clinical use. In addition, some of these studies may have to be performed in patient-derived CD34+ cells and thereof derived macrophages. As an alternative to this lentiviral-based gene therapy approach, precise genome editing tools using designer nucleases such as TALEN or CRISPR/Cas9 might also be employed to induce site-specific DNA repair. While the CRISPR/Cas9 methodology is frequently applied to induce gene disruption by the non-homologous end-joining machinery,30 recent studies using primary HSPCs have also proven its suitability to correct the disease-causing gene.31–33
As patients suffering from CSF2RA deficiency are very rare, CD34+ cells and macrophages derived from patient-specific induced pluripotent stem cells (iPSC) may be utilized as an intermediate step.34 The potential to use this technology has already been demonstrated in previous studies using hPAP patient-specific iPSC. Here, lentiviral vectors have been used in order to correct the hPAP phenotype in iPSC-derived macrophages.35,36 Similarly, iPSC technology has been used to gain insights into CSF2RB-deficiency using hematopoietic differentiation of murine iPSC from Csf2rb-deficient mice.37
In summary, here a SIN lentiviral vector (Lv.EFS.CSF2RAcoop) is introduced, which would be suited to express the CSF2RA transgene in CD34+ patient cells in order to restore GM-CSF-mediated downstream signaling. No adverse effect of CSF2RA expression in cell lines or in primary CD34+ cells were observed in this study, highlighting the suitability of the presented lentiviral architecture for future efforts to correct hematopoietic cells with CSF2RA-deficiency.
Supplementary Material
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Cluster of excellence REBIRTH: Exc 62/1 to T.M. and N.L., as well as by the individual grant: DFG LA 3680/2-1), Else Kröner-Fresenius-Stiftung (EKFS; 2015_A92 to N.L.), MHH Hannover (young academy to N.L.), National Heart, Lung and Blood Institutes (R01 HL085453, R21 HL106134, R01HL118342; to B.C.T.), American Thoracic Society Foundation (Unrestricted Research Grant; to T.S.), CCHMC Foundation Trustee Grant (to T.S.), and an the Translational Pulmonary Science Center (endowment; to B.C.T.). We thank Matthias Ballmaier (Cell Sorting Facility, Hannover Medical School) for assistance in cell sorting, and Doreen Lüttge and Theresa Buchegger (Institute of Experimental Hematology, Hannover Medical School) for excellent technical assistance.
Author Disclosure
The authors have no commercial, proprietary, or financial interest in the products or companies described in this article.
References
- 1.Suzuki T, Sakagami T, Rubin BK, et al. . Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J Exp Med 2008;205:2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Moczygemba M, Doan ML, Elidemir O, et al. . Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1. J Exp Med 2008;205:2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki T, Sakagami T, Young LR, et al. . Hereditary pulmonary alveolar proteinosis: pathogenesis, presentation, diagnosis, and therapy. Am J Respir Crit Care Med 2010;182:1292–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Liu CH, Roberts AI, et al. . Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don't know. Cell Res 2006;16:126–133 [DOI] [PubMed] [Google Scholar]
- 5.Nishinakamura R, Miyajima A, Mee PJ, et al. . Hematopoiesis in mice lacking the entire granulocyte-macrophage colony-stimulating factor/interleukin-3/interleukin-5 functions. Blood 1996;88:2458–2464 [PubMed] [Google Scholar]
- 6.Kleff V, Sorg UR, Bury C, et al. . Gene therapy of betac-deficient pulmonary alveolar proteinosis (betac-PAP): studies in a murine in vivo model. Mol Ther 2008;16:757–764 [DOI] [PubMed] [Google Scholar]
- 7.Szabolcs P, Cavazzana-Calvo M, Fischer A, et al. . Bone marrow transplantation for primary immunodeficiency diseases. Pediatr Clin North Am 2010;57:207–237 [DOI] [PubMed] [Google Scholar]
- 8.Michlitsch JG, Walters MC. Recent advances in bone marrow transplantation in hemoglobinopathies. Curr Mol Med 2008;8:675–689 [DOI] [PubMed] [Google Scholar]
- 9.Malatack JJ, Consolini DM, Bayever E. The status of hematopoietic stem cell transplantation in lysosomal storage disease. Pediatr Neurol 2003;29:391–403 [DOI] [PubMed] [Google Scholar]
- 10.Happle C, Lachmann N, Skuljec J, et al. . Pulmonary transplantation of macrophage progenitors as effective and long-lasting therapy for hereditary pulmonary alveolar proteinosis. Sci Transl Med 2014;6:250ra113. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T, Arumugam P, Sakagami T, et al. . Pulmonary macrophage transplantation therapy. Nature 2014;514:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willinger T, Rongvaux A, Takizawa H, et al. . Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc Natl Acad Sci U S A 2011;108:2390–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachmann N, Brennig S, Hillje R, et al. . Tightly regulated “all-in-one” lentiviral vectors for protection of human hematopoietic cells from anticancer chemotherapy. Gene Ther 2015;22:883–892 [DOI] [PubMed] [Google Scholar]
