Abstract
Improvements in health in the past decades have resulted in increased numbers of the elderly in both developed and developing regions of the world. Advances in therapy have also increased the prevalence of patients with chronic and degenerative diseases. Muscle wasting, a feature of most chronic diseases, is prominent in the elderly and contributes to both morbidity and mortality. A major research goal has been to identify the proteolytic system(s) that is responsible for the degradation of proteins that occurs in muscle atrophy. Findings over the past 20 years have clearly confirmed an important role of the ubiquitin proteasome system in mediating muscle proteolysis, particularly that of myofibrillar proteins. However, recent observations have provided evidence that autophagy, calpains and caspases also contribute to the turnover of muscle proteins in catabolic states, and furthermore, that these diverse proteolytic systems interact with each other at various levels. Importantly, a number of intracellular signaling pathways such as the IGF1/AKT, myostatin/Smad, PGC1, cytokine/NFκB, and AMPK pathways are now known to interact and can regulate some of these proteolytic systems in a coordinated manner. A number of loss of function studies have identified promising therapeutic approaches to the prevention and treatment of wasting. However, additional biomarkers and other approaches to improve early identification of patients who would benefit from such treatment need to be developed. The current data suggests a network of interacting proteolytic and signaling pathways in muscle. Future studies are needed to improve understanding of the nature and control of these interactions and how they work to preserve muscle function under various states of growth and atrophy.
Keywords: Muscle wasting, ubiquitin, proteasome, autophagy, calpains, caspases, IGF1, Akt, activin, myostatin, PGC1, cytokines, AMPK
Understanding muscle wasting - exploring the roles of proteolytic systems
Over the last 20 years the view of skeletal muscle as an inert peripheral tissue that performs voluntary contractile activity has been revised and replaced with the concept that muscle mass and muscle metabolism are highly dynamic and are modulated by multiple influences. Indeed one of the defining characteristics of skeletal muscle tissue is its plasticity in the face of changing metabolic and contractile demands, which in turn reflects natural growth and aging, as well as the impact of environmental influences including acute and chronic disease.
Both developed and developing countries have seen a sustained rise in the number of older people in their populations and an increase in the prevalence of people suffering from chronic and degenerative diseases such as cancer and chronic heart, lung, renal and rheumatological conditions1. Aging and these diverse diseases are associated with weight loss and muscle wasting2, and, moreover, low muscle mass is associated with increased morbidity and poorer survival3–6. The prognostic importance of muscle wasting may reflect simply an association with more active or debilitating disease; however there is evidence that muscle atrophy contributes directly to morbidity and mortality in a number of ways. For example, several studies suggest that those with lowest muscle mass are at highest risk of chemotherapy drug toxicity7. Also muscle wasting has functional consequences and contributes to reduced mobility, social dependence and specific complications such as falls8. In the extreme, muscle atrophy of the respiratory muscles can compromise respiratory function and predispose to pneumonia and death9. Thus there is increasing interest in developing ways to identify and limit muscle atrophy, so as to improve function in general and reduce specific symptoms and complications.
As muscle atrophy is an important prognostic sign in many diseases, the study of underlying mechanisms may offer insights into ways to improve outcomes for affected patients. This critical adaptation of muscle tissue is achieved largely through changes affecting the dynamic balance between protein synthesis and proteolysis in multinucleate myofibers, which form the bulk of muscle tissue, rather than by cellular apoptosis and replacement. Not only does the balance of net synthesis and proteolysis in muscle define overall skeletal muscle mass, but also there is differential control of this balance at the level of individual muscles that can allow one muscle to atrophy while another hypertrophies. To achieve this, there is exquisite control of expression of individual muscle proteins at the myofiber level, which in turn establishes the phenotypic features of the muscle tissue including its contractile and metabolic properties. In contrast, different mechanisms come into play in the response to muscle damage, where the proliferation and maturation of latent muscle satellite cells is more important10.
Because combating muscle atrophy may be a route to deliver significant improvements in outcomes for large numbers of patients, approaches to identify and treat affected individuals must be considered. Recent advances in accurate measurement of total muscle mass using computed tomography (CT) image analysis in some diseases, including cancer11, have become extremely important, but such approaches are still not widely applied and may not be the optimum way to identify the earliest changes in atrophying muscle tissue and thus to guide preventative therapies. As a result one priority is to develop more sophisticated biomarkers of impending pathological changes in muscle mass or phenotype. Candidate biomarkers include expression levels of molecules that reflect key aspects of the balance between protein synthesis and proteolysis within myofibers. To interpret such biomarkers and to rationally design new therapies to inhibit muscle wasting, there is a need for a better understanding of the different pathways underpinning the balance between protein synthesis and proteolysis for bulk protein and specific substrates, and an improved appreciation of the integration of the activities of these pathways.
This review will outline current knowledge on the proteolytic pathways in skeletal muscle, including autophagy, which has re-emerged as a highly regulated route for organelle degradation with specific importance in muscle. Changes in intramuscular protein synthesis and proteolysis pathways are initiated or modulated by a variety of extracellular signals, and the integration of these signaling pathways and their effects on different proteolytic pathways will be discussed. In particular the molecular control points which allow cross-talk between proteolytic and protein synthesis and the evidence of interactions between different proteolytic pathways will be highlighted.
Proteolytic systems in muscle wasting
Ubiquitin proteasome system
General features of the UPS
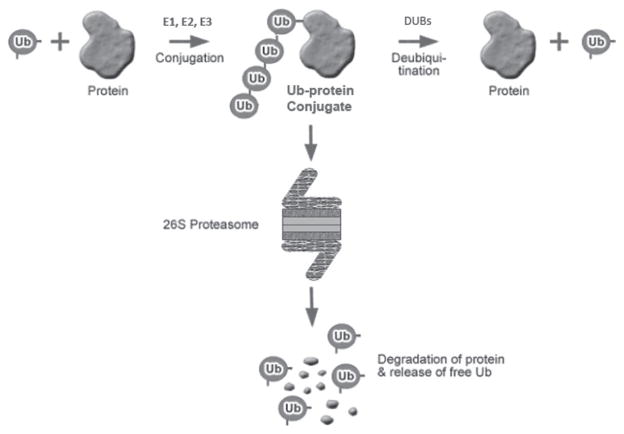
A large body of evidence indicating an important role for the ubiquitin proteasome system (UPS) in mediating muscle wasting has now accumulated. Ubiquitin, an abundant 8 kDa protein found in all eukaryotic cells, exerts its effects by serving in post-translational modification of other proteins (Figure 1) (reviewed in12). In this modification, the C-terminal end of ubiquitin is usually attached to the ε-amino group of the side chains of lysine residues within the target protein. Additional ubiquitin moieties can be added to produce a polyubiquitin chain in which each moiety is connected via an isopeptide bond between the C-terminal glycine of the distal ubiquitin and a lysine residue of the more proximal ubiquitin. Ubiquitin has seven lysine residues, and a proteomic analysis in yeast indicates that all seven lysines can be used to form ubiquitin–ubiquitin linkages, though to varying degrees13. Use of different lysine residues results in different conformations of the polyubiquitin chain, which are believed to confer distinct functions. Lys48-linked chains are well recognized to target substrates for degradation by the 26S proteasome14. However, recent evidence indicates that some other chain linkages can also target proteins to the proteasome15–17.
Figure 1.
The ubiquitin-proteasome system. Conjugation of ubiquitin (Ub) chains to proteins in which each ubiquitin is linked to the other via Lys 48 targets the protein substrate for recognition and degradation by the 26S proteasome. Conjugation is mediated by the sequential action of three enzymes - E1 (ubiquitin activating enzyme), which activates and transfers Ub to E2 (ubiquitin conjugating enzyme), which then works in concert with E3 (ubiquitin protein ligase) to mediate ubiquitination. E3s recognize substrates. The conjugation can also be reversed by deubiquitinating enzymes (DUBs).
Conjugation of ubiquitin to proteins involves a sequence of reactions18 catalyzed by three enzymes–ubiquitin activating enzyme (E1)19, ubiquitin conjugating enzyme (E2, UBC) (reviewed in20) and ubiquitin ligase (E3)21. For some substrates, a fourth enzyme, E4, lengthens short ubiquitin chains22. A single E1 gene appears to exist in somatic cells and supplies activated ubiquitin to a larger family of E2s23–24. Approximately 30 genes encode E2s in mammalian cells (reviewed in20). Each E2 appears to interact with distinct E3s and different E3s recognize distinct substrates. Where multiple E2s can interact with an E3, the different E2s can mediate formation of different types of ubiquitin chain linkages25. Thus, there are multiple pathways of ubiquitin conjugation leading to precise ubiquitination of specific proteins. E3s can be organized into two major classes. One class (~90 human genes) contains a conserved C-terminal HECT domain (Homologous to E6-AP Carboxy-Terminus - named after E6AP, the first E3 described in this class)26 and functions by first accepting ubiquitin from E2 onto a cysteine residue and then conjugating the ubiquitin to the substrate27. The other E3 class (~800 human genes) contains a conserved RING finger motif28–29 and functions by binding both substrate and the E221 and activating E2’s conjugating activity30–31. Ligases can exist as monomeric proteins or as multi-subunit complexes such as the family of cullin-RING ligases in which the substrate recognition and E2 binding functions are located on distinct subunits of the complex.
As in other post-translational modifications, ubiquitination is reversible and is mediated by ~100 deubiquitinating enzymes present in the mammalian genome (reviewed in32). Besides reversing the effects of ubiquitination and releasing free ubiquitin for re-use, these enzymes also play an essential role in processing the products encoded by the four ubiquitin genes. Two of these genes encode polyubiquitin (multiple copies of ubiquitin in tandem) while the other two encode ubiquitin-ribosomal protein fusions. Deubiquitinating enzymes can be subclassified into 5 families based on the presence of conserved sequence motifs32. The largest subfamily of deubiquitinating enzymes are the ubiquitin specific proteases (USP).
The 26S proteasome consists of a 20S cylindrical core bound on one or both ends by 19S caps (reviewed in33). The 20S core is comprised of four stacked rings of seven subunits each and contains in the interior of the rings, the active sites involved in peptide bond hydrolysis. The 19S caps serve to identify substrates for the proteasome and to translocate them to the proteolytic machinery in the 20S core complex. They recognize polyubiquitin chains either directly or via intermediary adaptor proteins which can bind ubiquitinated substrates as well as 19S subunits34. The 19S cap contains deubiquitinating enzyme activities. Rpn11 is an integral subunit of the 19S cap and appears to be able to cleave ubiquitin chains from the substrate35. Associated with the 19S cap are two other deubiquitinating enzymes USP14 and UCH37 (UCH, ubiquitin C-terminal hydrolase)36–37. The 19S cap also contains ATPase activities which are involved in opening the gate to the 20S core as well as unfolding the substrate for insertion into the channel of the 20S complex.
The UPS is activated in atrophying skeletal muscle
One of the earliest findings demonstrating a role for the UPS in muscle wasting was the observation of increased steady state levels of ubiquitinated proteins in atrophying muscle38–42. This suggested that ubiquitination is activated and increases flux through the pathway by increasing availability of ubiquitinated substrates for the proteasome. Supporting this model is increased expression of many components involved in ubiquitin conjugation in atrophying skeletal muscle (Table 1) (reviewed in43). Genes encoding ubiquitin are induced as are genes encoding several ubiquitin conjugating enzymes and ubiquitin ligases. The functional roles of these, particularly the ubiquitin ligases, will be discussed below.
Table 1.
List of genes in the UPS reported to be regulated in skeletal muscle wasting (examples of references in parentheses).
| Ubiquitin genes | Ubiquitin conjugating enzymes | Ubiquitin ligases | Deubiquitinating enzymes | Proteasome subunits |
|---|---|---|---|---|
| UbB (93, 200–201) | E214k/HR6B/UBC2 (46, 202) | MAFbx/Atrogin-1 (58, 60) | USP14 (44) | Psma1 (39, 208–209) |
| UbC (93, 200–201) | E220k (203) UBC4/UBC5 (44, 204) | MuRF-1 (58) | USP19 (55) | Psma2 (200, 210–211) |
| UBA52 (44) | Cbl-b (76) | Psma3 (39, 47, 200) | ||
| Ub S27A (44) | E4 (44) | Psma4 (39, 46, 209) | ||
| E3α/UBR1 (205–206) | Psma5 (44) | |||
| E3α-II/UBR2 UBR3 (77, 207) | Psmb1 (200, 212) | |||
| Psmb2 (212–213) | ||||
| Psmb3 (44) | ||||
| Psmb4 (44) | ||||
| Psmc1 (51) | ||||
| Psmc2 (214–215) | ||||
| Psmc4 (51) | ||||
| Psmc5 (48) (51) | ||||
| Psmd1 (48, 51) | ||||
| Psmd3 (48) | ||||
| Psmd4 (51) | ||||
| Psmd8 (51) (44) | ||||
| Psmd11 (44) | ||||
| Psme1 (48) | ||||
| Psme3 (48) |
Increased mRNA expression of many subunits of the 20S proteasome has been observed in atrophying skeletal muscle (Table 1)44–47. Increased expression of some 19S cap subunits have also been reported but is not as consistent a finding44,48–50. Proteasome activity in muscle, as measured by in vitro assays, has also been found to be increased in some catabolic conditions51–53. Overall the data are consistent with increased expression and activity of the proteasome in atrophying muscle.
To date only two deubiquitinating enzymes have been reported to be regulated in atrophying skeletal muscle. Both are up-regulated, indicating that they are not involved in mediating the overall increase in levels of ubiquitinated proteins seen in atrophy. Usp14 mRNA has been demonstrated to be up-regulated in muscle in rodents in response to fasting, uremia, diabetes and cancer44. This deubiquitinating enzyme is associated with the proteasome and so increased expression may be important for removing the ubiquitin chain prior to substrate degradation by the proteasome. However, mutational analysis of the yeast ortholog UBP6 suggest that its deubiquitinating activity may actually serve to protect proteins from degradation,54 so further studies are required to delineate more precisely the role of USP14 in muscle.
