Summary
Major depressive disorder (MDD) is a leading cause of disease burden worldwide. While the incidence, symptoms and treatment of MDD all point toward major sex differences, the molecular mechanisms underlying this sexual dimorphism remain largely unknown. Here, combining differential expression and weighted gene coexpression network analyses, we provide a comprehensive characterization of male and female transcriptional profiles associated with MDD across 6 brain regions. We overlap our human profiles with those from a mouse model of chronic variable stress and capitalize on converging pathways to define molecular and physiological mechanisms underlying the expression of stress susceptibility in males and females. Our results show a major rearrangement of transcriptional patterns, with male and female transcriptional profiles sharing very limited overlap, an effect seen in depressed humans and in stressed mice. We identify male and female hub genes and confirm their sex-specific impact as stress-susceptibility mediators. For example, downregulation of the female-specific hub gene DUSP6 in prefrontal cortex mimics stress susceptibility in females only by increasing ERK signaling and pyramidal neuron excitability. Such DUSP6 downregulation also recapitulates the transcriptional remodelling that occurs in PFC of depressed females. Together, our findings reveal dramatic sexual dimorphism at the transcriptional level in MDD and highlight the importance of studying sex-specific treatments for this disorder.
Introduction
Major depressive disorder (MDD) is a chronic debilitating illness, affecting yearly 350 million people worldwide, inducing major economic and medical burdens on societies1. While it affects both males and females, MDD is characterized by a strong sexual dimorphism: females are 2–3 times more likely to develop MDD2,3 and exhibit higher symptom severity, greater functional impairment, more atypical depressive symptoms and higher rates of co-morbid anxiety4–7. Furthermore, males and females with MDD respond differently to antidepressant treatment8–11.
There is likewise a wealth of data showing functional sex differences in MDD. For instance, males and females with MDD exhibit different magnitude, laterality and direction of volumetric changes observed across the brain12–17. Differences in serotonin (5-HT) synthesis as well as in the density of the 5-HT autoreceptor 1A (5-HT1A) have also been reported between males and females with MDD18–21. In addition, activation of several components of the hypothalamic-pituitary-adrenal (HPA) axis differs in males and females with MDD following stress22. Depressed women exhibit higher cortisol levels23–25 and higher corticotropin releasing factor sensitivity25–28 compared to their male counterparts. While these various features define sex differences in MDD, the underlying molecular mechanisms remain largely unknown.
Indeed, there have been few genome-wide characterizations and comprehensive comparisons of the transcriptional signatures defining MDD in males and females across the brain29. Global analyses of the male transcriptome in MDD have revealed alterations in genes related to the glutamatergic, GABAergic, serotonergic and polyaminergic systems across several cortical and subcortical brain regions30–36, lipid metabolism, immune response, ATP synthesis, regulation of transcription and translation, fibroblast growth factor signaling and cell proliferation33,37–41 in cortical regions, regulation of the HPA axis in hypothalamus42 and control of circadian rhythms in cortical and subcortical regions43.
On the other hand, many fewer studies have assessed female transcriptional regulation in MDD. With very few exceptions10,38,44,45, the large majority of these studies adopted candidate gene approaches showing alterations in brain-derived neurotrophic factor (BDNF), GABAergic, somatostatin (SST), cholinergic, serotonergic and glutamatergic systems as well as mitochondrial, energy metabolism and circadian rhythms in cortical and limbic regions46–54. Importantly, these findings have been complemented by transcriptional studies performed in various mouse models of stress. Although still focused predominantly on males, these studies have revealed interesting insight into the mechanisms underlying the effects of chronic social defeat stress (CSDS), chronic variable or unpredictable stress (CVS, CUS) or social isolation across several brain regions44,45,55–62.
With the still limited success of genome-wide association studies (GWAS) to identify genes associated with MDD in the general population63–65, systems-based approaches are emerging as additional tools for revealing pathogenic etiologies in complex neuropsychiatric syndromes. Combined with conventional differential gene expression analyses, network-based approaches offer a data-driven, molecular classification scheme for MDD while revealing previously unappreciated subtypes of this heterogeneous syndrome. While these approaches have provided novel insight in the study of different psychiatric conditions66,67,68–73, they have not yet been used to characterize the sexual dimorphism of MDD across different brain regions.
Here, we combined differential gene expression and network-based approaches to provide an integrative and unbiased characterization of male and female transcriptional signatures in human MDD. Because MDD is associated with abnormal functioning of numerous brain regions74, RNAseq was performed across 6 cortical and subcortical brain regions, making this the largest RNAseq dataset available to date for human MDD. To provide functional and mechanistic significance for our findings, we compared our sex-specific human MDD signatures with RNAseq transcriptional profiles generated from male and female mice following CVS. We identified sex-specific transcriptional networks in MDD, predicted network hubs that drive these sex-specific networks and, using viral-mediated gene transfer, validated the ability of key hub genes to drive stress susceptibility in a sex-specific fashion. We expanded the mechanistic characterization of these effects by focusing on two sexually dimorphic gene networks. We showed that the alteration of these gene networks induces a series of molecular and cellular changes in the prefrontal cortex in a sex specific fashion. Overall, this study provides a comprehensive molecular description of the transcriptional signatures associated with MDD in males and females and defines some of the mechanisms underlying this sexual dimorphism.
Results
Differential gene expression analysis reveals brain region- and sex-specific transcriptional profiles in MDD
We used RNAseq to examine the transcriptome of 6 brain regions of humans with MDD and extensively matched control subjects: ventromedial PFC (vmPFC; also known as subgenual PFC; Broadman area [BA] 25), OFC (BA11), dorsolateral PFC (dlPFC; BA8/9), anterior insula (aINS), nucleus accumbens (NAc) and ventral subiculum (vSUB). For each brain region, we examined 26 MDD postmortem samples (13 males; 13 females) and 22 controls (13 male; 9 females), and revealed significant transcriptional changes across every brain region in males and females with MDD compared to control subjects (Supplementary Tables 1 and 2). Besides a significant effect of phenotype (PC4, p<0.01), sex (PC3, p<1.0e-4) and brain region (PC1, p<5.0e-36), our analysis revealed a significant effect of RIN (PC1, p<1.0e-3), age (PC2, p<0.01) and alcohol abuse (PC2, p<0.05) on total variance and were included in our statistical model as covariates. Furthermore, given the well-known impact of antidepressant treatment on gene expression75, medication status was also accounted for in our analysis. Other covariates including drug abuse, cause of death, history of child abuse and toxicology were not significantly associated with global variation in gene expression.
We validated our RNAseq findings on a subset of regulated genes: 80% (16 out of 20) and 100% (20 out of 20) of genes differentially expressed by RNAseq in male or female vmPFC, respectively, also showed altered expression by NanoString analysis of RNA samples from the same brains (Supplementary Table 3). We further validated our findings in independent samples from different cohorts. We validated 85% (17 out of 20) of genes initially found to be differentially expressed in a second cohort of males with MDD, and validated 75% (15 out of 20) of genes initially found to be differentially expressed in a third cohort of females with MDD. Sociodemographic and clinical information for these cohorts is listed in Supplementary Tables 10, 11 and 12. Importantly all cohorts included samples from two geographically distant brain banks. Together, these findings confirm the validity of our results and suggest that our findings can be generalized to independent populations with different cultural and genetic backgrounds.
We next used an unbiased rank-rank hypergeometric overlap (RRHO) analysis to highlight shared transcriptional changes across brain regions in males and females with MDD. RRHO is a threshold-free assessment of statistical overlap76 which we used here to map both region-specific and sex-specific transcriptional signatures. The analysis in males showed a coordinated upregulation of transcriptional signatures across all cortical regions, including dlPFC, OFC and vmPFC (Fisher’s Exact Test [FET] p<1.0E-204, Figure 1a). A similar but weaker signal was found between the aINS and OFC and, to a lesser extent still, the NAc (Figure 1a). Such regional comparisons in female MDD were strikingly different (Figure 1b). While similar albeit weaker overlapping patterns were found across cortical regions, our analysis highlighted a coordinated upregulation of genes across vSUB and most of the cortical regions including OFC, vmPFC and aINS (FET p<1.0E-204; Figure 1b). vmPFC and aINS also showed concordant gene upregulation in females (Figure 1b). FETs restricted to the subsets of differentially expressed genes (DEGs) across brain regions confirmed overlaps in upregulated genes in both males and females with MDD (Extended Figure 1a and b).
Figure 1.
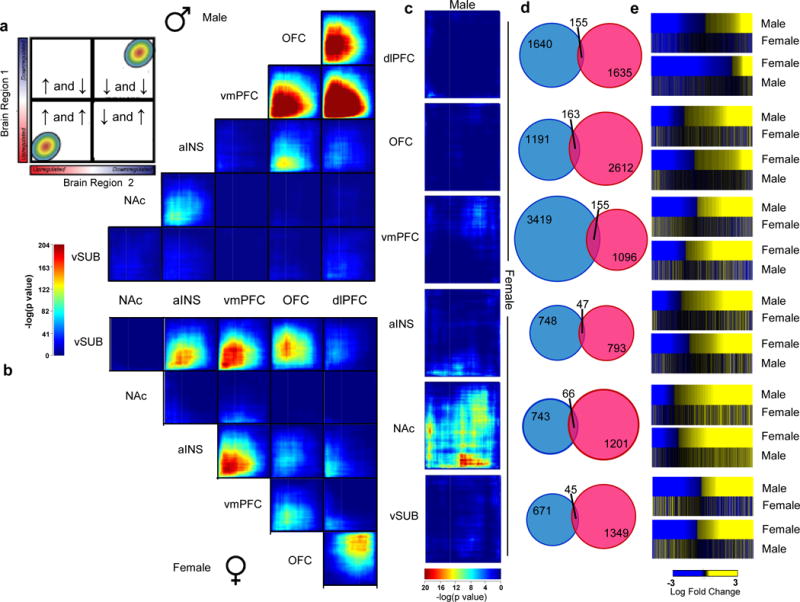
Differential expression profiles in humans with MDD reveal distinct sex-specific transcriptional signatures across brain regions. a,b, Rank-rank hypergeometric overlap (RRHO) maps comparing region to region transcriptional profiles in a, males and b, females with MDD. The upper left panel in a displays the overlap relationship across brain regions. The color bar between a and b represents degree of significance. c, RRHO maps directly comparing male and female transcriptional profiles across brain regions. Degree of significance is depicted in the color bar below the RRHO maps. d, Venn diagrams displaying low overlap between genes differentially expressed (p<0.05) in males (blue) and females (pink) across brain regions. e, Heatmaps comparing transcriptional changes (log fold change; below the heatmaps) in males and females with MDD compared to controls across brain regions.
When comparing directly the transcriptional patterns observed in male vs. female MDD across brain regions, we found surprisingly little overlap between male MDD and female MDD transcriptional signatures in any brain region analyzed (Figure 1c). The lack of overlap was further validated when restricting our analysis to DEGs. Indeed, direct comparison of DEGs in males vs. females with MDD revealed only 5–10% overlap (Figure 1d) across all six brain regions. Likewise, the directionality of the changes observed in males and females with MDD are not preserved as revealed by the heatmaps in Figure 1e. These analyses confirmed that the sex differences observed in MDD-associated transcriptional abnormalities do not reflect baseline sex differences in gene expression. Overall, these findings highlight a dramatic sexual dimorphism in the molecular signatures associated with MDD across multiple brain regions.
Although our subsequent analysis focused on protein-coding genes, our datasets reveal MDD derangements in several types of non-coding RNAs (Supplementary Tables 1 and 2). These data thus provide a valuable resource for future analyses.
Dramatic sex-specific transcriptional responses are recapitulated in mice after CVS
We next assessed whether similarly dramatic sex differences in transcriptional profiles are produced in mice in response to CVS. We found that 21 days of CVS induced a range of depression- and anxiety-related behavioral abnormalities to an equivalent degree in male vs. female mice (Figure 2a–d). These include increased latency to eat in the novelty-suppressed feeding test (F(1,40)=24.3, p<0.0001; Figure 2a), decreased time spent grooming in the splash test (F(1,40)=8.68, p<0.01; Figure 2b), a trend for increased time spent immobile in the forced swim test (F(1,40)=3.61, p=0.06; Figure 2c) and decreased sucrose preference (F(1,40)=10.04, p<0.005; Figure 2d).
Figure 2.
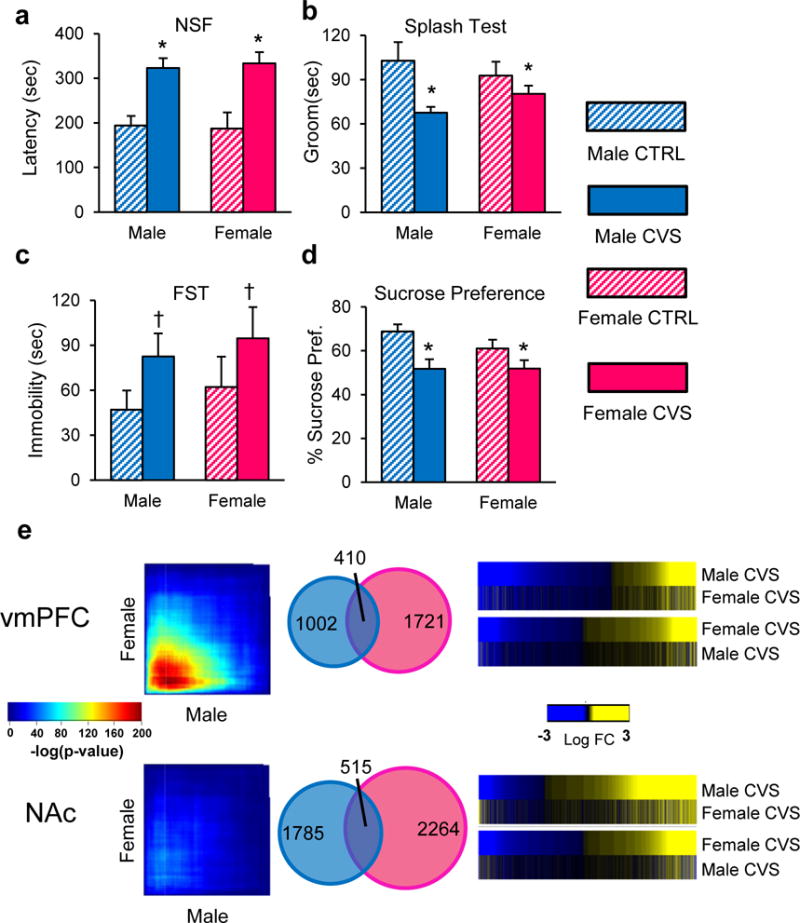
Chronic variable stress (CVS) induces an equivalent depressive-like phenotype in male and female mice despite the induction of largely distinct transcriptional profiles. a, Quantification of latency to eat in the novelty suppressed feeding (NSF) test, b time spent grooming in the splash test, c time swimming in the forced swim test (FST) and d sucrose preference in male (blue) and female (pink) mice. Bars, mean ± sem; * p<0.05; † p<0.1. e, RRHO maps comparing male and female stressed mice in the vmPFC and NAc. Degree of significance is depicted in the color bar in between the RRHO maps. Venn diagrams displaying overlap between genes differentially expressed (p<0.05) in male (blue) and female (pink) stressed mice in both brain regions. Heatmaps comparing transcriptional changes (log fold change; in between the heatmaps) in stressed male and female mice compared to controls across both brain regions. Significance in tests a–d was determined using two-way ANOVA with Tuckey correction. n =10/condition.
We used RNAseq to survey the transcriptome in two representative brain regions, vmPFC and NAc, from 20 males and 20 females (half stressed, half unstressed controls). Similar to what we found in humans, our analysis revealed sex-specific changes in gene expression between stressed and unstressed mice across both brain regions (Supplementary Tables 4 and 5). Our analysis shows that DEGs are found to be expressed across several different cell types (neurons, astrocytes, oligodendrocytes, microglia and endothelial cells). However, we found a larger proportion of astrocyte and neuronal genes being significantly downregulated in female vmPFC and male NAc as well as astrocytic and endothelial genes being significantly downregulated in male vmPFC (Extended Figure 2).
As found for human MDD, direct comparison of male vs. female transcriptional profiles in the NAc by RRHO revealed the absence of overlap between stressed males and females (Figure 2e). While genes upregulated in the vmPFC displayed some overlap between the sexes (Figure 2e), there was nevertheless a relatively small degree of overlap between DEGs in stressed males vs. females, with ~25% of DEGs being shared in the vmPFC and ~20% in the NAc (Figure 2e). Heatmaps in Figure 2e highlight this small overlap between DEGs in males and females. This 20–25% overlap in chronically stressed mice is greater than the 5–10% overlap in depressed humans and likely reflects the far greater homogeneity of the mouse system. Indeed, the fact that the same repeated stress, which induces equivalent behavioral impairments in the two sexes, only induces 20–25% of the same gene expression changes in genetically identical male and female mice underscores the major finding of this study, namely, profound sex differences in transcriptional responses to chronic stress. The findings also suggest that similar behavioral effects of stress in males and females are mediated by largely distinct molecular adaptations.
