Abstract
Optogenetics with microbial opsin genes has enabled high-speed control of genetically specified cell populations in intact tissue. However, it remains a challenge to independently control subsets of cells within the genetically targeted population. Although spatially precise excitation of target molecules can be achieved using two-photon laser-scanning microscopy (TPLSM) hardware, the integration of two-photon excitation with optogenetics has thus far required specialized equipment or scanning and has not yet been widely adopted. Here we take a complementary approach, developing opsins with custom kinetic, expression and spectral properties uniquely suited to scan times typical of the raster approach that is ubiquitous in TPLSM laboratories. We use a range of culture, slice and mammalian in vivo preparations to demonstrate the versatility of this toolbox, and we quantitatively map parameter space for fast excitation, inhibition and bistable control. Together these advances may help enable broad adoption of integrated optogenetic and TPLSM technologies across experimental fields and systems.
Microbial opsin genes1 can be introduced into neurons2,3 to provide control over defined circuit elements in intact tissue or organisms (reviewed in ref. 4). Most such studies have involved synchronous control of genetically targeted cell populations over millimeter-scale spatial domains3–19, for example, in studies of sleep-wake transitions5, parkinsonian circuitry6,7, gamma rhythms8–10, feeding behavior11,12, olfaction13, anxiety and fear14–17 and memory storage18,19. Yet methods for guiding spatial delivery of light excitation itself could allow improved precision and complexity in optogenetic modulation20–26, and indeed single-photon guided-light strategies have been used in mammalian tissue27,28 for optogenetic circuit mapping29,30 and dissection of anxiety circuitry14. Even more spatially precise control has been achieved using TPLSM31 methods for generating restricted excitation volumes, and such methods have advanced the study of circuit wiring, activity and plasticity (reviewed in ref. 32) in combination with caged compounds33–35. Although they are elegant, the TPLSM uncaging methods do not readily allow direct targeting of genetically specified cell populations, are difficult to apply in vivo in mammals and encounter challenges in controlling sustained, precisely timed spike trains. In contrast, optogenetics and two-photon excitation together could enable manipulation of single or multiple genetically and spatially targeted cells with high temporal resolution over sustained intervals and within intact volumes.
Prior groundbreaking studies have provided proof of principle for two-photon activation of neurons using the channelrhodopsin variant ChR2-H134R (ChR2HR). One pioneering study was able to overcome the low single-channel conductance of ChR2HR and produce spikes in cultured neurons using spiral scanning to open sufficient ChR2HR channels on individual neurons36. Two other reports of neuron activation in slice preparations relied on elegant hardware innovations and larger focal spots of nonscanning illumination37,38. Building on this past work and our own observations2,3,10,39–41, we found that, for timescales and excitation volumes typical of two-photon laser-scanning hardware, additional limitations of two-photon optogenetics include the off-kinetics and maximal photocurrents of optogenetic tools. By selecting and engineering new opsins with appropriate off-kinetics and photocurrent properties, we now bring a panel of depolarizing, hyperpolarizing and bistable capabilities from the one-photon realm into the two-photon realm for culture, slice and in vivo preparations; moreover, these tools require only standard raster-scanning two-photon microscopy hardware and protocols, thereby complementing earlier two-photon optogenetic studies.
RESULTS
Two-photon excitation of chimeric ChRs in cultured neurons
To design tools for activating neurons with standard two-photon raster scanning, we first sought to pinpoint limitations of ChR2HR. We began with cultured hippocampal neurons transduced with ChR2HR and used whole-cell recording during unidirectional raster-scanning two-photon excitation with a 40×/0.8–numerical aperture (NA) objective (940-nm light at the two-photon ChR2HR spectral peak with 237-femtosecond pulse length and 20-mW laser power; Supplementary Fig. 1a). Resulting depolarizations were found to first increase and then decrease over a single frame scan, and variation in the time taken to scan a cell altered peak depolarization (Supplementary Fig. 1b,c). We therefore hypothesized that not only improving expression and membrane targeting but also sufficiently slowing channel deactivation to allow two-photon–elicited photocurrents to accumulate during raster scanning could be useful for robust two-photon– driven spiking.
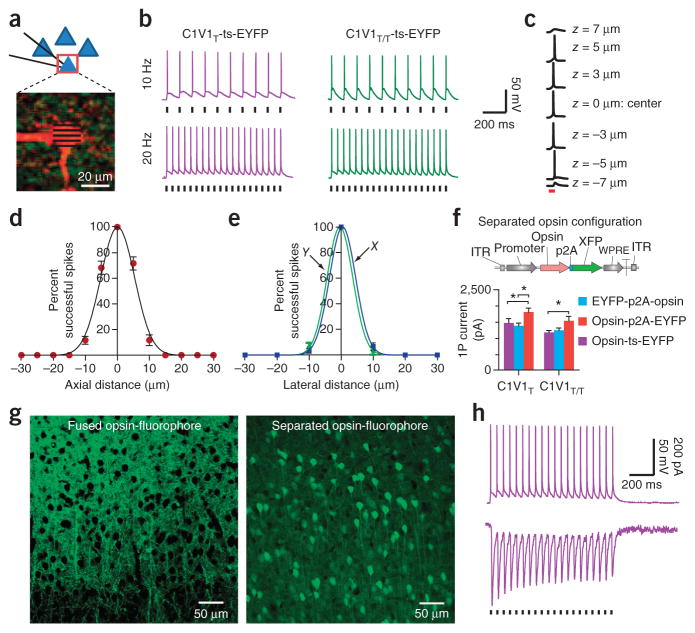
The red-shifted C1V1 channelrhodopsin family (based on chimeras between ChR1 and VChR1 and including the E162T (C1V1T) and E122T/E162T (C1V1T/T)10 variants as well as membrane-trafficking modifications such as the ‘ts’ signal39; Fig. 1a–c) displays off-kinetics (τoff, or time constant of deactivation after light exposure ends; Fig. 1d,e) two- to fivefold slower than those reported for ChR2HR41; moreover, photocurrents were greater for C1V1T than for ChR2HR with single-photon full-field illumination (Fig. 1b). As both properties suggested utility for two-photon application, we transfected the C1V1 opsins into cultured neurons and performed whole-cell recordings while raster scanning over the soma (Fig. 1c).
Figure 1.
Two-photon control of spiking with C1V1 variants in culture. (a) C1V1 mutants (white bands: mutations). (b) C1V1 and ChR2HR single-photon photocurrents (C1V1, 1,075 ± 74 pA (n = 7); C1V1T, 1,474 ± 130 pA (n = 18); C1V1T/T, 1,175 ± 58 pA (n = 18); ChR2HR, 1,075 ± 74 pA (n = 12), *P = 0.02; see Online Methods for parameters; Mann-Whitney two-tailed test; error bars, s.e.m.). (c) C1V1T-ts-EYFP–expressing hippocampal neuron. Purple square: typical ROI. (d) TPLSM photocurrents. Bars indicate scan times; typical ROIs were 15 × 15 μm to 20 × 20 μm. (e) τoff values (C1V1, 105 ± 1.1 ms (n = 6); C1V1T, 60 ± 1.9 ms (n = 6); C1V1T/T, 40 ± 1.3 ms (n = 5)). (f) TPLSM photocurrents for varying NA (C1V1: 40×/0.8, 239 ± 21 pA (n = 17), 20×/0.5, 490 ± 44 pA (n = 6), P = 0.00007; C1V1T: 40×/0.8, 244 ± 20 pA (n = 14), 20×/0.5, 582 ± 32 pA (n = 6), P = 0.00005; C1V1T/T: 40×/0.8, 269 ± 46 pA (n = 11), 20×/0.5, 585 ± 80 pA (n = 7), P = 0.001; * denotes significance at P < 0.05 level). (g) C1V1T TPLSM photocurrents elicited by different light wavelengths. (h) TPLSM action spectra (C1V1, n = 5; C1V1T, n = 5; C1V1T/T, n = 6). Values were normalized to the maximum photocurrent within the cell across wavelengths. (i) Laser intensity versus TPLSM photocurrent (C1V1, n = 8; C1V1T, n = 8; C1V1T/T, n = 13). Normalization as in h. (j) Left, 1,040-nm, TPLSM-evoked spiking. Right, spiking efficacy (10 Hz: C1V1 n = 6, 36.7% ± 6% versus C1V1T and versus C1V1T/T, both n = 6, 100% ± 0%, P = 0.002; 20 Hz: C1V1 n = 6, 18.3% ± 3.7% versus C1V1T n = 6, 56.7 ± 10%, P = 0.001 and versus C1V1T/T n = 6, 91 ± 4%, P = 0.01; * denotes significance at P < 0.05 level). Black squares: spiking of the same cells in response to direct current injection (400-pA, 10-ms current pulses provided at the indicated frequency). Resting membrane potential (Rm), −65 mV; 2P, two photon.