USP19 has also been shown to be up-regulated at the mRNA level in skeletal muscle in multiple catabolic conditions55. Silencing of USP19 in L6 muscle cells results in increased expression of myofibrillar proteins56. The finding that this is dependent on increased expression of myogenin implies that USP19 acts on signaling pathways that regulate myogenin and thereby a transcriptional program regulating myofibrillar protein genes. This suggests an additional novel mechanism in which the UPS can mediate muscle wasting - by suppressing the transcription and synthesis of myofibrillar proteins. Recently, mice lacking USP19 expression have been found to lose less muscle mass upon denervation,57 indicating that USP19 is indeed a gene required for the full catabolic response.
Inactivation of ubiquitin ligases confers resistance to muscle wasting
Although many UPS genes are induced in muscle in wasting conditions, only a limited number have been tested by genetic inactivation and found to be essential for mediating atrophy. Because most of these genes encode ubiquitin ligases, the subsequent discussion will focus on these enzymes. Deletion of the MuRF-1 (MuRF, muscle RING finger containing ligase) or MAFbx/Atrogin-1 (muscle atrophy F-box containing protein/Atrogin-1) ubiquitin ligases (58) results in ~35–50% sparing of muscle wasting following two weeks of denervation. These two ligases are muscle specific with expression in skeletal, cardiac and uterine (59) muscle. MuRF-1 is a monomeric ligase with a tripartite RING finger:B box:coiled-coiled motif. MuRF-2 and MuRF-3 are two other similar RING finger ligases with muscle and cardiac specific expression. MAFbx/Atrogin-1 is an F-box containing protein and, like other F-box subunits of ligases, is part of a cullin-RING ligase (CRL) complex (58, 60). The F-box containing subunit is the critical substrate recognition component of the complex. Multiple studies (see Table 1) have demonstrated marked induction (as much as 50-fold) of the mRNAs encoding MuRF-1 and MAFbx/Atrogin-1 in many conditions of muscle atrophy, usually either simultaneously or just prior to activation of wasting. In denervation or spinal cord isolation forms of disuse atrophy, MAFbx/Atrogin-1 is highly induced within a few days of loss of neural stimulation, but returns to baseline by two weeks61. MuRF-1 ligase is elevated and remains so as long as 4 weeks.
In vivo and in vitro studies have provided insights into possible mechanisms by which these two ligases mediate muscle wasting. Yeast two-hybrid studies using MuRF-1 or MuRF-2 as baits identified eight myofibrillar proteins as interactors and possible substrates– titin, nebulin, nebulin related protein (NRAP), troponin-I, troponin-T, MyLC2 (MyLC, myosin light chain), myotilin and T-cap62. Isolation of MuRF-1 from muscle extracts revealed co-binding of a number of thick filament proteins including myosin binding protein C (MyBP-C), MyLC1, MyLC2, myosin heavy chain (MyHC) isoforms 1, 4, 8, and filamin63. Incubation of these isolates with E1, E2, and ubiquitin demonstrated that these proteins can indeed be ubiquitinated. Furthermore MyBP-C, MyLC1 and MyLC2 are selectively lost upon denervation prior to the loss of MyHC and actin; however this does not occur in the mice expressing an inactive form of MuRF-1, which, like the MuRF-1 knock-out mice, show sparing of loss of muscle mass upon denervation63. Similarly there is less loss of MyHC in MuRF-1 deficient mice upon denervation or dexamethasone treatment, and MyHC can be ubiquitinated in vitro64. Troponin-I has similarly been shown to be a substrate of MuRF-1 in cardiac muscle65. In contrast though, levels of several thin filament proteins are not perturbed by loss of MuRF-163. These data support a model in which MuRF-1 plays a key role in ubiquitination and degradation of thick filament proteins.
Although MuRF-1 deficient mice have normal muscle structure or function under basal conditions, inactivation of an additional MuRF ligase leads to hypertrophy of both skeletal and cardiac muscle. Most MuRF-1/MuRF-2 double knock-out mice do not survive beyond the perinatal period. The few that do survive show marked cardiac hypertrophy associated with decreased ventricular function and altered sarcomeric and mitochondrial ultrastructure66. MuRF-1/MuRF-3 double knock-out mice manifest skeletal and cardiac hypertrophy67. In these mice, force generation is abnormal in both cardiac and skeletal muscle and is associated with accumulation of both b/slow myosin as well as MyHCIIa myosin. Thus, further loss of MuRF activity actually leads to myopathy with poorer muscle function despite increased mass.
The mechanisms by which the MAFbx/Atrogin-1 ligase might be involved in muscle wasting are less clear. MAFbx/Atrogin-1 was found to interact with myoblast determination protein (MyoD) in a yeast two-hybrid assay, and over-expression of MAFbx/Atrogin-1 in muscle cells results in degradation of MyoD and decreased muscle cell differentiation68. MAFbx/Atrogin-1 recognizes a LXXXLL motif in MyoD. A similar motif exists in myogenin and indeed MAFbx/Atrogin-1 can also modulate myogenin levels in C2C12 cells69. Finally, MAFbx/Atrogin-1 can target the eukaryotic initiation factor eIF3-f for degradation in C2C12 myotubes, which results in myotube thinning70. eIF3-f is a regulatory subunit of eIF3 which plays an important role in initiation of translation. Over-expression of eIF3-f in C2C12 cells increases myotube size, and electroporation of eIF3-f into mouse muscle results in fiber hypertrophy. All of these results are consistent with increased expression of MAFbx/Atrogin-1 causing decreased transcription and translation of muscle myofibrillar proteins. However, the precise relevance of these substrates in vivo remains unknown since they have not yet been carefully examined in MAFbx/Atrogin-1 knock-out mice.
Other substrates have been identified through studies of the role of MAFbx/Atrogin-1 in the development of cardiac hypertrophy. In cardiomyocytes, calcineurin interacts with calmodulin to promote the activity of NFAT, a transcriptional activator of genes involved in mediating cardiac hypertrophy. MAFbx/Atrogin-1 can target calcineurin for ubiquitination and degradation71. Accordingly, mice over-expressing this protein in the heart show resistance to hypertrophy induced by IGF1 (insulin-like growth factor 1)/growth hormone. In addition, MAFbx/Atrogin-1 can polyubiquitinate FOXO1 and FOXO3a (FoxO, forkhead box protein), which are transcriptional activators of MAFbx/Atrogin-1 and MuRF-1 72. However, the ubiquitin chains formed are via Lys63 linkages and do not target the FoxO isoforms for degradation, but rather increase the transcriptional activity of FoxO; this results in increased expression of these two ligases, which, in turn, leads to a feed forward amplification that promotes atrophy or resistance to hypertrophy.
Recent experiments have shown that MAFbx/Atrogin-1 is also expressed in statin-induced muscle injury and suggests similarities between atrophy and myopathy mechanisms73. MAFbx/Atrogin-1 mRNA levels were significantly higher in muscle biopsies from a set of statin-treated patients with muscle fatigue symptoms than in control biopsy samples (in age-matched patients undergoing knee replacement)73. Similarly, MAFbx/Atrogin-1 induction was found in both cultured muscle cells and in the muscles of whole zebrafish exposed to pharmacologic levels of statins but not in statin-treated muscle cells and fish lacking MAFbx/Atrogin-173. As well as inhibiting cholesterol synthesis, statins inhibit other biochemical reactions, including isoprenylation of small G-proteins (geranylgeranylation, farnesylation) and formation of coenzyme Q, which is required for oxidative phosphorylation. While it does not appear that statin inhibition of cholesterol or coenzyme Q biosynthesis is responsible for atrogin-1 induction, statin-induced MAFbx/Atrogin-1 expression and muscle damage can be prevented by geranylgeranol74. Furthermore, inhibitors of the transfer of geranylgeranyl isoprene units to protein targets cause statin-like muscle damage and MAFbx/Atrogin-1 induction in cultured cells and in fish74. These findings support the concept that dysfunction of small GTP-binding proteins lead to statin-induced muscle damage since these molecules require modification by geranylgeranyl moieties for their cellular localization and activity.
Another ligase that may be involved in disuse atrophy is Cbl-b (a ubiquitin ligase). This RING finger containing ligase is involved in the down-regulation of a number of plasma membrane receptor tyrosine kinases (reviewed in75) and results in decreased signaling. Atrophy induced by unloading has been observed to induce expression of Cbl-b and association of this ligase with IRS-1 (IRS, insulin receptor substrate), a key downstream signaling mediator of insulin and IGF176. This results in degradation of IRS-1, decreased growth factor signaling and activation of MuRF-1 and MAFbx/Atrogin-1 expression. Transduction of muscle with plasmid expressing Cbl-b induces atrophy. Finally, Cbl-b knock-out mice show sparing of muscle wasting and myofiber atrophy upon unloading, a finding that confirms an essential role of Cbl-b in this form of wasting76.
All three ligases involved in the N-end rule pathway of protein degradation (Ubr1, Ubr2, Ubr3) are up-regulated in atrophying skeletal muscle (44, 77). These ligases recognize specific amino acids at the N-termini of some of their substrates78. Ubr2, which has been characterized in more detail77, is induced in muscle of tumor-bearing animals as well as in muscle cells treated with TNF-α (tumor necrosis factor) or IL-6 (IL, interleukin), cytokines that are often increased in cancer. Over-expression of Ubr2 in muscle cells stimulates overall ubiquitin conjugation.
Although ubiquitination can clearly target proteins for degradation by the proteasome, it has been generally difficult to reconstitute degradation of a substrate in vitro simply with the enzymes involved in ubiquitination and purified proteasomes. This suggests the existence of factors that mediate delivery of ubiquitinated proteins to the proteasome. Indeed a number of such adaptor proteins have been identified. Members of the RAD23 family contain ubiquitin-like (Ubl) domains that can bind the Rpn10 subunit of the 19S cap of the proteasome as well as UBA (C-terminal ubiquitin-associated) domains that can bind polyubiquitinated proteins34. The adaptor protein, ubiquitin binding protein ZNF216, has been shown to be induced in atrophying muscle79. It contains a Zn finger A20 motif that binds polyubiquitin chains. The finding that mice lacking ZNF216 lose approximately 30% less muscle mass upon denervation confirms a role for this protein in muscle wasting.
Together, these studies provide compelling evidence for an essential role of the UPS in muscle wasting. A role in the targeting of thick filament proteins confirms as expected that the UPS is involved in degradation of myofibrillar proteins. Interestingly, recent evidence also suggests a role of the UPS in mediating the fall in protein synthesis seen in most catabolic conditions. The numerous components of the UPS that are involved suggest that the UPS mediates atrophy through multiple mechanisms; this may explain why inactivation of single genes in the UPS typically leads to only partial protection against wasting.
Autophagy
Lysosomal proteolysis
Lysosomes are membrane-bound organelles that contain a variety of hydrolases, including over a dozen proteases whose functions together are able to thoroughly digest proteins. The organellar sequestration of these hydrolases is critical in preventing autodigestion of the cell. Four major mechanisms exist to control the entry of substrates into lysosomes: endocytosis, by which plasma membrane proteins can be internalized into endosomes and trafficked to the lysosome for degradation80; a chaperone mediated direct import pathway that involves transport of selected proteins across the lysosomal membrane81; microautophagy, in which small amounts of cytoplasm are sequestered via budding and internalization of the lysosomal membrane82; and macroautophagy (quantitatively the most important mechanism), a process in which cytoplasmic contents including organelles such as mitochondria and endoplasmic reticulum are sequestered by double membrane vesicular structures termed autophagosomes83. Maturation of these autophagosomes involves fusion with endosomes which then traffic to lysosomes.
Mechanisms of macroautophagy
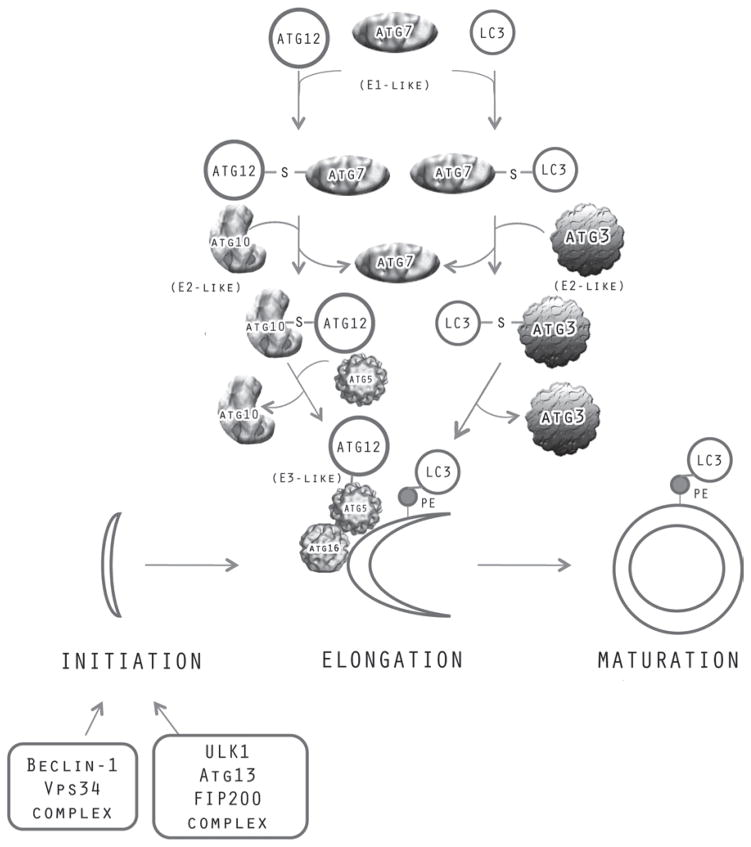
In the late 1960s and 1970s, formation of autophagosomes was shown to be activated in the liver under catabolic conditions such as glucagon stimulation and insulin or amino acid deficiency84–85. However, the molecular mechanisms by which this process occurred remained essentially unknown until the late 1990s, when Ohsumi and colleagues identified a large number of genes in yeast that were required for autophagy86. Surprisingly, molecular analysis of the genes indicated that a number of them were involved in a system of protein conjugation remarkably similar to the ubiquitin system (Figure 2)86. In yeast, two ubiquitin-like (Ubl) proteins, ATG12 and ATG8 (ATG, autophagy system protein), are activated by an activating enzyme ATG7 that, like E1, forms a high energy thiolester linkage between the activating enzyme and the carboxy terminus of the ubiquitin-like molecule. The Ubl proteins, ATG12 or ATG8, are then transferred to the cysteine residue of ATG10 or ATG3, respectively, which function analogously to ubiquitin conjugating enzymes (E2). ATG10 conjugates ATG12 to a lysine residue on its substrate ATG5. The ATG12/ATG5 then interacts with ATG16 to form oligomers that associate with membranes; this complex has E3-like activity, supporting the conjugation by ATG3 of ATG8 to membrane bound phosphatidylethanolamine. This conjugation pathway identified in yeast is highly conserved in mammalian cells. However, there are several orthologs of the Ubl protein ATG8 - LC3 (microtubule-associated proteins 1A/1B light chain 3A), GABARAP, GATE16, and conjugation of these onto phosphatidylethanolamine has been observed87.
