Abstract
The ubiquitin proteasome system plays a critical role in skeletal muscle atrophy. A large body of research has revealed that many ubiquitin ligases are induced and play an important role in mediating the wasting. However, relatively little is known about the roles of deubiquitinases in this process. Although it might be expected that deubiquitinases would be downregulated in atrophying muscles to promote ubiquitination and degradation of muscle proteins, this has not to date been demonstrated. Instead several deubiquitinases are induced in atrophying muscle, in particular USP19 and USP14. USP19, USP2 and A20 are also implicated in myogenesis. USP19 has been most studied to date. Its expression is increased in both systemic and disuse forms of atrophy and can be regulated through a p38 MAP kinase signaling pathway. In cultured muscle cells, it decreases the expression of myofibrillar proteins by apparently suppressing their transcription indicating that the ubiquitin proteasome system may be activated in skeletal muscle to not only increase protein degradation, but also to suppress protein synthesis. Deubiquitinases may be upregulated in atrophy in order to maintain the pool of free ubiquitin required for the increased overall conjugation and degradation of muscle proteins as well as to regulate the stability and function of proteins that are essential in mediating the wasting. Although deubiquitinases are not well studied, these early insights indicate that some of these enzymes play important roles and may be therapeutic targets for the prevention and treatment of muscle atrophy.
Keywords: Ubiquitin, Skeletal muscle, Myofibrillar proteins, Deubiquitination, Cachexia
1. Increased ubiquitination of muscle proteins in atrophic conditions
The first observations of activation of the ubiquitin–proteasome system (UPS) in conditions of muscle wasting were reported approximately twenty years ago (Haas and Riley, 1988; Medina et al., 1991). Amongst these findings were key observations that the increases in rates of proteolysis observed upon fasting or denervation – prototypic conditions of muscle wasting due to systemic regulatory factors or loss of neural activity respectively – were largely due to an ATP dependent mechanism (Furuno et al., 1990; Wing and Goldberg, 1993). Importantly, expression of ubiquitin and ubiquitinated proteins increased in skeletal muscle in parallel with these increases in rates of ATP dependent proteolysis (Medina et al., 1995; Wing et al., 1995) suggesting that the UPS was this ATP dependent proteolytic system. Furthermore, myofibrillar proteins were preferentially ubiquitinated (Wing et al., 1995) consistent with the preferential degradation of these proteins in the catabolic conditions (Furuno et al., 1990; Li and Wassner, 1984; Lowell et al., 1986).
2. Enzymes that mediate ubiquitination and deubiquitination of proteins
The increase in levels of ubiquitinated proteins in the atrophying muscles suggested that the increased flux through this pathway was due to activation of conjugation of ubiquitin to proteins in this tissue. Conjugation of ubiquitin is mediated by the sequential action of three enzymes (Hershko et al., 1983). Ubiquitin activating enzyme (E1/UBA) activates ubiquitin (Haas and Rose, 1982) and transfers it to one of a family of ubiquitin conjugating enzymes (E2/UBC/UBE) (rev. in (Pickart, 2001)). The E2 then conjugates ubiquitin to protein substrates in concert with a third protein, ubiquitin–protein ligase (E3) (Reiss et al., 1989) that plays the important role of binding the substrate. Two genes encode E1 enzymes (Ciechanover et al., 1984; Kulka et al., 1988; Pelzer et al., 2007), but UBA1 appears to be the dominant enzyme in somatic cells. Approximately 30 mammalian E2s exist and each can interact with a subset of E3s which number ~800 in the human genome (rev. in (Glickman and Ciechanover, 2002, Pickart, 2001)).