Next, we compared transcriptional profiles in the vmPFC and NAc between human MDD and mouse CVS cohorts using a combinatorial bioinformatic approach. We first identified a significant number of genes being commonly up- or downregulated in the vmPFC or NAc in depressed humans and in stressed mice. In males, depressed humans and stressed mice share 62 upregulated DEGs (fold-change [FC]: 1.2; p=0.08) and 90 downregulated DEGs (FC: 1.1; p=0.15) genes in vmPFC and 109 upregulated DEGs (FC: 3.1; p=2.3e-26) and 44 downregulated DEGs (FC: 3.7; p=3.73e-14) in NAc (Extended Figure 3a, Supplementary Table 6). Similarly, depressed human females and stressed female mice share 128 upregulated DEGs (FC: 1.8; p=2.3e-11) and 123 downregulated DEGs (FC: 5.6; p=9.8e-60) in vmPFC and 81 downregulated DEGs (FC: 2.1; p=7.6e-11) and 89 upregulated DEGs (FC: 1.5; p=4.9e-5) in NAc (Extended Figure 3b, Supplementary Table 6). A gene ontology overlap analysis identified several pathways commonly associated with MDD in humans and stress in mice. These several functional pathways in the vmPFC and NAc conserved across species in males or females (Extended Figure 4; Supplementary Table 7) may be commonly involved in stress-induced pathology in the two species.
Network-based approaches identify sex-specific transcriptional signatures associated with MDD
Previous work has demonstrated that systems-based network analyses reveal additional important transcriptional regulation in complex diseases compared with differential gene expression analysis alone66–73. Since neural circuits spanning several brain regions have been implicated in MDD74, it is imperative that the molecular pathogenesis of the syndrome be investigated at the network level. Thus, to provide a better resolution of sex-specific transcriptional signatures in brain and to gain novel insight into the molecular mechanisms involved in MDD, we constructed male and female multi-brain region coexpression networks that combined all 6 brain regions studied. Our analysis identified numerous male and female transcriptional modules, each named by an arbitrary color. We deduced 143 modules in males with MDD and 158 in control males each comprising between 10 to 6,687 genes. Likewise, our analysis uncovered 167 female MDD and 127 female control modules, each ranging from 10 to 10,395 genes.
We next used module differential connectivity (MDC) analysis to quantify changes in network connectivity between MDD and control conditions in both males and females (Extended Figure 5a,b). MDC is the ratio of the average connectivity for any pair of genes within MDD modules compared to the same gene pairs in control modules73 and as such informs on the structural reorganization of transcriptional profiles taking place between any two conditions (i.e., MDD vs. controls, males vs. females). The distribution of male and female MDC scores for MDD modules highlights several modules with a gain of connectivity (GOC), while only a small subset of male modules show a loss of connectivity (LOC) (Extended Figure 5b). Our analysis highlights several biological processes associated with male and female MDD modules that exhibit a GOC or LOC (Extended Figure 5c; Supplementary Tables 8 and 9). While several of these functions have previously been associated with MDD, our analysis provides novel evidence that several of these functions are affected in a sex-specific fashion.
We directly compared the sex-specific transcriptional organization of gene modules by measuring the level of homology between modules in males and females with MDD. Extended Figure 5d displays the modules in males and females with MDD sharing high levels of homology at the gene membership and functional levels (FET corrected p-value<0.05; fold enrichment>15). Interestingly, of the 55 male MDD modules conserved in female MDD, only 10 showed conserved connectivity with their female MDD homologues while the remaining 45 male MDD modules gained connectivity compared to their female MDD homologues. Extended Figure 5e displays the biological processes shared by males and females with MDD and characterized by a GOC. Together, this suggests that, even for modules conserved on a gene membership level, only a small subset of them displays conserved connectivity between males and females with MDD, which again highlights the strong sexual dimorphism characterizing transcriptional regulation in MDD.
Furthermore, consistent with previous network-based comparisons67,69,71,73,77, we identified several modules enriched for genes expressed predominantly in specific cell types such as neurons, microglia, oligodendrocytes, astrocytes or endothelial cells to be associated with a GOC or LOC in males or females with MDD (Extended Figure 6a,b). The results illustrate that MDD is associated with transcriptional alterations occurring within several cell types, which appear to be affected differently in males and females. Indeed, our analysis shows that modules associated with a GOC in males with MDD are enriched for genes expressed in several cell types (Extended Figure 6a), whereas in female MDD, GOC modules are enriched mainly for neuronal genes (Extended Figure 6b). Interestingly, female MDD modules that are enriched for other (non-neuronal) cell types show conserved connectivity (Extended Figure 6b).
Overlapping differential gene expression with module coexpression analysis identifies sex-specific hub genes
We next assessed the degree of enrichment for DEGs in male and female MDD modules. In total, we identified 77 modules in male MDD and 107 in female MDD enriched for DEGs in at least one brain region (FDR<0.1). The degree of enrichment in female modules is reported in Figure 3a–c, while the male enrichment is illustrated in Extended Figure 7a–c. Modules are ranked based on a composite measure considering the degree of enrichment for DEGs across brain regions and MDC against control conditions and, as such, allows prioritizing modules based on greater disruption (Figure 3c; Extended Figure 7c). In male MDD, the most highly ranked modules associate with transmission of nerve impulse, organic acid metabolism, nerve ensheathment and regulation of catecholamine metabolism (Supplementary Table 8), whereas the most highly ranked female MDD modules are associated with nerve ensheathment, mitogen-activated protein kinase (MAPK) activity, inflammatory response and synaptic transmission (Supplementary Table 9). While some of these terms are seen in both male and female models (e.g., nerve ensheathment), our analysis shows that both the patterns of MDC (see previous section; Extended Figure 5d,e) and DEG enrichment differs in males vs. females with MDD, further supporting the sexual dimorphism characterizing MDD.
Figure 3.
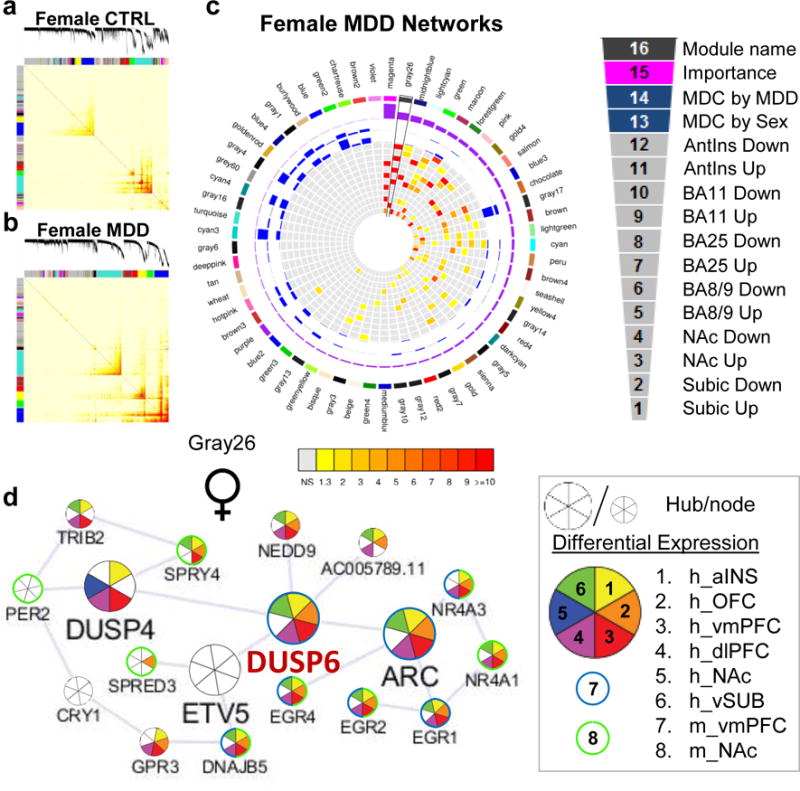
Gene co-expression modules in females with MDD are enriched for DEGs across brain regions. a, Topological overlap matrix (TOM) plots for control and b, MDD modules in females. Light color represents low topological overlap and progressively darker red color represents higher overlap. Each module is assigned by unique color. c, Circos plots displaying the degree of enrichment for DEGs (p<0.05) in female modules. Colors within squares of the plots represent the corrected FET p-value of the enrichment of DEGs across modules which are depicted in the color bar below the circos plot. Legend on the top right corner defines individual layers of the circos plot. d, Gray26 module in female MDD shows enrichment for DEGs across brain regions. Hubs and nodes are defined by the size of the circles with colors representing enrichment for DEGs across brain regions (depicted in the bottom right panel). DUSP6, labelled in red, was selected for sex-specific in vivo functional validation studies.
We used ARACNE to display the structure of male and female MDD networks and to identify intramodular hub genes within these networks (Figure 3d; Extended Figure 7d). Intramodular hub genes are genes highly connected with neighboring genes within a given module and have been shown to be significantly associated with disease traits in other experimental systems67,78–83. To gain insight into sex-specific modular organization, we focussed on two of the highest ranked gene modules associated with female or male MDD (Figure 3d and Extended Figure 7d). The female MDD-specific Gray26 module is involved in MAPK activity and is composed of 18 genes, of which 4 are hub genes. Gray26 has a marginal GOC compared to male MDD (MDC=2.66; FDR=0.3) but is highly enriched for genes downregulated across every brain region examined (aINS: FDR=4.32e-28, vmPFC: FDR=8.22e-23, OFC: FDR=1.48e-20, dlPFC: FDR=4.67e-19, and vSUB: FDR=3.03e-18) with the exception of the NAc (Figure 3c–d).
In contrast, the male-specific Peru module, which is enriched in genes associated with synaptic transmission, is composed of 311 genes, of which 43 are hub genes. Peru in male MDD has a GOC (MDC=2.54, FDR<0.01) compared to female MDD and is significantly enriched for genes upregulated, most strikingly, in vmPFC (FDR=5.68e-79) and to a lesser extent in vSUB (FDR=1.29e-15) and aINS (FDR=7.63e-5), and downregulated in NAc (FDR=9.82e-5; Extended Figure 7c–d).
Together, these results suggest that Gray26 and Peru modules, in female and male MDD, respectively, are subjected to sex-specific regulation, which may be distinctly associated with the expression of MDD-specific traits in the two sexes.
In vivo manipulation of male and female gene networks induces sex-specific behavioral alterations in mouse CVS
To validate these bioinformatics predictions, we used viral-mediated gene transfer in a subthreshold version of the CVS model. Unlike 21 days of CVS used above, shorter exposures to CVS (6 days males, 3 days females) by themselves are insufficient to induce behavioral abnormalities in male or female mice, but can reveal stress susceptibility in response to specific manipulations44,62.
We focused our validation efforts on two genes, DUSP6 and EMX1, based on their modular status as intramodular hub genes in female- or male-specific MDD modules and on their patterns of differential expression. The dual-specific phosphatase DUSP6, which is involved in several brain-related functions through inactivation of the ERK pathway84,85, is a highly connected hub gene within the female-specific Gray26 module and is downregulated in vmPFC in female MDD subjects and stressed female mice (Figure 3d). Likewise, the transcription factor EMX1, involved in the specification of neurons in the developing cortex86, is a highly connected hub gene within the male-specific Peru module and is upregulated in the vmPFC in male MDD subjects, in both our RNAseq and replication cohorts (Extended Figure 7d). We generated Herpes simplex virus (HSV) vectors to downregulate DUSP6 expression, and one to overexpress EMX1, and injected them into the vmPFC of mice subjected to subthreshold CVS.
We found that DUSP6 downregulation (t(1,7)=3.31, p<0.05; Figure 4a, b) in the female vmPFC followed by 3 days of CVS increased the latency to eat in the novelty-suppressed feeding test (F(1,25)=13.26; p<0.005; Figure 4c) compared to several other groups: GFP unstressed control (p<0.01), GFP stressed (p=0.053) and DUSP6 unstressed control (p<0.005) mice. DUSP6 downregulation also decreased sucrose preference (F(1,14)=7.07; p<0.05; Figure 4e) compared to GFP unstressed controls (p=0.076) and DUSP6 unstressed controls (p<0.05) but not GFP stressed mice. On the other hand, downregulation of DUSP6 in female vmPFC has no effect in the forced swim test, open field or elevated plus maze (Extended Figure 8a–d). In contrast, despite the equivalent downregulation, DUSP6 downregulation in male vmPFC failed to induce stress susceptibility in both the novelty-suppressed feeding and sucrose preference tests (Figure 4d,f), supporting the sex-specific role of DUSP6 in controlling stress responses. Conversely, overexpression of EMX1 (t(1,5) =2.40, p<0.05; Extended Figure 7e) in the male vmPFC followed by 6 days of stress increased the latency to initiate eating in the novelty-suppressed feeding test (F(1,33)=11.00, p<0.005; Extended Figure 7f) compared to GFP unstressed controls (p<0.05) and EMX1 unstressed controls (p<0.05) but not GFP-stressed mice. In contrast, EMX1 overexpression in the female vmPFC, despite an equivalent degree of overexpression, failed to induce stress susceptibility (Extended Figure 7g), supporting the sex-specific role of EMX1 in controlling stress responses. Finally, to confirm the role of DUSP6 in vmPFC in controlling stress susceptibility in females, we attempted to rescue the phenotype induced by stress in females. We generated an HSV vector to overexpress DUSP6 and injected it into the vmPFC of female mice subjected to 6 days of variable stress (Extended Figure 8e). DUSP6 overexpression in female vmPFC (t(1,6)=2.83, p<0.05; Extended Figure 8e) blocked the expression of the stress phenotype as measured with novelty-suppressed feeding (F(1,34)=19.35, p<0.0001; Extended Figure 8f) and sucrose preference (F(1,31)=5.56, p<0.05; Extended Figure 8g) compared to stressed mice expressing GFP alone.
Figure 4.
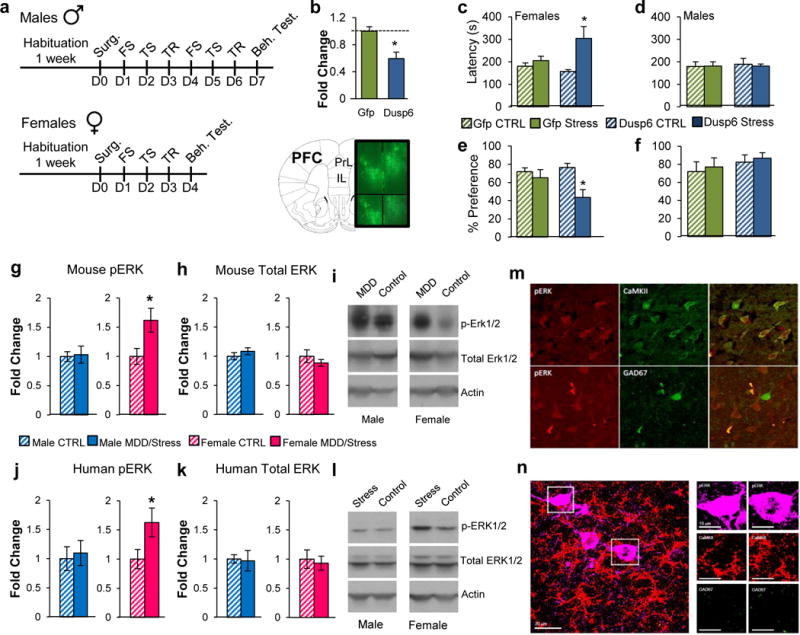
DUSP6 downregulation in vmPFC induces a sex-specific depressive-like phenotype associated with increased ERK signaling in females. a, Schematic representation of the behavioral paradigm used to assess the impact DUSP6 downregulation in males and females. b, HSV-mediated downregulation of DUSP6 covering the infra- and prelimbic regions of the vmPFC in mice. c, Behavioral consequence of DUSP6 viral downregulation in the novelty-supressed feeding test in females and d, males and in the sucrose preference test in e, females and f, males. Significance in b was assessed using independent sample t-test. Significance in tests c–f was determined using two-way ANOVA with Tuckey correction. b n=5/condition, c–d n =10/condition, e–f n=7/condition. Bars, mean ± sem; * p<0.05. g, Phospho-ERK1/2 levels assessed by Western blot in male (blue) and female (pink) mice in vmPFC with and without stress (21 days CVS). h, Total ERK1/2 protein levels assessed by Western blot in male (blue) and female (pink) mice in vmPFC with and without stress (21 days CVS). i, Representative blot of phospho-ERK1/2, total ERK1/2 and actin in male and female mice with and without stress (21 days CVS). j, Phospho-ERK1/2 levels assessed by Western blot in males (blue) and females (pink) with and without MDD in vmPFC. k, Total ERK1/2 levels assessed by Western blot in males (blue) and females (pink) vmPFC with and without MDD. l, Representative blot of phospho-ERK1/2, total ERK1/2 and actin in males and females with and without MDD. Significance in tests g–h and j–k was determined using independent sample t-test (two-tail in mice; one tail in humans). g–h n=12/stressed mice and n=10 in control conditions. j–k, Male CTRL n=18, Male MDD n=19, Female CTRL n=14, Female MDD n=18. Bars, mean ± sem; * p<0.05. m, Phospho-ERK1/2 (red) in females with MDD co-localizes in CaMKII reactive pyramidal cells (green; upper panel) but not in GAD67 reactive cells (green; lower panel). n, Phospho-ERK1/2 (purple) in female stressed mice co-localizes in CaMKII reactive pyramidal cells (red) but not in GAD67 reactive cells (green).