Figure 2.
Formation of autophagasomes in autophagy. Initiation of the phagaphore can be induced by two signaling complexes–one containing Beclin-1/Vps34 and the other Atg13/ULK1/FIP200. Elongation /maturation of the phagophore is mediated by two protein conjugation pathways–one in which Atg12 becomes conjugated to Atg5 through a pathway mediated by the Atg7 activating enzyme and the Atg10 conjugating enzyme. In the other pathway, LC3 or its paralogs, GABARAP and GATE16, become conjugated to phagophore bound phosphatidylethanolamine through a pathway mediated by the Atg7 activating enzyme, the Atg3 conjugating enzyme and the Atg12-Atg5/Atg16 ligase-like complex.
The mechanisms by which the proteins in these conjugation pathways and those encoded by other autophagy genes mediate autophagosome formation are becoming clearer (Figure 2). The autophagosome membranes appear to be derived mainly from the ER88 and the outer mitochondrial membrane89. Initial nucleation to form a phagophore can be induced by two signaling complexes–one that contains ULK1 kinase/ATG13/FIP200 and another that contains Beclin-1/ATG14/VPS34 class III phosphatidylinositol 3-kinase. This latter complex promotes the formation of PI3P which in turn can recruit the ATG5/ATG12/ATG16 complex to the phagophore membrane. Oligomerization of this complex promotes the formation of a coat and elongation of the autophagosome membrane. The conjugation of LC3 (ATG8) to phosphatidylethanolamine can then promote budding of the membrane and closure of the autophagosome90. Such closure results in release of the LC3 from the membrane by the ATG4 deconjugating enzyme. The autophagosome then appears to merge initially with endocytic compartments and ultimately with lysosomes91.
Macroautophagy in skeletal muscle
Until recently, lysosomal proteolysis was generally viewed as having a minor role in the degradation of skeletal muscle proteins. Studies in isolated muscles from denervated92 or fasted rodents93 indicated that although lysosomal proteolysis could be activated in these catabolic conditions, it appeared to make a minor contribution to the increase in proteolysis seen under these conditions. Most of the increase in muscle proteolysis could be ascribed to the UPS93–94 and so attention was focused on this pathway. However, more recent studies have clearly implicated a role for macroautophagy in the activation of protein breakdown. Firstly, many autophagy related genes–LC3, Gabarap, Bnip3, Atg12, Atg495–96 as well as the lysosomal protease, cathepsin L44,97, are induced in muscle in catabolic states. Secondly, suppression of the IGF1/AKT/FoxO (AKT, serine/threonine kinase involved in insulin/IGF1 signaling) pathway, a major signaling pathway that activates protein breakdown (see below), also activates autophagic pathways. Indeed transduction of constitutively active FoxO3 into muscle myofibers is sufficient to activate autophagosome formation95. Transgenic mice that express GFP-LC3 fusion proteins in which activation of macroautophagy is associated with increased GFP signal from autophagosomal membranes has revealed that there is marked increase in autophagasome formation in skeletal muscle upon fasting and that this is more prominent in the fast glycolytic muscles which manifest a larger degree of atrophy98. Finally and most importantly, transduction of myofibers by electroporation with plasmids expressing shRNA against LC3 is able to partially block the atrophy induced by FoxO3 expression, indicating that at least part of the muscle wasting occurs via an autophagic mechanism95).
However, chronic inactivation of autophagy does not render mice resistant to muscle wasting. In fact, skeletal muscle specific loss of the activating enzyme ATG7 leads to a form of myopathy in which there is myofibrillar disorganization, accumulation of cytoplasmic and sarcomeric aggresomes, abnormal mitochondria and myofiber atrophy99. Thus, autophagy is required for maintenance of normal muscle fiber function. From studies in other cellular systems (reviewed in100), it is clear that autophagy plays a variety of homeostatic functions–clearance of protein aggregates, turnover of ribosomes, mitochondria and endoplasmic reticulum, and a role in programmed cell death. Inactivation of autophagy can lead to disorganized sarcomeric structure101, and abnormal accumulation of damaged organelles and protein aggregates, which in turn lead to cellular toxicity. Autophagy also plays a normal role in cellular energy economy. Low levels of amino acids result in decreased mTOR (mammalian target of rapamycin) activity which results in activation of macroautophagy and consequently hydrolysis of proteins and fatty acids to generate ATP in the cell. Therefore, it is not surprising that chronic blockade of this pathway will lead to myofiber damage.
The activation of macroautophagy in muscle wasting is now well established. However, the relative role of autophagy in relation to other proteolytic systems, in particular the UPS, remains to be established. Importantly, the substrates that are targeted by macroautophagy in muscle remain unclear. One established target is mitochondria. Recent studies102 indicate that fasting, denervation or expression of constitutively active FoxO3 result in altered organization of the mitochondrial network with possibly increased turnover. Furthermore, inhibiting loss of mitochondria from fission can offer partial protection from muscle wasting due to fasting. Finally, inducing a decrease in mitochondrial number and function can induce muscle wasting. Thus, activation of macroautophagy may lead to wasting not just by targeting proteins for degradation, but also by targeting organelles such as mitochondria.
Calpains
The calpain (CAPN) superfamily of Ca2+-dependent cytosolic cysteine proteases is encoded by 14 genes103 and can be categorized into different subgroups. mu(μ)-calpain (CAPN1) and m-calpain (CAPN2) differ in their sensitivity to Ca2+. ‘Typical’ calpains (1, 2, 3, 8a, 9, 11, 12, and 13) have a canonical domain structure on the 80 kDa catalytic subunit and three of these (CAPN1, 2 and 9) can form heterodimers with a smaller 30 kDa regulatory subunit. The ‘atypical’ calpains (CAPN5, 6, 7, 8b, 10, and 15) have altered domain structure including absent domain IV containing an EF-hand motif. Finally some calpains are ubiquitous (1, 2, 5, 7, 10, 13, and 15), whereas others are more tissue-restricted in expression [3a (striated muscle), 6 (placenta), 8 (stomach), 9 (digestive tract), 11 (testis), and 12 (hair follicle cells)]104.
Calpains undergo autolytic activation though the unautolysed proteins still retain some proteolytic activity105. Localization and autolytic activation seem to be important for controlling calpain activity. In some cases binding to membranes or other proteins may stabilize the calpains, limit autolysis and help direct activity to specific substrates, but other evidence suggests membrane binding and specifically exposure to membrane phospholipids reduces the Ca2+ activation threshold103. In addition, activity of dimeric calpains (CAPN1, 2 and 9) is also specifically inhibited by calpastatin104, a monomeric protein that has recently been shown to bind calpain in the presence of Ca2+ and assumes a structure which allows inhibition of the calpain active site while avoiding proteolytic cleavage itself106–107.
Ubiquitous calpains are modulated by contractile activity
Approximately 7 different calpains are expressed in muscle, and post-mortem calpain mediated proteolysis in livestock muscle is a major determinant of meat tenderness, which is of importance to the food industry and the subject of intense research108. However, CAPN expression and activity are recognized as an important component of adaptive changes in muscle related to contractile activity. Thus myofibrillar-bound CAPN1 activity is increased early in electrical low frequency stimulation-induced fast-to-slow myofiber type transition, whereas preferential increases in CAPN2 myofibrillar-bound activity are seen in response to direct muscle injury109. Human gene expression studies showed no change in mRNAs encoding CAPN1, 2 and calpastatin as a result of disuse, but increases in all three genes were observed during remobilizing110. In addition other studies that have demonstrated increased calpain activity in human muscle after eccentric, i.e. muscle-damaging, exercise, particularly in the myofibrillar fraction, suggest that calpain-mediated proteolysis is involved in the myofibrillar disruption observed after this type of exercise111.
The endogeous calpain inhibitor, calpastatin, which is active against CAPN1 and 2 but not CAPN3 in muscle, is also highly regulated in this tissue. For example, during myoblast fusion calpastatin levels are dramatically reduced, allowing calpain-dependent degradation of certain membrane-associated muscle proteins112. Muscle-specific calpastatin over-expression also reduced muscle fiber atrophy by 30% and inhibited slow-to-fast myosin isoform transition in an unloading model113. Few studies have looked at the post-translational control of calpastatin activity, but other proteases, such as CASP1 (caspase-1), can degrade or inactivate calpastatin114.
Calpain 3 is muscle-specific and required for muscle homeostasis
CAPN3a (p94) has muscle-specific expression and mutations lead to limb-girdle muscular dystrophy type 2A (LGMD2A). In contrast to data on CAPN1 and 2, mRNA levels for CAPN3 are down-regulated acutely in animal models of muscle wasting such as cancer115, and denervation116, and in human disuse atrophy110. However CAPN3 down-regulation may not be a universal occurrence in atrophying muscle as CAPN3 was not identified as a consistently regulated atrogene in other studies of gene expression in muscle atrophy44. In contrast, CAPN3 activation is increased in response to eccentric exercise117.
The mechanism by which CAPN3 mutation leads to dystrophy is still not entirely clear but the dystrophy phenotype is due to reduced CAPN3 activity rather than hyperactivation. CAPN3 binds to the giant titin molecule in the backbone of the myofibril; many studies have suggested it has a wide range of roles including remodeling of contractile proteins, control of apoptosis and modulation of gene expression118.
As described below, there is evidence that the transcription factor NFκB (nuclear factor kappa-B) is important in mediating muscle atrophy in different conditions, particularly those associated with effects of cytokines such as TNF. NFκB has pleotropic effects and also promotes transcription of inhibitor of apoptosis proteins that counteract the pro-apoptotic effects of cytokines119. CAPN3 substrates include IκBα (NFκB inhibitor alpha) and cardiac ankyrin repeat protein (CARP) which bind NFκB. Lower levels of active CAPN3 can lead to increased myonuclear apoptosis indirectly by increasing NFκB cytoplasmic sequestration through binding to IκBα120 or CARP118 with consequent reduced anti-apoptotic effects of NFκB.
Inhibiting calpain activity in muscle can reduce muscle wasting
Despite some evidence for a general up-regulation of calpain activity in muscle in disease models such as acute diabetes121, most of the earlier studies could demonstrate only a small fraction of overall proteolysis as being calpain-dependent. Many of these studies used isolated muscle preparations and compared the effects of inhibitors of different classes of proteases to demonstrate the role of specific proteolytic pathways. These studies were limited by their in vitro nature, the lack of specificity of some of the protease inhibitors used, and a failure to appreciate the inter-dependence or cross-talk between different proteolytic pathways. This may explain why, despite prior evidence for little effect of calpain-inhibition on bulk proteolysis, calpain inhibition does appear to have useful functional effects in a variety of clinically-relevant models of muscle wasting. These findings suggest that calpain-inhibition may still be a potentially useful therapeutic approach. Thus over-expression of calpastatin or use of exogenous calpain inhibitors has been shown to ameliorate or reduce: a) sepsis-induced muscle (diaphragm122 and heart123) weakness, b) post-MI cardiac muscle remodeling124, c) disuse atrophy125, and d) hyperglycemia-related cardiac muscle dysfunction126.
Calpain proteolysis links with other proteolytic pathways
The success of calpain inhibitors in slowing muscle wasting in many experimental conditions suggests a sequential process of coordinated protein cleavages by different classes of proteases, including calpain-dependent steps. There is now further experimental evidence for such interaction between calpains and other pathways related to proteolysis and protein synthesis in muscle. Thus in studies of proteolysis in rat diaphragms, calpain activation increased UPS proteolysis and inhibited AKT and mTOR phosphorylation, whereas proteasome inhibition prevented the effects of calpain-activation on overall proteolysis127. Other studies of interaction between calpains and UPS include those in CAPN3 knock-out mice which have smaller muscle fibers, slower rate of atrophy in response to unloading, but also markedly impaired recovery from unloading compared to wild type. CAPN3-deficient animals, which lack the normal marked increase in CAPN3 on reloading, also failed to demonstrate the normal accumulation of high molecular weight ubiquitin-protein conjugates in muscle during reloading. This suggests that CAPN3 has an important role in remodeling during recovery from disuse and that the overall process involves UPS degradation of products of CAPN3 cleavage128. Protein cleavage, including calpain-dependent steps upstream of UPS, may also have a role in enhancing proteasome degradation by providing protein fragments with NH2-terminus amino acid residues that are particularly favored for ubiquitination by the Ubr family of ubiquitin ligases and targeted for proteasomal degradation129.
