Abstract
The aim of this study is to optimize laboratory preparation of teeth for DNA identification. By sectioning the tooth topographically into two different radicular portions, it was analyzed whether these portions of mineralized tissue differ in the quantity and quality of DNA they contain. 25 teeth were subject to different experimental conditions and total DNA was quantified for each individual tooth's radicular portion: apical and remaining root, according to a 2003 study by Gaytemenn and Sweet. We verified, with statistically significant figures, that the apical portion of the tooth is that which contains the greatest quantity of DNA. Different analytical procedures were studied for various polymorphic markers to evaluate the quality of the DNA. We concluded that the tooth is topographically distinct in both DNA quantity and quality. The tooth's apical portion is the preferential choice in sample preparation of dental mineralized tissue for molecular analysis and identification.
KEYWORDS: tooth, apical root, identification, genetic profile
Introduction
The dentition is an organ, made up of different types of tissues which are structurally disposed for a specific function (1, 2). Enamel, the protective tissue is acellular, avascular and non-enervated and therefore unimportant for genetic analysis. Cement is responsible for tooth anchoring and suffers constant remodeling. Cement is composed of cement cells located in lacunae and can characteristically invaginate the interior of apical canaliculus and canaliculi (3). Dentin, (2, 3) characterized by its histological organization can be designated secondary or tertiary (which comprises sclerotic dentin).
The choice of a tooth as a sample for genetic analysis occurs only in situations of extreme degradation when other biological tissues are not considered suitable for sampling (4-10). In practice, dental pulp, due to being highly vascularized connective tissue, was the first to be studied. However, for that same reason it is also the first part of the tooth to be degraded, therefore in this study mineralized dental tissues are being studied.
Through molecular analysis of the tooth, specifically mineralized tissues, we aimed to establish individual identification and genetic profile typing using DNA analysis.
Our specific objective was to study the apical radicular portion and the remaining radicular portion to analyze whether these portions differed in DNA quantity and quality.
MATERIALS AND METHODS
25 teeth belonging to individuals between the age of 18 and 87 were studied after informed consent was obtained. Equal proportions of intact teeth and those with caries or restorations were present. The teeth were exposed to different experimental conditions, some were buried in different mediums: pH=6 (n=7); pH=5 (n=3); pH=7 (n=5); buried in sand(n=7) for a period of 7 months while others subject to various atmospheric conditions (Iberian Peninsula) for two years (n=3). The material used for sampling was in different stages of genetic alteration after cell death (6, 11, 12).
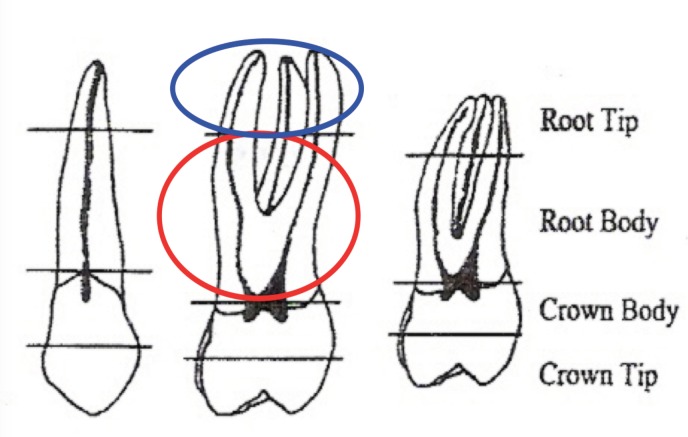
Each tooth was chemically cleaned, with hypochlorite, and mechanically, with a rotary device, it was then pulpectomized (fig.1), and only the root portion kept. The root portion was then divided further into two portions, the apex and the remaining portion, in conformity to the topographic drawings of Gaytmenn and Sweet’s 2003 study (13) (fig.2).
Fig. 1.
Sequence of the used tooth, cuts by Gaytmenn and Sweet
Fig. 2.
The Gaytmenn and Sweet guidelines
DNA was extracted using the ArchivePure commercial™ kit (5Prime®). Total DNA quantification was performed by real time PCR, by using the Human Quantifiler kit (Applied Biosystems®).
Total DNA quantification value was associated with the study viability of the polymorphisms which are most relevant for identification, namely autosomal STRs (14, 15), with the aid of AmpFℓSTR® Identifiler™ and AmpFℓSTR® Minifiler™ commercial kits, both by Applied Biosystems.