Because the C1V1 action spectrum is red shifted, we began with the longest illumination wavelength compatible with widely used titanium-sapphire (Ti-S) lasers (1,040 nm), and we found that standard raster scanning with 0.5-NA objectives gave rise to photocurrents in the range of 500–600 pA (Fig. 1f). All C1V1-family two-photon raster-scanning illumination-evoked photocurrents were significantly larger than corresponding currents for ChR2HR (Supplementary Fig. 2a), even when normalized to saturating-range single-photon full-field illumination-evoked photocurrents in each cell (Supplementary Fig. 2b), which further suggests utility for C1V1 variants in two-photon excitation. C1V1T narrowly outperformed other members of the C1V1 family by this measure, consistent with high functional expression (Fig. 1b) and an approximately threefold-increased τoff relative to that of ChR2HR (Fig. 1d,e)41, although all C1V1 variants outperformed ChR2HR.
After this result, we mapped illumination parameters. We first explored an experimental parameter governing laser exposure time by comparing peak photocurrent to maximal charge transfer (that is, the area under the photocurrent-versus-time curve) for a raster scan under conditions in which pixel dwell time was held constant but resolution of each line scan was increased; peak photocurrents depended heavily upon this practical scan-resolution parameter, which influences both scan-line time and scan-line number (Supplementary Fig. 2c–h). Next, cellular photocurrent recovery between scans, a process of practical importance, was tracked and found to proceed with kinetics such that essentially full recovery was attained with a 30-s interscan interval (Supplementary Fig. 3a). We next found that the greatest photocurrents under these conditions occurred at 1,040 nm (Fig. 1g–i; less widely available, longer-wavelength illumination approaches are likely to exceed this level for the red-shifted C1V1 family, as photocurrents had not peaked at 1,040 nm); when laser power at 1,040 nm was varied between 1 and 30 mW (Fig. 1i), data were found to fit the expected quadratic function for low laser-intensities—indicative of a two-photon process (Supplementary Fig. 3b). When we varied only the parameter of dwell time (the duration the laser remained on each pixel, which influences total scan-line time while holding number of scan lines constant), we noted an inverted-U pattern in photocurrent amplitude (Supplementary Fig. 3c,d); only for certain regimes and certain C1V1 variants could higher peak photocurrents be achieved by increasing time per scan line via increasing dwell time, and in some cases lower peak photocurrents actually resulted, again consistent with off-kinetics (Supplementary Fig. 3c–f).
It is of practical importance to note that under the same pixel dwell time and scan resolution, using a 20×/0.5-NA objective with a greater illumination point-spread function and thus excitation volume (Supplementary Fig. 4a–c) will elicit significantly larger photocurrents than using (for example) a 40×/0.8-NA objective (Fig. 1f); this expected effect of the illumination point-spread function depends on (among other parameters) the degree to which the back aperture of the objective is filled as described42, and influences ChR variants similarly (Supplementary Fig. 4). In a final practical test of opsin performance in culture, we found that faster opsin off-kinetics (Fig. 1d,e) predicted improved high-frequency performance, with C1V1T/T generating the best-resolved high-frequency 1,040-nm-elicited photocurrents in voltage clamp (Supplementary Fig. 5) as well as the highest- frequency sustained spike trains in current clamp (Fig. 1j). Notably, for all C1V1 family members, standard raster-scan illumination at 1,040 nm generated sustained, precisely timed spike trains (Fig. 1j).
Two-photon C1V1-driven activation of neurons in slices
Whereas use of a simple two-photon diffraction-limited spot in combination with optogenetics to generate spikes has been successful in culture36, spike generation in tissue slices (‘in slice’) has been accomplished using (for example) advanced methodologies to shape two-photon spots37,38. To explore C1V1-variant functionality in slice, we targeted pyramidal cells in slices from prefrontal cortex or hippocampus of adult mice via injection of adeno-associated virus41 (AAV5) bearing C1V1-family opsins under the control of the CaMKIIα promoter, and we performed whole-cell recordings (20×/0.5-NA objective, 32 °C) 5–6 weeks after transduction during standard raster scanning with 1,040-nm illumination (Fig. 2a). We tested C1V1T and C1V1T/T in slice, given their improved frequency response (Fig. 1j) compared with that of C1V1, and confirmed the potent responses to 1,040-nm light that had been seen in culture (Supplementary Fig. 6). At high frame-scan frequencies, C1V1T/T (with faster off-kinetics) showed greater photocurrent recovery than C1V1T between light pulses (that is, reduced current persisting between scans) in voltage clamp (Supplementary Fig. 6a,b); in current clamp, we were able to drive action potentials in C1V1T-transduced cells reliably at 20 Hz and in C1V1T/T-transduced cells at up to 40 Hz (Fig. 2b and Supplementary Fig. 6c).
Figure 2.
Two-photon control of spiking with C1V1 variants in acute brain slices. (a) Top, targeted neurons during TPLSM excitation (red box, ROI). Bottom, two-photon image of patched/Alexa 594–filled neuron expressing C1V1T-EYFP; lines schematize TPLSM pattern. Throughout, ROIs in slice were ~10 × 10 μm to 15 × 15 μm. (b) TPLSM spike generation under 1,040-nm illumination in neurons expressing C1V1T-ts-EYFP or C1V1T/T-ts-EYFP (C1V1T-ts-EYFP in hippocampus (n = 15) and prefrontal cortical (n = 51) neurons; C1V1T/T-ts-EYFP in hippocampus (n = 7) and prefrontal cortical (n = 8) neurons). Resting membrane potential (Rm), −65 to −70 mV; black ticks, scan onset. (c) TPLSM activation of a representative C1V1T-ts-EYFP–expressing cell from targeting different axial (z) positions. Rm, −65 mV. (d) Percent successful spikes versus axial position in C1V1T-ts-EYFP–expressing cortical neurons. Rm, −65 mV; n = 4; error bars, s.e.m. (e) Percent successful spikes versus lateral distance from the center of a representative cortical cell expressing C1V1T-ts-EYFP. Blue, X; green, Y; Rm, −65 mV; n = 7; error bars: s.e.m. (f) Top, p2A strategy: opsin separated from fluorophore (XFP). Bottom, single-photon (1P) photocurrent for different configurations of C1V1, fluorophore and p2A (C1V1T: EYFP-p2A-opsin 1,383 ± 80 pA (n = 8), opsin-ts-EYFP 1,474 ± 130 pA (n = 12), opsin-p2A-EYFP 1,811 ± 125 pA (n = 18; P = 0.04 versus two comparison groups); C1V1T/T: EYFP-p2A-opsin 1,245 ± 67 pA (n = 8), opsin-ts-EYFP 1,175 ± 58 pA (n = 9), opsin-p2A-EYFP 1,541 ± 135 pA (n = 17, P = 0.009 versus opsin-ts-EYFP)). P values, Mann-Whitney two-tailed test for non-Gaussian distribution. (g) Left, cortical neurons expressing C1V1T-ts-EYFP. Right, cortical neurons expressing C1V1T-p2A-EYFP. (h) Top, current clamp: TPLSM-stimulation of C1V1T-p2A-EYFP (20 Hz; Rm, −65 mV). Bottom, voltage clamp.