Together, these results demonstrate the role of DUSP6 and EMX1 in mediating sex-specific stress susceptibility. From a more global perspective, the findings validate our bioinformatics analyses and confirm the functional importance of sex-specific transcriptional profiles in the expression of stress susceptibility associated with human MDD.
Defining the molecular mechanisms underlying the sex-specific effect of DUSP6 in female mice
DUSP6 has previously been shown to induce its effects through the phosphorylation of ERK1/2 specifically84,85. Consistent with the downregulation of DUSP6 found in the vmPFC of chronically stressed female mice, we found a significant increase in phosphorylated ERK1/2 levels in this region of stressed female mice compared to control females (t(1,19)=2.33, p<0.05; Figure 4g, i), but no difference in males. Importantly, this effect was not associated with a change in total ERK1/2 levels (Figure 4h, i). Our analysis revealed very similar findings in humans with MDD. Phospho-ERK1/2 levels were increased in females with MDD compared to healthy controls (t(1,27)=1.92; p<0.05; Figure 4j, l), with no difference seen in males. Here again, no difference was found in total ERK1/2 levels in females or males with MDD (Figure 4k, l).
We used immunohistochemistry (IHC) to confirm our findings and locate the cellular substrate in which these effects occur. IHC revealed elevated phospho-ERK1/2 levels in the vmPFC of stressed (21 days of CVS) compared to unstressed female mice (t(1,11)=2.39, p<0.05; Extended Figure 9a), with no effect seen in males. Analysis of human postmortem tissue also confirmed elevated phospho-ERK1/2 levels in the PFC of females with MDD (t(1,12)=2.96, p<0.05; Extended Figure 9b, c). This effect was not specific to any cortical layer, as increased phospho-ERK1/2 was found uniformly across cortical layers (layerII/III: t(1,22)=2.35, p<0.05; layerV/VI: t(1,22)=2.19, p<0.05; Extended Figure 9d).
We further characterized these effects by defining the subcellular component in which the elevated levels of phospho-ERK1/2 are found in the PFC of depressed females. Our analysis shows that phospho-ERK1/2 immunoreactivity localizes predominantly in CaMKII-expressing pyramidal cells, but not in GAD67-expressing GABAergic cells nor in cells not labeled by either marker (Figure 4m). A parallel analysis of stressed female mice revealed equivalent localization patterns, with phospho-ERK1/2 immunoreactivity localizing in CaMKII- but not GAD67-expressing cells (Figure 4n).
Sex-specific modifications of synaptic activity by DUSP6 and EMX1
Given the role of ERK1/2 in regulating cellular activity87,88, our demonstration of increased phospho-ERK1/2 levels in pyramidal neurons suggest a potential impact on excitatory synaptic transmission in the vmPFC of depressed females and stressed mice. Thus, we assessed the impact of DUSP6 downregulation on the excitability of pyramidal neurons in this region of female and male mice (Figure 5a). Downregulation of DUSP6 in female vmPFC significantly increased the frequency of spontaneous excitatory postsynaptic currents (sEPSCs; F(2,151)=6.31, p<0.005); Figure 5b,c) without changing their amplitude (Figure 5b,d) in infected (DUSP6 downregulation) compared to control GFP-expressing (p<0.005) and uninfected (p<0.01) pyramidal neurons. Interestingly, these effects were specific to females as no change in sEPSC frequency (Figure 5e,f) or amplitude (Figure 5e,g) was seen in male vmPFC.
Figure 5.
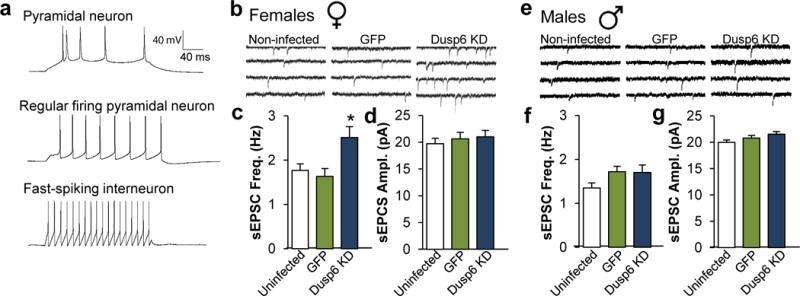
DUSP6 downregulation alters the physiological properties of vmPFC pyramidal neurons in a sex-specific fashion. a, Representative action potential traces of burst firing pyramidal neuron (upper), regular firing pyramidal neuron (middle) and interneuron (lower). b, Representative traces of neuronal activity in non-infected, GFP-infected and DUSP6 KD-infected pyramidal neurons of vmPFC of female mice. c, DUSP6 KD-induced changes of spontaneous excitatory postsynaptic current (sEPSC) frequency in Hertz (Hz) and d, amplitude in picoAmpere (pA) in female vmPFC. e, Representative traces of neuronal activity in non-infected, GFP-infected and DUSP6 KD-infected pyramidal neurons of vmPFC of male mice. f, DUSP6 KD effects on sEPSC frequency and g, amplitude in male vmPFC. Significance in tests b–c and e–f was determined using two-way ANOVA with Tuckey correction. c–d Female non-infected: n=10 mice and n=25 cells, Female GFP-infected: n=4 mice and n=17 cells, Female DUSP6 KD-infected: n=5 mice and n=24 cells. f–g, Male non-infected: n=9 mice and n=32 cells, Male GFP-infected: n=5 mice and n=29 cells, Male DUSP6 KD-infected: n=4 and n=30 cells. Bars, mean ± sem; * p<0.05.
We confirmed that these effects of DUSP6 downregulation were associated with elevated phospho-ERK1/2 levels. Cells infected by the DUSP6 downregulating HSV also showed elevated phospho-ERK1/2 levels (Extended Figure 10a,b). Elevated phospho-ERK1/2 immunoreactivity was found predominantly in CaMKII-expressing pyramidal neurons but not GAD67-expressing GABAergic neurons nor in cells not stained by either marker (Extended Figure 10a,b,d). Further investigation revealed that cells exhibiting elevated phospho-ERK1/2 levels following DUSP6 downregulation also coexpress the vesicular glutamate transporter, VGLUT1, confirming their identify as glutamatergic neurons (Extended Figure 10c). Interestingly, phospho-ERK1/2 immunoreactivity was found in the cell soma and dendrites (Extended Figure 10a, c). Importantly, these changes induced by the viral downregulation of DUSP6 were found both in males (t(1,6)=14.51, p<0.0001; Extended Figure 10e) and females (t(1,6)=8.23, p<0.0005; Extended Figure 10e), consistent with equivalent downregulation of DUSP6 in the two sexes. The results show that this equivalent biochemical effect induces very different downstream physiological actions in females vs. males.
We performed a similar assessment of the physiological impact of EMX1 overexpression in the vmPFC of both males and females. Overexpression of EMX1 in male vmPFC significantly increased the frequency of sEPSCs (F(2,152)=7.92, p<0.005; Extended Figure 11a,b) without changing their amplitude (Extended Figure 11a,c) in infected (EMX1 overexpressing) compared to GFP-expressing (p<0.005) and uninfected (p<0.0001) pyramidal neurons. These effects were specific to males as no change in EPSC frequency (Extended Figure 11d,e) or amplitude (Extended Figure 11d,f) was seen in female vmPFC. While these results highlight the sex-specific contribution of both DUSP6 and EMX1 in regulating synaptic excitability, they also underline the convergence of these effects toward common functional outputs, namely increased neuronal excitability in the vmPFC following prolonged stress.
DUSP6 and EMX1 control the transcriptional organization of their respective gene networks
We next assessed how the downregulation of DUSP6 would change gene expression profiles in females and how the structure of the Gray26 gene network would be affected by this manipulation. The downregulation of DUSP6 followed by 3 days of stress (Figure 6a) in female vmPFC induced a massive reorganization of the Gray26 gene network (Figure 6b). Indeed, our analysis shows that 5 and 6 genes within the Gray26 module (out of 18 total genes) were up- and downregulated, respectively, by DUSP6 downregulation plus sub-threshold stress. Such DUSP6 downregulation plus sub-threshold stress also reproduces a large proportion of the transcriptional changes induced by 21 days of stress in female mice. Indeed, using RRHO analysis, we found a significant overlap for genes commonly upregulated in the vmPFC after DUSP6 downregulation and 21 days of stress (FET p<1.0E-32; Figure 6c). Analysis of the gene ontology pathways commonly upregulated revealed several relevant terms related to ERK signaling such as Ras protein signaling, phosphorylation metabolism, and ATPase coupled activity (Figure 6d). Furthermore, our analysis shows that 36% (n=781) of the genes differentially expressed in female vmPFC after 21 days of stress are also differentially expressed in the vmPFC of female after DUSP6 downregulation followed by 3 days of stress (Figure 6e). These results demonstrate directly that downregulation of DUSP6 in vmPFC reproduces a significant proportion of the transcriptional profile associated with prolonged stress and provide important in vivo validation of our gene network analysis. The transcriptional regulation observed upon DUSP6 downregulation is consistent with the prominent role played by ERK in controlling gene expression89.
Figure 6.
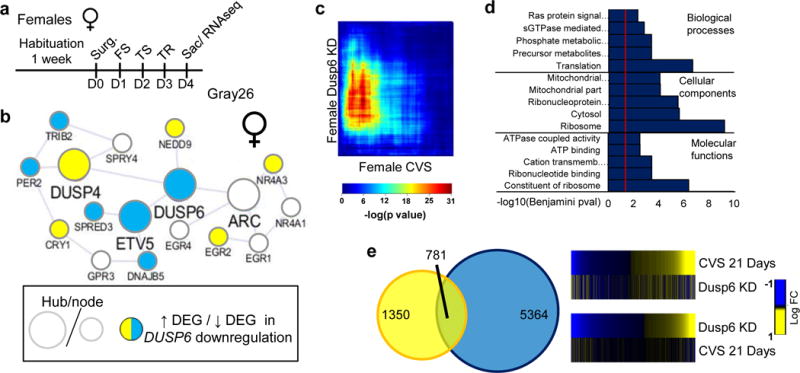
DUSP6 downregulation in female vmPFC reproduces the transcriptional and network alterations induced by 21 days of CVS in female vmPFC. a, Schematic representation of the behavioral paradigm used to assess the impact of DUSP6 downregulation in female mice. b, Transcriptional reorganization of the female-specific Gray26 gene network by DUSP6 downregulation in female vmPFC. Hubs and nodes are defined by the size of the circles with colors representing directionality of differential expression in the vmPFC (depicted in the bottom left panel). c, RRHO map directly comparing transcriptional profiles of females after DUSP6 downregulation with 21 days of CVS in the female vmPFC. Degree of significance is depicted in the color bar below the RRHO map. d, Top ontological terms enriched for downregulated genes following DUSP6 downregulation in female vmPFC. e, Venn diagram displaying the overlap between genes differentially expressed (p<0.05) in females after 21 days of CVS (yellow) and females after DUSP6 downregulation (blue) in the vmPFC. Heatmaps on the right compare transcriptional changes (log fold change; right to the heatmaps) in females after 21 days of CVS and females after DUSP6 downregulation in the vmPFC.
Our analysis of the male-specific Peru module also revealed a global reorganization following EMX1 overexpression in vmPFC. 18 and 3 hub genes within the Peru gene network (out of 43 total hub genes) were up and downregulated, respectively, by EMX1 overexpression followed by 6 days of stress (Extended Figure 12a,b). However, unlike our findings in females following DUSP6 downregulation, we found small overlap between genes commonly up or downregulated in the vmPFC following EMX1 overexpression and 21 days of stress in males (Extended Figure 12c). Furthermore, our analysis shows that only 14% (n=195) of genes differentially expressed following 21 days of stress in males are also differentially expressed following EMX1 overexpression (Extended Figure 12d). Thus, while EMX1 overexpression in the vmPFC increases stress susceptibility and neuronal excitability in males, its impact in reproducing the transcriptional alterations induced by prolonged stress in males appears limited, likely underlying a more subtle role of this gene.
Discussion
The heterogeneity of depressive disorders is one of the major impediments to better understanding them and developing improved treatments90. While males and females with MDD exhibit both similar and different symptomology, our molecular characterization of sex-specific transcriptional signatures shows that MDD is in fact widely divergent in males and females. In this study, we started with a combination of bioinformatics approaches in humans and stressed mice to characterize the transcriptional signatures associated with MDD in males and females. We then used a mouse model of variable stress to assess the role of sex-specific gene networks in mediating stress susceptibility and define the molecular and cellular mechanisms underlying these effects in a sex-specific fashion. Together, our results provide a comprehensive characterization of sex-specific transcriptional signatures defining MDD in males and females and highlight potential new avenues to develop more targeted therapeutic strategies for the treatment of MDD in males and females.
Transcriptional changes associated with MDD have been the focus of several investigations. As a result, several functional pathways have been associated with MDD in males30–43 and females46–54. While these studies provided interesting insight into the mechanisms underlying MDD in the two sexes, prior studies have not consistently compared male and female profiles and, as such, the extent to which the transcriptional signatures defining MDD differs in males vs. females remains unknown. Leveraging the intrinsic properties of gene coexpression network analysis78–81, we highlighted the fundamental differences in stress responses in males vs. females and provided a framework for better understanding the molecular basis of the sexual dimorphism characterizing MDD. Our findings suggest that MDD in males and females may arise in part from the actions of similar gene modules, which share cellular and biological specificity, but which are organized and expressed differently across brain regions in the two sexes. Alterations in this precise transcriptional structure may then interfere with the coordinated activity of several brain regions in males and in females and consequently disrupt the strategies normally used for coping with stress.
Our network-based approach not only allowed us to identify sex-specific gene coexpression modules significantly associated with MDD, but also provides valuable insight into candidate genes which likely contribute to depression in males and females. Indeed, for every module constructed in our analysis, we identified a series of hub genes, which by definition78–82 are more likely to carry important functional roles. In females, we provided compelling evidence for a role of DUSP6 acting in vmPFC in modulating stress susceptibility. Indeed, DUSP6 downregulation induced stress susceptibility while its overexpression blocked it. Our results suggest that DUSP6 downregulation increases stress susceptibility by increasing the excitability of a population of glutamatergic pyramidal neurons in this brain region via the activation of ERK signaling in females specifically. This is in contrast with previous studies in rodents showing impairment of LTP91,92 and retraction of pyramidal dendrites93 in the PFC after chronic unpredictable stress (CUS). Similar postmortem findings reported reduction of various synaptic proteins including AMPA and NMDA subunits91,94,95 in the dorsolateral PFC of suicide completers with MDD. Antidepressant-like effects were also obtained upon the optogenetic stimulation of the PFC in a mouse model of chronic social defeat stress96. It is important to note that our study was conducted in the vmPFC, the functional homologue of BA25, while most previous studies focused on different cortical regions97 or on NAc98. Furthermore, in humans, BA25 showed significant increased baseline activity in treatment resistant MDD patients compared to controls, the activity of which was reduced to control levels after 3–6 weeks of treatment with deep brain stimulation74.
Our results suggest that these effects were mediated through activation of the ERK signalling pathway. Previous studies in depressed humans showed lower expression of several signalling proteins in PFC, such as PKA, ERK, Ras, mTOR and CREB (reviewed in99). In humans, decreased Raf-ERK1/2 signaling and increased expression of MAPK phosphatases, negative regulators of ERK, was reported in the dorsolateral PFC of male suicide completers with MDD100–102. Importantly, we did not observe any change in total ERK1/2 levels, but rather increased phosphorylation levels of ERK1/2 which we causally linked to DUSP6 downregulation seen in the vmPFC of stressed female mice and depressed human females. Even though DUSP6 downregulation activates the ERK pathway to an equivalent degree in female vs. male vmPFC, the downstream consequences are different: such downregulation increases pyramidal neuron excitability in females but not males. These findings reveal fundamental differences between the sexes in the control of neuronal excitability, which defines important directions for future research.