Studies from many groups over the past twenty years have indeed identified many components in the ubiquitin conjugating apparatus that are induced in atrophying skeletal muscle. Levels of the ubiquitin conjugating enzyme HR6b (Ube2b) mRNA are increased in many conditions of muscle wasting (Lorite et al., 2001; Temparis et al., 1994; Wing and Banville, 1994). However, mice lacking this gene are not protected from muscle wasting of fasting indicating that this gene is not essential (Adegoke et al., 2002). Single reports have also indicated increased expression of E2–20k (Ube2h) (Li et al., 2003), UBC4/UBC5 (Ube2g) isoform (Chrysis and Underwood, 1999) or Ubc6 (Ube2j1) (Lecker et al., 2004) in some conditions of atrophy.
A number of ubiquitin ligases are induced, including atrogin-1 (Fbxo32) (Bodine et al., 2001; Gomes et al., 2001), MuRF-1 (Trim63) (Bodine et al., 2001), Cbl-b (Cblb) (Nakao et al., 2009), UBR1 (Fischer et al., 2000; Lecker et al., 1999), UBR2, UBR3 (Kwak et al., 2004; Kwon et al., 2001), and TRAF6 (Paul et al., 2010). Atrogin-1 and MuRF1 are most upregulated with levels of expression increasing by as much as 100 fold at the mRNA level, suggesting important roles for these ligases. Indeed, mice lacking either one of these genes show sparing of muscle wasting in response to denervation (Bodine et al., 2001). However, other ligases can also contribute to the atrophic process. Cbl-b knockout mice also show protection against denervation induced wasting (Nakao et al., 2009) and TRAF6 knockout mice are protected from multiple causes of atrophy including cancer cachexia (Paul et al., 2010), fasting (Paul et al., 2012) and denervation (Paul et al., 2010).
These studies clearly established that activation of ubiquitin conjugation can promote muscle protein degradation and contribute to the increase in ubiquitinated proteins in atrophying muscles. However, ubiquitination can also be modulated by deubiquitinases (rev. in (Komander et al., 2009; Reyes-Turcu et al., 2009)). There are ~95 human genes encoding enzymes that remove ubiquitin (Nijman et al., 2005). These can be subdivided into five major families based on conserved protein motifs. The largest family (~60 genes) is the USP (ubiquitin specific protease) family containing a conserved catalytic core domain bearing classical Cys, His, Asp domains which cooperate to render the thiol group on the cysteine residue an effective nucleophile for catalyzing peptide bond hydrolysis. The second largest class is the Otubain class (14 genes), members of which contain an OTU domain originally described in a Drosophila gene required for ovarian development (Balakirev et al., 2003). UCH (Ubiquitin C-terminal hydrolase) enzymes were the first deubiquitinases to be described. There are four of these in the human genome and one of these is BAP1 which binds the BRCA1 breast cancer susceptibility gene and can function as a tumor suppressor (Jensen et al., 1998). The four MJD (Machado–Joseph Disease) enzymes contain N-terminal Josephine domains and the best studied of these is ataxin3 which when mutated causes a form of ataxia with Parkinsonian features (Burnett et al., 2003). Finally, there are up to 14 JAMM (JAB1/MPN/Mov34) domain containing proteins with four clearly having hydrolytic activity against ubiquitin or ubiquitin like proteins. One of these, Rpn11/POH1, is an integral subunit of the 19S cap of the proteasome and plays a critical role in cleaving the ubiquitin chain from substrates prior to their degradation (Yao and Cohen, 2002). Unlike the other four classes of deubiquitinases, the JAMM domain enzymes are not cysteine proteases, but metalloproteinases in which a Zn2+ atom in the active site coordinates a water molecule which serves as the nucleophile to attack the peptide bond.