We next tested the spatial resolution of two-photon spike elicitation: during whole-cell recording from prefrontal slices, we positioned the scanning region of interest (ROI) at the soma center and moved the objective in the lateral and axial planes (Supplementary Fig. 7a,b). For suitable spatial resolution, we found that reliable spike generation, required centering the ROI on the soma (Fig. 2c; typical cell size was ~10 × 10 × 15 μm). The resolution of spiking revealed a full-width half-maximum value of 12.5 μm axially (Fig. 2d) and 9.3–9.5 μm laterally (Fig. 2e) around the soma, consistent with photocurrent measurements for the same axial and lateral positions (Supplementary Fig. 7).
It was challenging to define ROIs around tightly packed cells visualized via this membrane-delimited opsin-fluorophore fusion. Therefore, to enable robust targeting of individual neurons, we separated fluorophore from opsin to fill targeted cells with the fluorophore while maintaining opsin expression at the membrane. Using the previously described 9-amino-acid picornavirus p2A motif for expression of two proteins from a single promoter39,43, we separated C1V1T from EYFP (Fig. 2f) and injected AAV5 bearing this separated-opsin configuration into prefrontal cortex. The p2A motif was designed to remain on the opsin C terminus so that immunostaining for p2A still allows subcellular tracking of opsins (Supplementary Fig. 8a). In slices prepared 5–6 weeks after injection, this configuration enabled robust discrimination and definition of single cells, crucial for these two-photon methods (Fig. 2g and Supplementary Fig. 8b). Finally, we verified functionality: in a comparison of three different configurations (opsin-ts-EYFP, opsin-p2A-EYFP and EYFP-p2A-opsin), the highest photocurrents were observed in the opsin-p2A-EYFP configuration (Fig. 2f) along with robust raster-scanning 1,040-nm-driven spike generation in slice (Fig. 2h).
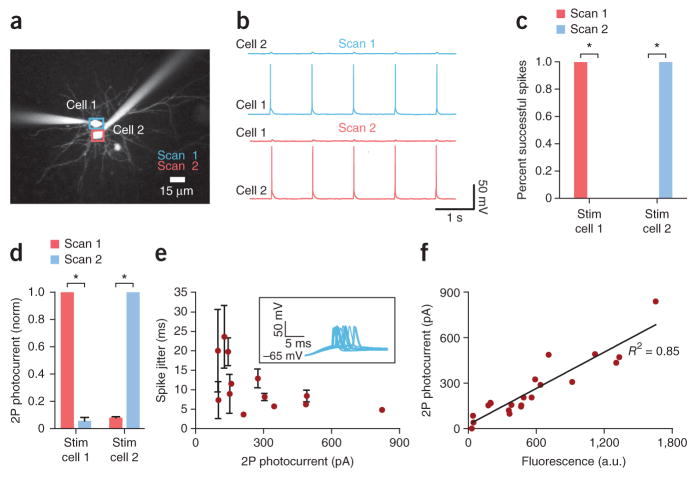
To confirm that intact-tissue, single cell–resolution, raster-scanning two-photon spike generation was possible, we performed dual whole-cell recordings from neurons separated by no more than 1–2 μm (Fig. 3a) and attempted to independently fire each cell (Fig. 3b,c). We did not observe spikes in adjacent nontargeted cells while the targeted cell was driven to spike (Fig. 3b), a property also reflected in the small photocurrents recorded from adjacent nontargeted cells (Fig. 3d). As expected, increased photocurrent was associated with shortened latency to spike, lower jitter (Fig. 3e) and greater fluorescence intensity (Fig. 3f). Finally, to test optogenetic two-photon stimulation of dendritic spines in slice, we filled patched cells with Alexa 594 dye to ensure that the stimulated spine would be part of the patch-clamped neuron, and we found that we could readily induce photocurrents averaging 7.3 pA (Supplementary Fig. 8c; n = 10 cells, 29 spines; 1,040-nm raster scans over single spines; 40×/0.8-NA objective).
Figure 3.
Spatial resolution of C1V1-mediated two-photon excitation in acute brain slices. (a) Two-photon image of dual whole-cell recording from Alexa 594–filled pyramidal cells expressing C1V1T-p2A-EYFP. (b) Current-clamp traces during TPLSM activation of cell 1 or cell 2. Resting membrane potential (Rm), −65 mV. (See Online Methods for stimulation parameters.) Typical ROI sizes were as in Figure 2a. (c) Spike probability for TPLSM stimulation (stim) of cell 1 or 2. Rm, −65 mV; similar results were obtained in n = 4 cell pairs (*P = 0.03) wherein no spikes were seen in adjacent cells despite 100% fidelity in driving the targeted cell at 1 Hz, regardless of which cell was targeted. (d) TPLSM-elicited photocurrents in voltage-clamp mode. Values were normalized to the maximum photocurrent observed between cell 1 and cell 2 during stimulation of either cell 1 or cell 2 (n = 4 cell pairs, *P = 0.03; error bars, s.e.m.; P values, Mann-Whitney two-tailed test for non-Gaussian distribution). (e) Spike jitter (s.d. of spike timing stimulated at 1 Hz) versus TPLSM photocurrent for pyramidal cells expressing C1V1T-p2A-EYFP (n = 12 cells). Inset, representative current clamp traces showing jitter. Rm, −65 mV; error bars, s.d. of mean time to spike. (f) Plot of 1,040-nm TPLSM photocurrent versus EYFP fluorescence of pyramidal cells expressing C1V1T-p2A-EYFP. Cells were selected here not for high expression but to cover a broad range of expression, to assess relationship between fluorescence and photocurrent; linear fit R2 = 0.85. a.u., arbitrary units.
In vivo optogenetic activation of single neurons
We next turned to the mammalian in vivo preparation (Fig. 4a). We injected AAV5 bearing CaMKIIα::C1V1T-p2A-EYFP into superficial somatosensory cortex of adult mice and observed robust expression in superficial layers (Fig. 4b). The cell-filling configuration sufficed to allow not only selection of individual cells but also visualization of individual dendritic processes and spines in vivo (Fig. 4c). After visualizing and targeting individual neurons in anesthetized mice, we performed loose patch recordings under two-photon–guided patch-clamp to test stimulation of individual neurons with appropriate spatial resolution (Fig. 4a,d). By placing the ROI at the center of single pyramidal cells, we were able to drive precise spike trains using 1,040-nm light and standard raster-scanning light delivery to the single-cell ROI in the living mouse (Fig. 4d). We noted that moving in either axial or lateral directions as in slice reliably eliminated spike generation, indicating high spatial (Fig. 4d) as well as temporal precision in vivo.
Figure 4.