Interestingly, our results suggest that the expression of stress susceptibility in males and females, although induced by different pathways and mechanisms, resolves into some common physiological outputs. Indeed, both DUSP6 downregulation and EMX1 overexpression resulted in higher frequency of sEPSCs in females and males, respectively. While our findings support the involvement of the ERK pathway in mediating DUSP6’s behavioral and physiological effects, the mechanisms mediating EMX1’s effects are less clear. EMX1 is a transcription factor critical in determining neuronal fate, differentiation of cortical neurons and GABAergic cells in the hippocampus and amygdala and contributing to the early development of the corpus callosum86,103,104. However, the transcriptional overlap between EMX1 overexpressing male mice and 21 days CVS males is fairly small. This may suggest a more subtle role of EMX1 in mediating stress susceptibility. Being highly expressed in early cortical developmental phases, EMX1 is believed to be involved in early functional circuitry103,104. As such, its acute overexpression during adulthood may be sufficient to activate critical pathways involved in synaptic plasticity but may require longer incubation periods before reproducing to a significant level the transcriptional pattern induced by 21 days of stress. As a transcription factor, one may also hypothesize that its binding to DNA requires precise chromatin structure that may be obtained only after prolonged periods of stress. Furthermore, compared to DUSP6, EMX1 is part of a larger gene network and may work in collaboration with other members of the network to induce stronger behavioral alterations. More work will be required to understand the complex impact of EMX1 on synaptic plasticity and stress susceptibility in males.
MDD remains a highly heterogeneous and complex disorder constraining our understanding of its molecular etiologies. Previous attempts to decipher the genetic bases of MDD yielded limited success. More recently, studies performed on larger populations have been more successful64,65. Interestingly, several gene targets identified in these studies are found within the top gene networks in our studies, which further validates our bioinformatics analyses and highlights the importance of using combinatorial approaches when studying the genetic and transcriptional mechanisms underlying the expression of complex disorders. Several of these networks associate with neurotransmission, nerve ensheathment, neuronal differentiation, calcium signaling, cathecholamine metabolism, transcription factors and inflammatory response in males and females with MDD and highlight the heterogeneity of the disorder, not only from a clinical but also from a molecular and mechanistic point of view. Importantly, while these studies reported no MDD-associated genetic sex difference64,65, we found these gene targets within highly sexually dimorphic gene networks, suggesting that the sexual dimorphism defining MDD may be taking place at the transcriptional rather than at the genetic levels although more work is required to understand this complex relationship.
To conclude, we provide molecular, physiological and behavioral evidence supporting the strong sexual dimorphism characterizing the clinical features of MDD. Given the complexity of MDD, we leveraged the use of network-based approaches combined with differential gene expression and provided an unbiased and comprehensive characterization of the transcriptional sexual dimorphism in MDD. We further characterized the effects of stress in males and females by identifying specific functional pathways preserved in a mouse model of variable stress. Besides defining the transcriptional profiles of males and females with MDD, we also provided solid evidence uncovering the molecular and cellular mechanisms underlying sex-specific stress susceptibility. Together, this study represents a great resource for the study of sex-differences in mood disorders and the identification of molecular pathways which may be significantly involved in mediating the effect of stress in males and females.
While our interpretations are constrained by the intrinsic limitations associated with the study of postmortem brain tissue and our relatively small sample size, we succeed in illustrating robust differences in gene regulation in MDD with clear sex differences, findings validated in a chronic stress model in mice. It will be interesting in future studies to extend this work to additional cohorts of MDD subjects and to assess the potential associations between brain and blood gene networks, with the promise that the latter might reveal gene signatures related to diagnostic subtypes or treatment responses. Nevertheless, by showing here that MDD in males and females originates from the alterations of mostly distinct genes converging on partly similar functional pathways, we provide a strong basis on which to build the foundation for a better understanding of the molecular mechanisms taking place in the brain of males and females with MDD, insights which can be used to develop treatment strategies targeted selectively to the two sexes.
Methods
Human subjects
Brain tissue was obtained from the Douglas Bell Canada Brain Bank (DBCBQ; Douglas Mental Health Institute, Verdun, Québec). All subjects were Caucasians of French–Canadian descent, a population with a well identified founder effect105. Sociodemographic and clinical information is listed in Supplementary Table 10. Males and females were group-matched for age, pH and postmortem intervals (PMI). Other information included presence of comorbid disorders, treatment history, smoking history, history of early life adversity, cause of death and presence of drug and/or alcohol abuse (Supplementary Table 10). Inclusion criteria for both cases and controls were the following: the subject had to be Caucasian and of French Canadian origin and the subject had to die suddenly without prolonged agonal state. Forty-eight subjects (26 MDD: 13 males, 13 females and 22 controls [CTRLs]: 13 males, 9 females) were recruited for this study. Tissue from six brain regions—orbitofrontal cortex (OFC; BA11), dorsolateral PFC (BA8/9; dlPFC), cingulate gyrus 25 (BA25; cg25; vmPFC), anterior insula (aINS); nucleus accumbens (NAc) and ventral subiculum (vSUB)—was carefully dissected at 4°C after having been flash-frozen in isopentene at −80°C. An additional group of 32 male samples (15 MDD and 17 CTRL) from The University of Texas Southwestern Medical Center brain bank was used for the validation of male DEGs. A third cohort composed of 18 female samples (6 MDD and 12 CTRL) was used for the validation of female DEGs. Sociodemographic and clinical information for the second (males) and third (females) cohorts is listed in Supplementary Tables 11 and 12, respectively. Tissue dissection was performed by histopathologists using reference neuroanatomical maps106,107. The human study was approved by the research ethics boards of the McGill University and the University of Texas Southwestern Medical Center. Written informed consent was obtained from all participants.
Psychological Autopsies and Psychopathology
Information concerning psychiatric history and socio-demographics was obtained by way of psychological autopsies performed by trained clinicians with the informants best-acquainted with the deceased as described elsewhere4 and which has been shown to yield highly valid information108–114. Both cases and controls were characterized by the same psychological autopsy methods, therefore avoiding the occurrence of systematic biases. Diagnoses were obtained using DSM-IV115 criteria by means of SCID-I interviews116 adapted for psychological autopsies.
Animal Experiments
Animals
Eight-week-old C57BL/6J male or female mice (Jackson Laboratory) were used. All mice were habituated to the animal facility for 1 week before experimental manipulations and were maintained at 23–25°C on a 12 h light/dark cycle (lights on from 7:00 A.M.) with ad libitum access to food and water. All behavioral experiments were conducted once. Experiments were conducted in accordance with guidelines of the Society for Neuroscience and the institutional animal care and use committee (IACUC) at Mount Sinai.
Chronic variable stress (CVS)
CVS, modified from LaPlant et al., 200944, consists of three different stressors repeated over a period of 21 days in both male and female mice. Stressors are alternated during the 21 days to prevent stress habituation. Stressors were administered in the following order: 100 random mild foot shocks at 0.45 mA for 1 h (10 mice to a chamber), a tail suspension stress for 1 h, and a restraint stress placed inside of a 50 mL falcon tube for 1 h within the home cage. The three stressors were then repeated for the next 21 days in the same order. 40 mice (20 males: 10 stressed, 10 unstressed; 20 females: 10 stressed and 10 unstressed) were used for the behavioral experiments. Mice were housed five per cage over the period of stress and rehoused one mouse per cage before behavioral assessment. For both males and females, a group of unstressed grouped-house mice were used as controls. Statistical outliers were detected and removed if sample values exceeded twice the standard deviation. The impact of stress on males and females was assessed using a generalized linear model with stress (stress or control) and sex (male or female) as main factors. Significance was fixed at p<0.05. A second cohort of mice (20 males: 10 stressed, 10 control and 20 females: 10 stressed, 10 control) went at the same time through the same stress paradigm but were killed on the day after the last stressor to collect vmPFC and NAc tissue for RNAseq using 12 and 14 gauge punches, respectively. The same protocol was applied to generate tissue for protein and immunohistochemistry (IHC) experiments. No statistical outliers were removed from any of these cohorts.
Splash test
Behavioral testing was based on a published protocol117. The test was performed under red light (230V, 15W). Mice were habituated to the room for 1 h prior to testing. Mice were sprayed on the back with a 10% sucrose solution 3 times and then placed into an empty housing cage and behavior was recorded for 5 min via videotape. The total amount of time grooming over the 5 min period was recorded and hand-scored by an observer blind to experimental conditions.
Novelty suppressed feeding
Behavioral testing was adapted from a published protocol118. Mice were food restricted overnight prior to testing. On day of testing mice habituated to the testing room for 1 h. Under red light conditions, mice were then placed into a plastic box 50 × 50 × 20 cm with bedding. A single pellet of food was placed in the center of the box. Mice were placed in the corner of the box and the latency to eat was scored up to 10 min during testing. Mice were then immediately transferred to their home cage in standard lighting conditions and the latency to eat was recorded.
Sucrose preference
Behavioral testing was conducted according to previously published protocols55,119. Immediately after splash test, mice were separated and individually housed. They were given 2 bottles filled with water for a 24 h habituation period. The following day, immediately after NSF testing, one of the two 50 ml bottles was replaced with a 2% sucrose bottle for 24 h. The 2 bottles were then weighed and position was switched for an additional 24 h. The total duration of the test was 48 h. Sucrose was calculated by determining the percentage of total sucrose consumption divided by total liquid consumption (sucrose + water).
Forced swim test (FST)
Behavioral testing was conducted according to previously published protocols120. 24 h after the NSF test, animals were placed in the test room for an h before behavioral testing. Mice were tested in a 4 L Pyrex glass beaker, containing 2.5 L of water at 25±1°C for 6 min. Behavior was videotaped and hand scored using a 5 sec sampling method for % time spent immobile by an observer blind to experimental conditions.
Open field test (OFT)
Mice were acclimated to the testing facility for 1 h before testing. Under red light conditions, mice were then placed into a plastic box 50 × 50 × 20 cm with bedding. Mice were placed in the corner of the box and the latency to eat was scored up to 10 min during testing. Mice were then immediately transferred to their home cage in standard lighting conditions. Time spent in the periphery and center was calculated using an automated system (Noldus Ethovision; Noldus Interactive Technologies).
Elevated plus maze (EPM)
Mice were acclimated to the testing facility for 1 h before testing. Animals were placed in the EPM under red light conditions for 5 min. Each arm of the maze measured 12 × 50 cm. The black Plexiglas cross-shaped maze consisted of two open arms with no walls and two closed arms (40 cm high walls) and was on a pedestal 1 m above floor level. Behavior was tracked using an automated system (Noldus Ethovision; Noldus Interactive Technologies). Behavior was measured as total time spent in combined open arms and total time in combined closed arms.
Sub chronic variable stress (sCVS)
A modified version of the CVS was used for functional behavioral cellular and molecular studies which, by itself, is insufficient to induce stress-related behavioral abnormalities in males and females but when combined with viral-mediated modification of gene expression can reveal altered stress susceptibility62. In males, the same stressors described above were repeated over a period of 6 days, while in females stressors were repeated for 3 days after surgery. On day 7 in males and 3 in females (post-surgery), behavior was assessed in the NSF, sucrose preference, OFT, FST and EPM test as described above. Mice were killed the following day. The conventional sCVS protocol was used to assess the functional in vivo behavioral effect of DUSP6 over expression as described before62. In females, 6 days of the same stressors described above are sufficient to induce a stress phenotype. On day 7 (post-surgery), behavior was assessed in the NSF, sucrose preference, and grooming test as described above. Mice were killed the following day.
Viral constructs
Knockdown (KD) constructs designed to target DUSP6 mRNA were cloned using BLOCK-iT Pol II miR RNAi kit (Invitrogen). Briefly, four artificial miRNAs oligos were designed using Invitrogen’s RNAi Designer (www.invitrogen.com/rnai) to KD DUSP6 and cloned into pcDNA6.2-GW. Mouse neuroblastoma (N2a) cells were transfected using lipofectamine 2000 (Invitrogen) with the different plasmids designed to target DUSP6 or LacZ as control. The level of DUSP6 mRNA KD was assessed using qRT-PCR. The miRNA causing the most efficient down regulation was further Gateway cloned (Invitrogen) into the p1005+ HSV vector. DUSP6 and EMX1 coding regions were excised from DUSP6 (Origene cat# MR222688) and EMX1 (Origene cat# RC208006) expression clones and cloned into a p1005+ HSV vector to induce overexpression in the brain.
Stereotaxic surgeries and viral gene overexpression and knockdown
EMX1 and DUSP6 overexpression and DUSP6 knockdown in the vmPFC was achieved by injecting Herpes Simplex Virus (HSV) vectors expressing EMX1, DUSP6 or an siRNA targeting DUSP6, as well as green fluorescent protein (HSV–EMX1/DUSP6/siDUSP6–GFP). HSV–GFP was used as a control. Briefly, mice were anesthetized using a mixture of ketamine (10 mg/kg) and xylazine (1 mg/kg), and the following stereotaxic coordinates were used for viral delivery: +1.8 mm (anterior/posterior), +0.8 (lateral), −2.75 (dorsal/ventral) at an angle of 15° from the midline (relative to Bregma). A total of 0.5 μl of HSV solution was delivered bilaterally over a 5 min period (0.1 μl/min), followed by 5 min of rest.
The effect of DUSP6 downregulation (siDUSP6) and EMX1 overexpression was initially assessed on a total 40 mice (10 stressed males, 10 stressed females, 10 unstressed males, 10 unstressed females) for each virus through the modified sCVS paradigm. Male and female mice were tested 7 and 4 days after surgery, respectively, and viral injection sites were confirmed using standard histological methods. Statistical outliers were detected and removed if sample values exceeded twice the standard deviation. For every tested gene, the impact of stress on males and females was assessed using a generalized linear model with stress (stress or control) and virus (tested gene or GFP control) as main factors. Significance was fixed at p<0.05 with Tuckey post-hoc test.
The same paradigm was used to generate tissue for the IHC validation experiments and molecular RNAseq experiments. For IHC validation of viral mediated DUSP6 downregulation, 40 mice (10 stressed males, 10 stressed females, 10 unstressed males, 10 unstressed females) were stressed through the same paradigm and sacrificed a day following the last stressor. The same tissue was used for IHC co-staining experiments. For RNAseq on viral mediated DUSP6 downregulation and EMX1 overexpression, 10 female/male mice were used (5 DUSP6/EMX1 stressed and 5 GFP stressed) went through the same protocol as described above and sacrificed a day following the last stressor.
The effect of DUSP6 overexpression was assessed on the conventional sCVS model on 20 female mice (10 stressed and 10 unstressed females) for both DUSP6 and GFP viruses. Female mice were tested 7 days after surgery, respectively, and viral injection sites were confirmed using standard histological methods. Statistical outliers were detected and removed if sample values exceeded twice the standard deviation. For every tested gene, the impact of stress on females was assessed using a generalized linear model with stress (stress or control) and virus (DUSP6 or GFP control) as main factors with Tuckey post-hoc test. Significance was fixed at p<0.05 with Tuckey post-hoc test.
Perfusion, tissue processing, immunohistochemistry and confocal microscopy
Mice were anaesthetized with a mixture of ketamine (10 mg/kg) and xylazine (1 mg/kg) and perfused with cold 4% paraformaldehyde in PBS. Brains were removed, post-fixed, cryoprotected in PBS + 15% sucrose for 2–3 hours followed by immersion in PBS + 30% sucrose for 24 h, and then sectioned coronally (35 μm) on a microtome. Free-floating sections were permeabilized with blocking buffer (PBS + 3% normal donkey serum, 0.3% Triton X-100) for 2 hours and then incubated with primary antibodies (GAD67, Santa Cruz (sc-7512), 1:250; CaMKII, Santa Cruz (sc-5306), 1:400; phospho-ERK1/2, Cell Signaling (4370s), 1:200; VGlut1, Millipore (AB5905), 1:10000) overnight at 4°C. Slices were then washed 4×15 min with PBS, incubated with corresponding secondary antibodies (donkey anti-mouse Cy3 1:400, donkey anti-rabbit Cy5 1:400, or donkey anti-goat Cy2 1:400, Jackson ImmunoResearch) for 2 hours, washed 4 × 15 min with PBS, mounted on microscope slides followed by counterstaining with DAPI and coverslipped for confocal imaging.
Slices from mice brain were imaged using a Zeiss LSM 780 microscope. For analysis of GAD67 and GFP co-localization with phospho-ERK1/2, 5-um z-stacks were taken at ×63 magnification in the vmPFC of male or female mice. A total of 8-21 phospho-ERK1/2-positive cells per animal were analyzed using NIH ImageJ. Cell body mean optical density (O.D.) was measured for phospho-ERK1/2-positive cells in each channel (GAD67-Cy5, phospho-ERK1/2-Cy3, GFP) and compared to mean O.D. values of a reference positive cell.