3. Potential roles of deubiquitinases in muscle atrophy
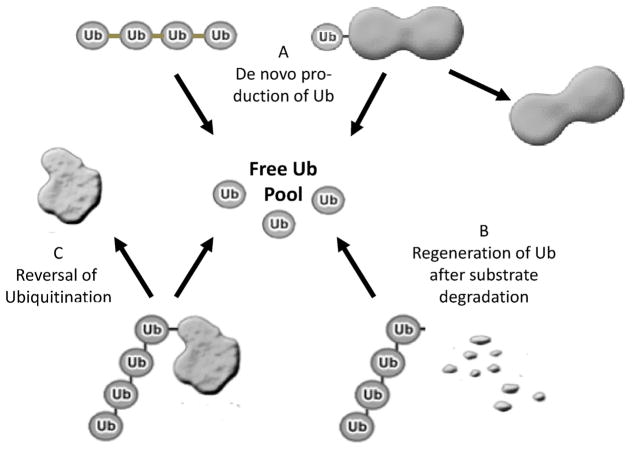
The most obvious mechanism by which deubiquitination could be involved in muscle wasting would be downregulation of deubiquitinases to promote ubiquitination and degradation of proteins. However, surprisingly, to date no deubiquitinase has been reported to be downregulated in atrophying muscle. In contrast, there is evidence for induction of expression of some deubiquitinases. Although seemingly counterintuitive, a number of possibilities exist as to how upregulation of a deubiquitinase could contribute to enhancement of muscle proteolysis (Fig. 1). Deubiquitinases play essential roles in maintaining cellular levels of free ubiquitin. Several deubiquitinases, USP14, UCH37, POH1 are associated with the 19S regulatory cap of the 26S proteasome. POH1 appears to cleave the chain at the junction of linkage of ubiquitin with the target protein (Yao and Cohen, 2002). USP14 and UCH37 can trim ubiquitin chains (Crosas et al., 2006; Lam et al., 1997). Thus, these enzymes could promote proteolysis by regenerating free ubiquitin from the ubiquitin chains to ensure an adequate supply of the peptide for further conjugation of proteins destined for degradation. It is also well established now that some forms of ubiquitination of plasma membrane proteins such as monoubiquitination or lysine 63 linked ubiquitin chains can promote internalization and trafficking of these membrane proteins via endosomes and multivesicular bodies into lysosomes for degradation. Again, the ubiquitin is removed from the cargo prior to its degradation. The deubiquitinases isopeptidase T/USP5 (Papa and Hochstrasser, 1993) and/or AMSH (McCullough et al., 2004) play important roles in recycling ubiquitin in this compartment. Failure to do so in yeast bearing mutants of Doa4, the ortholog of USP5, results in depletion of free ubiquitin (Swaminathan et al., 1999).
Fig. 1.
Potential roles for deubiquitinases in skeletal muscle atrophy. Deubiquitinases may be activated to support or promote proteolysis by maintaining the pool of free ubiquitin that is required for the increased overall conjugation to substrates targeted for degradation. Deubiquitinases generate free ubiquitin by processing the products of the polyubiquitin genes Ubb and Ubc and the ubiquitin fusion genes Uba52 and Rps27a (A) as well as by removing ubiquitin from proteins committed to degradation by the proteasome or by the lysosome (B). Deubiquitinases may also act on specific protein substrates to reverse the functions of ubiquitination (C). In this way, they not only contribute to the pool of free ubiquitin, but importantly may promote muscle atrophy by stabilizing proteins that promote wasting (see text for more detailed discussion). If ubiquitination of these specific proteins does not target for degradation but confers other functions, deubiquitinases may promote atrophy by enhancing the functions of activators or by inhibiting the functions of repressors of pathways that promote wasting. Muscle atrophy could also be promoted by downregulation of deubiquitinases that normally stabilize substrates targeted for degradation or stabilize repressors of pathways that promote wasting. At this time, all of these possibilities remain speculative.
Ubiquitin is encoded by four genes in the mammalian genome. Two of these genes, Ubb and Ubc, are polyubiquitin genes (Finley et al., 1987; Ozkaynak et al., 1987) encoding multiple copies of ubiquitin arranged in tandem. The other two genes, Uba52 and Rps27a, encode ubiquitin fused to one of two proteins – the L40 or S27a subunits of the 60S ribosomal complex, respectively (Finley et al., 1989; Redman and Rechsteiner, 1989). All of these genes can be induced in atrophying muscles (Lecker et al., 2004). Enhanced deubiquitinases could play a role in processing the increased production of these polyproteins to their individual proteins. In this way, the pool of free ubiquitin is also enhanced.