A different type of cross-talk was demonstrated in a recent study in cancer cell lines: IκBα is a calpain and UPS substrate but proteasome inhibition led to a counter-regulatory increase in calpain-dependent degradation of IκBα and increased NFκB signaling, rather than the expected stabilization of IκBα130. Calpains can also interact with caspase proteases in a variety of different ways to activate131–132 or inhibit apoptosis, depending on conditions133.
In summary calpains contribute to proteolytic cleavage of substrates prior to proteasomal degradation in muscle atrophy and have an additional important role in responses to muscle injury and remodeling after disuse. Calpain activity may be especially important to accelerate release of components of large multiprotein structures such as the myofibril. CAPN inhibition shows promise as a means to reduce muscle wasting in certain types of acute illness, but longer term inhibition of CAPN3 may lead to impaired remodeling and muscle dysfunction or dystrophy.
Caspases
In humans, the caspases (CASP) comprise a family of 12 cysteine-dependent proteases which cleave proteins preferentially after aspartic acid residues. Caspases have a variety of roles but as a group are best known for their involvement in programmed cell death pathways (apoptosis). Classical caspase-dependent apoptosis involves initiator caspases (CASP2, 8, 9, 10) which activate effector caspases (CASP 3, 6, 7) through proteolytic cleavage. The effector caspases then cleave a variety of substrates as part of an orchestrated process of cellular destruction134. However some caspases (CASP1, 4, 5) are not thought to be directly involved in apoptosis, but rather to play roles in inflammatory cell and cytokine maturation135.
In muscle atrophy early experimental results showing a dominant role for the ATP-dependent UPS in accelerated proteolysis suggested little or no role for caspase-dependent proteolysis (reviewed in136). However there is evidence from experimental models linking caspase activity with specific types of muscle atrophy, e.g. in aging137 and in cardiac muscle remodeling after myocardial infarct138. Myonuclear apoptosis appears to be a feature of many models of muscle wasting, particularly those involving denervation. Other data in such diseases as chronic obstructive pulmonary disease139 and congestive heart failure140 have suggested that myonuclear apoptosis is also increased in association with muscle atrophy. Whether this limited form of apoptosis of nuclei without cell death reflects a response to muscle atrophy to reduce the number of nuclei and preserve the size of myonuclear domains within smaller (atrophied) myofibrils, or indicates that apoptosis is a driving pathological process in atrophy in chronic disease is still uncertain.
Caspases may also affect muscle wasting through non-apoptotic pathways. In muscles of diabetic and uremic rats, CASP3 activation is increased and is associated with increased muscle levels of a 14 kDa actin cleavage fragment141. Inhibiting caspase activity in isolated muscles and cultured muscle cells reduces production of the 14 kDa fragment and inhibits the accelerated proteolysis due to diabetes. Moreover insulin and IGF1 inhibit the production of the 14 kDa fragment through a PI3K-dependent mechanism (PI3K, phosphoinositide-3-kinase), suggesting that these hormones have a direct effect on CASP3 in addition to their inhibition of the UPS via FoxO as described below141. The nature of the activation of UPS proteolysis by CASP3 in catabolic conditions has been studied further; in addition to potentially cleaving and releasing substrates for the UPS, there is evidence that in muscle tissue CASP3 cleavage of two ATPase-containing subunits on the 19S proteasome regulatory particle [PSMC1 (RPT2) and PSMC5 (RPT6)] amplifies proteasome activity142.
Genetic inactivation of CASP3 in mice results in partial protection from denervation-induced muscle atrophy. However, CASP3-deficient mice have similar levels of the 14 kDa actin fragment and activation of the UPS, suggesting that the initial actin degradation step may be performed by other caspases or indeed other proteases. However pro-apoptotic signaling and myonuclear apoptosis was reduced after denervation in muscle from CASP3 knock-out animals. This suggests the primary role for CASP3 in denervation atrophy may be in promoting apoptosis rather than in actin cleavage143. The functional impact of CASP3 activity was further demonstrated by CASP3 knock-down in rat heart muscle, which led to inhibition of sepsis-induced changes in contractile properties, an increase in selected contractile proteins, and a reduction in markers of DNA damage144.
The precise roles and targets of caspase activation are unclear. Like calpains, caspases appear to work upstream of the UPS to facilitate the release of myofibrillar proteins, and in some studies there is evidence of parallel activation of both caspases and calpains contributing to muscle weakness and wasting122. In addition, caspase cleavage promotes atrophy by initiating myonuclear apoptosis. Caspases interact with other proteolytic systems including calpains as described above, and can also enhance proteasome activity142. Furthermore there is now evidence from studies in other cell types suggesting that caspase activity may have a role in actively directing a cell away from autophagy in favor of apoptosis through caspase cleavage of Beclin-1145.
Major signaling pathways regulating muscle proteolysis
The IGF1/PI3K/AKT/FoxO pathway
Great strides have been made in the last decade in uncovering the intracellular signaling pathways that control muscle fiber size. A central pathway involves IGF1 and insulin, which are most important in promoting net protein accumulation and resulting growth/hypertrophy of adult muscle fibers. In skeletal muscle, the binding of IGF1 or insulin to the IGF1 receptor activates two major signaling pathways: the Ras-Raf-MEK-ERK pathway and the PI3K/AKT pathway. The Ras-Raf-MEK-ERK pathway affects fiber type composition but has little effect on muscle fiber size146, while activation of the PI3K/AKT pathway induces muscle hypertrophy by increasing translation (protein synthesis) through phosphorylation/activation of GSK (glycogen synthase kinase) and mTOR kinases58,147. In addition to its activation through AKT phosphorylation, mTOR is also controlled by nutrient supply148. During fasting or nutrient insufficiency, low levels of amino acids lead directly to inhibition of mTOR. This inhibition is augmented by the fall in insulin and IGF1 in these states, which decrease nutrient transport into muscle, further amplifying the reduction in mTOR activity. mTOR can thus integrate signals from the IGF1/PI3K/AKT pathway with information about the cell’s nutritional status, both of which then influence mTOR activity. The latter is mediated by two multi-subunit mTOR complexes, the major one being mTORC1, in which mTOR complexes with raptor, and the less important one, mTORC2, in which mTOR complexes with rictor149. The TORC1 complex is primarily the controller of protein synthesis through its phosphorylation of S6 kinase, while mTORC2 is postulated to regulate cytoskeletal organization. Interestingly, skeletal muscle lacking raptor (but not rictor) develops a dystrophic phenotype with smaller, damaged fibers 150. This phenotype seems to stem from a complex combination of dysregulated pathways downstream of mTORC1 that includes decreased translation resulting from reduced S6K activity, suppression of PGC-1α (PGC-1, peroxisome proliferator-activated receptor gamma coactivator), and loss of feedback inhibition of IRS-1 that leads to hyperphosphorylation of AKT.
Several recent findings suggest that decreased activity of the IGF1/PI3K/AKT signaling pathway leads to muscle atrophy. Inhibition of PI3K and expression of dominant negative (inhibitory) forms of AKT reduce myotube diameter in culture151, and muscles from mice lacking AKT1 or AKT2 are smaller than their wild type littermates13. Conversely, stimulation of AKT can prevent denervation atrophy152–153. As noted above, much of the IGF1-stimulated growth in muscle results from an increase in protein synthesis. Reduced AKT activity when IGF1 is low presumably causes decreases in translation that contribute to the loss of mass seen in muscle atrophy. However there is now evidence that strongly suggests the IGF1/PI3K/AKT pathway not only promotes protein synthesis but also induces reciprocal changes in protein degradation. This is demonstrated in part by evidence from experiments in tissue culture showing insulin and IGF1 inhibition of glucocorticoid-induced activation of proteolysis154. These experiments strongly suggest that the IGF1/PI3K/AKT pathway, in addition to affecting translation mechanisms, suppresses protein degradation.
One downstream target of the IGF1/PI3K/AKT pathway that could mediate these IGF1 effects on protein degradation is the FoxO class of transcription factors, a subfamily of the large group of forkhead transcription factors. Mammalian cells contain three members of this family, FoxO1 (FKHR), FoxO3 (FHKRL1), and FoxO4 (AFX)155. AKT blocks the function of all three transcription factors by phosphorylation of three conserved residues, leading to their sequestration in the cytoplasm away from target genes156. Dephosphorylation of FoxO factors leads to nuclear entry and transcription of downstream genes that promote growth-suppression or apoptosis157. FoxO gene expression is also tightly regulated. Fasting and glucocorticoids induce the expression of FoxO factors in muscle, while refeeding suppresses FoxO transcription158. In seminal papers by both the Goldberg and the Glass groups159–160, manipulation of FoxO1 and FoxO3 levels was shown to have dramatic effects on muscle fiber size: expression of constitutively active forms led to dramatic atrophy while dominant negative (inhibitory) forms prevented denervation-induced atrophy. Dephosphorylation/activation of FoxO leads to induction of a conserved set of atrophy-specific genes (atrogenes) including the muscle-specific ubiquitin ligases, MAFbx/Atrogin-1 and MuRF-1 as well as the FoxO factors themselves, and promotes increased rates of protein degradation. Recent data has further demonstrated that activation of FoxO factors also promotes autophagy in skeletal muscle95–96. The parallel activation of ubiquitin-mediated and autophagic protein degradation may promote the concerted dismantling of muscle during atrophic states in which different proteolytic pathways target different cellular constituents; the UPS for soluble and myofibrillar proteins and the autophagy system for organelles such as mitochondria. Thus, suppression of IGF1/PI3K/AKT signaling, leading to reduced levels of AKT and FoxO phosphorylation, FoxO nuclear translocation and increased FoxO-dependent transcription, appears to be the dominant atrophy signaling pathway in skeletal muscle.
The PGC-1α transcriptional coactivator suppresses FoxO transcription
The PGC-1 family of transcriptional coactivators has been recently identified as promoters of the transcriptional program for mitochondrial biogenesis and oxidative metabolism that is critical to the maintenance of energy homeostasis in muscle161–162. Transgenic expression of PGC-1α in fast-twitch, glycolytic muscles transforms the type IIb muscle fibers into a more oxidative, mitochondria-rich phenotype163–164. PGC-1α therefore appears to be an important mediator in regulating mitochondrial metabolic properties in skeletal muscle and is a prime candidate as a target regulator. Interestingly, it has been found that PGC-1α mRNA falls dramatically in various types of atrophying muscle165. Furthermore, in transgenic mice over-expressing PGC-1α, denervation and fasting cause a much smaller decrease in muscle fiber diameter and a smaller induction of MAFbx/Atrogin-1 than in control mice165. Importantly, when the levels of PGC-1α are maintained artificially by electroporation of PGC-1α cDNA into adult muscle fibers, muscles are protected from the atrophy induced by denervation, fasting, or expression of FoxO3. This protective effect can explain how exercise, which induces expression of PGC-1α, maintains muscle mass and retards atrophy, even in the face of circulating catabolic factors166. Furthermore, recent findings using chromatin immunoprecipitation (ChIP) assays indicate that PGC-1α directly or indirectly inhibits FoxO-dependent binding to and transcription of the atrogin-1 promoter165. Mechanistically, an alternative explanation for the protection afforded by PGC-1α is that it may lead to expression of an inhibitor of FoxO transcription. PGC-1α therefore is a prime anti-atrophy target, the activity of which might be enhanced to protect against muscle atrophy through its ability to augment mitochondrial number and function.
AMPK also acts on the IGF1 signaling pathway and promotes atrophy signaling
Another intracellular signaling molecule that seems, at least in part, to feed into the IGF1/AKT/FoxO pathway is AMPK (AMP activated kinase). AMPK phosphorylation and activation are triggered by falling energy content (increased AMP/ATP ratio)167, a state often present in catabolic conditions in which muscles atrophy. Recent studies have shown that an increase in muscle p-AMPK stimulates MAFbx/Atrogin-1 expression in mice and in cultured muscle cells102,168. In mice treated with the AMPK agonist, AICAR, the agonist raises phospho-AMPK and increases MAFbx/Atrogin-1 expression, which is blocked by the AMPK inhibitor, Compound C. AMPK activation promotes increased FoxO expression as well as its dephosphorylation and activation; this provides a mechanism by which AMPK leads to atrogene expression102,169. This pathway provides a potential means to link mitochondrial dysfunction and its consequent decrease in ATP production with atrophy development and to further amplify atrophy-promoting signaling pathways. Energy insufficiency can lead to FoxO activation directly via increased AMPK activity, and in addition, FoxO-mediated autophagy promotes mitochondrial loss and further energy imbalance through increased dependence on glycolytic as opposed to oxidative metabolism.
NFκB and p38: other signaling pathways that act in parallel to the IGF1 signaling pathway
The secreted cytokine TNF170 is well known as a cachectic factor that, along with the related factor TWEAK (TNF-related weak inducer of apoptosis)171 promotes muscle atrophy through multiple mechanisms172. Fundamentally, TNF binding to its receptor leads to activation of NFκB and its downstream targets173. Recent studies have demonstrated the role of NFκB in cytokine-induced loss of myofibrillar proteins such as myosin174. Since NFκB is also activated by sepsis175 and disuse176, it likely plays a general role in development of muscle atrophy. Indeed, in culture models, blocking NFκB lowers rates of protein degradation,177 and conversely TNF treatment can attenuate insulin-stimulated protein synthesis178. TNF-mediated NFκB activation can also inhibit myocyte differentiation through the suppression of myogenic transcription factors like MyoD174,179–180. This added effect of NFκB on myogenesis may suppress the ability of muscle precursor, or satellite, cells to be recruited into atrophying muscle, amplifying the loss of muscle mass179. Recent studies have demonstrated that the cytokine, TWEAK, has even more robust effects in causing muscle wasting than its related cousin, TNF171. In cultured cells or mice treated with TWEAK and in muscle-specific TWEAK transgenic animals, dramatic reductions in fiber size were seen. These effects may be due to the concomitant stimulation of multiple processes contributing to atrophy.