Human IHC and DAB
Phospho-ERK1/2 immunohistochemistry analysis was performed in postmortem human brain (n=4/group) to assess phospho-ERK1/2 cellular expression in orbitofrontal cortex (BA11). This tissue was selected for the human IHC because BA25 was not available for immunochemistry and because the module differential expression enrichment pattern in females was exactly the same in BA25 and BA11. All tissue slots were fixed in 4% PFA for 3 weeks, then cryoprotected gradually in10%, 20% and 30% sucrose in 0.1M PBS, and snap frozen in cold methylbutane. Non-serial sections (30 μm thick, 60 μm apart) were cut on a cryostat (Leica CM3050S) and mounted on superfrost plus slides for phospho-ERK1/2 IHC.
All slides were subjected to microwave antigen retrieval (citra solution, BioGenex; 95°C for 10 min), quenched free of endogenous peroxidases in 0.3% H2O2, and then follow the instruction of ImmPRESS Excel staining kit (Vector Laboratories). Briefly, sections were blocked in normal horse serum prior to overnight incubation of phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) rabbit monoclonal antibody (1:200; Cell Signaling). Primary antibody was detected by sequential incubation with goat anti-mouse IgG and HRP-conjugated horse anti-goat IgG. Diaminobenzidine chromogen was used to detect immunoperoxidase signal, and hematoxylin was used as the counterstain. Negative controls were performed by omitting primary antibody from procedure and by using primary antibody preincubated with phosphor-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) blocking peptide (Cell Signaling).
For double staining, primary antibodies were simultaneously incubated (phospho-ERK1/2 1:50/CaMKII 1:400, phospho-ERK1/2 1:50/GAD67 1:800). Cy3-conjugated donkey anti-rabbit IgG and cy2-conjugated donkey anti-mouse IgG (1:200, Jackson Immunoresearch) were used to detect primary antibodies.
Stereology method
All slides were coded prior to blinding, and the observer was blinded to subgroups. Unbiased estimates for phospho-ERK1/2+ and phospho-ERK1/2− cells in each group were obtained using stereology on an Olympus BX51 System Microscope (Optronics, Goleta, Calif., USA). Cell counts were collected using the Optical Fractionator Probe within the Stereo Investigator software (MBF Bioscience, MicroBrightField, Inc., Williston, Vt., USA). Region of Interest (ROI) of all cortical layers and cortical sublayers (II/III and V/VI layers) was identified with the help of hematoxylin counterstain. Cell counting was performed using a 40x/0.63 NA lens. Phospho-ERK1/2+ cells were identified as the cells containing a brown reaction product of sufficient intensity within the whole cell body, and phospho-ERK1/2− cells were identified as the cells only containing a blue reaction of hematoxylin within the nucleus (the cells that related to blood vessels were excluded from counting). The optical dissector height was 15 μm. The area-sampling fraction was 1/20 for all layers and 1/12.5 for sublayers counting. 2 sections were counted for all layers and 3 sections were counted for sublayers statistics analysis.
Protein extraction, SDS-PAGE and Western blotting
For analysis in humans, tissue from vmPFC from our initial RNAseq cohort was combined with samples from our third cohort (total sample: Male CTRL n=18, Male MDD n=19, Female CTRL n=14, Female MDD n=18). Two male CTRL, 1 male MDD and 2 female CTRL samples were excluded from the analysis because of outliers using the Grubb’s test. For the analysis of mouse vmPFC, 10 male CTRL, 12 male stressed, 10 female CTRL and 12 female stressed samples were processed. No sample was removed from the animal cohort. Tissue from human and mouse vmPFC was carefully dissected and sonicated in RIPA buffer (pH=7.4, plus protease and serine/threonine and tyrosine phosphatase inhibitors). The homogeneous mixture was centrifuged at 10,000 × g for 10-minutes at 4°C and the supernatant was kept for protein analysis. Quantification of samples was determined using a DC protein assay. Samples (25 μg/lane) were mixed with reducing sample buffer containing SDS and DTT, and heated to 95°C for 5 min. Samples were resolved by SDS-PAGE on pre-cast BioRad gels and transferred to PVDF membranes. Membranes were blocked in 5% BSA in TBS-tween (TBST) for 1 h and incubated at 4°C overnight with primary antibodies (pERK, Cell Signaling (4370s), 1:1000; ERK, Cell Signaling (4696s), 1:1000; Actin, MP Biomedicals (691002), 1:20,000) diluted in 5% bovine serum albumin in TBST. After thorough washing, membranes were incubated with peroxidase conjugated secondary antibodies (Vector Labs) in blocking solution at room temperature for 2 h, washed thoroughly, and developed using chemiluminescent substrate (Thermo Scientific). Quantification of protein expressed in each lane was determined by densitometry using Image J (US National Institutes of Health). Phospho-ERK1/2 and total ERK1/2 expression levels were normalized to Actin to gain an accurate quantification. Variation in phospho-ERK1/2 and total ERK1/2 levels, in both mice and humans, was assessed by means of independent sample t-test comparing male MDD with male CTRL or Female MDD and Female CTRL. T-tests in mice were two tailed and one-tailed in humans. Significance was fixed at p<0.05.
In vitro electrophysiology
Recordings were restricted to pyramidal neurons with large tear-drop shaped somas and conspicuous apical dendrites. Interneurons, which exhibited small rounded somas and the lack of the apical process, were avoided. Besides the morphological criteria, the action potential firing patterns were also used to differentiate between pyramidal neurons and interneurons121. Mice were injected with siDUSP6, EMX1 or GFP expressing HSVs two days before electrophysiological recordings. Recordings were performed on 23 females (10 non-infected, 4 GFP infected, 5 siDUSP6 infected and 4 EMX1 infected) and 22 males (9 non infected, 5 GFP infected, 4 EMX1 infected and 4 siDUSP6 infected). No outliers were removed. Virally infected neurons were distinguished from non-infected neurons by their GFP signals under epifluorescence microscopy. Spontaneous EPSCs (sEPSCs) of pyramidal neurons were recorded for 3 min in the voltage-clamp mode, with a K-based internal solution (in mM: 130 K-methanesulfate, 10 KCl, 10 HEPES, 0.4 EGTA, 2.0 MgCl2, 3 MgATP, 0.5 Na3GTP, 7.5 phosphocreatine, pH 7.4; 285 mOsm). The data (frequency and amplitude of sEPSCs) was analyzed with Minianalysis Program (Synaptosoft Inc.). For siDUSP6 and EMX1, the sex-specific impact of DUSP6 and EMX1 on pyramidal neurons excitability was assessed using a generalized linear model with sex (Male or Female) and virus (siDUSP6-infected, GFP-infected or non-infected) as main factors with Tuckey post-hoc test. Significance was fixed at p<0.05.
RNA sequencing
RNA from every brain sample (humans and mice) was extracted using RNAeasy micro kit using trizol, followed by DNAse 1 treatment, as described by the manufacturer (Qiagen). RNA integrity (RIN) and concentration were assessed using a bioanalyzer (Agilent). ERCC spike-in controls were added at a concentration of 1:1000 to every RNA sample. Libraries were constructed using the ScriptSeq Complete Gold Kit (Epicentre, Illumina) proceeded by ribosomal RNA depletion starting with 1 μg of RNA in humans, and 100 ng of vmPFC and 300 ng of NAc in mice. PFC samples from virally infected mice (siDUSP6, EMX1 and GFP) were processed the same way using 300ng RNA. Samples were barcoded for multiplexing and sequenced at 50 bp paired-end on Illumina HiSeq2500. Samples were pooled 8 per lane and sequenced twice at a depth of 50 million reads per sample.
Data analysis and differential expression analysis
FASTQ and FASTX-toolkit were used to examine the sequencing quality and trim reads. Paired-end reads for the human data were aligned to the GENCODE 2014 version of the human genome while the mouse data were mapped to the GENCODE vM2 mm10 annotation using TopHat. Both datasets were mapped using the comprehensive gene annotation on the reference chromosome only. In humans, all samples passed quality control (QC) with the exception of 6 samples (1 female MDD in OFC, 1 male control and 1 male MDD in vmPFC, 1 female control in dlPFC, 1 male control in aINS and 1 female MDD in vSUB) that had excessively high levels of rRNA or extremely low mapped read % and thus removed from the analysis. In mice, 4 samples were removed (1 stressed male and 2 stressed females in the vmPFC, 1 stressed female in the NAc) due to excessively high levels of rRNA or extremely low mapped read %. Using the same criteria in the virally infected mice RNAseq samples, 1 sample was removed from the female siDUSP6 and none from the GFP female mice while no sample was removed from the male EMX1 and 1 from the male GFP expressing mice. These samples were excluded from further analysis. ERCC spike-in controls were used to assess the quality of each sequencing run and to insure replicability of runs 1 and 2 for every sample as previously described122. Reads from run 1 and 2 for every sample were merged and counted using HTSeq. A gene was considered the union of all its exons in any known isoforms, based on GENCODE annotation. Any reads that fell in multiple genes were excluded from the analysis. Threshold for filtering out low expressed genes was set to >5 reads in at least 80% of the samples as previously described123.
Human and mouse samples were processed separately. Differential gene expression between groups of samples was determined in several stages to ensure both statistical significance and biological relevance. In the first stage, linear models implemented in the ‘limma’ package124 of Bioconductor125 were used to compute the variance of gene expression across all groups. Gene expression was transformed and normalized using voom in the limma package. In humans, models were adjusted for age, RIN, alcohol abuse, and medication status. Among the extensive information collected for the individuals in this study, these variables were selected for adjustment based on a combination of domain knowledge and variance analysis of the RNAseq data. Eigen-R2126 was used to estimate the amount of variance in RNAseq data explained by each variable. The estimate for each variable is similar to taking the average of the correlations between the variable and the expression values for each gene. Correlation averages are vulnerable to technical artifacts such as stochastic noise for genes with little or no expression values so Eigen-R2 uses principal component analysis to reduce the contribution of these and other problematic genes. Using Eigen-R2 we found evidence that phenotype (PC4: p<0.01), sex (PC3: p<1.0e-4) and brain regions (PC1: p<5.0e-36) explained the most variance of any variable. Differential gene expression was assessed through a generalized linear model implemented in limma, with phenotype (MDD vs CTRL) and sex (male and female) as main factors for every brain region. Our analyses were adjusted for age (PC2: p<0.01), RIN (PC1: p<1.0e-3) and alcohol abuse (PC2; p<0.05) because these contributed to the variance according to the Eigen-R2 analyses, and we chose to adjust for medication status because medication is well known to change gene expression in the brain75. Overall, our model allowed us to generate a comprehensive evaluation of gene expression changes in males and females with MDD relative to controls, while controlling for the effects of major covariates and considering baseline differences in gene expression between males and females. An individual gene was called differentially expressed if the p-value of its t-statistic was at most 0.05. The exact same procedure was followed for the CVS mouse data analysis with phenotype (stress vs CTRL) and sex (male and female) as main factors and for the virally infected mice with phenotype (stress vs CTRL) and virus (EMX1 or siDUSP6 vs GFP). For the latter, male and female mice were analyzed separately given the difference in stress paradigm. RIN was the only factor we accounted for in our statistical analysis of our different mouse cohorts.
The Rank Rank Hypergeometric Overlap (RRHO) test76,127 was used to evaluate degree of overlap in gene signatures across brain regions in males and in females with MDD separately and directly in males and females for every brain region. The RRHO analysis was also used to assess the overlap between virally infected and CVS mice. Only genes found across all brain regions in both males and females with and without MDD (intersection) were used in this analysis. A one-sided version of the test only looking for over-enrichment was used. RRHO difference maps were produced by calculating for each pixel the normal approximation of difference in log odds ratio and standard error of overlap with expression data in the intersection list. This Z score was then converted to a p-value and corrected for multiple comparisons across pixels128. For display, the p-value was multiplied by the sign of the effect to show both stronger and weaker matching between in vitro systems. Sex-specific regional overlap was further refined using Fisher’s exact test on gene across brain regions that were significantly differentially expressed (p<0.05) in males and in females.
Comparisons between human and mouse transcriptional profiles were performed using the human and mouse orthologues which we gathered using the Biomart package in R. The interspecies statistical overlap in the vmPFC and NAc for males and females was quantified using Fisher’s exact test with significance set at p<0.05. Interspecies assessment of gene ontology (GO) terms overlap was performed using an in house R script. GO terms from both species being enriched for genes differentially expressed (p<0.05) after multiple testing correction were first identified in the vmPFC and NAc. Only terms being significantly enriched after multiple testing correction from both species were considered overlapping.
NanoString assay
RNA from the vmPFC of all human brain cohorts was processed through the NanoString-n-Counter system (Life Science). Up to 125 ng of RNA was submitted to the qPCR CORE at Mount Sinai for analysis. The code set was designed by the company with unique sequences. We included a set of 3 male and 3 female housekeeping genes identified using our RNAseq data as genes showing no difference in cases vs. controls in both males and females (Fold Change +/− 0; p=0.99). For the original discovery cohort (RNAseq cohort), normalization of NanoString read counts was performed using housekeeping genes. Normalization of the replication cohorts was accomplished using NormaGene. No outliers were removed. Data were analyzed using one tail independent samples t-test with significance set a p<0.05.
Multi-tissue network analysis
We generated coexpression modules with RNA-sequencing expression data, using weighted gene coexpression analysis79. We constructed multi-tissue coexpression networks that simultaneously captured the intra- and inter-tissue gene-gene interactions between the MDD and normal states in both males and females. These networks can be characterized only when multiple tissues are monitored in a population of individuals covering all tissues analyzed (i.e., aINS, OFC, vmPFC, dlPFC, NAc, vSUB).
The weighted network analysis began with a matrix of the Pearson correlations between all gene pairs, and then converted the correlation matrix into an unsigned adjacency matrix using a power function, so that the resulting adjacency matrix, i.e., the weighted coexpression network, is approximately scale-free. To explore the modular structures of the coexpression network, the adjacency matrix was further transformed into a topological overlap matrix (TOM)79. Because topological overlap between two genes reflects both their direct interaction and their indirect interactions through all other genes in the network, this approach helps create more cohesive and biologically more meaningful modules. To identify modules of highly coregulated genes, we used average linkage hierarchical clustering to group genes based on the topological overlap of their connectivity, followed by a dynamic cut-tree algorithm to dynamically separate clustering dendrogram branches into gene modules. To distinguish between modules, each module was assigned a unique (and arbitrary) color identifier. The gene with the highest intramodular connectivity was considered the “hub gene” (most connected gene). We used DAVID v6.7129 to assess the functional annotation (GO and KEGG) for each module, and we report FDR p-values and fold enrichment, as provided by DAVID.
To quantify differences in transcript network organization between MDD and control subjects in both males and females, as well as male MDD vs female MDD, we employed a modular differential connectivity (MDC) metric73. In brief, MDC represents the ratios of the connectivity of all gene pairs in a module from MDD subjects to that of the same gene pairs from control subjects. MDC is a continuous measure ranging from 0 to infinity. MDC > 1 indicates gain of connectivity or enhanced co-regulation between genes, whereas MDC < 1 indicates loss of connectivity or reduced co-regulation between genes. The statistical significance of the MDC metrics was computed through the FDR procedure. The significance or FDR of the MDC statistic can be accessed by permuting the data underlying the two networks. We estimated FDR based both on shuffled samples (i.e., networks with nonrandom nodes but random connections) and shuffled gene labels (i.e., networks with random nodes but nonrandom connections), and then selected the larger value as the final FDR estimate.
To further identify key regulator (driver) genes of the modules identified by WGCNA, we applied the key driver analysis31 to the module-based unweighted coexpression networks derived from ARACNE (Algorithm for the Reconstruction of Accurate Cellular Networks)130. ARACNE was used first to identify significant interactions between genes in each module based on their mutual information, and then remove indirect interactions through data processing inequality (DPI). For each ARACNE-derived unweighted network, we further identified the key regulators by examining the number of N-hob neighborhood nodes (NHNN) for each gene. For a given network, let μ be the numbers of N-hop downstream nodes and d be the out degrees for all the genes. Genes with a number of NHDN greater than ū+σ(μ) were nominated as causal regulators. The regulators with degree above d̄+2σ(d), where d denotes the number of downstream genes, become key causal regulators of a corresponding network module associated with MDD differential connectivity31. This criterion identified genes with number of NHNNs significantly above the corresponding average value.
The assessment of module enrichment for genes expressed in specific cell types was performed using a published RNAseq dataset131. We focused our analysis on neuronal, astrocytic, myelinating oligodendrocytes, microglial and endothelial cell types. We first excluded all genes with no human orthologues and filtered out genes expressed at values below 1 FPKM. We then created sublists for genes expressed at least 5 times more in one cell type vs. the average of the other cells type and used Fisher’s exact test to quantify the significance of the enrichment with significance fixed at p<0.05.