Two-photon optogenetic control of spike firing in vivo in adult mammals. (a) Schematic of the experimental setup for in vivo two-photon control of superficial layer 2 and 3 (2/3) somatosensory neurons transduced with C1V1T-p2A-EYFP. (b) Left, two-photon image of layer 2/3 pyramidal neurons transduced with C1V1T-p2A-EYFP in somatosensory cortex (150–250 μm below pia). Right, two-photon image of layer 1 pyramidal neurons transduced with C1V1T-p2A-EYFP in somatosensory cortex (50–150 μm below pia). (c) Two-photon image of dendritic spines on pyramidal cells transduced with C1V1T-p2A-EYFP in layer 2/3 of somatosensory cortex. (d) Upper left, in vivo two-photon image of layer 2/3 pyramidal cells transduced with C1V1T-p2A-EYFP (imaged during loose patch). Lower left, trace showing precise spike-train control with 5-Hz 1,040-nm raster-scanning illumination; the amplitude and waveform of these evoked spikes recorded in cell-attached mode matched the spontaneous spikes in each cell. Upper right, axial resolution of two-photon optogenetic control of spiking in vivo. Blue triangles indicate pyramidal neurons and red boxes illustrate ROI positioning. Spiking of layer 2/3 cells as a function of raster scan position is shown (20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel, 3.2 μs, scan resolution, 0.6 μm per pixel; line scan speed, 0.19 μm μs−1; laser intensity, 20 mW). Typical ROI sizes were as in Figure 2a. Lower right, lateral resolution of two-photon optogenetic control of spiking in vivo. Similar results were observed in n = 3 neurons in vivo, all 150–250 μm below pia in layer 2/3 in somatosensory cortex.
Bistable two-photon optogenetic control
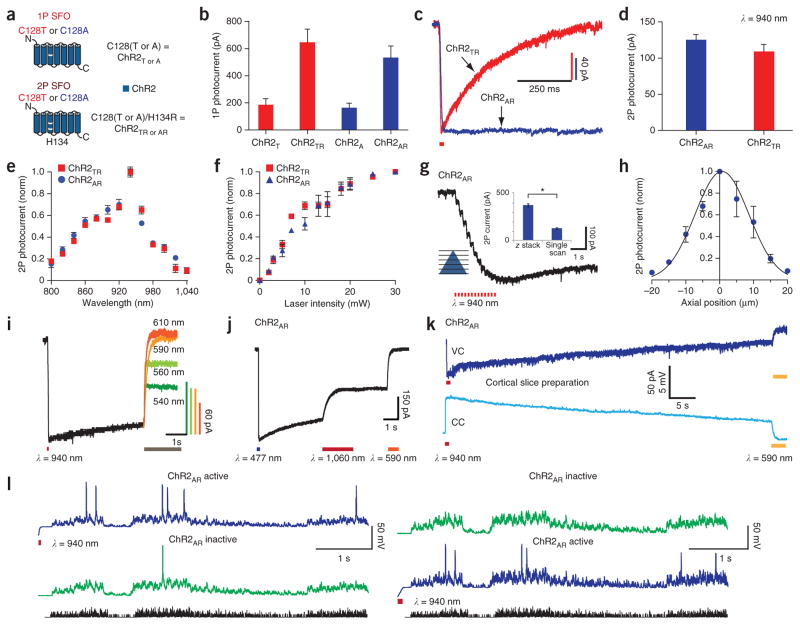
A distinct kind of optogenetic control relevant to two-photon methods involves bistable or step-function opsin (SFO) genes, which give rise to prolonged photocurrents far outlasting the initiating pulses of light10,40. Blue light–activated SFOs can be inactivated with yellow light10,40, and SFOs have been useful for delivering prolonged but still precisely temporally delimited excitability (rather than defined spike trains) to targeted cell populations in behaving mouse neocortex10. This allows elicitation of sparse, distributed, asynchronous and endogenously timed activity in targeted cell populations (rather than exogenously coordinated spiking)—suited to presumed sparse coding in populations, such as principal cells in neocortex, that are often studied with two-photon imaging. However, no study has reported bistable photocurrents using two-photon illumination owing to the challenge of generating photocurrents in the >50-pA range typically required to modulate circuit physiology and behavior with currently published SFOs10,40. For example, although the stabilized SFOs (SSFOs)10 are the most kinetically stable among the opsins used in optogenetic experiments, SSFO currents (like earlier C128 SFO currents) are already on the small end of the useful range (~50–200 pA) even with maximal single-photon full-field excitation, thereby posing challenges for less-potent raster- scanning two-photon approaches.
We therefore developed a high-photocurrent SFO for two-photon raster-scanning excitation. Adding the H134R mutation44 onto the background of two earlier SFO variants40, ChR2-C128T (ChR2TR) and ChR2-C128A (ChR2AR) (Fig. 5a) increased one-photon photocurrents threefold (Fig. 5b) while retaining the prolonged off-kinetics of the original SFOs (Fig. 5c); ChR2AR and ChR2TR displayed mean τoff values of 53 s and 3.1 s, respectively (Fig. 5c and Supplementary Fig. 9a). We next found that these novel variants allowed single-scan two-photon (940-nm) elicitation of photocurrents in the required 100- to 150-pA range (Fig. 5d), and we denote these novel variants as 2PSFOs (though of course these tools can also be controlled with single-photon methods, as demonstrated below).
Figure 5.
Bistable two-photon optogenetic control in culture and slice with novel ChR variants. (a) Earlier SFOs (1P SFO) and novel multiple-mutant opsins (2P SFO). (b–j) Experiments in cultured neurons. (b) Single-photon photocurrents. ChR2A: 161 ± 33 pA, n = 7; ChR2AR: 532 ± 87 pA, n = 7, P = 0.0003 versus ChR2A; ChR2T: 185 ± 45 pA, n = 4; ChR2TR: 645 ± 98 pA, n = 7, P = 0.0002 versus ChR2T. (c) TPLSM-stimulated 2PSFO (ChR2TR τoff = 3.2 ± 0.05 s (n = 8), ChR2AR τoff = 53.3 ± 2.8 s (n = 13)). (d) TPLSM photocurrents. ChR2AR: 125 ± 7 pA (n = 15), ChR2TR: 109 ± 10 pA (n = 8). Error bars, s.e.m. (e) Action spectra. Values were normalized to the maximum photocurrent within the cell across wavelengths; ChR2AR (n = 5), ChR2TR (n = 4). ROIs were 15 × 15 to 20 × 20 μm. (f) Laser intensity versus 940-nm, TPLSM-photocurrent (ChR2TR (n = 3), ChR2AR (n = 3)); normalization as in e. (g) ChR2AR TPLSM photocurrents for multiple–axial-plane stimulation. Inset, photocurrents from multiple- or single-plane stimulation (multiplane: 366 ± 15 pA (n = 2), single plane: 125 ± 15 pA (n = 15, *P = 0.01 versus multiplane)). P values, Mann-Whitney two-tailed test for non-Gaussian distribution. (h) TPLSM photocurrent versus axial position (n = 4). (i) Single-photon deactivation of TPLSM-elicited ChR2AR photocurrents. Gray bar, timing of deactivating pulses. (j) TPLSM deactivation of 477-nm-elicited ChR2AR photocurrents. (k) Current clamp (bottom, CC) and voltage clamp (top, VC) of ChR2AR TPLSM-stimulation in cortical slice. TPLSM scan was applied across soma (n = 3). (l) Left, single hippocampal neuron in acute slice with (top) or without (bottom) TPLSM activation of ChR2AR, stimulated with simulated excitatory postsynaptic current (sEPSC) train (black). Right, separate cell with sequence of conditions reversed (n = 4).