Important recent studies in mice highlight the importance of NFκB activation in promoting muscle atrophy. Using transgenic mice that constitutively activate NFκB (MIKK mice, for muscle specific expression of IKK, which phosphorylates IκBα and leads to activation of NFκB) or repress NFκB (MISR mice, for muscle specific “super-repressor”, which expresses a dominant negative form of IκB that prevents NFκB activation), Shoelson and coworkers demonstrated corresponding atrophied and hypertrophied muscles respectively181. Interestingly, the expression of the ubiquitin-ligase MuRF-1, but not MAFbx/Atrogin-1, was up-regulated in the MIKK skeletal muscle, a finding that provided the first functional dissociation of the signals that induce MuRF-1 and MAFbx/Atrogin-1181. The fact that MAFbx/Atrogin-1 is up-regulated in essentially all types of atrophying muscle is strong support for activation of a second parallel pathway, distinct from NFκB, (e.g. FoxO activation), during atrophy. Further experiments showed that NFκB is also involved in mediating atrophy and remodeling due to unloading; NFκB1 knock-out animals demonstrated less atrophy following disuse (unloading) as well as a blockade of the fiber-type switching usually associated with atrophying muscle182.
In addition to activating NFκB, TNF can also mediate signaling through p38. Interestingly, in cultured muscle cells, TNF acts to induce MAFbx/Atrogin-1 expression, and this up-regulation can be blocked by pharmacologic inhibitors of p38183. Whether or not p38 has any effect on FoxO or MuRF-1 induction or on the NFκB pathway is not known.
Myostatin
Myostatin is a member of the TGFα family that may also promote atrophy. Loss of function mutations in myostatin lead to skeletal muscle hypertrophy in multiple species including humans184–187, whereas infusion of cells expressing myostatin into adult mice leads to muscle wasting188. Forced over-expression of myostatin in skeletal muscle, however, does not show quite the same robust effects as the genetic mutants and infusion experiments above. In muscle-specific myostatin expressing transgenic mice, only males have reduced muscle and myofiber size189 and in experiments in which myostatin was electroporated into skeletal muscle, only 10–20% reduction in fiber cross-sectional area was seen190. Despite this, anti-myostatin peptibodies are capable of blocking muscle atrophy in mice with chronic kidney disease191. Myostatin binds to the high affinity activin type 2 receptor (ActRIIB) and oligomerizes with the co-receptors ALK4 or ALK5, which mediate its signaling through phosphorylation of Smad2 and 3 (Smad, mothers against decapentaplegic homolog)185,192. Expression of dominant negative ActRIIB in transgenic mice results in skeletal muscle hypertrophy and suggests a role of the ActRIIB pathway in limiting muscle growth185. Exciting new experiments demonstrate that antagonizing this receptor can dramatically prevent cancer-induced cachexia193. Administration of an ActRIIB decoy receptor (sActRIIB) in multiple cancer cachexia models prevents further wasting, but also fully reverses skeletal muscle loss and atrophy of the heart, thereby dramatically prolonging survival of the tumor-bearing animals. The potent effects of the ActRIIB decoy may be augmented by inhibition of other circulating ligands like activin that bind to and stimulate ActRIIB. Thus, the ActRIIB pathway inhibition appears to have therapeutic potential for treating various types of muscle wasting, especially cancer cachexia.
Integrated control of signaling pathways that promote atrophy
As described above, in atrophying muscle, suppression of IGF1 signaling and activation of AMPK, NFκB and Smad signaling through ActRIIB all contribute to the loss of muscle mass (Figure 3). It appears quite likely that these pathways act in concert with cross-talk between them. For example, AMPK activation has been shown to suppress, through dephosphorylation, the FoxO transcription factors102. The cytokine TWEAK both activates NFκB and suppresses IGF1 signaling171, and signaling through ActRIIB may also contribute to reduced activity of mTOR192. Recent experiments also demonstrate that FoxO activation leads to IRS-2-mediated stimulation of the MEK/ERK signaling pathway and promotes expression of poly-ubiquitin194. Yet another example of cross-talk between signaling pathways is a recent study that shows glucocorticoid-induced glucocorticoid receptor sequestration of PI3K promoting reduction of IGF1 signaling in acutely diabetic mice195. Finally, the muscle wasting associated with inflammation involves IL-6 and serum amyloid A protein-induced impairment of IGF1 signaling196.
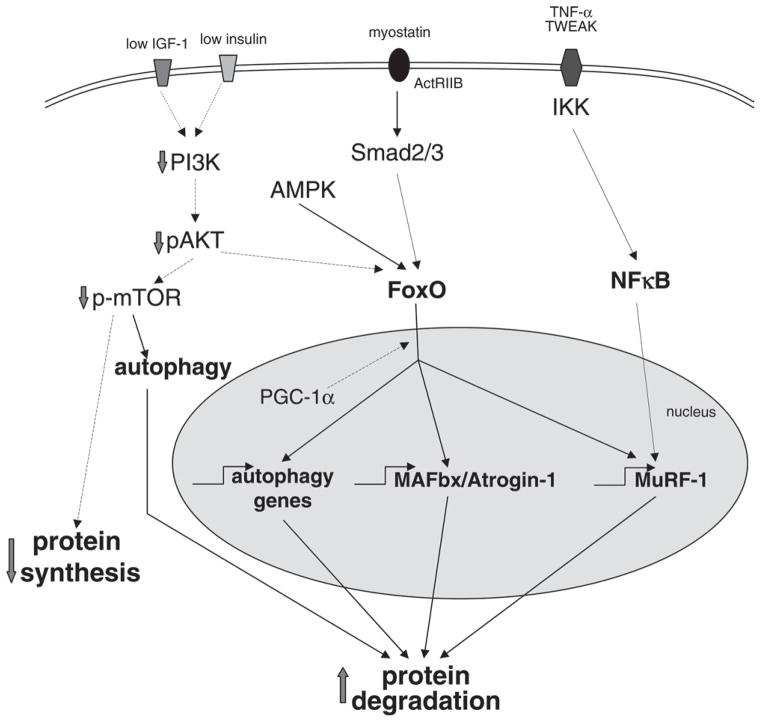
Figure 3.
Signaling pathways involving MAFbx/Atrogin-1 and MuRF-1 that promote muscle wasting. Three major intracellular signaling pathways act to increase muscle protein degradation and suppress synthesis, leading to the loss of mass seen in atrophying muscle. Suppression of signaling through PI3K and AKT, caused by reduced insulin, insulin resistance or reduced IGF1, leads to FoxO translocation to the nucleus and transcription of the atrogenes MAFbx/Atrogin-1 and MuRF-1 as well as activation of autophagy. Myostatin binding to the ActRIIB leads to signaling through Smad intermediates that stimulates FoxO and other downstream targets. Cytokines such as TNF and TWEAK promote NFκB activation and lead to transcriptional activation of atrogenes such as MuRF-1. AMPK activation can augment atrophy signaling through phosphorylation of FoxO while PGC-1α can suppress these pathways also at the level of FoxO. (dashed lines-inhibition; solid lines - stimulation).
The contribution of multiple signaling pathways to overall control of muscle mass is also consistent with the requirement of muscle to be able to respond to multiple extra-cellular signals. These include circulating factors (e.g. insulin, IGF1, thyroxine, corticosteroids and pro-inflammatory cytokines), paracrine molecules (e.g. mechano-growth factor (MGF) and myostatin), the type and concentration of metabolic substrates (e.g. branched-chain amino acids), environmental conditions (e.g. oxygen, acidosis), neuronal stimulation frequency or pattern and tension or workload. The integration of these multiple inputs and signaling pathways in muscle defines muscle mass, energy and protein metabolism, and fiber phenotype. In conditions leading to muscle atrophy, the balance among these different factors is disturbed, and changes typically occur in multiple components of this complex network with stimulation or inhibition of overlapping intracellular signaling pathways. In this way it is clear that net muscle wasting will show wide variation among individuals with the same disease, due to the combined effects of the many factors which impact on muscle mass and function.
Biomarkers and barriers to their use
The improved molecular understanding of the process of muscle wasting has the potential to produce better biomarkers for diagnosis and to gauge treatment. Clinically, the release of muscle enzymes like creatine kinase, aldolase and aspartate aminotransferase into the bloodstream is commonly used to diagnose muscle damage (myopathy) as opposed to atrophy or wasting, in which elevated concentrations of these substances are typically not found. In fact circulating factors that correlate with atrophy are lacking. Reduced concentrations of IGF1 might be expected in atrophy conditions, but since most IGF1 is produced locally, levels in the circulation correlate poorly with disease. Likewise, cytokines such as IL-1 and IL-16 and other factors such as TNF-α and myostatin have potential as biomarkers, but to date have not been well correlated with atrophying muscle. On the other hand, gene expression data has been used successfully, at least in research studies, to define the atrophy process. Muscle-specific ubiquitin protein ligases such as MAFbx/Atrogin-1 and MuRF-1 that are dramatically up-regulated as atrophy occurs have been used in a large number of animal and human197 studies as sensitive markers of atrophy. The major barrier to their more generalized clinical use is the invasiveness of the muscle biopsy that is required to obtain the material necessary to measure gene expression. To date, attempts to identify trace levels of MAFbx/Atrogin-1 or MuRF-1 in the circulation of patients or animals with atrophying muscle have been unsuccessful (S.L., unpublished). One additional potential biomarker mentioned above is the caspase-derived 14 kDa actin fragment identified from atrophying muscles by Mitch and coworkers141. Though this fragment has recently been observed in atrophying muscles from hemodialysis and burn patients198, its broad use suffers from the same pitfalls as the gene expression markers above, namely, that it also requires a muscle biopsy to be performed.
Current potential drug targets and strategies
It is clear from the above that we now have a more comprehensive understanding of muscle proteolytic pathways, their functions and their regulation. Loss of function studies have identified a number of genes within these pathways whose inactivation reduces muscle wasting (Table 2). Because most of these are enzymes, the development of pharmacological inhibitors to them is an attractive and feasible approach towards the prevention and treatment of muscle atrophy. These targets presently include the ubiquitin ligases MuRF-1, MAFbx/Atrogin-1, Cbl-b, the deubiquitinating enzyme USP19, and CASP3. MuRF-1 and MAFbx/Atrogin-1 are particularly interesting because their expression is restricted to striated muscle tissues and they show very high induction of expression in a broad array of catabolic conditions that include local disuse as well as systemic diseases such as cancer 197. Thus, even modest inhibition of these muscle-specific E3 ligases may be sufficient to have a significant protective effect and their tissue-specific expression may limit the risk of systemic toxicity. In fact, strong and prolonged inhibition of these enzymes may result in myopathy, a suggestion that is supported by the observation of muscle damage and decreased function upon genetic inactivation of two of the three MuRF ligases66–67.
Table 2.
Genetic gain- and loss-of-function studies in muscle wasting.
| Molecule | Function | Effect on muscle atrophy | References |
|---|---|---|---|
| MAFbx/Atrogin-1 | Muscle-specific E3 ligase | Promote | (58) |
| MuRF-1 | Muscle-specific E3 ligase | Promote | (58) |
| Cbl-b | E3 ligase | Promote | (76) |
| Calpastatin | Endogenous calpain inhibitor | Protect | (113, 125) |
| Caspase 3 | Protease | Promote | (143) |
| AKT | Kinase | Protect | (147, 153) |
| TORC1 | Kinase | Promote | (150) |
| FoxO | Transcription factor | Promote | (95–96, 159–160, 216) |
| PGC-1α | Transcriptional co-activator | Protect | (165) |
| JunB | Transcription factor | Protect | (217) |
| NFκB | Transcription factor | Promote | (181–182) |
| TWEAK | Cytokine | Promote | (171) |
| ActRIIB | Cytokine receptor | Promote | (193) |
| Myostatin | Cytokine | Promote | (184–189) (190) |
Other candidates as drug targets include genes with more general tissue expression. These include Cbl-b and USP19, which also regulate receptor tyrosine kinase trafficking28 and cell cycle199, respectively, but the general lack of a major phenotype in mouse knock-outs of those genes indicate that they may still be viable drug targets. Whole body inactivation of CASP3, an effector caspase in apoptosis, leads to a high proportion of embryonic death, a finding which makes this a less attractive candidate for inhibition. As discussed above, acute inhibition of autophagy can also inhibit muscle wasting, but chronic inhibition such as occurs in gene inactivation studies of an important autophagy gene leads to severe myopathy. Furthermore inhibition of CAPN1 and 2 through calpastatin over-expression also ameliorates muscle atrophy, so calpains may be useful therapeutic targets113,125. Despite concerns regarding inhibition of broadly expressed drug targets with multiple cellular functions, there is evidence that inhibition of such pathways in adults or mature tissues may prove less toxic than expected. For example, given the universal importance of the UPS for protein turnover and regulation of multiple cellular pathways in all eukaryotic cells, it is surprising that the proteasome inhibitor Velcade (Bortezemib), the only drug inhibitor of the UPS that is approved for clinical use (in the treatment of multiple myeloma), is well tolerated. However, sadly, to date there is no evidence that Velcade has beneficial effects on human muscle wasting.
An inhibitor of any of these targets could possibly be effective if it possessed a pharmacokinetic profile that lowered its toxicity and/or was administered for only short periods of time when catabolic stimuli were maximal (e.g. episode of sepsis, short periods of fasting, peri-operatively). Such limited treatment could avoid toxicity arising from inhibition of other important functions of the targeted enzymes (e.g. muscle recovery, remodeling, or quality control of proteins and organelles).