We tested conservation of MDD modules between males and females with MDD by analyzing overlap of module members. Fisher’s exact test was used to quantify significant overlap and enrichment. Modules in male MDD that show significant overlap (FDR p-value of 0.05) and a fold change of greater than 15 with a module in female MDD were considered conserved, while the others were considered sex-specific. For enrichment analysis using gene sets from our differential expression analysis, Fischer’s exact test was performed. Fold enrichment and FDR corrected p-value are reported in all cases.
Statistical analyses
Sample size calculation was not performed. However, we justified every experiments sample size based on several previously published reports using similar or even smaller sample size and showing the power to detect significant statistical differences. For every statistical assessment, estimate of variance was calculated and even variance was reached. RNAseq gene expression data for differential expression was normalized. Because of the limited sample size for behavioral experiments, normality was not always reached. Network analyses, including network construction, module differential connectivity, GO enrichment, module conservation and module differential expression enrichment were adjusted for multiple testing. Differential expression, NanoString validation and cell type enrichment analyses were not corrected for multiple testing. RRHO analysis was corrected using the Benjamini-Hochberg method. Behavioral and electrophysiological results data were corrected using Tuckey posthoc test. Protein expression and IHC quantification was performed using two tailed or one tailed unpaired samples t-test. Data were not randomized. For the human studies, experimenters were blinded to the experimental design.
Extended Data
Extended Figure 1.
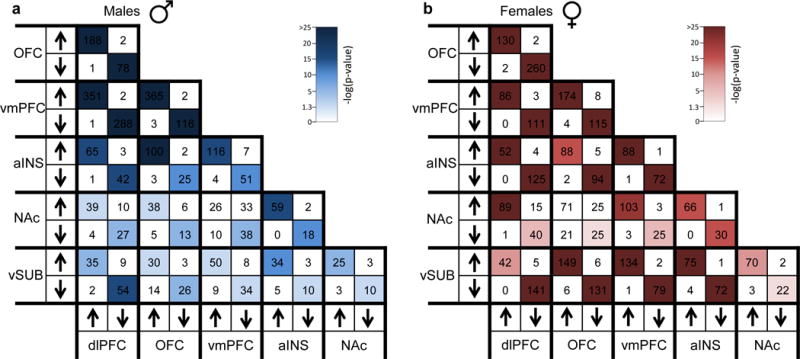
Degree of overlap for genes significantly differentially expressed in a, males with MDD and b, females with MDD across brain regions. Arrows on x and y axis correspond to directionality of gene expression changes across brain regions and numbers within squares highlight the number of overlapping genes. Colors within squares represent the −log(corrected Fisher’s Exact test p value) of the enrichment across brain regions which is depicted in the color key.
Extended Figure 2.
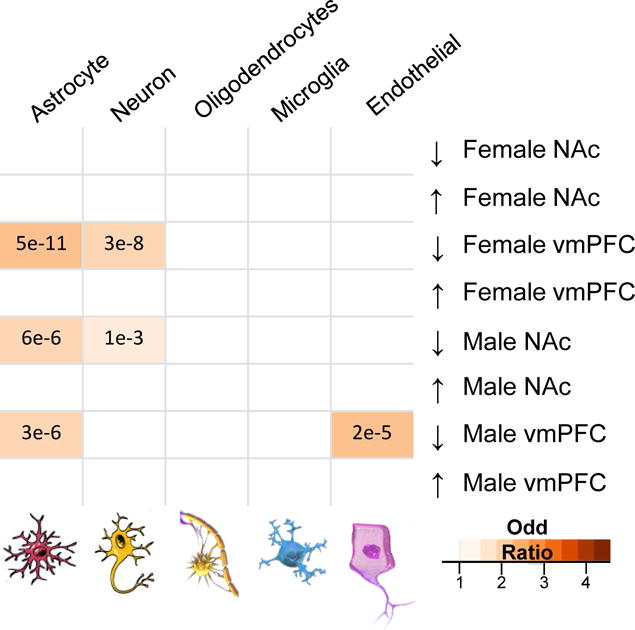
Cell-type specific enrichment for genes differentially expressed in males and females after 21 days of CVS. Numbers in boxes represent the corrected Fisher’s Exact Test p-values of the enrichment. Colors of boxes represent the odds ratio of the cell type enrichment and is depicted in the color bar. Cell types considered in this analysis include astrocytes, neurons, myelinated oligodendrocytes, microglia and endothelial cells.
Extended Figure 3.
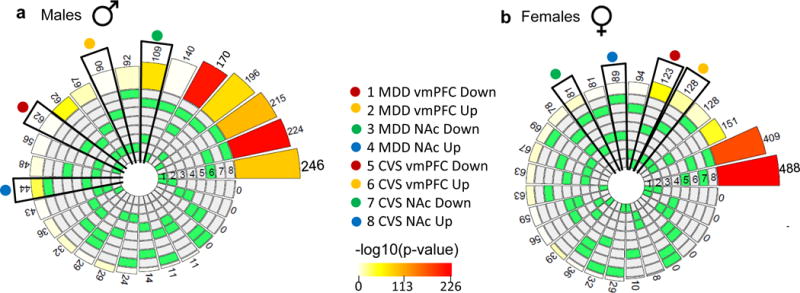
Degree of overlap in the vmPFC and NAc for genes differentially expressed in humans with MDD and mice subjected to 21 days of CVS in a, males and b, females. Green squares in the circos plots represent the two conditions that are compared which are listed below each plot. The size of the bars surrounding the circos plots corresponds to the number of overlapping genes which are also listed for every comparison, and the intensity of the color corresponds to the −log(corrected Fisher’s Exact Test p-value) of the enrichment across conditions as depicted in the color key between circos plots.
Extended Figure 4.
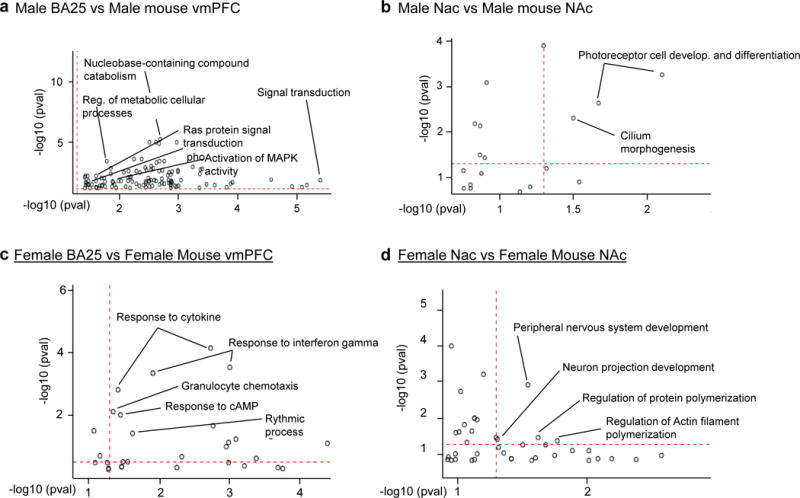
Degree of overlap for gene ontology (GO) terms significantly associated with MDD in humans and 21 days of CVS in mice in the vmPFC and NAc. a, GO terms significantly overlapping in the vmPFC of males with MDD and stressed male mice. b, GO terms significantly overlapping in the NAc of males with MDD and stressed male mice. c, GO terms significantly overlapping in the vmPFC of females with MDD and stressed female mice. d, GO terms significantly overlapping in the NAc of females with MDD and stressed female mice. For each graph, the y axis represents the level of significance (−log pvalue) of the enrichment for GO terms in humans and the x axis represents the level of significance (−log pvalue) of the enrichment for GO terms in mice. The dotted red line marks the threshold of significance (p<0.05).
Extended Figure 5.
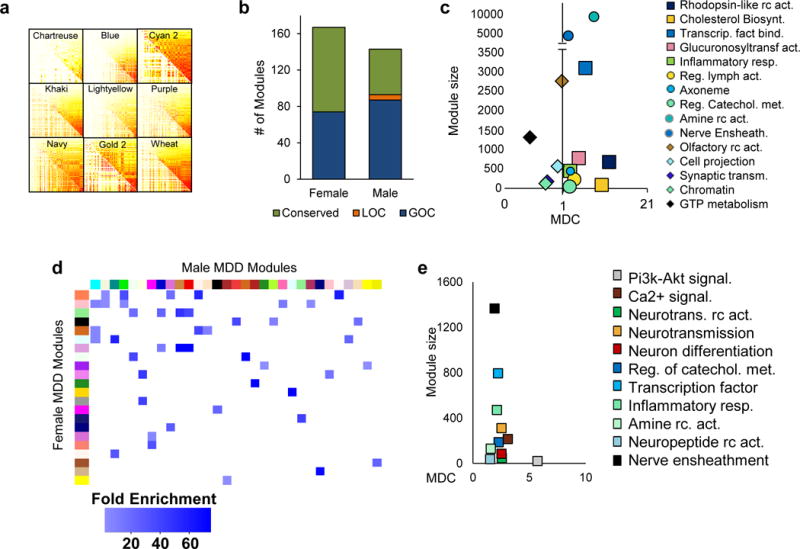
Multi brain region gene co-expression network analysis in humans reveals distinct sex-specific transcriptional signatures in males and females with MDD. a, Individual topological overlap covariance matrices of the top 9 differentially connected modules (FDR<0.1) in males and females with MDD (upper right triangle of each module) vs. that in the control state (lower left triangle of each module). b, Proportion of modules differentially connected (FDR<0.1) (blue: gain of connectivity [GOC], green: conserved connectivity, and orange: loss of connectivity [LOC]) in males and females with MDD vs. controls. The x axis represents the number of modules in every category. c, Most significant enrichment of functional categories in modules showing GOC (square) or LOC (circle) in males with MDD, and GOC (diamond) in females with MDD. The y axis denotes the number of genes per module, whereas the x axis represents module differential connectivity (MDC) values. d, Modules in male MDD that share high levels (corrected Fisher’s Exact Test<0.05; fold enrichment>15) of conservation in females with MDD. Modules are defined by an arbitrary color in female MDD (y axis) and male MDD (x axis). Enrichment (fold enrichment) is defined by the darkness of the blue color with paler blue representing lower enrichment (minimum 15) and darker blue representing higher enrichment. e, Gene ontology (GO) terms associated with male MDD modules sharing high levels of homology with female MDD modules showing a GOC (square). The y axis denotes the number of genes per module, whereas the x axis represents MDC values.
Extended Figure 6.
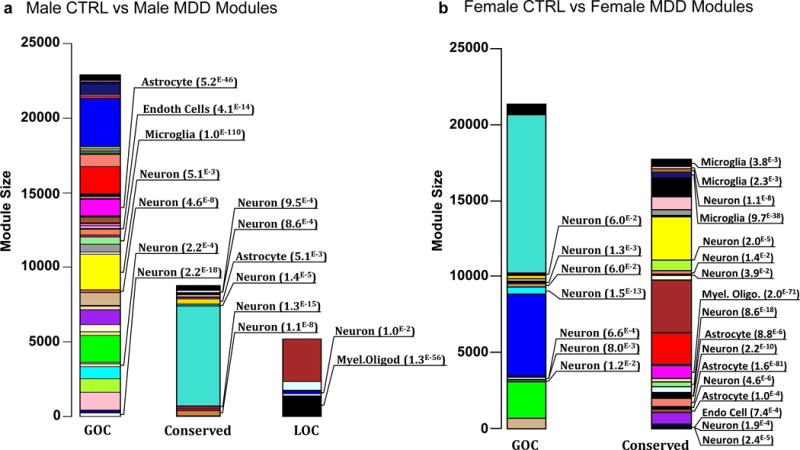
Modules in males and females with MDD are enriched for cell-type specific genes. Stacked bar graphs showing modules enriched for genes expressed in particular cell types in a, male MDD and b, female MDD. The y axis represents module size defined by number of genes within each module. Modules are defined by an arbitrary color name and are clustered by module differential connectivity (MDC) as gain of connectivity (GOC), loss of connectivity (LOC) and conserved. There is no module associated with a LOC in females with MDD. Enrichment is determined by corrected Fisher’s Exact Test p-value and is listed only for modules that reached significance (FDR p<0.05).
Extended Figure 7.
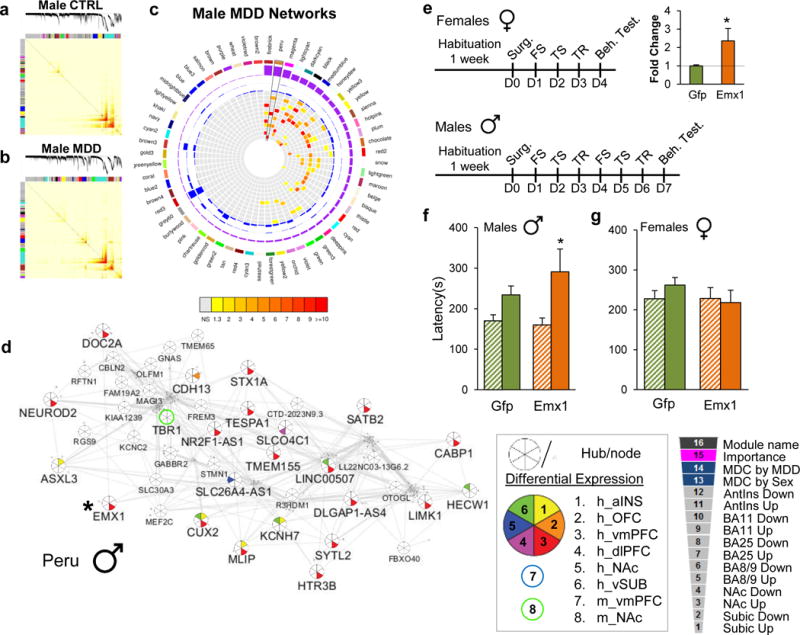
Gene co-expression modules in males with MDD are enriched for DEGs across brain regions. a, Topological overlap matrix (TOM) plots for control and b, MDD modules in males. Light color represents low topological overlap and progressively darker red color represents higher overlap. Each module is assigned by a unique color. c, Circos plots displaying the degree of enrichment for DEGs (p<0.05) in male modules. Colors within squares of the plots represent the corrected Fisher’s Exact Test p-value of the enrichment across modules which are depicted in the color bar below the circos plot. Legend on the bottom right corner defines individual layers of the circos plot. d, Peru module in male MDD shows enrichment for DEGs across brain regions. Hubs and nodes are defined by the size of the pie charts with colors representing enrichment for DEGs across brain regions (depicted in the bottom right panel). The star highlights EMX1, which was selected for sex-specific in vivo phenotypic validation studies. e, Schematic representation of the behavioral paradigm used to assess the impact of EMX1 overexpression in the vmPFC of males and females. Bar graph shows the HSV-mediated overexpression of EMX1 covering the infra- and prelimbic region of vmPFC in male mice. f, Behavioral consequence of EMX1 overexpression in the novelty-supressed feeding (NSF) test in males and g, females. Significance in tests e was determined using student independent sample t-test and in f–g using two-way ANOVA with Tuckey correction. e n=5/condition, f–g n=10/condition. Bars, mean ± sem; * p<0.05.
Extended Figure 8.
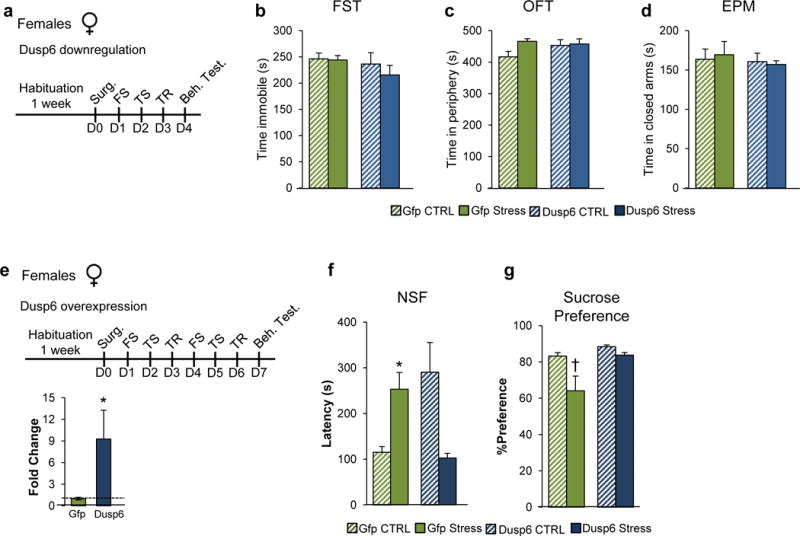
a, Schematic representation of the behavioral paradigm used to assess the impact of DUSP6 downregulation in the vmPFC of females. Behavioral consequences of DUSP6 downregulation in female vmPFC in the b, forced swim test (FST), c, open field test (OFT), and d, elevated plus maze (EPM). e, Schematic representation of the behavioral paradigm used to assess the impact of DUSP6 overexpression in females following sub-chronic variable stress. Bottom graph shows the HSV-mediated overexpression of DUSP6 covering the infra- and prelimbic region of the vmPFC in mice. Behavioral consequences of DUSP6 viral overexpression in the f, novelty-supressed feeding test (NSF) and g, sucrose preference test. Significance in tests e was determined using independent sample t-test while significance in b–d and f–g was determined using two-way ANOVA with Tuckey correction. b–d n =7/condition, e n=5/condition, f–g n=10/condition. Bars, mean ± sem; * p<0.05.