Mitochondrial DNA analysis was performed on samples over which validation of the 7 markers or any autosomal marker was not viable (16-19), and in which it was not possible to obtain an identifying autosomal profile.
The results of the quantitative study of DNA, performed on the apical and remaining tooth radicular portion, were analyzed from 25 paired samples. Quantification results were then statistically analyzed with SPSS (Statistical Package Social Science) software. Such analysis began by studying normality of the variable quantification through the Kolmogorov-Smirnov test. The non-parametric inferential median test was applied for the absence of normality within the variable quantification.
RESULTS
The results for the parameter total DNA quantification (ng/μL) obtained for each of the portions analyzed, the conservation medium and characterization of the analyzed polymorphisms are shown in Figure 3 and Table 1.
Fig. 3.
Quantification results of the apical radicular portion and the remaining radicular portion, statistically analyzed with SPSS.
Table1. Characterization of samples according to conservation medium, total DNA identification and analyzed polymorphisms in 25 paired samples.
| SAMPLES | MEDIUMS | RADICULAR TOOTH | total DNA quantification (ng/μL) | STR Profile | MITOCHONDRIAL |
|---|---|---|---|---|---|
| 1 | 2YEARS | REMAINING | 0,006 | * | ANALYZED |
| APICAL | 1,09 | 15 POLYMORPHISMS | ** | ||
| 2 | REMAINING | 0,1 | * | ANALYZED | |
| APICAL | 1,17 | 15 POLYMORPHISMS | ** | ||
| 3 | REMAINING | 0,311 | 15 POLYMORPHISMS | ** | |
| APICAL | 1,88 | 15 POLYMORPHISMS | ** | ||
| 4 | pH6 | REMAINING | 0,019 | * | ANALYZED |
| APICAL | 0,022 | * | ANALYZED | ||
| 5 | REMAINING | 0,003 | * | ANALYZED | |
| APICAL | 1,53 | 15 POLYMORPHISMS | ** | ||
| 6 | REMAINING | 0,0000 | * | ANALYZED | |
| APICAL | 0,03534 | * | ANALYZED | ||
| 7 | REMAINING | 0,024 | * | ANALYZED | |
| APICAL | 0,2396 | 14 POLYMORPHISMS | ** | ||
| 8 | REMAINING | 0,0605 | * | ANALYZED | |
| APICAL | 0,5172 | 15 POLYMORPHISMS | ** | ||
| 9 | REMAINING | 0,0061 | * | ANALYZED | |
| APICAL | 0,1027 | * | ANALYZED | ||
| 10 | REMAINING | 0,017 | * | ANALISADO | |
| APICAL | 0,113 | * | ANALYZED | ||
| 11 | pH5 | REMAINING | 0,0158 | * | ANALYZED |
| APICAL | 0,157 | * | ANALYZED | ||
| 12 | REMAINING | 0,033 | * | ANALYZED | |
| APICAL | 0,0846 | * | ANALYZED | ||
| 13 | REMAINING | 0,031 | * | ANALYZED | |
| APICAL | 0,033 | * | ANALYZED | ||
| 14 | pH7 | REMAINING | 0,0250 | * | ANALYZED |
| APICAL | 0,2955 | 12 POLYMORPHISMS | ** | ||
| 15 | REMAINING | 0,002 | * | ANALYZED | |
| APICAL | 0,0502 | * | ANALYZED | ||
| 16 | REMAINING | 0,0045 | * | ANALYZED | |
| APICAL | 0,047 | * | ANALYZED | ||
| 17 | REMAINING | 0,2491 | 15 POLYMORPHISMS | ** | |
| APICAL | 0,2756 | 15 POLYMORPHISMS | ** | ||
| 18 | REMAINING | 0,0012 | * | ANALYZED | |
| APICAL | 0,004 | * | ANALYZED | ||
| 19 | SAND | REMAINING | 0,0096 | * | ANALYZED |
| APICAL | 0,0158 | * | ANALYZED | ||
| 20 | REMAINING | 0,0077 | * | ANALYZED | |
| APICAL | 0,031 | * | ANALYZED | ||
| 21 | REMAINING | 0,0605 | * | ANALYZED | |
| APICAL | 0,1084 | * | ANALYZED | ||
| 22 | REMAINING | 0,007 | * | ANALYZED | |
| APICAL | 0,004 | * | ANALYZED | ||
| 23 | REMAINING | 0,001 | * | ANALYZED | |
| APICAL | 0,039 | * | ANALYZED | ||
| 24 | REMAINING | Indeterminated | * | ANALYZED | |
| APICAL | 0,0069 | * | ANALYZED | ||
| 25 | REMAINING | Indeterminated | * | ANALYZED | |
| APICAL | 0,0020 | * | ANALYZED |
Legend: * Validated less than 7 markers ** Not been studied
Discussion and Conclusions
Various types of teeth were sampled for this study: intact, carious, endodontically treated and restored. Teeth without caries were not included as referred to in the studies by Schwartz and colleagues (1991) (20) Chen, Sun e Wu (1994) (21) ou Alvarez-Garcia (1995) (7), López (1996) (22) and Utsuno e Minaguchi (2004) (23). Over the years, the pulp has been considered the ideal source of DNA. Pulp degradation in extreme forensic situations, endodontic clinical procedures and difficulty anatomically in removing pulp tissue, makes, pulp tissue unsuitable for sampling. In this study the teeth were cleaned and pulpectomized and the mineralized tissues of two root portions were analysed.