The discussion in this review also suggests strongly that drug manipulation of signaling pathways such as the IGF1/AKT, cytokine-NFκB, and myostatin/activin pathways may be a feasible approach to the treatment of muscle wasting. Many of these pathways modulate both protein degradation and protein synthesis in a reciprocal manner to mediate changes in muscle size. As a result, targeting signaling pathways may be more effective than attempts at inhibiting proteolysis alone. However, once again signaling pathways which modulate overall protein synthesis and proteolysis are also broadly utilized in most cell types and, as discussed above, targeting such fundamental processes to control muscle atrophy may be limited by systemic toxicity. In the case of the IGF1/AKT pathway, one would be concerned about the stimulatory effects of the administration of growth hormone, IGF1 or activators of downstream signaling molecules on growth in other tissues or in cancerous cells. Finally from a drug design perspective, identifying specific activators of enzymes is much more challenging than identifying specific inhibitors. However, the recent report of dramatic protection against cancer cachexia in mice by administration of a decoy receptor for activins demonstrates that targeting signaling pathways is indeed promising and that systemic toxicities may be avoided when administered for a limited period of time.
Conclusions - from pathways to networks and beyond
The last 20 years have witnessed major advances in our understanding of proteolysis in general as well as in skeletal muscle in particular. The application of gain of function and loss of function approaches to evaluating the roles of specific genes in proteolytic systems has clearly demonstrated that the UPS and the autophagic/lysosomal systems play important roles in mediating muscle protein catabolism. More limited studies suggest roles for calcium-dependent proteases and caspases, but further studies are required to better define the mechanisms by which these proteolytic systems may exert their effects on muscle protein balance. Nonetheless, the involvement of multiple proteolytic systems is concordant with the notion that as the myofiber atrophies, multiple cellular components (e.g. myofibrillar proteins, mitochondria, sarcoplasmic reticulum, nuclei) need to be degraded. Since all proteolytic systems have some degree of substrate specificity, a requirement for the coordinate involvement of multiple proteolytic systems is not surprising. Interestingly, as described in this review, the proteolytic systems interact with each other directly–e.g. the ubiquitination of some damaged proteins can lead to binding to the p62 adaptor protein and targeting to the autophagy pathway; calpains may cleave a subunit of the proteasome and thereby activate the enzyme.
The recent advances in our understanding of the signaling pathways that modulate skeletal muscle fiber growth and atrophy also support the concept of coordinate regulation of multiple proteolytic systems. As described in this review, the IGF1/AKT/FoxO pathway modulates in a coordinate fashion both the ubiquitin proteasome system and the autophagic pathway. Other pathways such as myostatin /activin or AMPK signaling can exert at least some of its stimulatory effects on protein degradation via inhibition of AKT and activation of FoxO. PGC-1α can antagonize FoxO action at the promoters of its target genes, and recent evidence indicates that stress activated MAP kinases can stimulate export of FoxO from the nucleus. The ability of diverse signaling pathways to cross-talk at various levels between the initial signal and its target proteolytic system indicates clearly that proteolytic systems function within complex networks rather than as independent pathways.
The analysis of the functions of such signaling pathways as the AKT pathway also suggests a coordinate control of protein synthesis and protein degradation. Intriguingly, though, recent evidence suggests that coordinate control of protein synthesis and degradation also occurs through more intricate mechanisms in which induction of the MAFbx/Atrogin-1 ligase can lead to degradation of myoD, myogenin and eIF3-f that result in decreased rates of transcription and translation of important muscle cell proteins.
Despite the progress made in the last 20 years, a number of deficits in our understanding remains: (a) The structure of the network that is becoming apparent needs to be characterized more completely. In particular, more of the key nodes and the signalling molecules in the network need to be identified; (b) The functions of each proteolytic system need to be better characterized–e.g. what are all of the substrates (or organelles in the case of autophagy) of each proteolytic system? (c) How is the degradation of the various myofiber components regulated to maintain correct proportions of each of the components? (d) How important is myofiber loss in atrophy as well as new myofiber formation, or myoblast fusion to existing fibers, in hypertrophy or recovery from atrophy? (e) What are the contributions of the various proteolytic systems and protein synthesis to overall protein balance at various time points in the muscle wasting process? What are the implications of this with respect to the usefulness of therapies targeted to specific proteolytic systems? (f) Can longer term treatments to limit or prevent muscle atrophy be devised which enhance muscle function and preserve muscle tissue physiological plasticity? Although many questions remain, advances in recent years offer significant hope that effective treatment to prevent or reverse muscle wasting and sarcopenia can be found.
Abbreviations
- ActRIIB
activin type 2 receptor
- AKT
serine/threonine kinase involved in insulin/IGF1 signaling
- AMPK
AMP activated kinase
- Atg or ATG
autophagy system gene or protein
- CASP
caspase
- CAPN
calpain, Ca2+-dependent cytosolic cysteine protease
- Cbl-b
a ubiquitin ligase, functions as a negative regulator of many signaling pathways downstream of receptor tyrosine kinases
- CRL
cullin-RING ligase
- CT
computed tomography
- E1
ubiquitin activating enzyme
- E2
ubiquitin conjugating enzyme, UBC
- E3
ubiquitin protein ligase
- E4
ubiquitin chain elongating enzyme
- eIF3
eukaryotic initiation factor for translation
- FoxO
forkhead box protein, transcriptional activator
- GSK
glycogen synthase kinase
- IGF1
insulin-like growth factor 1
- IκBα
NFκB inhibitor alpha
- IL
interleukin
- IRS
insulin receptor substrate
- LC 3
microtubule-associated proteins 1A/1B light chain 3A, protein involved in elongation of autophagosome
- MAFbx/Atrogin-1
muscle atrophy F-box containing protein/Atrogin-1, ubiquitin protein ligase
- mTOR
mammalian target of rapamycin
- MuRF
muscle RING finger containing ligase
- MyBP-C
myosin binding protein C
- MyHC
myosin heavy chain
- MyLC
myosin light chain
- MyoD
myoblast determination protein, muscle differentiation factor
- NFκB
nuclear factor kappa-B, pleiotropic transcription factor involved in many biological processes including inflammation, immunity, differentiation, cell growth, tumorigenesis and apoptosis
- PGC-1
peroxisome proliferator-activated receptor gamma coactivator, stimulator of transcription factors and nuclear receptors activities
- PI3K
phosphoinositide-3-kinase
- Smad
mothers against decapentaplegic homolog, intracellular signal transducer and transcriptional modulator activated by TGF-beta (transforming growth factor) and activin type 1 receptor kinases
- TNF
tumor necrosis factor, inflammatory cytokine
- TWEAK
TNF-related weak inducer of apoptosis, inflammatory cytokine
- Ubl
ubiquitin-like
- UCH
ubiquitin C-terminal hydrolase
- UPS
ubiquitin proteasome system
- USP
ubiquitin specific protease
Footnotes
Referee William E. Mitch, Professor of Medicine and Chief of Nephrology, Baylor College of Medicine, Houston, TX, USA
Declarations of interest
The authors report no declarations of interest.
References
- 1.Trends in aging - United States and worldwide. CDC. MMWR Morb Mortal Wkly Rep. 2003;52:101–104. 106. [PubMed] [Google Scholar]
- 2.Evans WJ, Morley JE, Argilés J, Bales C, Baracos V, Guttridge D, et al. Cachexia: a new definition. Clin Nutr. 2008;27:793–799. doi: 10.1016/j.clnu.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Marquis K, Debigaré R, Lacasse Y, LeBlanc P, Jobin J, Carrier G, et al. Midthigh muscle cross-sectional area is a better predictor of mortality than body mass index in patients with chronic obstructive pulmonary disease. Amer J Resp Crit Care. 2002;166:809–813. doi: 10.1164/rccm.2107031. [DOI] [PubMed] [Google Scholar]
- 4.Anker SD, Coats AJ. Cardiac cachexia: a syndrome with impaired survival and immune and neuroendocrine activation. Chest. 1999;115:836–847. doi: 10.1378/chest.115.3.836. [DOI] [PubMed] [Google Scholar]
- 5.Giles JT, Bartlett SJ, Andersen RE, Fontaine KR, Bathon JM. Association of body composition with disability in rheumatoid arthritis: impact of appendicular fat and lean tissue mass. Arthritis Rheum. 2008;59:1407–1415. doi: 10.1002/art.24109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan B, Birdsell L, Martin L, Baracos V, Fearon K. Sarcopenia in an overweight or obese patient is an adverse prognostic factor in pancreatic cancer. Clin Cancer Res. 2009;15:6973–6979. doi: 10.1158/1078-0432.CCR-09-1525. [DOI] [PubMed] [Google Scholar]
- 7.Prado CM, Baracos VE, McCargar LJ, Reiman T, Mourtzakis M, Tonkin K, et al. Sarcopenia as a determinant of chemotherapy toxicity and time to tumor progression in metastatic breast cancer patients receiving capecitabine treatment. Clin Cancer Res. 2009;15:2920–2926. doi: 10.1158/1078-0432.CCR-08-2242. [DOI] [PubMed] [Google Scholar]
- 8.Janssen I, Heymsfield SB, Ross R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J Am Geriatr Soc. 2002;50:889–896. doi: 10.1046/j.1532-5415.2002.50216.x. [DOI] [PubMed] [Google Scholar]
- 9.Arora NS, Rochester DF. Effect of body weight and muscularity on human diaphragm muscle mass, thickness, and area. J Appl Physiol. 1982;52:64–70. doi: 10.1152/jappl.1982.52.1.64. [DOI] [PubMed] [Google Scholar]
- 10.Srikuea R, Pholpramool C, Kitiyanant Y, Yimlamai T. Satellite cell activity in muscle regeneration after contusion in rats. Clin Exp Pharmacol Physiol. 2010;37:1078–1086. doi: 10.1111/j.1440-1681.2010.05439.x. [DOI] [PubMed] [Google Scholar]
- 11.Prado CM, Birdsell LA, Baracos VE. The emerging role of computerized tomography in assessing cancer cachexia. Curr Opin Supp Palliative Care. 2009;3:269–275. doi: 10.1097/SPC.0b013e328331124a. [DOI] [PubMed] [Google Scholar]
- 12.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 13.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 14.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Shang F, Deng G, Liu Q, Guo W, Haas AL, Crosas B, et al. Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. J Biol Chem. 2005;280:20365–20374. doi: 10.1074/jbc.M414356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 19.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. J Biol Chem. 1982;257:10329–10337. [PubMed] [Google Scholar]
- 20.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 21.Reiss Y, Heller H, Hershko A. Binding sites of ubiquitin-protein ligase. Binding of ubiquitin- protein conjugates and of ubiquitin-carrier protein. J Biol Chem. 1989;264:10378–10383. [PubMed] [Google Scholar]
- 22.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 23.Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- 24.Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, et al. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J Biol Chem. 1988;263:15726–15731. [PubMed] [Google Scholar]
- 25.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 26.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thiolester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 28.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-Type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 29.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deffenbaugh AE, Scaglione KM, Zhang L, Moore JM, Buranda T, Sklar LA, et al. Release of ubiquitin-charged Cdc34-S - Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell. 2003;114:611–622. doi: 10.1016/s0092-8674(03)00641-x. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa M, Ohta T, Xiong Y. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J Biol Chem. 2002;277:15758–15765. doi: 10.1074/jbc.M108565200. [DOI] [PubMed] [Google Scholar]
- 32.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Madura K. Rad23 and Rpn10: perennial wallflowers join the melee. Trends Biochem Sci. 2004;29:637–640. doi: 10.1016/j.tibs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 36.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 37.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 38.Wing SS, Haas AL, Goldberg AL. Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and denervation atrophy. Biochem J. 1995;307:639–645. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baracos VE, DeVivo C, Hoyle DH, Goldberg AL. Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Amer J Physiol Endo Metab. 1995;268:E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- 40.Lorite MJ, Thompson MG, Drake JL, Carling G, Tisdale MJ. Mechanism of muscle protein degradation induced by a cancer cachectic factor. Br J Cancer. 1998;78:850–856. doi: 10.1038/bjc.1998.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llovera M, Garcia-Martinez C, Agell N, Lopez-Soriano FJ, Argiles JM. Muscle wasting associated with cancer cachexia is linked to an important activation of the ATP-dependent ubiquitin-mediated proteolysis. Int J Cancer. 1995;61:138–141. doi: 10.1002/ijc.2910610123. [DOI] [PubMed] [Google Scholar]
- 42.Tiao G, Fagan J, Roegner V, Lieberman M, Wang JJ, Fischer JE, et al. Energy-ubiquitin-dependent muscle proteolysis during sepsis in rats is regulated by glucocorticoids. J Clin Invest. 1996;97:339–348. doi: 10.1172/JCI118421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wing SS. Control of ubiquitination in skeletal muscle wasting. Int J Biochem Cell Biol. 2005;37:2075–2087. doi: 10.1016/j.biocel.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 45.Taillandier D, Aurousseau E, Meynial-Denis D, Bechet D, Ferrara M, Cottin P, et al. Coordinate activation of lysosomal, Ca2+-activated and ATP- ubiquitin-dependent proteinases in the unweighted rat soleus muscle. Biochem J. 1996;316:65–72. doi: 10.1042/bj3160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, et al. Increased ATP-ubiquitin-dependent proteolysis in skeletal muscles of tumor-bearing rats. Cancer Res. 1994;54:5568–5573. [PubMed] [Google Scholar]