Extended Figure 9.
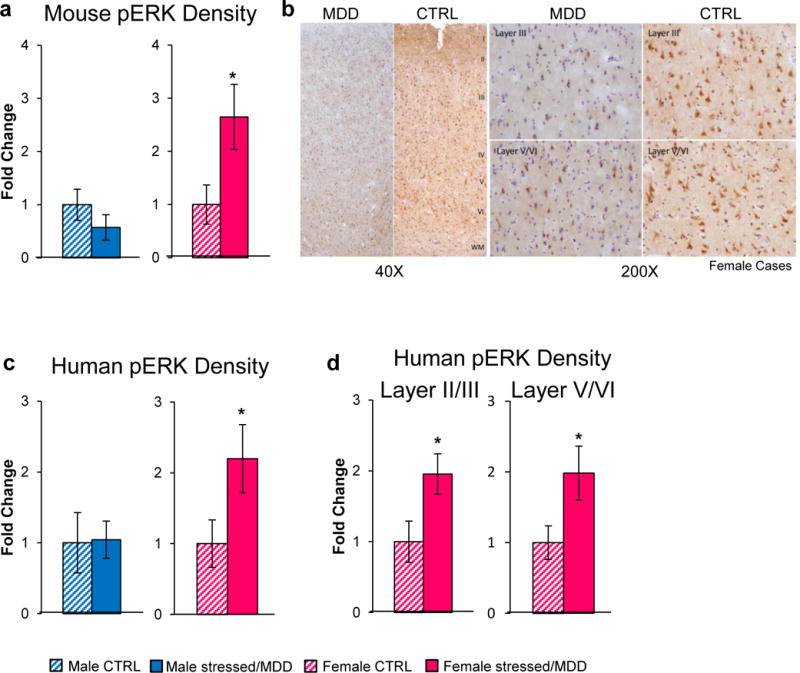
Females with MDD show elevated levels of phospho-ERK1/2 in PFC. a, Quantitative assessment of phospho-ERK1/2 levels measured by IHC in male (blue) and female (pink) mice with and without stress (21 days CVS). b, Representative image of phospho-ERK1/2 DAB staining across PFC cortical layers in postmortem brains (BA11) from females with MDD and healthy control (40×). Right panels show the layer specificity of the effects at a larger magnification (200×). c, Quantification of phospho-ERK1/2-reactive cell density in males (blue) and females (pink) with and without MDD. d, Layer specific (layers II/III and layers V/VI) quantification of phospho-ERK1/2-reactive cell density in females with and without MDD. Significance in tests a, c–d was determined using independent sample t-tests. a n=6–7 cells/condition, c–d n=4/condition. Bars, mean ± sem; * p<0.05.
Extended Figure 10.
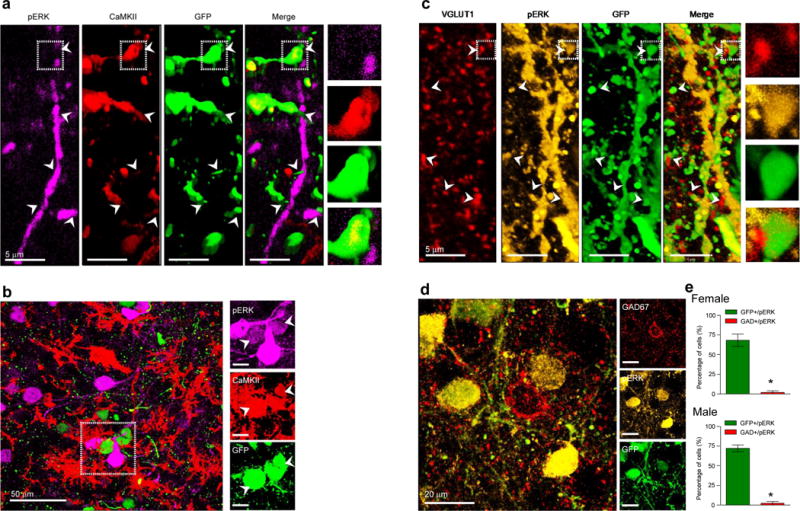
Viral downregulation of DUSP6 in vmPFC increases phospho-1/2 levels in glutamatergic but not GABAergic cells in both males and females. Phospho-ERK1/2 staining in both male and female mice co-localizes in the dendrites of a, CaMKII (red) and c, VGlut1-reactive pyramidal cells. Outer boxes highlight the degree of co-localization of phospho-ERK1/2 in a, CaMKII and c, VGlut1 expressing cells at the synaptic level within virally DUSP6 KD-infected pyramidal cells. b, phospho-ERK1/2 staining in both male and female mice co-localizes in the soma of b, CamKII (red) but not d, GAD67 cells. Outer boxes in b highlight the degree of somatic co-localization of phospho-ERK1/2 in CaMKII expressing cells within virally DUSP6 KD-infected pyramidal cells. e, Quantification of HSV-DUSP6 KD-infected cells in the vmPFC showing GAD67 and phospho-ERK1/2 reactivity in females (upper panel) and males (lower panel). Significance in tests e was determined using student independent sample t-test. e n=4/condition. Bars, mean ± sem; * p<0.05.
Extended Figure 11.
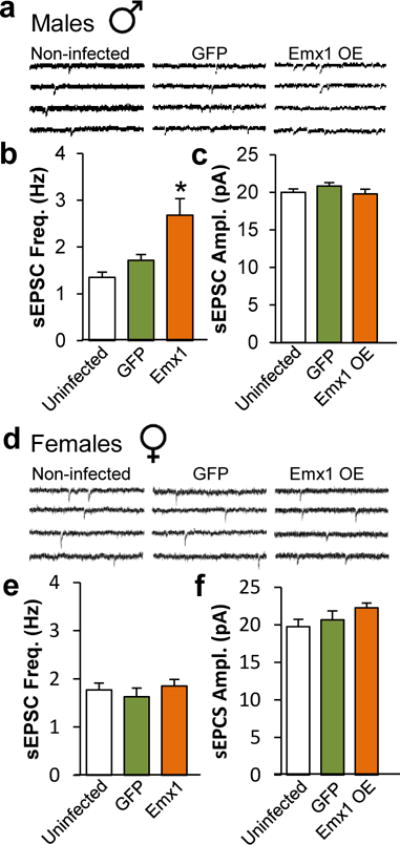
EMX1 overexpression alters the physiological properties of vmPFC pyramidal neurons in a sex-specific fashion. a, Representative traces of neuronal activity in non-infected, GFP and EMX1-infected pyramidal neurons in male mice vmPFC. b, Quantification of EMX1-induced changes of spontaneous excitatory post-synaptic current (sEPSC) frequency in hertz (Hz) and c, amplitude in picoAmpere (pA) in male vmPFC. d, Representative traces of neuronal activity in non-infected, GFP and EMX1-infected pyramidal neurons in female mice vmPFC. e, Quantification of EMX1 effects on sEPSC frequency and f, amplitude in female vmPFC. Significance in tests b–c and e–f was determined using two-way ANOVA with Tuckey correction. b–c, Male non-infected: n=9 mice and n=32 cells, Male GFP-infected: n=5 mice and n=29 cells, Male EMX1-infected: n=4 and n=27 cells. e–f, Female non-infected: n=10 mice and n=25 cells, Female GFP-infected: n=4 mice and n=17 cells, Female EMX1-infected: n=4 mice and n=28 cells. Bars, mean ± sem; * p<0.05.
Extended Figure 12.
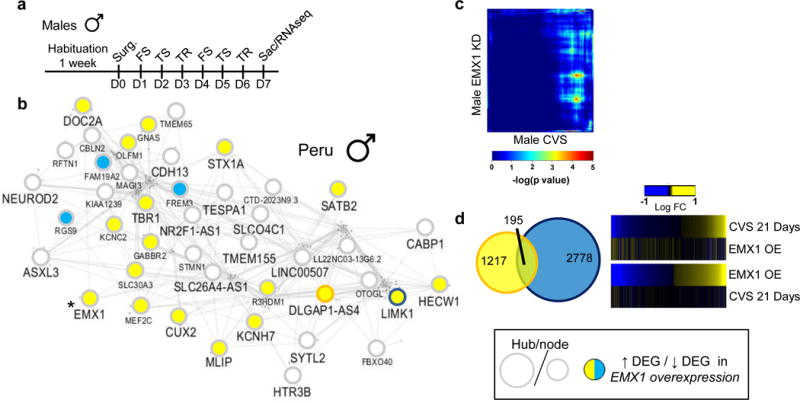
EMX1 overexpression in male vmPFC modifies the transcriptional and network structures induced by stress. a, Schematic representation of the behavioral paradigm used to assess the impact of EMX1 overexpression in male mice. b, Transcriptional reorganization of the male-specific Peru gene network by EMX1 viral overexpression in male vmPFC. Hubs and nodes are defined by the size of the circles with colors representing directionality of differential expression in the vmPFC (depicted in the bottom right panel). c, RRHO map directly comparing transcriptional profiles of males after EMX1 overexpression with 21 days CVS males in the vmPFC. Degree of significance is depicted in the color bar below the RRHO map. d, Venn diagrams displaying the overlap between genes differentially expressed (p<0.05) in males after 21 days of CVS (yellow) and males after EMX1 overexpression (blue) in the vmPFC. Heatmaps on the right compare transcriptional changes (log fold change; over the heatmaps) in males after 21 days of CVS and males after EMX1 overexpression in the vmPFC.
Supplementary Material
Acknowledgments
We thank Dr. Vincent Vialou for his in vivo expertise, Kelly Gleason at The University of Texas Southwestern Medical Center, and Josée Prud’homme and Danielle Cecyre at the Douglas Bell-Canada Brain Bank, for their help with procuring human brain samples and Dr. Omar Jabado for his expert advice on RNAseq.
Footnotes
Supplementary Information
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions
B.L. and E.J.N. conceived the project, designed the experiments, and wrote the manuscript. B.L. also generated and analyzed all the data. B.Z. and L.S. oversaw all bioinformatics analyses. A.K., Y.D., C.T., S.R., N.M. and G.T. also contributed to the study design. O.E., G.M., Z.L., P.J.H., E.C., O.I., H.K., and A.O. contributed to the cloning, in vivo surgeries and behavioral experiments. G.H. M.P., G.M. and A.O. contributed to the behavioral experiments. C.M., C.T., G.M., and M.C. helped with the protein and IHC experiments. J.W. and Y.D. generated the electrophysiological data. I.P., J.R.S., Z.L. and E.L. contributed to the differential expression and network analyses. R.N. packaged the viruses. C.T., N.M. and G.T. contributed brain samples. All authors contributed to the preparation of the manuscript.
References
- 1.Organization., W. H. The Global burden of Disease 2004 update. 2008 [Google Scholar]
- 2.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuehner C. Gender differences in unipolar depression: an update of epidemiological findings and possible explanations. Acta Psychiatr Scand. 2003;108:163–174. doi: 10.1034/j.1600-0447.2003.00204.x. [DOI] [PubMed] [Google Scholar]
- 4.Breslau N. Gender differences in trauma and posttraumatic stress disorder. The journal of gender-specific medicine: JGSM: the official journal of the Partnership for Women’s Health at Columbia. 2002;5:34–40. [PubMed] [Google Scholar]
- 5.Kessler RC, McGonagle KA, Swartz M, Blazer DG, Nelson CB. Sex and depression in the National Comorbidity Survey. I: Lifetime prevalence, chronicity and recurrence. J Affect Disord. 1993;29:85–96. doi: 10.1016/0165-0327(93)90026-g. [DOI] [PubMed] [Google Scholar]
- 6.Kessler RC. Epidemiology of women and depression. J Affect Disord. 2003;74:5–13. doi: 10.1016/s0165-0327(02)00426-3. [DOI] [PubMed] [Google Scholar]
- 7.Kornstein SG, et al. Gender differences in chronic major and double depression. J Affect Disord. 2000;60:1–11. doi: 10.1016/s0165-0327(99)00158-5. [DOI] [PubMed] [Google Scholar]
- 8.Kornstein SG, et al. Gender differences in treatment response to sertraline versus imipramine in chronic depression. Am J Psychiatry. 2000;157:1445–1452. doi: 10.1176/appi.ajp.157.9.1445. [DOI] [PubMed] [Google Scholar]
- 9.Martenyi F, Dossenbach M, Mraz K, Metcalfe S. Gender differences in the efficacy of fluoxetine and maprotiline in depressed patients: a double-blind trial of antidepressants with serotonergic or norepinephrinergic reuptake inhibition profile. Eur Neuropsychopharmacol. 2001;11:227–232. doi: 10.1016/s0924-977x(01)00089-x. [DOI] [PubMed] [Google Scholar]
- 10.Khan A, Brodhead AE, Schwartz KA, Kolts RL, Brown WA. Sex differences in antidepressant response in recent antidepressant clinical trials. J Clin Psychopharmacol. 2005;25:318–324. doi: 10.1097/01.jcp.0000168879.03169.ce. [DOI] [PubMed] [Google Scholar]
- 11.Young EA, et al. Sex differences in response to citalopram: a STAR*D report. J Psychiatr Res. 2009;43:503–511. doi: 10.1016/j.jpsychires.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botteron KN, Raichle ME, Drevets WC, Heath AC, Todd RD. Volumetric reduction in left subgenual prefrontal cortex in early onset depression. Biol Psychiatry. 2002;51:342–344. doi: 10.1016/s0006-3223(01)01280-x. [DOI] [PubMed] [Google Scholar]
- 13.Frodl T, et al. Hippocampal changes in patients with a first episode of major depression. Am J Psychiatry. 2002;159:1112–1118. doi: 10.1176/appi.ajp.159.7.1112. [DOI] [PubMed] [Google Scholar]
- 14.Hastings RS, Parsey RV, Oquendo MA, Arango V, Mann JJ. Volumetric analysis of the prefrontal cortex, amygdala, and hippocampus in major depression. Neuropsychopharmacology. 2004;29:952–959. doi: 10.1038/sj.npp.1300371. [DOI] [PubMed] [Google Scholar]
- 15.Kong L, et al. Sex differences of gray matter morphology in cortico-limbic-striatal neural system in major depressive disorder. J Psychiatr Res. 2013;47:733–739. doi: 10.1016/j.jpsychires.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacerda AL, et al. Anatomic evaluation of the orbitofrontal cortex in major depressive disorder. Biol Psychiatry. 2004;55:353–358. doi: 10.1016/j.biopsych.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am J Psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- 18.Frey BN, Skelin I, Sakai Y, Nishikawa M, Diksic M. Gender differences in alpha-[(11)C]MTrp brain trapping, an index of serotonin synthesis, in medication-free individuals with major depressive disorder: a positron emission tomography study. Psychiatry Res. 2010;183:157–166. doi: 10.1016/j.pscychresns.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Sakai Y, et al. Cortical trapping of alpha-[(11)C]methyl-l-tryptophan, an index of serotonin synthesis, is lower in females than males. Neuroimage. 2006;33:815–824. doi: 10.1016/j.neuroimage.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman J, et al. Quantification of the Serotonin 1A Receptor Using PET: Identification of a Potential Biomarker of Major Depression in Males. Neuropsychopharmacology. 2015;40:1692–1699. doi: 10.1038/npp.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underwood MD, et al. Neuron density and serotonin receptor binding in prefrontal cortex in suicide. Int J Neuropsychopharmacol. 2012;15:435–447. doi: 10.1017/s1461145711000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bangasser DA, Valentino RJ. Sex differences in stress-related psychiatric disorders: neurobiological perspectives. Front Neuroendocrinol. 2014;35:303–319. doi: 10.1016/j.yfrne.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chopra KK, et al. Sex differences in hormonal responses to a social stressor in chronic major depression. Psychoneuroendocrinology. 2009;34:1235–1241. doi: 10.1016/j.psyneuen.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Peeters F, Nicholson NA, Berkhof J. Cortisol responses to daily events in major depressive disorder. Psychosomatic medicine. 2003;65:836–841. doi: 10.1097/01.psy.0000088594.17747.2e. [DOI] [PubMed] [Google Scholar]
- 25.Young EA, Altemus M. Puberty, ovarian steroids, and stress. Ann N Y Acad Sci. 2004;1021:124–133. doi: 10.1196/annals.1308.013. [DOI] [PubMed] [Google Scholar]
- 26.Gallucci WT, et al. Sex differences in sensitivity of the hypothalamic-pituitary-adrenal axis. Health psychology: official journal of the Division of Health Psychology, American Psychological Association. 1993;12:420–425. doi: 10.1037//0278-6133.12.5.420. [DOI] [PubMed] [Google Scholar]
- 27.Heuser IJ, et al. Age-associated changes of pituitary-adrenocortical hormone regulation in humans: importance of gender. Neurobiology of aging. 1994;15:227–231. doi: 10.1016/0197-4580(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 28.Kunugi H, et al. Assessment of the dexamethasone/CRH test as a state-dependent marker for hypothalamic-pituitary-adrenal (HPA) axis abnormalities in major depressive episode: a Multicenter Study. Neuropsychopharmacology. 2006;31:212–220. doi: 10.1038/sj.npp.1300868. [DOI] [PubMed] [Google Scholar]
- 29.Mehta D, Menke A, Binder EB. Gene expression studies in major depression. Curr Psychiatry Rep. 2010;12:135–144. doi: 10.1007/s11920-010-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard R, et al. Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychiatry. 2011;16:634–646. doi: 10.1038/mp.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudary PV, et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci U S A. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duric V, et al. Altered expression of synapse and glutamate related genes in postmortem hippocampus of depressed subjects. Int J Neuropsychopharmacol. 2013;16:69–82. doi: 10.1017/s1461145712000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klempan TA, et al. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry. 2009;14:175–189. doi: 10.1038/sj.mp.4002110. doi:4002110 [pii] 10.1038/sj.mp.4002110 [doi] [DOI] [PubMed] [Google Scholar]
- 34.Sequeira A, et al. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. doi:4001969 [pii] 10.1038/sj.mp.4001969 [doi] [DOI] [PubMed] [Google Scholar]
- 35.Sequeira A, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sequeira A, et al. Gene expression changes in the prefrontal cortex, anterior cingulate cortex and nucleus accumbens of mood disorders subjects that committed suicide. PLoS One. 2012;7:e35367. doi: 10.1371/journal.pone.0035367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans SJ, et al. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci U S A. 2004;101:15506–15511. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- 39.Kang HJ, et al. Gene expression profiling in postmortem prefrontal cortex of major depressive disorder. J Neurosci. 2007;27:13329–13340. doi: 10.1523/jneurosci.4083-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lalovic A, Klempan T, Sequeira A, Luheshi G, Turecki G. Altered expression of lipid metabolism and immune response genes in the frontal cortex of suicide completers. J Affect Disord. 2010;120:24–31. doi: 10.1016/j.jad.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Tochigi M, et al. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neuroscience research. 2008;60:184–191. doi: 10.1016/j.neures.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Wang SS, Kamphuis W, Huitinga I, Zhou JN, Swaab DF. Gene expression analysis in the human hypothalamus in depression by laser microdissection and real-time PCR: the presence of multiple receptor imbalances. Mol Psychiatry. 2008;13:786–799. 741. doi: 10.1038/mp.2008.38. [DOI] [PubMed] [Google Scholar]