Two-photon action spectra were similar to that of ChR2HR, peaking at 940 nm (Fig. 5e); laser intensity dependence fit the quadratic function (at low laser intensities) indicative of a two-photon process (Fig. 5f and Supplementary Fig. 9b). Photocurrent measures were also obtained while varying the practical dwell-time and scan-resolution parameters; results are summarized in Supplementary Figure 9c,d. We next performed whole-cell recordings in hippocampal neurons transduced in culture with ChR2AR, with the scanning ROI at the center of the cell. We observed a profound decrement in 940-nm-evoked photocurrent when moving off the neuron (Fig. 5g,h) consistent with the anticipated spatial resolution. We conjectured that 2PSFO activation would be reversible using red-shifted light (via a single-photon mechanism); indeed, we were able to deactivate the 2PSFOs fully with 590-nm light and partially with more blue-shifted light (Fig. 5i). We also found that continuous raster scanning with 1,060-nm ultrashort pulsed illumination allowed successful deactivation of 2PSFOs in the targeted focal plane (Fig. 5j). Because the raster-scanning two-photon activation is spatially constricted to a subset of opsins, deactivation is partial, whereas full-field one-photon application affects all opsins (Supplementary Fig. 9e); applications may thus be enabled wherein the experimenter can selectively activate all, and then partially deactivate subpopulations of, targeted cells or compartments. Notably, the same wavelength that deactivates SFOs can also activate SFOs, depending on state of the photocycle, because their activation and deactivation spectra overlap10 (Supplementary Fig. 9f).
Finally, we tested for induction of bistable photocurrents in slice. A 2PSFO lentivirus (CaMKIIα::EYFP-p2A-ChR2AR) was injected into adult mouse cortex. In slices prepared 4–5 weeks later, even a single 940-nm raster scan using a 40×/0.8-NA objective sufficed to induce suitable bistable photocurrents (mean = 71 pA, n = 8 cells) in pyramidal neurons (Fig. 5k); in current clamp, this two-photon activation consistently depolarized neurons to subthreshold levels (mean = 8.2 mV, n = 3 cells) just as in hippocampal culture (mean = 9.7 mV, n = 9 cells), increasing the probability of spiking to injected current pulse trains as typically desired for SFOs (Fig. 5l).
Two-photon optogenetic inhibition
Experimental advantages from genetic and optical selection of targeted elements with two-photon–based methods would also apply to optogenetic loss of function. Molecular strategies have enhanced hyperpolarizing tools (Fig. 6a)39,41, and the longer τoff of the engineered proton pump eArch3.0 (refs. 41,45–47) relative to the faster chloride pump eNpHR3.0 (refs. 39,41), along with large photocurrents, could allow two-photon–based raster-scanning inhibition. We transduced cultured hippocampal neurons with eArch3.0 and found that a single raster scan through an ROI around the soma (Fig. 6b) produced suitable outward current (Fig. 6c) depending, as expected, on time per scan line and number of scan lines (Supplementary Fig. 10a,b). Though not clearly at peak, the greatest current with widely used Ti-S lasers occurred at 1,040 nm and saturated at approximately 20 mW (Fig. 6d,e); low–laser intensity data fit the expected quadratic function indicative of a two-photon process (Supplementary Fig. 10c). Suitable spatial resolution was also observed (Supplementary Fig. 10d), and we found that using the 20×/0.5-NA objective increased current amplitude (Fig. 6f) as expected. Finally, although single raster scans provided robust peak currents, repetitive fast scanning, even with a delay between scans, still enabled prolonged outward photocurrents (Fig. 6g) that stably inhibited spiking (Fig. 6h).
Figure 6.
Two-photon optogenetic inhibition. (a) Version 3.0 of opsins for enhancing photocurrent magnitude. (b) Two-photon image of cultured hippocampal neuron transduced with eArch3.0-EYFP; red square: ROI borders. (c) Voltage-clamp traces showing outward currents for 1,040 nm, TPLSM-scanned eArch3.0-expressing neurons at different scan resolutions (blue: n = 7, photocurrent:176.1 ± 20.6 pA; yellow: n = 7 cells, photocurrent 278.7 ± 41.6 pA). (d) Action spectrum of eArch3.0 (n = 3; error bars: s.e.m.). Values were normalized to the maximum photocurrent within the cell across all wavelengths. (e) Laser intensity versus 1,040-nm, TPLSM photocurrent (n = 6). Normalization as in d. (f) TPLSM-evoked eArch3.0 photocurrents elicited with different NA objectives (40×/0.8 NA: n = 7; 20×/0.5 NA; n = 6, *P = 0.03 versus 40×/0.8 NA). P values, Mann-Whitney two-tailed test for non-Gaussian distribution. (g) Trace of continuous TPLSM scan of eArch3.0-expressing neuron. Trace within dashed box at left expanded at right. (h) Trace of TPLSM-mediated spike inhibition in a cultured neuron expressing eArch3.0; spiking evoked by 300-pA current injection. Rm, −65 mV. (i) Two-photon image of pyramidal prefrontal neurons expressing eArch3.0. (j) TPLSM activation of eArch3.0 showing spike-inhibition dependence on TPLSM axial (z) position targeting. Rm, −65 mV. (k) Axial position versus successful spikes. Rm, −65 mV; n = 4. Values were normalized to the maximum number of spikes within the cell across all axial positions. (l) TPLSM inhibition over long timescales with eArch3.0. Top, traces showing key temporal windows. Bottom, spike rate over time. Rm, −65 mV; n = 5. TPLSM scan initiated at 15 s and terminated at 75 s.
We finally turned to acutely prepared slices injecting AAV5 into prefrontal cortex of adult mice to deliver CaMKIIα::eArch3.0-ts-EYFP and observing robust expression 4–6 weeks after injection (Fig. 6i). We defined ROIs centered on pyramidal somata and, while injecting current to drive spiking, applied repetitive raster scans to the ROI. We observed potent inhibition of spiking (Fig. 6j): as we moved the ROI axially from the center of the cell, inhibition of spiking steeply diminished (Fig. 6j,k), indicating suitable spatial resolution in this preparation. Temporal stability was also suitable: in whole-cell recordings with spiking elicited by injected 5-Hz current, continuous 1,040-nm raster scanning centered on the cell produced stable inhibition of spiking (Fig. 6l).
DISCUSSION
Optogenetic tools have been applied across many preparations to determine causal contributions of genetically specified cell types to behavioral and physiological outputs of neuronal circuits. Beyond genetic specificity, further precision can be obtained with optogenetics by activating or inhibiting specific projections from subsets of neurons or neurons defined by specific projections14,30,39. Prior work has been instrumental in demonstrating two-photon optogenetic control and in identifying limitations of two-photon methods in combination with optogenetics. By modifying and validating distinct microbial opsins, here we sought to allow basic raster-scanning two-photon microscopes, hardware and protocols (Supplementary Fig. 11 and Supplementary Table 1) to optogenetically modulate neuronal circuitry at the level of single cells in preparations ranging from culture to the living mammal. The tools demonstrated here (i) address this practical goal, (ii) maintain the temporal precision required for fast spike generation, bistable modulation and spike inhibition (Supplementary Table 2) and (iii) maintain use of a diffraction-limited two-photon spot (thereby eliminating potential power loss due to divided beams or diffraction gratings). In addition, the flexibility of a single diffraction-limited spot to activate or silence can be combined with other elegant spatial light-modulation technologies to probe millisecond-timescale processing48,49.