There are several techniques for tooth preparation before DNA extraction (7, 20-23), the most common is the crushing or grinding, which implies sample destruction. In this paper the tooth was preserved for morphological study, crown and restorations description. The main purpose of this research is to reduce the damage of archaeological specimens, maintain the crown morphology and validate access through the pulp, when this can yield potential samples (25, 26).
The apical portion of the tooth was compared to the remaining radicular portion, and a topographic molecular analysis was made considering the two different portions of the root: the apical portion and remaining root. The values obtained in the quantification of total DNA were higher for the apical portion than for the remaining root (table 1)the same applies to the number of genetic profiles that were obtained in each radicular portion. The type of the tooth did not affect the results obtained. Due to the tooth’s anatomy, the pulp chamber communicates with the surrounding environment through the tooth’s apical foramen located near the apex, and through which mineralized tissues of the erupted tooth maintain their activity. Elements resulting from cell death are disseminated through the apex: potential elements of DNA degradation and contamination.The pulp that fills the dentinal canalicules is degraded after death in a centripetal manner. Recent studies have focused on mineralized tissuesas an alternative source of DNA. One such study, published in 2003 by Gaytemenn and colleagues, shows that the mid portion of the root is the ideal mineralized tissue for genetic analysis.
Our results contradict their findings (Table 1 and Fig 1).
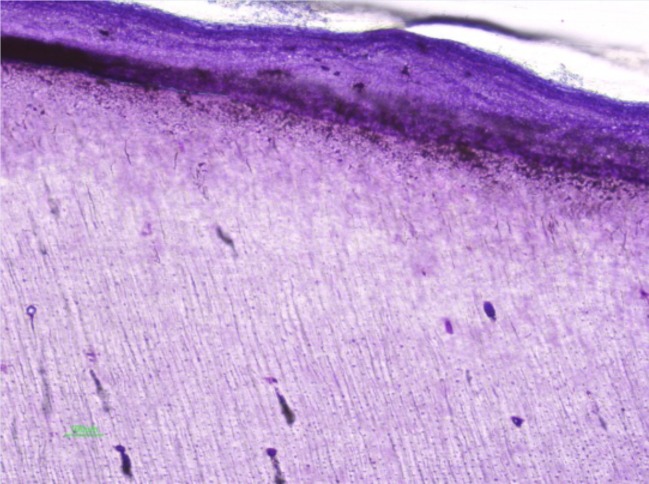
Our results can be explained based on the fact that apical canicular obliteration through the formation of tertiary dentin can preserve some of the pulpar contents in the topographic region, resembling a "mosquito in amber"; on the other hand cementogenesis in the apical portion can occur by invagination into the canaliculi in a rapid and disorganized fashion enabling the trapping of cementocites in its lacunae (fig. 4). In our study it was the apical portion that through its cellular content presented Ageing processes increase the quantity of collected DNA, which can be explained by the increased numbers of cell repairs and regenerative processes with increased sequestration of cells in the mineralized matrix (fig. 4).
Fig. 4.
Microscopic photograph of cementogenesis in the apical root portion.