Another important mechanism by which induction of deubiquitinases can promote muscle wasting is through modulation of proteins that regulate atrophy. For example, increased expression of deubiquitinases could stabilize transcription factors that activate genes required for atrophy such as the ubiquitin ligases previous discussed. A number of deubiquitinases are associated with ligases (see (Kee et al., 2005; Lu et al., 2009; Mouchantaf et al., 2006; Wu et al., 2004) for examples) and so it is possible that increased deubiquitinases could stabilize ubiquitin ligases that are involved in muscle wasting. Deubiquitinases could also modulate signaling pathways that regulate proteolysis. They could act to stabilize repressors of signaling pathways that suppress proteolysis or stabilize activators of signaling pathways that stimulate proteolysis. It is important to note that ubiquitination may serve non-proteolytic functions such as regulation of chromatin structure and gene expression, DNA repair, cell signaling, and cellular localization. These effects are due to the ubiquitination either modifying the structure and thereby function of the protein or promoting the recruitment of other proteins. Deubiquitinases could decrease such non-proteolytic functions which could through indirect mechanisms promote muscle protein degradation. Increased deubiquitination may also stimulate degradation by promoting substrate availability. For example, the A20 deubiquitinase removes lysine 63 chains from RIP1 allowing it to be ubiquitinated with lysine 48 chains that target it for degradation (Wertz et al., 2004).
4. USP19 and skeletal muscle
The most intensively studied deubiquitinase to date with respect to skeletal muscle is USP19. It is induced in muscle wasting, but the changes are modest rendering it generally not detected in microarray analyses as a differentially expressed gene. Increased mRNA levels of this enzyme have been seen in multiple conditions including fasting, diabetes, three different tumor bearing models, high dose glucocorticoid therapy, denervation and rats exposed to cigarette smoke (Combaret et al., 2005; Liu et al., 2011; Ogawa et al., 2012; Robert et al., 2012). This induction can be mediated through p38 MAP kinase signaling (Liu et al., 2011). This upregulation in a broad array of conditions suggests that USP19 may be involved in an important common process in the pathogenesis of wasting. Antioxidants such as β-carotene can suppress muscle loss as well as the induction of USP19 (Ogawa et al., 2012). In vitro studies in cultured myotubes have demonstrated that catabolic stimuli such as glucocorticoids (Sundaram et al., 2009), cigarette smoke extract (Liu et al., 2011), and hydrogen peroxide (Ogawa et al., 2012) can induce USP19 expression. Evidence for a functional role arises from studies in myotubes which demonstrate that silencing of USP19 can increase expression of myofibrillar proteins (Sundaram et al., 2009). Since such proteins make up the bulk of muscle protein content, it suggests that induction of USP19 levels indeed promote atrophy. In addition, such silencing can reverse the catabolic effects of dexamethasone in myotubes suggesting inhibition of this enzyme as a potential therapy for wasting (Sundaram et al., 2009). Interestingly, the increase in levels of myofibrillar proteins was associated with increased expression of mRNA for these proteins suggesting that USP19 regulates a myogenic program. Indeed, we observed that depletion of USP19 induced the expression of the myogenic regulatory factor myogenin and this increase in myogenin was required for the increase in myofibrillar protein expression (Sundaram et al., 2009). Thus, induction of USP19 appears to play a role in protein balance through regulation of synthesis of myofibrillar proteins promoting the concept that the UPS is activated in muscle to suppress protein synthesis as well as activate protein degradation. This ability of USP19 to modulate muscle mass appears to be relevant in vivo as our preliminary studies indicate that mice lacking the enzyme have sparing of muscle loss in response to denervation atrophy (Sundaram et al., 2010).