Potential applications include mapping of neuronal connectivity, which could give rise to a wealth of information on both normal circuitry and pathophysiological processes. For example, animal models of psychiatric disease states might show differences in local connectivity patterns among neurons. To collect these data, multiple paired recordings from ex vivo preparations typically must be conducted, but the technology presented here may allow faster data collection and a more complete picture of network connectivity. By combining this technology with behavioral, physiological and imaging tools currently available50, dissection of the causal contribution of arbitrarily defined sets of elements to circuit physiology and behavior becomes possible.
ONLINE METHODS
Molecular cloning
ChRs and ChR point mutants were human codon optimized, and point mutations were produced by site-directed mutagenesis (QuickChange, Agilent). pLenti-CaMKIIα::opsin-EYFP-WPRE constructs were designed with BamHI between the promoter and the opsin, NotI between the opsin and the fluorophore, and EcoRI between the fluorophore and the WPRE. Cell-filling versions of all the opsins were constructed using a ribosomal skip site (p2A) between the opsin and fluorophore. All opsins were flanked by BamHI and AscI, and the p2A-FYP by AscI and EcoRI, to facilitate modular cloning. Opsins were cloned into the BamHI/EcoRI site of the AAV-CaMKIIα vector, and DNA was packaged by the UNC Vector Core as AAV5. Constructs were sequenced for accuracy of cloning and AAV vectors were tested for in vitro expression before viral production.
Hippocampal neuron culture preparation and transfection
Primary cultured hippocampal neurons were prepared from P0 Sprague-Dawley rat pups (Charles River). CA1 and CA3 were isolated, digested with 0.4 mg ml−1 papain (Worthington), and plated onto glass coverslips precoated with 1:30 Matrigel (Becton Dickinson Labware). Cultures were maintained in a 5% CO2 humid incubator with Neurobasal-A medium (Invitrogen) containing 1.25% FBS (HyClone), 4% B-27 supplement (Gibco), 2 mM Glutamax (Gibco) and 2 mg ml−1 fluorodeoxyuridine (FUDR) (Sigma), and were grown on coverslips in a 24-well plate at a density of 65,000 cells per well.
For transfecting each well, a DNA-CaCl2 mix was prepared with 2 μg DNA (Qiagen endotoxin-free preparation) and 1.875 μl 2 M CaCl2 (final Ca2+ concentration 250 mM) in 15 μl H2O. To DNA-CaCl2 we added 15 μl of 2× HEPES-buffered saline (pH 7.05). After 20 min at room temperature (20–22 °C), the mix was added dropwise into each well (from which the growth medium had been removed and replaced with prewarmed minimal essential medium (MEM)), and transfection proceeded for 45–60 min at 37 °C, after which each well was washed three times with 1 ml warm MEM before the original growth medium was returned.
Animal care
All experiments were conducted under the protocols approved by the Stanford Administrative Panel on Laboratory Animal Care. Male C57/BL6 mice were virus-injected at 3–4 weeks of age as previously described41. Slice physiology was performed 5–7 weeks post-injection.
Stereotactic injections
Adeno-associated virus (AAV) serotype 5 was used for C1V1(E162T or E122T/E162T)-ts-EYFP, C1V1(E162T)-p2A-EYFP, and eArch3.0, all produced by the University of North Carolina at Chapel Hill Vector Core; lentivirus for ChR2(C128A/H134R) in vivo injection was produced as described (http://www.stanford.edu/group/dlab/optogenetics/). One microliter of virus was stereotactically injected bilaterally into either the medial prefrontal cortex (+1.7 mm anteroposterior, 0.3 mm mediolateral and 2.8 mm dorsoventral (from bregma)), somatosensory cortex (−0.5 mm anteroposterior, 2.9 mm mediolateral and 0.3 mm dorsoventral (from bregma)), or hippocampus (−1.0 mm anteroposterior, 1.0 mm mediolateral and 2.0 mm dorsoventral (from bregma)) of 3- to 4-week-old mice.
Microscope hardware
All two-photon experiments were conducted on a customized microscope (Prairie Technologies) fitted on an Olympus BX51. As described, either 40×/0.8-NA or 20×/0.5-NA LUMPlanFL/N (Olympus) objectives were used for all experiments. For two-photon experiments, raster scanning was performed through a defined ROI in a unidirectional, sequential line manner (Supplementary Fig. 11). Typical ROI dimensions were 15 × 15 μm to 20 × 20 μm in culture preparations and 10 ×10 μm to 15 × 15 μm in slice preparations but were adapted on the basis of individual soma morphology. For this scan size and typical scan speeds, there was 8–9% spatial overscan per scan line (Supplementary Fig. 7e) and illumination-time duty cycle of ~30%; illumination was restricted to the ROI, shuttered on and off with the Pockels cells. The time taken (defined here as off-sample time; Supplementary Fig. 7e) to return from the end of the exposure of one scan line (the last exposed pixel of the scan line) to the start of the exposure of next scan line (the first exposed pixel of the scan line) was 2–3 times longer than exposure time on-sample per line. The total scan line time is composed of the off-sample time and the exposure time. Total scan frame time for a cell (or ROI) was typically 10–15 ms, which is likewise a combination of the total exposure time (laser irradiation on cell or ROI) and the total off-sample time. In order to estimate typical exposure times, three parameters are employed: scan speed, scan resolution and size of the ROI. The exposure time per cell (or ROI) is estimated as the irradiation time per line times the number of lines; irradiation time per line is (size of ROI in x dimension)/(scan speed), whereas number of lines corresponds to (size of ROI in y dimension)/(scan resolution). In cultured hippocampal neurons, for optimized scanning, the total exposure time was typically ~4–5 ms. For both slice and in vivo experiments, the total exposure time was typically ~3–4 ms. Note also that the fill factor allowed efficient stimulation, and a Pockels cell was used to blank the laser outside the ROI.
Light delivery
One-photon
For ChR2(H134R) and ChR2 (C128T or C128A/H134R), blue-light pulses were elicited using the XCite halogen light source (EXFO) with a blue 470/30-nm filter (Semrock) coupled to a Uniblitz shutter. For inactivation of ChR2 (C128A/H134R), 520/15-nm, 540/20-nm, 560/25-nm, 590/20-nm, or 610/20-nm band-pass filters were used with light power density of 5 mW mm−2. For single-photon–to–two-photon comparison of photocurrents, a saturating blue pulse was used at 15 mW mm−2. For C1V1, current comparisons were conducted with a 540/20-nm filter and Olympus 40×/0.8-NA LUMPlanFL/N objective.
Two-photon
A Coherent Ultra II Ti-Sapphire laser was used for all experiments. Supplementary Table 1 shows pulse width for each wavelength. In cases where EYFP and Alexa 594 were visualized, an FF560 dichroic with filters 630/69 and 542/27 (Semrock) was used to separate emissions.
For representation of TPLSM-stimulus timing in all figures, black tick marks are aligned to the signal-controlling voltage applied to the Pockels cell (via a Conoptics modulation box) that shutters the two-photon laser on at the very first pixel of a single line of an ROI. Likewise, light-off is defined as the place and time where the Pockels cell shutters the laser to an off position after the last pixel of the final line in an ROI.