Forensic laboratory practice shows that success in DNA typing depends on the conditions under which bodies are maintained and their capacity to degrade (6, 11, 12, 24). In order to promote extreme degradation of the pulp we have mimicked extreme forensic conditions as in prior studies by Pfeiffer and colleagues (1999), Burger and colleagues (1999) and in studies by Alonso and colleagues (2001). To evaluate DNA quality, different polymorphisms were studied: 15 STRs and Amelogenin, the full profile for identification according to the European Standard.
We have verified that sand and pH=5 were the mediums that produced the poorest results in DNA analysis: less than seven nuclear markers were validated, only permitting the characterization of maternal lineage through the mitochondrial polymorphisms chosen for this study. We verified that DNA quantity and quality are correlated (27). For samples under 0,179ng/µL (tab. 1) mitochondrial DNA was used to obtain an ID. For the other samples we were capable of detecting more than seven autosomal markers, which are the preferred markers for DNA identification (16, 17). In samples 1,2,5,7,8 and 14, (Table 1) autosomal polymorphic validation for an ID (15 STRs) was obtained in the apical radicular portion while this was not possible in the remaining radicular portion. To increase the chances of compiling a complete profile from the samples (table1), we amplified with the more sensitive next generation kits, such as AmpFℓSTR® NGMTM PCR amplification kit (Applied Biosystems (D3S1358, vWA, D16S539, D2S1338, D8S1179, D21S11, D18S51, D19S433, TH01, FGA, D1S1656, D12S391, D10S1248, D2S441 and the gender determination locus amelogenin) and the PowerPlex® ESI 17 System (Promega) (D22S1045, D2S1338, D19S433, D3S1358, Amelogenin, D2S441, D10S1248, D1S1656, D18S51,D16S539, D12S391, D21S11, vWA, TH01, SE33, FGA AND D8S1179) (28).
We believe that these preliminary results warrant a further study with a larger sample size.
This study emphasizes the value of the genetic analysis of mineralized tooth tissues as an alternative to pulp, especially in extreme forensic conditions.
Footnotes
The authors declare that they have no conflict of interest.
References
- 1.Mjor IA, Sveen OB, Heyeraas KJ. Pulp-dentin biology in restorative dentistry. Part 1: normal structure and physiology. Quintessence Int. 2001. Jun;32(6):427–46. [PubMed] [Google Scholar]
- 2.Berkovitz BVB, Holland GR, Moxham BJ. A colour atlas and textbook of oral anatomy, histology and embryology. London, 2nd ed. Wolfe; 1992 [Google Scholar]
- 3.Mjor IA, Smith MR, Ferrari M, et al. The structure of dentine in the apical region of human teeth. Int Endod. 2001;5:346–53. 10.1046/j.1365-2591.2001.00393.x [DOI] [PubMed] [Google Scholar]
- 4.Pötsch L, Meyer U, Rothschild S, et al. Application of DNA techniques for identification using human dental pulp as a source of DNA. Int J Legal Med. 1992;05:139–43. 10.1007/BF01625165 [DOI] [PubMed] [Google Scholar]
- 5.Smith BS, Weedn VW, Warnock GR, et al. A systematic-approach to the sampling of dental DNA. J Forensic Sci. 1993;38:1194–209. [PubMed] [Google Scholar]
- 6.Pfeiffer H, Mornstad H, Teivens A. Influence of soil storage and exposure period on DNA revorery from teeth. Int J Legal Med. 1999;112:142–4. 10.1007/s004140050219 [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-García A, Munoz I, Pestoni C, et al. Effect of environmental factors on PCR-DNA analysis from dental pulp. Int J Legal Med. 1996;109:125–9. 10.1007/BF01369671 [DOI] [PubMed] [Google Scholar]
- 8.Pfeiffer H, Brinkmann B, Huhne J, et al. Mitochondrial DNA extraction and typing from isolated dentin-experimental evaluation in a Korean population. Int J Legal Med. 1998;111:309–13. 10.1007/s004140050177 [DOI] [PubMed] [Google Scholar]
- 9.Pfeiffer H. The Kaiser’s tooth. Int J Legal Med. 2003;117:118–20. [DOI] [PubMed] [Google Scholar]