- 47.Voisin L, Breuille D, Combaret L, Pouyet C, Taillandier D, Aurousseau E, et al. Muscle wasting in a rat model of long-lasting sepsis results from the activation of lysosomal, Ca2+-activated, and ubiquitin- proteasome proteolytic pathways. J Clin Invest. 1996;97:1–8. doi: 10.1172/JCI118586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attaix D, Taillandier D, Combaret L, Ralliere C, Larbaud D, Aurousseau E, et al. Expression of subunits of the 19S complex and of the PA28 activator in rat skeletal muscle. Mol Biol Rep. 1997;24:95–98. doi: 10.1023/a:1006806103675. [DOI] [PubMed] [Google Scholar]
- 49.Combaret L, Tilignac T, Claustre A, Voisin L, Taillandier D, Obled C, et al. Torbafylline (HWA 448) inhibits enhanced skeletal muscle ubiquitin-proteasome-dependent proteolysis in cancer and septic rats. Biochem J. 2002;361:185–192. doi: 10.1042/0264-6021:3610185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heng AE, Ventadour S, Jarzaguet M, Pouch-Pelissier MN, Guezennec CY, Bigard X, et al. Coordinate expression of the 19S regulatory complex and evidence for ubiquitin-dependent telethonin degradation in the unloaded soleus muscle. Int J Biochem Cell Biol. 2008;40:2544–2552. doi: 10.1016/j.biocel.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Combaret L, Taillandier D, Dardevet D, Bechet D, Ralliere C, Claustre A, et al. Glucocorticoids regulate mRNA levels for subunits of the 19 S regulatory complex of the 26 S proteasome in fast-twitch skeletal muscles. Biochem J. 2004;378:239–246. doi: 10.1042/BJ20031660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bossola M, Muscaritoli M, Costelli P, Grieco G, Bonelli G, Pacelli F, et al. Increased muscle proteasome activity correlates with disease severity in gastric cancer patients. Ann Surg. 2003;237:384–389. doi: 10.1097/01.SLA.0000055225.96357.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minnaard R, Wagenmakers AJ, Combaret L, Attaix D, Drost MR, van Kranenburg GP, et al. Ubiquitin-proteasome-dependent proteolytic activity remains elevated after zymosan-induced sepsis in rats while muscle mass recovers. Int J Biochem Cell Biol. 2005;37:2217–2225. doi: 10.1016/j.biocel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 55.Combaret L, Adegoke OA, Bedard N, Baracos V, Attaix D, Wing SS. USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am J Physiol Endocrinol Metab. 2005;288:E693–E700. doi: 10.1152/ajpendo.00281.2004. [DOI] [PubMed] [Google Scholar]
- 56.Sundaram P, Pang Z, Miao M, Lu Y, Wing SS. USP19 deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells. Am J Physiol Endocrinol Metab. 2009;297:E1283–E1290. doi: 10.1152/ajpendo.00409.2009. [DOI] [PubMed] [Google Scholar]
- 57.Sundaram P, Pang Z, Miao M, Bedard N, Moore T, Wing SS. Loss of USP19 increases transcription of myofibrillar proteins in L6 muscle cells and decreases muscle wasting in response to denervation in mice. J Cachexia Sarcopenia Musc. 2010;1:87. [Google Scholar]
- 58.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 59.Bdolah Y, Segal A, Tanksale P, Karumanchi SA, Lecker SH. Atrophy-related ubiquitin ligases atrogin-1 and MuRF-1 are associated with uterine smooth muscle involution in the postpartum period. Am J Physiol Regul Integr Comp Physiol. 2007;292:R971–R976. doi: 10.1152/ajpregu.00617.2006. [DOI] [PubMed] [Google Scholar]
- 60.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 62.Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J Mol Biol. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 63.Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6:376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117:2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem. 2005;280:2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- 69.Jogo M, Shiraishi S, Tamura TA. Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Lett. 2009;583:2715–2719. doi: 10.1016/j.febslet.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 70.Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, et al. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, et al. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanai JI, Cao P, Tanksale P, Imamura S, Koshimizu E, Zhao J, et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J Clin Invest. 2007;117:3940–3951. doi: 10.1172/JCI32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao P, Hanai J, Tanksale P, Imamura S, Sukhatme VP, Lecker SH. Statin-induced muscle damage and atrogin-1 induction is the result of a geranylgeranylation defect. FASEB J. 2009;23:2844–2854. doi: 10.1096/fj.08-128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 76.Nakao R, Hirasaka K, Goto J, Ishidoh K, Yamada C, Ohno A, et al. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol Cell Biol. 2009;29:4798–4811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwak KS, Zhou X, Solomon V, Baracos VE, Davis J, Bannon AW, et al. Regulation of protein catabolism by muscle-specific and cytokine-inducible ubiquitin ligase E3a-II during cancer cachexia. Cancer Res. 2004;64:8193–8198. doi: 10.1158/0008-5472.CAN-04-2102. [DOI] [PubMed] [Google Scholar]
- 78.Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 2006;25:554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 81.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 83.Mehrpour M, Esclatine A, Beau I, Codogno P. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- 84.Lardeux BR, Mortimore GE. Amino acid and hormonal control of macromolecular turnover in perfused rat liver. Evidence for selective autophagy. J Biol Chem. 1987;262:14514–14519. [PubMed] [Google Scholar]
- 85.Schworer CM, Mortimore GE. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc Natl Acad Sci USA. 1979;76:3169–3173. doi: 10.1073/pnas.76.7.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 87.Tanida I, Komatsu M, Ueno T, Kominami E. GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun. 2003;300:637–644. doi: 10.1016/s0006-291x(02)02907-8. [DOI] [PubMed] [Google Scholar]
- 88.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 91.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Furuno K, Goodman MN, Goldberg AL. Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. J Biol Chem. 1990;265:8550–8557. [PubMed] [Google Scholar]
- 93.Wing SS, Goldberg AL. Glucocorticoids activate the ATP-ubiquitin-dependent proteolytic system in skeletal muscle during fasting. Am J Physiol Endo Metab. 1993;264:E668–E676. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- 94.Price SR, Bailey JL, Wang X, Jurkovitz C, England BK, Ding X, et al. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome proteolytic pathway by a mechanism including gene transcription. J Clin Invest. 1996;98:1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 96.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 97.Deval C, Mordier S, Obled C, Bechet D, Combaret L, Attaix D, et al. Identification of cathepsin L as a differentially expressed message associated with skeletal muscle wasting. Biochem J. 2001;360:143–150. doi: 10.1042/0264-6021:3600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 100.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 101.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 102.Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zatz M, Starling A. Calpains and disease. N Engl J Med. 2005;352:2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- 104.duVerle D, Takigawa I, Ono Y, Sorimachi H, Mamitsuka H. CaMPDB: a resource for calpain and modulatory proteolysis. Genome Inform. 2010;22:202–213. [PubMed] [Google Scholar]
- 105.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 106.Hanna RA, Campbell RL, Davies PL. Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature. 2008;456:409–412. doi: 10.1038/nature07451. [DOI] [PubMed] [Google Scholar]
- 107.Moldoveanu T, Gehring K, Green DR. Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature. 2008;456:404–408. doi: 10.1038/nature07353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kemp CM, Sensky PL, Bardsley RG, Buttery PJ, Parr T. Tenderness--an enzymatic view. Meat Sci. 2010;84:248–256. doi: 10.1016/j.meatsci.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 109.Sultan KR, Dittrich BT, Pette D. Calpain activity in fast, slow, transforming, and regenerating skeletal muscles of rat. Am J Physiol Cell Physiol. 2000;279:C639–C647. doi: 10.1152/ajpcell.2000.279.3.C639. [DOI] [PubMed] [Google Scholar]
- 110.Jones SW, Hill RJ, Krasney PA, O’Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- 111.Raastad T, Owe SG, Paulsen G, Enns D, Overgaard K, Crameri R, et al. Changes in calpain activity, muscle structure, and function after eccentric exercise. Med Sci Sports Exerc. 2010;42:86–95. doi: 10.1249/MSS.0b013e3181ac7afa. [DOI] [PubMed] [Google Scholar]
- 112.Barnoy S, Glaser T, Kosower NS. The calpain-calpastatin system and protein degradation in fusing myoblasts. Biochim Biophys Acta. 1998;1402:52–60. doi: 10.1016/s0167-4889(97)00144-4. [DOI] [PubMed] [Google Scholar]
- 113.Tidball JG, Spencer MJ. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J Physiol. 2002;545:819–828. doi: 10.1113/jphysiol.2002.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barnoy S, Kosower NS. Caspase-1-induced calpastatin degradation in myoblast differentiation and fusion: cross-talk between the caspase and calpain systems. FEBS Lett. 2003;546:213–217. doi: 10.1016/s0014-5793(03)00573-8. [DOI] [PubMed] [Google Scholar]
- 115.Busquets S, Garcia-Martinez C, Alvarez B, Carbo N, Lopez-Soriano FJ, Argiles JM. Calpain-3 gene expression is decreased during experimental cancer cachexia. Biochim Biophys Acta. 2000;1475:5–9. doi: 10.1016/s0304-4165(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 116.Stockholm D, Herasse M, Marchand S, Praud C, Roudaut C, Richard I, et al. Calpain 3 mRNA expression in mice after denervation and during muscle regeneration. Am J Physiol Cell Physiol. 2001;280:C1561–C1569. doi: 10.1152/ajpcell.2001.280.6.C1561. [DOI] [PubMed] [Google Scholar]
- 117.Murphy RM, Goodman CA, McKenna MJ, Bennie J, Leikis M, Lamb GD. Calpain-3 is autolyzed and hence activated in human skeletal muscle 24 h following a single bout of eccentric exercise. J Appl Physiol. 2007;103:926–931. doi: 10.1152/japplphysiol.01422.2006. [DOI] [PubMed] [Google Scholar]
- 118.Laure L, Danièle N, Suel L, Marchand S, Aubert S, Bourg N, et al. A new pathway encompassing calpain 3 and its newly identified substrate cardiac ankyrin repeat protein is involved in the regulation of the nuclear factor-κB pathway in skeletal muscle. FEBS J. 2010;277:4322–4337. doi: 10.1111/j.1742-4658.2010.07820.x. [DOI] [PubMed] [Google Scholar]
- 119.Salvesen G, Duckett C. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 120.Baghdiguian S, Martin MM, Richard I, Pons F, Astier C, Bourg N, et al. Calpain 3 deficiency is associated with myonuclear apoptosis and profound perturbation of the IkappaB alpha/NF-kappaB pathway in limb-girdle muscular dystrophy type 2A. Nat Med. 1999;5:503–511. doi: 10.1038/8385. [DOI] [PubMed] [Google Scholar]
- 121.Kettelhut IC, Pepato MT, Migliorini RH, Medina R, Goldberg AL. Regulation of different proteolytic pathways in skeletal muscle in fasting and diabetes mellitus. Braz J Med Biol Res. 1994;27:981–993. [PubMed] [Google Scholar]
- 122.Supinski GS, Wang WX, Callahan LA. Caspase and calpain activation both contribute to sepsis-induced diaphragmatic weakness. J Appl Physiol. 2009;107:1389–1396. doi: 10.1152/japplphysiol.00341.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Y, Li Y, Feng Q, Arnold M, Peng T. Calpain activation contributes to hyperglycaemia-induced apoptosis in cardiomyocytes. Cardiovasc Res. 2009;84:100–110. doi: 10.1093/cvr/cvp189. [DOI] [PubMed] [Google Scholar]
- 124.Mani SK, Balasubramanian S, Zavadzkas JA, Jeffords LB, Rivers WT, Zile MR, et al. Calpain inhibition preserves myocardial structure and function following myocardial infarction. Am J Physiol Heart Circ Physiol. 2009;297:H1744–H1751. doi: 10.1152/ajpheart.00338.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salazar JJ, Michele DE, Brooks SV. Inhibition of calpain prevents muscle weakness and disruption of sarcomere structure during hindlimb suspension. J Appl Physiol. 2010;108:120–127. doi: 10.1152/japplphysiol.01080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li X, Li Y, Shan L, Shen E, Chen R, Peng T. Over-expression of calpastatin inhibits calpain activation and attenuates myocardial dysfunction during endotoxaemia. Cardiovasc Res. 2009;83:72–79. doi: 10.1093/cvr/cvp100. [DOI] [PubMed] [Google Scholar]
- 127.Smith IJ, Dodd SL. Calpain activation causes a proteasome-dependent increase in protein degradation and inhibits the Akt signalling pathway in rat diaphragm muscle. Exp Physiol. 2007;92:561–573. doi: 10.1113/expphysiol.2006.035790. [DOI] [PubMed] [Google Scholar]
- 128.Kramerova I, Kudryashova E, Venkatraman G, Spencer MJ. Calpain 3 participates in sarcomere remodeling by acting upstream of the ubiquitin-proteasome pathway. Hum Mol Genet. 2005;14:2125–2134. doi: 10.1093/hmg/ddi217. [DOI] [PubMed] [Google Scholar]
- 129.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med. 2010;207:983–997. doi: 10.1084/jem.20092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rami A, Agarwal R, Spahn A. Synergetic effects of caspase 3 and mu-calpain in XIAP-breakdown upon focal cerebral ischemia. Neurochem Res. 2007;32:2072–2079. doi: 10.1007/s11064-007-9361-6. [DOI] [PubMed] [Google Scholar]
- 132.Nakagawa T, Yuan J. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chua BT, Guo K, Li P. Direct cleavage by the calcium-activated protease calpain can lead to inactivation of caspases. J Biol Chem. 2000;275:5131–5135. doi: 10.1074/jbc.275.7.5131. [DOI] [PubMed] [Google Scholar]