Illumination parameters and methodological notes for main figure panels
Figure 1b. Temperature (T): 23–24 °C; objective: 40×/0.8 NA; ChR2HR λexcitation: 477 nm; C1V1 variant λexcitation: 540 nm; light power density: 7 mW mm−2; n represents number of cells. Figure 1c. ‘ts’ denotes trafficking sequence. Figure 1d. T: 23–24 °C; objective: 40×/0.8 NA; λ: 1,040 nm; dwell time per pixel: 4.0 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. Figure 1e. T: 23–24 °C; objective: 40×/0.8 NA; λ: 1,040 nm; dwell time per pixel: 4.0 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Figure 1f. T: 23–24 °C; λ: 1,040 nm; laser intensity: 20 mW; for the 40×/0.8-NA objective, dwell time per pixel was 4.0 μs and scan resolution was 0.6 μm per pixel; for the 20×/0.5-NA objective, dwell time per pixel was 4.0 μs and scan resolution was 0.6 μm per pixel at line scan speed of 0.15 μm μs−1. n represents number of cells. Figure 1g. T: 23–24 °C; objective: 40×/0.8 NA; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1, laser intensity: 20 mW. Figure 1h. T: 23–24 °C; objective: 40×/0.8 NA; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1, laser intensity: 20 mW, n represents number of cells. Figure 1i. T: 23–24 °C; objective: 40×/0.8 NA; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1. n represents number of cells. Figure 1j. T: 23–24 °C; objective: 20×/0.5 NA; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. Electrical stimulation: 400-pA current injections for 10 ms at 5 Hz, 10 Hz, 20 Hz and 40 Hz. Spikes evoked by current injection (black) shown for the same cells. n represents number of cells.
Figure 2b. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. Figure 2c. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. Figure 2d. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. n represents the number of cells. Figure 2e. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs; laser intensity: 20 mW. n represents the number of cells. Figure 2f. λ: 540 nm; light power density: 7 mW mm−2. n represents the number of cells. Figure 2h. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW.
Figure 3b. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. Figure 3c. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. Figure 3d. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. Figure 3e. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW. There were 20 scans per cell stimulated at 1 Hz. Figure 3f. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW.
Figure 4d. 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 3.2 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.19 μm μs−1; laser intensity: 20 mW.
Figure 5b. λ: 477 nm; light power density: 7 mW mm−2. n represents number of cells. Figure 5c. T: 23–24 °C; 40×/0.8-NA objective; λ: 940 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. Figure 5d. T: 23–24 °C; 40×/0.8-NA objective; λ: 940 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Figure 5e. T: 23–24 °C; objective: 40×/0.8 NA; dwell time per pixel: 4.0 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Figure 5f. T: 23–24 °C; objective: 40×/0.8 NA; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1. Figure 5g. T: 23–24 °C; 40×/0.8-NA objective; λ: 940 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Measurements performed on rat hippocampal cultured pyramidal neurons. Figure 5h. T: 23–24 °C; 40×/0.8-NA objective; λ: 940 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Measurements were performed on rat hippocampal cultured pyramidal neurons. Figure 5i. TPLSM-stimulation: T: 23–24 °C; 40×/0.8-NA objective; λ: 940 nm (red bar); dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. One-photon stimulation: different wavelengths shown, all at 5 mW mm−2. Measurements performed on rat hippocampal cultured pyramidal neurons. Figure 5j. A 10-ms full-field pulse was applied (blue bar; λ: 477 nm; light power density: 7 mW mm−2) followed by a single-plane two-photon raster scan (dark red bar; 40×/0.8-NA objective; λ: 1,060 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 15 mW) and a full-field amber pulse (orange bar; λ: 610 nm; light power density: 5 mW mm−2). Measurements were performed on rat hippocampal cultured pyramidal neurons. Figure 5k. TPLSM stimulation (T: 32 °C; 40×/0.8-NA objective; λ: 940 nm (red bar); dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW) followed by a single-photon deactivating pulse (λ = 590 nm, yellow bar; light power density: 5 mW mm−2). n represents number of cells. Measurements were performed on prefrontal cortical pyramidal cells. Figure 5l. T: 23–24 °C; 40×/0.8-NA objective; λ: 940 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Measurements were performed on rat hippocampal cultured pyramidal neurons.
Figure 6a. Note positioning of the endoplasmic reticulum export motif and trafficking signal (ER and ts, respectively). Figure 6c. T: 23–24 °C; 20×/0.5-NA objective; λ: 1,040 nm; laser intensity: 20 mW. For the blue trace, dwell time per pixel was 4.0 μs, scan resolution (full field of view) was 1.2 μm per pixel, and line scan speed was 0.30 μm μs−1. For the yellow trace, dwell time per pixel was 4.0 μs, scan resolution was 0.6 μm per pixel, and line scan speed was 0.15 μm μs−1. Figure 6d. T: 23–24 °C; 40×/0.8-NA objective; dwell time per pixel: 4.0 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents the number of cells. Figure 6e. T: 23–24 °C; 40×/0.8-NA objective; λ: 1,040 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1. n represents the number of cells. Figure 6f. 40×/0.8-NA objective; T: 23–24 °C; λ: 1,040 nm; laser intensity: 20 mW; dwell time per pixel: 4.0 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; and 20×/0.5-NA objective: T: 23–24 °C; λ: 1,040 nm; laser intensity: 20 mW; dwell time per pixel: 4.0 μs, scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1. n represents number of cells. Figure 6g. T: 23–24 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1, laser intensity: 20 mW. Figure 6h. T: 23–24 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. Figure 6j. Current clamp traces showing spike inhibition resulting from 1,040-nm illumination of a pyramidal neuron in medial prefrontal cortex expressing eArch3.0, (T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW). Figure 6k. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells. Figure 6l. T: 32 °C; 20×/0.5-NA objective; λ: 1,040 nm; dwell time per pixel: 4.0 μs; scan resolution: 0.6 μm per pixel; line scan speed: 0.15 μm μs−1; laser intensity: 20 mW. n represents number of cells.
Slice and culture electrophysiology
Electrophysiological data were collected using Multiclamp 700B (Molecular Devices) for amplification and digitized using a Digidata 1440A. Cultured-neuron physiology. Whole-cell recordings were performed from cultured neurons 6–7 days after transfection in Tyrode solution (320 mOsm): 125 mM NaCl, 2 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose and 25 mM HEPES, titrated to pH 7.3–7.4 with NaOH. Tyrode was perfused and kept at room temperature (23–24 °C). Intracellular solution for these recordings contained (in mM): 130 potassium gluconate, 10 KCl, 10 HEPES, 10 EGTA and 2 MgCl2, titrated to pH 7.3 with KOH. All photocurrent recordings were in bath-applied tetrodotoxin (1 μM; Sigma-Aldrich), whereas recordings for spiking were in the presence of synaptic blockers 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; 10 μM; Sigma-Aldrich) and D-(−)-2-amino-5-phosphonovaleric acid (APV; 25 μM, Sigma-Aldrich). Stimulation bouts were separated by 1 min to allow cell and opsin recovery. Acute slice physiology. Coronal slices were prepared from either hippocampus or medial prefrontal cortex 5–7 weeks post-injection. Two-photon imaging/ stimulation and electrophysiological recordings were under the constant perfusion of aCSF, which contained (in mM): 126 NaCl, 26 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 1 MgCl2, 2 CaCl and 10 glucose. All recordings were at 32 °C. Patch electrodes (4–6 MΩ) were filled (in mM) with: 10 HEPES, 4 Mg-ATP, 0.5 MgCl2, 0.4 Na3-GTP, 10 NaCl, and 140 potassium gluconate. For experiments involving axial/lateral resolution, dual–whole-cell recordings, or subcellular stimulation, the pipette was filled with Alexa Fluor 594 hydrazide (Molecular Probes) in order to visualize the cell body.
For two-photon resolution measurements, stimulation bouts (at different axial and lateral position) were separated by 1 min to allow for cell and opsin recovery. For spike resolution measurements, spikes were evoked at 0.5 Hz. For two-photon photocurrent-driven spike jitter measurements, 20 spikes were evoked at 1 Hz. For two-photon spine stimulation, a 40×/0.8-NA objective was employed and spines were visualized using both EYFP and Alexa 594.