The ability of USP19 to modulate expression of myogenin suggests that it might also be involved in myogenesis. In support of this, estrogen acting via the ERα receptor can suppress differentiation and increase expression of USP19 in cultured muscle cells (Ogawa et al., 2011). This effect of estrogen on myogenesis is critically dependent on USP19 as silencing its expression blocked the effect on differentiation (Ogawa et al., 2011). Consistent with this, our recent studies also demonstrate that overexpression of USP19 can impair muscle cell differentiation in vitro (unpublished data).
The mechanisms by which USP19 might mediate these effects in muscle remain unknown. Previous studies in HEK293 cells have shown that USP19 can deubiquitinate and stabilize cIAP proteins (Mei et al., 2011). These proteins can promote TNFα signaling in several cell types (Varfolomeev et al., 2012) and can also inhibit myogenesis in primary muscle cells (Enwere et al., 2012). So an attractive hypothesis is that USP19 might promote muscle atrophy by stabilizing cIAPs and activating the NFκB pathway. However, to date, we have not been able to demonstrate such an effect on cIAPs in muscle cells.
5. Other deubiquitinases and skeletal muscle
Usp14 was observed to be highly induced on microarray analysis in several catabolic conditions including fasting, uremia, diabetes and cancer (Lecker et al., 2004), but exploration of this observation has not been reported. This induction is somewhat surprising as studies of its ortholog in yeast have suggested that USP14 functions to negatively regulate ubiquitination and degradation of proteasome substrates (Crosas et al., 2006; Hanna et al., 2006). Its function in skeletal muscle may be different and instead it may be required for recycling of ubiquitin to maintain the free ubiquitin pool as discussed earlier. If deubiquitination of substrates is required prior to degradation, then induction of USP14 could allow a larger flux of ubiquitinated proteins to be processed and degraded by the proteasome. It is possible that USP14 can promote or inhibit degradation depending on the individual substrate in question. Naturally occurring Usp14 mutant mice have been described (Wilson et al., 2002). The primary abnormality in these mice is ataxia which can progress to muscle paralysis, but this appears to be explained by a defect in neurotransmission in both the central and peripheral nervous system rather than a defect in muscle. A muscle specific inactivation of Usp14 has not been reported to date.
USP2 is expressed as two distinct isoforms each possessing an identical C-terminal catalytic core domain but distinct N-termini (Gousseva and Baker, 2003; Lin et al., 2000). These two isoforms can modulate muscle cell differentiation in vitro, but interestingly each with opposing effects (Park et al., 2002). USP2a mRNA expression increases transiently before membrane fusion, but USP2b mRNA increases as the cells fuse. Fusion and differentiation are stimulated by overexpression of USP2a whereas overexpression of USP2b inhibits these processes. However, it appears that USP2 is not essential for myogenesis as skeletal muscle appears grossly normal in mice lacking USP2, albeit both isoforms ((Bedard et al., 2011) and data not published).
A20 is an intriguing enzyme that possesses both a deubiquitinating activity localized to its OTU N-terminal domain and a ligase activity in its C-terminal seven tandem C2/C2 zinc finger domain (Wertz et al., 2004). As discussed earlier, it plays a critical role in regulating NFκB signaling through regulating the deubiquitination and subsequent ubiquitination and degradation of RIP1. A20 is highly expressed in regenerating muscle fibers in dystrophic mice and its silencing impairs muscle differentiation in vitro (Charan et al., 2012). It is suggested that A20 promotes regeneration by inhibiting the classical NFkB pathway and upregulating the alternate pathway.
6. Perspectives/future directions
As is evident from the brevity of this review, the potential role of deubiquitinases in muscle wasting remains largely unexplored. Although at this time, there are no published reports describing deubiquitinases that are suppressed to promote muscle protein ubiquitination and degradation, this possibility has not been completely excluded. The search for regulated deubiquitinases in muscle wasting has largely taken place at the level of expression of mRNA, so it remains possible that there are decreased activities of deubiquitinases occurring through post-transcriptional mechanisms. Levels of deubiquitinase proteins may be regulated at a translational level by factors such as miRNAs or other non-coding RNAs. In addition, it remains possible that there are deubiquitinase activities in skeletal muscle inhibited through mechanisms such as post-translational modifications, allosteric regulation, substrate activation or regulated localization in complexes or specific compartments. Regulation of specific deubiquitinases by all of these mechanisms have been observed (rev. in (Komander et al., 2009)).