- 134.Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 135.Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 136.Jagoe RT, Goldberg AL. What do we really know about the ubiquitin-proteasome pathway in muscle atrophy? Curr Opin Clin Nutr Metab Care. 2001;4:183–190. doi: 10.1097/00075197-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 137.Ogata T, Machida S, Oishi Y, Higuchi M, Muraoka I. Differential cell death regulation between adult-unloaded and aged rat soleus muscle. Mech Ageing Dev. 2009;130:328–336. doi: 10.1016/j.mad.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 138.Yarbrough WM, Mukherjee R, Stroud RE, Meyer EC, Escobar GP, Sample JA, et al. Caspase inhibition modulates left ventricular remodeling following myocardial infarction through cellular and extracellular mechanisms. J Cardiovasc Pharmacol. 2010;55:408–416. doi: 10.1097/FJC.0b013e3181d4ca66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Agusti AG, Sauleda J, Miralles C, Gomez C, Togores B, Sala E, et al. Skeletal muscle apoptosis and weight loss in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;166:485–489. doi: 10.1164/rccm.2108013. [DOI] [PubMed] [Google Scholar]
- 140.Vescovo G, Volterrani M, Zennaro R, Sandri M, Ceconi C, Lorusso R, et al. Apoptosis in the skeletal muscle of patients with heart failure: investigation of clinical and biochemical changes. Heart. 2000;84:431–437. doi: 10.1136/heart.84.4.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE. Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J Clin Invest. 2004;113:115–123. doi: 10.1172/JCI200418330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang XH, Zhang L, Mitch WE, LeDoux JM, Hu J, Du J. Caspase-3 cleaves specific 19 S proteasome subunits in skeletal muscle stimulating proteasome activity. J Biol Chem. 2010;285:21249–21257. doi: 10.1074/jbc.M109.041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Plant PJ, Bain JR, Correa JE, Woo M, Batt J. Absence of caspase-3 protects against denervation-induced skeletal muscle atrophy. J Appl Physiol. 2009;107:224–234. doi: 10.1152/japplphysiol.90932.2008. [DOI] [PubMed] [Google Scholar]
- 144.Chopra M, Das P, Sharma AC. Caspase-3 knock-down reverses contractile dysfunction induced by sepsis in adult rat ventricular myocytes. Br J Pharmacol. 2010;160:93–100. doi: 10.1111/j.1476-5381.2010.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Murgia M, Serrano AL, Calabria E, Pallafacchina G, Lomo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- 147.Aguilar V, Alliouachene S, Sotiropoulos A, Sobering A, Athea Y, Djouadi F, et al. S6 kinase deletion suppresses muscle growth adaptations to nutrient availability by activating AMP kinase. Cell Metab. 2007;5:476–487. doi: 10.1016/j.cmet.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 148.Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. J Biochem (Tokyo) 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- 149.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 150.Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 151.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 152.Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci USA. 2002;99:9213–9218. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 154.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–E601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 155.Tran H, Brunet A, Griffith EC, Greenberg ME. The many forks in FOXO’s road. Sci STKE. 2003;172:RE5. doi: 10.1126/stke.2003.172.re5. [DOI] [PubMed] [Google Scholar]
- 156.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 157.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 158.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 160.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 162.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 163.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 164.Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, et al. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 165.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Goldberg AL. Role of insulin in work-induced growth of skeletal muscle. Endocrinology. 1968;83:1071–1073. doi: 10.1210/endo-83-5-1071. [DOI] [PubMed] [Google Scholar]
- 167.Hardie DG, Sakamoto K. AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiol. 2006;21:48–60. doi: 10.1152/physiol.00044.2005. [DOI] [PubMed] [Google Scholar]
- 168.Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. AMP-activated protein kinase agonists increase mRNA content of the muscle-specific ubiquitin ligases MAFbx and MuRF1 in C2C12 cells. Am J Physiol Endocrinol Metab. 2007;292:E1555–E1567. doi: 10.1152/ajpendo.00622.2006. [DOI] [PubMed] [Google Scholar]
- 169.Nakashima K, Yakabe Y. AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Biosci Biotechnol Biochem. 2007;71:1650–1656. doi: 10.1271/bbb.70057. [DOI] [PubMed] [Google Scholar]
- 170.Beutler BA, Milsark IW, Cerami A. Cachectin/tumor necrosis factor: production, distribution, and metabolic fate in vivo. J Immunol. 1985;135:3972–3977. [PubMed] [Google Scholar]
- 171.Dogra C, Changotra H, Wedhas N, Qin X, Wergedal JE, Kumar A. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 2007;21:1857–1869. doi: 10.1096/fj.06-7537com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Argiles JM, Lopez-Soriano FJ. The role of cytokines in cancer cachexia. Med Res Rev. 1999;19:223–248. doi: 10.1002/(sici)1098-1128(199905)19:3<223::aid-med3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 173.von Haehling S, Genth-Zotz S, Anker SD, Volk HD. Cachexia: a therapeutic approach beyond cytokine antagonism. Int J Cardiol. 2002;85:173–183. doi: 10.1016/s0167-5273(02)00245-0. [DOI] [PubMed] [Google Scholar]
- 174.Ladner KJ, Caligiuri MA, Guttridge DC. Tumor necrosis factor-regulated biphasic activation of NF-kappa B is required for cytokine-induced loss of skeletal muscle gene products. J Biol Chem. 2003;278:2294–2303. doi: 10.1074/jbc.M207129200. [DOI] [PubMed] [Google Scholar]
- 175.Penner CG, Gang G, Wray C, Fischer JE, Hasselgren PO. The transcription factors NF-kappab and AP-1 are differentially regulated in skeletal muscle during sepsis. Biochem Biophys Res Commun. 2001;281:1331–1336. doi: 10.1006/bbrc.2001.4497. [DOI] [PubMed] [Google Scholar]
- 176.Hunter RB, Stevenson E, Koncarevic A, Mitchell-Felton H, Essig DA, Kandarian SC. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. FASEB J. 2002;16:529–538. doi: 10.1096/fj.01-0866com. [DOI] [PubMed] [Google Scholar]
- 177.Li YP, Reid MB. NF-kappaB mediates the protein loss induced by TNF-alpha in differentiated skeletal muscle myotubes. Am J Physiol Regul Integr Comp Physiol. 2000;279:R1165–R1170. doi: 10.1152/ajpregu.2000.279.4.R1165. [DOI] [PubMed] [Google Scholar]
- 178.Williamson DL, Kimball SR, Jefferson LS. Acute treatment with TNF-alpha attenuates insulin-stimulated protein synthesis in cultures of C2C12 myotubes through a MEK1-sensitive mechanism. Am J Physiol Endocrinol Metab. 2005;289:E95–E104. doi: 10.1152/ajpendo.00397.2004. [DOI] [PubMed] [Google Scholar]
- 179.Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 180.Langen RC, Schols AM, Kelders MC, Wouters EF, Janssen-Heininger YM. Inflammatory cytokines inhibit myogenic differentiation through activation of nuclear factor-kappaB. FASEB J. 2001;15:1169–1180. doi: 10.1096/fj.00-0463. [DOI] [PubMed] [Google Scholar]
- 181.Cai D, Frantz JD, Tawa NE, Jr, Melendez PA, Oh BC, Lidov HG, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 182.Hunter RB, Kandarian SC. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J Clin Invest. 2004;114:1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li YP, Chen Y, John J, Moylan J, Jin B, Mann DL, et al. TNF-alpha acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 185.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 188.Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, et al. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]
- 189.Reisz-Porszasz S, Bhasin S, Artaza JN, Shen R, Sinha-Hikim I, Hogue A, et al. Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am J Physiol Endocrinol Metab. 2003;285:E876–E888. doi: 10.1152/ajpendo.00107.2003. [DOI] [PubMed] [Google Scholar]
- 190.Durieux AC, Amirouche A, Banzet S, Koulmann N, Bonnefoy R, Pasdeloup M, et al. Ectopic expression of myostatin induces atrophy of adult skeletal muscle by decreasing muscle gene expression. Endocrinology. 2007;148:3140–3147. doi: 10.1210/en.2006-1500. [DOI] [PubMed] [Google Scholar]
- 191.Zhang L, Rajan V, Lin E, Hu Z, Han HQ, Zhou X, et al. Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J. 2011;25:1653–1663. doi: 10.1096/fj.10-176917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, et al. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:C1248–C1257. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- 193.Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, et al. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 2010;142:531–543. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 194.Zheng B, Ohkawa S, Li H, Roberts-Wilson TK, Price SR. FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. FASEB J. 2010;24:2660–2669. doi: 10.1096/fj.09-151480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hu Z, Wang H, Lee IH, Du J, Mitch WE. Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice. J Clin Invest. 2009;119:3059–3069. doi: 10.1172/JCI38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang L, Du J, Hu Z, Han G, Delafontaine P, Garcia G, et al. IL-6 and serum amyloid A synergy mediates angiotensin II-induced muscle wasting. J Am Soc Nephrol. 2009;20:604–612. doi: 10.1681/ASN.2008060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Murton AJ, Constantin D, Greenhaff PL. The involvement of the ubiquitin proteasome system in human skeletal muscle remodelling and atrophy. Biochim Biophys Acta. 2008;1782:730–743. doi: 10.1016/j.bbadis.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 198.Workeneh BT, Rondon-Berrios H, Zhang L, Hu Z, Ayehu G, Ferrando A, et al. Development of a diagnostic method for detecting increased muscle protein degradation in patients with catabolic conditions. J Am Soc Nephrol. 2006;17:3233–3239. doi: 10.1681/ASN.2006020131. [DOI] [PubMed] [Google Scholar]
- 199.Lu Y, Adegoke OAJ, Nepveu A, Nakayama KI, Bedard N, Wing SS. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin protein ligase for p27Kip1. Mol Cell Biol. 2009;29:547–558. doi: 10.1128/MCB.00329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Medina R, Wing S, Goldberg AL. Increase in levels of polyubiquitin and proteasome mRNA in skeletal muscle during starvation and denervation atrophy. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Llovera M, Garcia-Martinez C, Agell N, Marzabal M, Lopez-Soriano FJ, Argiles JM. Ubiquitin gene expression is increased in skeletal muscle of tumour-bearing rats. FEBS Letters. 1994;338:311–318. doi: 10.1016/0014-5793(94)80290-4. [DOI] [PubMed] [Google Scholar]
- 202.Wing SS, Banville D. 14-kDa ubiquitin-conjugating enzyme: structure of the rat gene and regulation upon fasting and by insulin. Amer J Physiol. 1994;267:E39–E48. doi: 10.1152/ajpendo.1994.267.1.E39. [DOI] [PubMed] [Google Scholar]
- 203.Li YP, Lecker SH, Chen Y, Waddell ID, Goldberg AL, Reid MB. TNF-alpha increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. FASEB J. 2003;17:1048–1057. doi: 10.1096/fj.02-0759com. [DOI] [PubMed] [Google Scholar]
- 204.Chrysis D, Underwood LE. Regulation of components of the ubiquitin system by insulin-like growth factor I and growth hormone in skeletal muscle of rats made catabolic with dexamethasone. Endocrinology. 1999;140:5635–5641. doi: 10.1210/endo.140.12.7217. [DOI] [PubMed] [Google Scholar]
- 205.Fischer D, Sun X, Gang G, Pritts T, Hasselgren PO. The gene expression of ubiquitin ligase E3alpha is upregulated in skeletal muscle during sepsis in rats - potential role of glucocorticoids. Biochem Biophys Res Commun. 2000;267:504–508. doi: 10.1006/bbrc.1999.1987. [DOI] [PubMed] [Google Scholar]
- 206.Lecker SH, Solomon V, Price SR, Kwon YT, Mitch WE, Goldberg AL. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J Clin Invest. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ilian MA, Forsberg NE. Gene expression of calpains and their specific endogenous inhibitor, calpastatin, in skeletal muscle of fed and fasted rabbits. Biochem J. 1992;287:163–171. doi: 10.1042/bj2870163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Price SR, England BK, Bailey JL, Van Vreede K, Mitch WE. Acidosis and glucocorticoids concomitantly increase ubiquitin and proteasome subunit mRNAs in rat muscle. Am J Physiol Cell Physiol. 1994;267:C955–C960. doi: 10.1152/ajpcell.1994.267.4.C955. [DOI] [PubMed] [Google Scholar]
- 210.Tiao G, Hobler S, Wang JJ, Meyer TA, Luchette FA, Fischer JE, et al. Sepsis is associated with increased mRNAs of the ubiquitin-proteasome proteolytic pathway in human skeletal muscle. J Clin Invest. 1997;99:163–168. doi: 10.1172/JCI119143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Taillandier D, Aurousseau E, Meynial-Denis D, Bechet D, Ferrara M, Cottin P, et al. Coordinate activation of lysosomal, Ca 2+-activated and ATP-ubiquitin-dependent proteinases in the unweighted rat soleus muscle. Biochem J. 1996;316:65–72. doi: 10.1042/bj3160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Williams A, Sun X, Fischer JE, Hasselgren PO. The expression of genes in the ubiquitin-proteasome proteolytic pathway is increased in skeletal muscle from patients with cancer. Surgery. 1999;126:744–749. [PubMed] [Google Scholar]
- 213.Fang CH, Li BG, Fischer DR, Wang JJ, Runnels HA, Monaco JJ, et al. Burn injury upregulates the activity and gene expression of the 20 S proteasome in rat skeletal muscle. Clin Sci (Lond) 2000;99:181–187. [PubMed] [Google Scholar]
- 214.Combaret L, Ralliere C, Taillandier D, Tanaka K, Attaix D. Manipulation of the ubiquitin-proteasome pathway in cachexia: pentoxifylline suppresses the activation of 20S and 26S proteasomes in muscles from tumor-bearing rats. Mol Biol Rep. 1999;26:95–101. doi: 10.1023/a:1006955832323. [DOI] [PubMed] [Google Scholar]
- 215.Lorite MJ, Smith HJ, Arnold JA, Morris A, Thompson MG, Tisdale MJ. Activation of ATP-ubiquitin-dependent proteolysis in skeletal muscle in vivo and murine myoblasts in vitro by a proteolysis-inducing factor (PIF) Br J Cancer. 2001;85:297–302. doi: 10.1054/bjoc.2001.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 217.Raffaello A, Milan G, Masiero E, Carnio S, Lee D, Lanfranchi G, et al. JunB transcription factor maintains skeletal muscle mass and promotes hypertrophy. J Cell Biol. 2010;191:101–113. doi: 10.1083/jcb.201001136. [DOI] [PMC free article] [PubMed] [Google Scholar]