In vivo electrophysiology
Mice were anesthetized with isoflurane (1%–2% in oxygen). A craniotomy above the somatosensory cortex was prepared as described previously41. The dura was carefully removed, and the exposed cortex was superfused with normal rat Ringer (NRR) solution (135 mM NaCl, 5.4 mM KCl, 5 mM HEPES, 1.8 mM CaCl2; pH 7.2 with NaOH). To dampen heartbeat- and breathing-induced motion, the cranial window was filled with agarose (type III-A, Sigma; 1% in NRR) and covered with an immobilized glass coverslip. Loose patch recordings were made by visually targeting EYFP+ pyramidal cells in layer 2/3 of the somatosensory cortex. Patch electrodes (4–6 MΩ) were filled (in mM): 10 HEPES, 4 Mg-ATP, 0.5 MgCl2, 0.4 Na3-GTP, 10 NaCl, 140 potassium gluconate and 80 Alexa Fluor 594 for pipette visualization.
Immunohistochemistry
Neuronal cultures were fixed on coverslips for 15 min with ice-cold 4% PFA, washed in PBS with glycine (0.1 M), permeabilized for 30 min with 0.4% saponin in 2% NDS, and incubated overnight at 4 °C with primary antibody in 2% NDS (Mouse anti-CaMKIIα 1:500, Abcam (ab22609); rabbit anti-2A 1:500, Millipore (ABS321)). Coverslips were washed and were incubated for 3 h at RT with secondary antibodies (donkey anti-mouse (715-166-151) or donkey anti-rabbit (711-165-152) conjugated to Cy3, 1:1,000, Jackson Laboratories). Coverslips were washed, incubated with DAPI (1:50,000) and mounted on slides with Fluoromount (Sigma). Fluorescence images were acquired on a confocal laser scanning microscope using a 40× oil- immersion objective.
For histochemistry, mice were anesthetized and perfused transcardially with cold PBS followed by 4% paraformaldehyde (PFA) in PBS; brains were removed and post-fixed in 4% PFA for 3 h at 4 °C and then equilibrated in 30% sucrose in PBS. Forty- micrometer-thick frozen coronal sections were stored in 25% glycerol and 30% ethylene glycol in PBS at 4 °C. Free-floating sections were washed in PBS, incubated for 30 min in 0.2% Triton X-100 (Tx100) and 2% normal donkey serum (NDS), and incubated overnight with primary antibody in 2% NDS (mouse anti-CaMKIIα 1:500, Abcam (ab22609); rabbit anti-2A 1:500, Millipore, (ABS321)). Sections were washed with PBS and incubated for 2 h at room temperature (RT) with secondary antibodies (donkey anti-mouse (715-166-151) or donkey anti-rabbit (711-165-152) conjugated to Cy3, 1:1000, Jackson Laboratories). Sections were then washed, incubated with DAPI (1:50,000) and mounted on slides with PVA-Dabco (Sigma). Fluorescence images were acquired on a laser-scanning confocal microscope using a 5× or 10× air objective or a 40× oil-immersion objective.
Statistics and data analysis
Physiology data were analyzed using Clampfit software (Axon Instruments). Statistical analysis was performed using Microsoft Excel (for t tests). For linear and Gaussian regression fits, GraphPad Prism (GraphPad Software) was used. All image analysis was performed using ImageJ (NIH). All comparisons were made allowing for non-Gaussian distributions using a Mann-Whitney two-tailed test.
Supplementary Material
Acknowledgments
We thank R. Pashaie and the Deisseroth laboratory members for helpful discussions. We thank Prairie Technologies (T. Keifer, M. Szulczewsk and A. Statz) for discussions and work with the two-photon microscope. R.P. is supported by the US National Institute of Mental Health (F30 MH095468). K.D. is supported by the US National Institutes of Health, the Gatsby Foundation and the Defense Advanced Research Program Agency REPAIR Program.
Footnotes
AUTHOR CONTRIBUTIONS
R.P., B.G. and K.D. contributed to study design. C.R., O.Y. and R.P. contributed to cloning of constructs. C.R. cultured primary neurons, performed transfections and managed viral production. R.P. and N.W. contributed to viral injections. R.P. performed all two-photon experiments in both culture and slice preparations. R.P. and B.G. contributed to in vivo two-photon experiments supervised by K.D. and M.J.S. I.G. performed histological processing and fluorescence imaging. R.P. performed all data analysis. A.M.P., D.S.P. and R.Y. contributed to design, analysis and interpretation. K.D. supervised all aspects of the work. R.P. and K.D. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Note: Supplementary information is available in the online version of the paper.
References
- 1.Zhang F, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 4.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domingos AI, et al. Leptin regulates the reward value of nutrient. Nat Neurosci. 2011;14:1562–1568. doi: 10.1038/nn.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi GB, et al. Driving opposing behaviors with ensembles of piriform neurons. Cell. 2011;146:1004–1015. doi: 10.1016/j.cell.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tye KM, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haubensak W, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciocchi S, et al. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468:277–282. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- 17.Johansen JP, et al. Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci USA. 2010;107:12692–12697. doi: 10.1073/pnas.1002418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goshen I, et al. Dynamics of retrieval strategies for remote memories. Cell. 2011;147:678–689. doi: 10.1016/j.cell.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 20.Jerome J, Foehring RC, Armstrong WE, Spain WJ, Heck DH. Parallel optical control of spatiotemporal neuronal spike activity using high-speed digital light processing. Front Syst Neurosci. 2011;5:70. doi: 10.3389/fnsys.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blumhagen F, et al. Neuronal filtering of multiplexed odour representations. Nature. 2011;479:493–498. doi: 10.1038/nature10633. [DOI] [PubMed] [Google Scholar]
- 22.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyart C, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel ADT. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stirman JN, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai S, Ueno K, Ishizuka T, Yawo H. Parallel and patterned optogenetic manipulation of neurons in the brain slice using a DMD-based projector. Neurosci Res. 2012 Mar 24; doi: 10.1016/j.neures.2012.03.009. published online. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci USA. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arenkiel BR, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing Channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adesnik H, Scanziani M. Lateral competition for cortical space by layer-specific horizontal circuits. Nature. 2010;464:1155–1160. doi: 10.1038/nature08935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 31.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 32.Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 33.Pettit DL, Wang SSH, Gee KR, Augustine GJ. Chemical two-photon uncaging: a novel approach to mapping glutamate receptors. Neuron. 1997;19:465–471. doi: 10.1016/s0896-6273(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 34.Fino E, et al. RuBi-Glutamate: two-photon and visible-light photoactivation of neurons and dendritic spines. Front Neural Circuits. 2009;3:2. doi: 10.3389/neuro.04.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuzaki M, Hayama T, Kasai H, Ellis-Davies GCR. Two-photon uncaging of γ-aminobutyric acid in intact brain tissue. Nat Chem Biol. 2010;6:255–257. doi: 10.1038/nchembio.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci USA. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papagiakoumou E, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci USA. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 41.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 43.Osborn MJ, et al. A picornaviral 2A-like sequence-based tricistronic vector allowing for high-level therapeutic gene expression coupled to a dual-reporter system. Mol Ther. 2005;12:569–574. doi: 10.1016/j.ymthe.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 44.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 45.Ihara K, et al. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J Mol Biol. 1999;285:163–174. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- 46.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaziri A, Emiliani V. Reshaping the optical dimension in optogenetics. Curr Opin Neurobiol. 2012;22:128–137. doi: 10.1016/j.conb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Nikolenko V, et al. SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators. Front Neural Circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerr JND, Greenberg D, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci USA. 2005;102:14063–14068. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.