Much work also remains to be done with the deubiquitinases that have been implicated to date in regulating muscle size. Although USP19 has been studied the most, many questions remain. In particular, the substrates of the enzyme and the molecular mechanisms by which USP19 mediates these effects in muscle remain unknown. The specific functions of USP14 in muscle atrophy also remain unknown as do the potential roles of USP2 in muscle regeneration or recovery from hypertrophy. Nonetheless, the insights to date with USP19 confirm that it plays a key role in skeletal muscle atrophy and so other deubiquitinases are also likely to be involved in important ways.
Acknowledgments
Results from the author’s laboratory described in this review were supported by the Canadian Institutes of Health Research, the Terry Fox Research Institute and the Fonds de recherche du Quebec – Santé.
References
- Adegoke OA, Bedard N, Roest HP, Wing SS. Ubiquitin-conjugating enzyme E214k/HR6B is dispensable for increased protein catabolism in muscle of fasted mice. American Journal of Physiology Endocrinology and Metabolism. 2002;283:E482–9. doi: 10.1152/ajpendo.00097.2002. [DOI] [PubMed] [Google Scholar]
- Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Reports. 2003;4:517–22. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard N, Yang Y, Gregory M, Cyr DG, Suzuki J, Yu X, et al. Mice lacking the USP2 deubiquitinating enzyme have severe male subfertility associated with defects in fertilization and sperm motility. Biology of Reproduction. 2011;85:594–604. doi: 10.1095/biolreprod.110.088542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–8. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Human Molecular Genetics. 2003;12:3195–205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Charan RA, Hanson R, Clemens PR. Deubiquitinating enzyme A20 negatively regulates NF-kappaB signaling in skeletal muscle in mdx mice. FASEB Journal. 2012;26:587–95. doi: 10.1096/fj.11-189829. [DOI] [PubMed] [Google Scholar]
- Chrysis D, Underwood LE. Regulation of components of the ubiquitin system by insulin-like growth factor I and growth hormone in skeletal muscle of rats made catabolic with dexamethasone. Endocrinology. 1999;140:5635–41. doi: 10.1210/endo.140.12.7217. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Combaret L, Adegoke OA, Bedard N, Baracos V, Attaix D, Wing SS. USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. American Journal of Physiology Endocrinology and Metabolism. 2005;288:E693–700. doi: 10.1152/ajpendo.00281.2004. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–13. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Enwere EK, Holbrook J, Lejmi-Mrad R, Vineham J, Timusk K, Sivaraj B, et al. TWEAK and cIAP1 regulate myoblast fusion through the noncanonical NF-kappaB signaling pathway. Science Signaling. 2012;5:ra75. doi: 10.1126/scisignal.2003086. [DOI] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–46. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fischer D, Sun X, Gang G, Pritts T, Hasselgren PO. The gene expression of ubiquitin ligase E3alpha is upregulated in skeletal muscle during sepsis in rats-potential role of glucocorticoids. Biochemical and Biophysical Research Communications. 2000;267:504–8. doi: 10.1006/bbrc.1999.1987. [DOI] [PubMed] [Google Scholar]
- Furuno K, Goodman MN, Goldberg AL. Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. Journal of Biological Chemistry. 1990;265:8550–7. [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiological Reviews. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A. Atrogin-1 Goldberg AL a muscle-specific F-box protein highly expressed during muscle atrophy. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14440–5. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousseva N, Baker RT. Gene structure, alternate splicing, tissue distribution, cellular localization, and developmental expression pattern of mouse deubiquitinating enzyme isoforms Usp 2–45 and Usp 2–69. Gene Expression. 2003;11:163–79. doi: 10.3727/000000003108749053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Riley D. The dynamics of ubiquitin pools within skeletal muscle. In: Schlesinger M, Hershko A, editors. The ubiquitin system, current communications in molecular biology. New York: Cold Spring Harbor Laboratory Press; 1988. pp. 178–85. [Google Scholar]
- Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. Journal of Biological Chemistry. 1982;257:10329–37. [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin–protein ligase system. Journal of Biological Chemistry. 1983;258:8206–14. [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO Journal. 2005;24:2414–24. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature Reviews Molecular Cell Biology. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, et al. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. Journal of Biological Chemistry. 1988;263:15726–31. [PubMed] [Google Scholar]
- Kwak KS, Zhou X, Solomon V, Baracos VE, Davis J, Bannon AW, et al. Regulation of protein catabolism by muscle-specific and cytokine-inducible ubiquitin ligase E3a-II during cancer cachexia. Cancer Research. 2004;64:8193–8. doi: 10.1158/0008-5472.CAN-04-2102. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Molecular and Cellular Biology. 2001;21:8007–21. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–40. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB Journal. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Solomon V, Price SR, Kwon YT, Mitch WE, Goldberg AL. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. Journal of Clinical Investigation. 1999;104:1411–20. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Wassner SJ. Effects of food deprivation and refeeding on total protein and actomyosin degradation. American Journal of Physiology. 1984;246:E32–7. doi: 10.1152/ajpendo.1984.246.1.E32. [DOI] [PubMed] [Google Scholar]
- Li YP, Lecker SH, Chen Y, Waddell ID, Goldberg AL, Reid MB. TNF-alpha increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. FASEB Journal. 2003;17:1048–57. doi: 10.1096/fj.02-0759com. [DOI] [PubMed] [Google Scholar]
- Lin H, Keriel A, Morales CR, Bedard N, Zhao Q, Hingamp P, et al. Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures. Molecular and Cellular Biology. 2000;20:6568–78. doi: 10.1128/mcb.20.17.6568-6578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu WG, Luo Y, Han FF, Yao XH, Yang TY, et al. Cigarette smoke-induced skeletal muscle atrophy is associated with up-regulation of USP-19 via p38 and ERK MAPKs. Journal of Cellular Biochemistry. 2011;112:2307–16. doi: 10.1002/jcb.23151. [DOI] [PubMed] [Google Scholar]
- Lorite MJ, Smith HJ, Arnold JA, Morris A, Thompson MG, Tisdale MJ. Activation of ATP–ubiquitin-dependent proteolysis in skeletal muscle in vivo and murine myoblasts in vitro by a proteolysis-inducing factor (PIF) British Journal of Cancer. 2001;85:297–302. doi: 10.1054/bjoc.2001.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, Ruderman NB, Goodman MN. Regulation of myofibrillar protein degradation in rat skeletal muscle during brief and prolonged starvation. Metabolism: Clinical and Experimental. 1986;35(12):1121–7. doi: 10.1016/0026-0495(86)90025-9. [DOI] [PubMed] [Google Scholar]
- Lu Y, Adegoke OA, Nepveu A, Nakayama KI, Bedard N, Cheng D, et al. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Molecular and Cellular Biology. 2009;29:547–58. doi: 10.1128/MCB.00329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. Journal of Cell Biology. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Wing SS, Goldberg AL. Increase in levels of polyubiquitin and proteasome mRNA in skeletal muscle during starvation and denervation atrophy. Biochemical Journal. 1995;307(Pt 3):631–7. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Wing SS, Haas A, Goldberg AL. Activation of the ubiquitin–ATP-dependent proteolytic system in skeletal muscle during fasting and denervation atrophy. Biomedica Biochimica Acta. 1991;50:347–56. [PubMed] [Google Scholar]