- 10.Marjanovic D, Durmic-Pasic A, Kakal N, et al. DNA identification of skeletal remains from world war II mass graves uncovered in Sloveniale. Croat Med J. 2007;4:513–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Burger S, Hummel B, Hermann, et al. DNA preservation: A microsatellite-DNA study on ancient skeletal remains. Electrophoresis. 1999;20:1722–8. [DOI] [PubMed] [Google Scholar]
- 12.Alonso A, Andelinovic S, Martin P, et al. DNA typing from skeletal remains: evaluation of multiplex and megaplex STR systems on DNA isolated from bone and teeth samples. Croat Med J. 2001;3:260–6. [PubMed] [Google Scholar]
- 13.Gaytmenn RM, Sweet SD. Quantification of forensic DNA from various regions of human teeth. J Sci. 2003;3:622–5. [PubMed] [Google Scholar]
- 14.Prinz M, Carracedo A, Mayr WR, et al. DNA Commission of the International Society for Forensic Genetics (ISFG): Recommendations regarding the role of forensic genetics for disaster victim identification (DVI). For Sci Int. Genetics. 2007;•••:13–12. [DOI] [PubMed] [Google Scholar]
- 15.Parsons TJ, Huel R, Davoren J et al. Application of novel “mini-amplicons” STR multiplexes to high volume casework one degraded skeletal remains. For Sci Int Genetics 2007;1:175-17 [DOI] [PubMed]
- 16.Butler JM. Genetics and genomics of core short tandem repeat loci used in human identity testing. J Sci. 2006. March;51:253–65. [DOI] [PubMed] [Google Scholar]
- 17.Castella V, Dimo-Simonim N, Brandt-Casadevall C et al. Forensic evaluation of the QIAshredder/QIAamp DNA extraction procedure. For Sci Int 2006;156:70-73 [DOI] [PubMed]
- 18.Brandstatter A, Parsons TJ, Parsons W. A rapid screening of mtDNA coding region SNPs for identification of west European Caucasian haplogroups. Int J Legal Med. 2003;117:291–8. 10.1007/s00414-003-0395-2 [DOI] [PubMed] [Google Scholar]
- 19.Parsons W, Tendt L, Ballard D et al.Identification of West Eurasian mitochondrial haplogroups by mtDNA SNP screening: Results of the 2006-2007 EDNAP collaborative exercise. For Sci Int Genet 2008;1:61-68 [DOI] [PubMed]
- 20.Schwartz TR, Schwartz EA, Miezerski L, et al. Characterization of deoxyribonucleic-acid (DNA) obtained from teeth subjected to various environmental conditions. J Sci. 1991;4:979–90. [PubMed] [Google Scholar]
- 21.Chen L, Sun G, Wu M. Influence exerted by environmental and hysicochemical factors on the results of sex identification of human dental pulp by polymerase chain reaction. Hua Hsi l Ko Ta Hsueh Hsueh Pao. 1994;25(3):253–8. [PubMed] [Google Scholar]
- 22.López J. Identificación de cadáveres calcinados y en grandes catástrofes: aplicación de métodos odontológicos actuales. Importancia de marcadores genéticos en tejido dental. Dissertation, 1996,Tese Doctoral, University Madrid [Google Scholar]
- 23.Utsuno H, Minaguchi K. Influence of template DNA degradation on the genotyping of SNPs and STR polymorphisms from forensic materials by PCR. Bull Tokyo Dent Coll. 2004;45:33–46. 10.2209/tdcpublication.45.33 [DOI] [PubMed] [Google Scholar]
- 24.Holland MM, Cave CA, Holland CA, et al. Development of a quality, high throughput DNA analysis procedure for skeletal samples to assist with the identification of victims from the World Trade Center attacks. Croat Med J. 2003;44:264–72. [PubMed] [Google Scholar]
- 25.Pinchi V, Torricelli F, Nutini AL, Conti M, Iozzi S. G. Norelli Tecnhiques of dental DNA extraction: some operative experiences. Forensic Sci Int. 2011;204:111–4. 10.1016/j.forsciint.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 26.Kitayama T, Ogawa Y, Fujii K, et al. Evaluation of a new experimental kit for the extraction DNA from bones and teeth using a non-power method. Leg Med (Tokyo). 2010;12:84–9. 10.1016/j.legalmed.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 27.Gill P, Whitaker J, Flaxman C, et al. An investigation of the rigor of interpretation rules for STRs derived from less than 100pg of DNA. J Sci. 2000;91:41–53. [DOI] [PubMed] [Google Scholar]
- 28.Senge T, Madea B, Junge A, et al. STRs miniSTRs and SNPs – a comparative study for typing degraded DNA. Leg Med (Tokyo). 2011;13:68–74. 10.1016/j.legalmed.2010.12.001 [DOI] [PubMed] [Google Scholar]