- 14.Shibata Y, Berclaz PY, Chroneos ZC, et al. . GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity 2001;15:557–567 [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eder M, Ernst TJ, Ganser A, et al. . A low affinity chimeric human alpha/beta-granulocyte-macrophage colony-stimulating factor receptor induces ligand-dependent proliferation in a murine cell line. J Biol Chem 1994;269:30173–30180 [PubMed] [Google Scholar]
- 17.Suzuki T, Sakagami T, van der Loo JC, et al. . A novel cell line (mAM-hGM-R) for measuring the bioactivity of GM-CSF and neutralizing capacity of GM-CSF autoantibodies in human serum. Am J Resp Crit Care Med 2010;181:A2983 [Google Scholar]
- 18.Yao Y, Vent-Schmidt J, McGeough MD, et al. . Tr1 cells, but not Foxp3+ regulatory T cells, suppress NLRP3 inflammasome activation via an IL-10-dependent mechanism. J Immunol 2015;195:488–497 [DOI] [PubMed] [Google Scholar]
- 19.Lachmann N, Czarnecki K, Brennig S, et al. . Deoxycytidine-kinase knockdown as a novel myeloprotective strategy in the context of fludarabine, cytarabine or cladribine therapy. Leukemia 2015;29:2266–2269 [DOI] [PubMed] [Google Scholar]
- 20.Bardenheuer W, Lehmberg K, Rattmann I, et al. . Resistance to cytarabine and gemcitabine and in vitro selection of transduced cells after retroviral expression of cytidine deaminase in human hematopoietic progenitor cells. Leukemia 2005;19:2281–2288 [DOI] [PubMed] [Google Scholar]
- 21.Hacein-Bey-Abina S, Pai SY, Gaspar HB, et al. . A modified gamma-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med 2014;371:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Ravin SS, Wu X, Moir S, et al. . Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med 2016;8:335ra357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott MG, Schmidt M, Schwarzwaelder K, et al. . Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 2006;12:401–409 [DOI] [PubMed] [Google Scholar]
- 24.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. . LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003;302:415–419 [DOI] [PubMed] [Google Scholar]
- 25.Biffi A, Montini E, Lorioli L, et al. . Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:1233158. [DOI] [PubMed] [Google Scholar]
- 26.Aiuti A, Biasco L, Scaramuzza S, et al. . Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeil JA, Hacein-Bey-Abina S, Payen E, et al. . Gene therapy in a patient with sickle cell disease. N Engl J Med 2017;376:848–855 [DOI] [PubMed] [Google Scholar]
- 28.Carbonaro DA, Zhang L, Jin X, et al. . Preclinical demonstration of lentiviral vector-mediated correction of immunological and metabolic abnormalities in models of adenosine deaminase deficiency. Mol Ther 2014;22:607–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene MR, Lockey T, Mehta PK, et al. . Transduction of human CD34+ repopulating cells with a self-inactivating lentiviral vector for SCID-X1 produced at clinical scale by a stable cell line. Hum Gene Ther Methods 2012;23:297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal PK, Ferreira LM, Collins R, et al. . Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 2014;15:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeWitt MA, Magis W, Bray NL, et al. . Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med 2016;8:360ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Ravin SS, Li L, Wu X, et al. . CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci Transl Med 2017;9. [DOI] [PubMed] [Google Scholar]
- 33.Dever DP, Bak RO, Reinisch A, et al. . CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 2016;539:384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lachmann N, Ackermann M, Frenzel E, et al. . Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports 2015;4:282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T, Mayhew C, Sallese A, et al. . Use of induced pluripotent stem cells to recapitulate pulmonary alveolar proteinosis pathogenesis. Am J Respir Crit Care Med 2014;189:183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lachmann N, Happle C, Ackermann M, et al. . Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis. Am J Respir Crit Care Med 2014;189:167–182 [DOI] [PubMed] [Google Scholar]
- 37.Mucci A, Kunkiel J, Suzuki T, et al. . Murine iPSC-derived macrophages as a tool for disease modeling of hereditary pulmonary alveolar proteinosis due to Csf2rb deficiency. Stem Cell Reports 2016;7:292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.