- Mei Y, Hahn AA, Hu S, Yang X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. Journal of Biological Chemistry. 2011;286:35380–7. doi: 10.1074/jbc.M111.282020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. Journal of Biological Chemistry. 2006;281:38738–47. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- Nakao R, Hirasaka K, Goto J, Ishidoh K, Yamada C, Ohno A, et al. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Molecular and Cellular Biology. 2009;29:4798–811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kariya Y, Kitakaze T, Yamaji R, Harada N, Sakamoto T, et al. The preventive effect of beta-carotene on denervation-induced soleus muscle atrophy in mice. British Journal of Nutrition. 2012:1–10. doi: 10.1017/S0007114512003297. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yamaji R, Higashimura Y, Harada N, Ashida H, Nakano Y, et al. 17beta-estradiol represses myogenic differentiation by increasing ubiquitin-specific peptidase 19 through estrogen receptor alpha. Journal of Biological Chemistry. 2011;286:41455–65. doi: 10.1074/jbc.M111.276824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO Journal. 1987;6:1429–39. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa FR, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–9. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Park KC, Kim JH, Choi EJ, Min SW, Rhee S, Baek SH, et al. Antagonistic regulation of myogenesis by two deubiquitinating enzymes, UBP45 and UBP69. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9733–8. doi: 10.1073/pnas.152011799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Molecular and Cellular Biology. 2012;32:1248–59. doi: 10.1128/MCB.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. Journal Cell Biology. 2010;191:1395–411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid HP, Scheffner M, et al. UBE1L2, a novel E1 enzyme specific for ubiquitin. Journal of Biological Chemistry. 2007;282:23010–4. doi: 10.1074/jbc.C700111200. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Redman KL, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438–40. doi: 10.1038/338438a0. [DOI] [PubMed] [Google Scholar]
- Reiss Y, Heller H, Hershko A. Binding sites of ubiquitin–protein ligase. Binding of ubiquitin–protein conjugates and of ubiquitin–carrier protein. Journal of Biological Chemistry. 1989;264:10378–83. [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual Review of Biochemistry. 2009;78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Mills JR, Agenor A, Wang D, DiMarco S, Cencic R, et al. Targeting protein synthesis in a Myc/mTOR-driven model of anorexia–cachexia syndrome delays its onset and prolongs survival. Cancer Research. 2012;72:747–56. doi: 10.1158/0008-5472.CAN-11-2739. [DOI] [PubMed] [Google Scholar]
- Sundaram P, Pang Z, Miao M, Bedard N, Moore T, Wing SS. Loss of USP19 increases transcription of myofibrillar proteins in L6 muscle cells and decreases muscle wasting in response to denervation in mice. Journal of Cachexia, Sarcopenia and Muscle. 2010;1:87. [Google Scholar]
- Sundaram P, Pang Z, Miao M, Yu L, Wing SS. USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells. American Journal of Physiology Endocrinology and Metabolism. 2009;297:E1283–90. doi: 10.1152/ajpendo.00409.2009. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Molecular Biology of the Cell. 1999;10:2583–94. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, et al. Increased ATP–ubiquitin-dependent proteolysis in skeletal muscles of tumor-bearing rats. Cancer Research. 1994;54:5568–73. [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Maecker H, Zobel K, Komuves LG, Deshayes K, et al. Cellular inhibitors of apoptosis are global regulators of NF-kappaB and MAPK activation by members of the TNF family of receptors. Science Signaling. 2012;5:ra22. doi: 10.1126/scisignal.2001878. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nature Genetics. 2002;32:420–5. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- Wing SS, Banville D. 14-kDa ubiquitin-conjugating enzyme: structure of the rat gene and regulation upon fasting and by insulin. American Journal of Physiology. 1994;267:E39–48. doi: 10.1152/ajpendo.1994.267.1.E39. [DOI] [PubMed] [Google Scholar]
- Wing SS, Goldberg AL. Glucocorticoids activate the ATP–ubiquitin-dependent pro-teolytic system in skeletal muscle during fasting. American Journal of Physiology. 1993;264:E668–76. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- Wing SS, Haas AL, Goldberg AL. Increase in ubiquitin–protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and atrophy denervation. Biochemical Journal. 1995;307(Pt 3):639–45. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Molecular and Cellular Biology. 2004;24:7748–57. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]