- 43.Li JZ, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. 2013;110:9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaPlant Q, et al. Role of nuclear factor kappaB in ovarian hormone-mediated stress hypersensitivity in female mice. Biol Psychiatry. 2009;65:874–880. doi: 10.1016/j.biopsych.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malki K, et al. Pervasive and opposing effects of Unpredictable Chronic Mild Stress (UCMS) on hippocampal gene expression in BALB/cJ and C57BL/6J mouse strains. BMC Genomics. 2015;16:262. doi: 10.1186/s12864-015-1431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripp A, et al. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. Am J Psychiatry. 2012;169:1194–1202. doi: 10.1176/appi.ajp.2012.12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guilloux JP, et al. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Mol Psychiatry. 2012;17:1130–1142. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol Psychiatry. 2015;20:1139. doi: 10.1038/mp.2015.114. [DOI] [PubMed] [Google Scholar]
- 49.Goswami DB, May WL, Stockmeier CA, Austin MC. Transcriptional expression of serotonergic regulators in laser-captured microdissected dorsal raphe neurons of subjects with major depressive disorder: sex-specific differences. J Neurochem. 2010;112:397–409. doi: 10.1111/j.1471-4159.2009.06462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szewczyk B, et al. Gender-specific decrease in NUDR and 5-HT1Areceptor proteins in the prefrontal cortex of subjects with major depressive disorder. Int J Neuropsychopharmacol. 2009;12:155–168. doi: 10.1017/s1461145708009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boldrini M, Underwood MD, Mann JJ, Arango V. Serotonin-1A autoreceptor binding in the dorsal raphe nucleus of depressed suicides. J Psychiatr Res. 2008;42:433–442. doi: 10.1016/j.jpsychires.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bassi S, Seney ML, Argibay P, Sibille E. Elevated Hippocampal Cholinergic Neurostimulating Peptide precursor protein (HCNP-pp) mRNA in the amygdala in major depression. J Psychiatr Res. 2015;63:105–116. doi: 10.1016/j.jpsychires.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seney ML, Tripp A, McCune S, Lewis DA, Sibille E. Laminar and cellular analyses of reduced somatostatin gene expression in the subgenual anterior cingulate cortex in major depression. Neurobiol Dis. 2015;73:213–219. doi: 10.1016/j.nbd.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin LC, Lewis DA, Sibille E. A human-mouse conserved sex bias in amygdala gene expression related to circadian clock and energy metabolism. Mol Brain. 2011;4:18. doi: 10.1186/1756-6606-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 56.Lisowski P, et al. Effect of chronic mild stress on hippocampal transcriptome in mice selected for high and low stress-induced analgesia and displaying different emotional behaviors. Eur Neuropsychopharmacol. 2011;21:45–62. doi: 10.1016/j.euroneuro.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Wallace DL, et al. CREB regulation of nucleus accumbens excitability mediates social isolation-induced behavioral deficits. Nat Neurosci. 2009;12:200–209. doi: 10.1038/nn.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15:1621–1623. doi: 10.1038/nn.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seney ML, et al. The Role of Genetic Sex in Affect Regulation and Expression of GABA-Related Genes Across Species. Frontiers in psychiatry. 2013;4:104. doi: 10.3389/fpsyt.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin LC, Sibille E. Somatostatin, neuronal vulnerability and behavioral emotionality. Mol Psychiatry. 2015;20:377–387. doi: 10.1038/mp.2014.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soumier A, Sibille E. Opposing effects of acute versus chronic blockade of frontal cortex somatostatin-positive inhibitory neurons on behavioral emotionality in mice. Neuropsychopharmacology. 2014;39:2252–2262. doi: 10.1038/npp.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodes GE, et al. Sex differences in nucleus accumbens transcriptome profiles associated with susceptibility versus resilience to sub-chronic variable stress. Journal of Neuroscience. 2015 doi: 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81:484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.consortium, C. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyde CL, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016 doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belzeaux R, et al. Predisposition to treatment response in major depressive episode: A peripheral blood gene coexpression network analysis. J Psychiatr Res. 2016;81:119–126. doi: 10.1016/j.jpsychires.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 67.Bagot RC, et al. Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron. 2016;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breen MS, et al. Gene networks specific for innate immunity define post-traumatic stress disorder. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang P, et al. A systems approach identifies networks and genes linking sleep and stress: implications for neuropsychiatric disorders. Cell reports. 2015;11:835–848. doi: 10.1016/j.celrep.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narayanan M, et al. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Molecular systems biology. 2014;10:743. doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayberg HS, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45:651–660. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 75.Vialou V, Feng J, Robison AJ, Nestler EJ. Epigenetic mechanisms of depression and antidepressant action. Annual review of pharmacology and toxicology. 2013;53:59–87. doi: 10.1146/annurev-pharmtox-010611-134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stein JL, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang B, Horvath S. Statistical Applications in Genetics and Molecular Biology 4. 2005 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 80.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS computational biology. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horvath S. Applications in Genomics and Systems Biology. Springer; 2011. [Google Scholar]
- 82.Ivliev AE, t Hoen PA, Sergeeva MG. Coexpression network analysis identifies transcriptional modules related to proastrocytic differentiation and sprouty signaling in glioma. Cancer Res. 2010;70:10060–10070. doi: 10.1158/0008-5472.can-10-2465. [DOI] [PubMed] [Google Scholar]
- 83.Langfelder P, Mischel PS, Horvath S. When is hub gene selection better than standard meta-analysis? PLoS One. 2013;8:e61505. doi: 10.1371/journal.pone.0061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Groom LA, Sneddon AA, Alessi DR, Dowd S, Keyse SM. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- 85.Muda M, et al. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271:27205–27208. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- 86.Schuurmans C, Guillemot F. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol. 2002;12:26–34. doi: 10.1016/s0959-4388(02)00286-6. [DOI] [PubMed] [Google Scholar]
- 87.Mazzucchelli C, et al. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron. 2002;34:807–820. doi: 10.1016/s0896-6273(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 88.Mazzucchelli C, Brambilla R. Ras-related and MAPK signalling in neuronal plasticity and memory formation. Cellular and molecular life sciences: CMLS. 2000;57:604–611. doi: 10.1007/pl00000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Busca R, Pouyssegur J, Lenormand P. ERK1 and ERK2 Map Kinases: Specific Roles or Functional Redundancy? Frontiers in cell and developmental biology. 2016;4:53. doi: 10.3389/fcell.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rush AJ. The varied clinical presentations of major depressive disorder. J Clin Psychiatry. 2007;68(Suppl 8):4–10. [PubMed] [Google Scholar]
- 91.Quan MN, Zhang N, Wang YY, Zhang T, Yang Z. Possible antidepressant effects and mechanisms of memantine in behaviors and synaptic plasticity of a depression rat model. Neuroscience. 2011;182:88–97. doi: 10.1016/j.neuroscience.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 92.Mailliet F, et al. Protection of stress-induced impairment of hippocampal/prefrontal LTP through blockade of glucocorticoid receptors: implication of MEK signaling. Exp Neurol. 2008;211:593–596. doi: 10.1016/j.expneurol.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 93.Martin KP, Wellman CL. NMDA receptor blockade alters stress-induced dendritic remodeling in medial prefrontal cortex. Cerebral cortex (New York, NY: 1991) 2011;21:2366–2373. doi: 10.1093/cercor/bhr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gourley SL, Kedves AT, Olausson P, Taylor JR. A history of corticosterone exposure regulates fear extinction and cortical NR2B, GluR2/3, and BDNF. Neuropsychopharmacology. 2009;34:707–716. doi: 10.1038/npp.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li N, et al. Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biol Psychiatry. 2011;69:754–761. doi: 10.1016/j.biopsych.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Covington HE, 3rd, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. 2010;30:16082–16090. doi: 10.1523/jneurosci.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ito H, Nagano M, Suzuki H, Murakoshi T. Chronic stress enhances synaptic plasticity due to disinhibition in the anterior cingulate cortex and induces hyper-locomotion in mice. Neuropharmacology. 2010;58:746–757. doi: 10.1016/j.neuropharm.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 98.Wang W, et al. Deficiency in endocannabinoid signaling in the nucleus accumbens induced by chronic unpredictable stress. Neuropsychopharmacology. 2010;35:2249–2261. doi: 10.1038/npp.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marsden WN. Synaptic plasticity in depression: molecular, cellular and functional correlates. Prog Neuropsychopharmacol Biol Psychiatry. 2013;43:168–184. doi: 10.1016/j.pnpbp.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 100.Dwivedi Y, et al. Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. J Neurochem. 2001;77:916–928. doi: 10.1046/j.1471-4159.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- 101.Dwivedi Y, Rizavi HS, Conley RR, Pandey GN. ERK MAP kinase signaling in post-mortem brain of suicide subjects: differential regulation of upstream Raf kinases Raf-1 and B-Raf. Mol Psychiatry. 2006;11:86–98. doi: 10.1038/sj.mp.4001744. [DOI] [PubMed] [Google Scholar]
- 102.Dwivedi Y, et al. Aberrant extracellular signal-regulated kinase (ERK)1/2 signalling in suicide brain: role of ERK kinase 1 (MEK1) Int J Neuropsychopharmacol. 2009;12:1337–1354. doi: 10.1017/s1461145709990575. [DOI] [PubMed] [Google Scholar]
- 103.Cocas LA, et al. Emx1-lineage progenitors differentially contribute to neural diversity in the striatum and amygdala. J Neurosci. 2009;29:15933–15946. doi: 10.1523/jneurosci.2525-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Piper M, et al. Emx and Nfi genes regulate cortical development and axon guidance in the telencephalon. Novartis Foundation symposium. 2007;288:230–242. discussion 242-235, 276-281. [PubMed] [Google Scholar]
- 105.Labuda M, et al. Linkage disequilibrium analysis in young populations: pseudo-vitamin D-deficiency rickets and the founder effect in French Canadians. Am J Hum Genet. 1996;59:633–643. [PMC free article] [PubMed] [Google Scholar]
- 106.Haines D. Neuroanatomy: An Atlas of Structures, Sections, and Systems. 5th. Lippincott Williams & Wilkins; 2000. [Google Scholar]
- 107.Nolte J. The Human Brain: An Introduction to Its Functional Neuroanatomy. 5th. Mosby-Year Book Inc; 2002. [Google Scholar]
- 108.Brent DA, et al. The validity of diagnoses obtained through the psychological autopsy procedure in adolescent suicide victims: use of family history. Acta Psychiatr Scand. 1993;87:118–122. doi: 10.1111/j.1600-0447.1993.tb03341.x. [DOI] [PubMed] [Google Scholar]
- 109.Brewin CR, Andrews B, Gotlib IH. Psychopathology and early experience: a reappraisal of retrospective reports. Psychol Bull. 1993;113:82–98. doi: 10.1037/0033-2909.113.1.82. [DOI] [PubMed] [Google Scholar]
- 110.Conner KR, Conwell Y, Duberstein PR. The validity of proxy-based data in suicide research: a study of patients 50 years of age and older who attempted suicide. II. Life events, social support and suicidal behavior. Acta Psychiatr Scand. 2001;104:452–457. doi: 10.1034/j.1600-0447.2001.00091.x. [DOI] [PubMed] [Google Scholar]
- 111.Dumais A, et al. Risk factors for suicide completion in major depression: a case-control study of impulsive and aggressive behaviors in men. Am J Psychiatry. 2005;162:2116–2124. doi: 10.1176/appi.ajp.162.11.2116. doi: 162/11/2116 [pii] 10.1176/appi.ajp.162.11.2116 [doi] [DOI] [PubMed] [Google Scholar]
- 112.Hawton K, et al. The psychological autopsy approach to studying suicide: a review of methodological issues. J Affect Disord. 1998;50:269–276. doi: 10.1016/s0165-0327(98)00033-0. [DOI] [PubMed] [Google Scholar]
- 113.Kelly TM, Mann JJ. Validity of DSM-III-R diagnosis by psychological autopsy: a comparison with clinician ante-mortem diagnosis. Acta Psychiatr Scand. 1996;94:337–343. doi: 10.1111/j.1600-0447.1996.tb09869.x. [DOI] [PubMed] [Google Scholar]
- 114.McGirr A, et al. Risk factors for completed suicide in schizophrenia and other chronic psychotic disorders: a case-control study. Schizophr Res. 2006;84:132–143. doi: 10.1016/j.schres.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 115.Association, A. P. Diagnostic and statistical manual of mental disorders. (4th) 1994 [Google Scholar]
- 116.Spitzer RL, Williams JB, Gibbon M, First MB. The Structured Clinical Interview for DSM-III-R (SCID). I: History, rationale, and description. Arch Gen Psychiatry. 1992;49:624–629. doi: 10.1001/archpsyc.1992.01820080032005. [DOI] [PubMed] [Google Scholar]
- 117.Isingrini E, et al. Association between repeated unpredictable chronic mild stress (UCMS) procedures with a high fat diet: a model of fluoxetine resistance in mice. PLoS One. 2010;5:e10404. doi: 10.1371/journal.pone.0010404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Santarelli L, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 119.Vialou V, et al. Serum response factor promotes resilience to chronic social stress through the induction of DeltaFosB. J Neurosci. 2010;30:14585–14592. doi: 10.1523/jneurosci.2496-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang CR, Seamans JK, Gorelova N. Electrophysiological and morphological properties of layers V-VI principal pyramidal cells in rat prefrontal cortex in vitro. J Neurosci. 1996;16:1904–1921. doi: 10.1523/JNEUROSCI.16-05-01904.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Akula N, et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry. 2014;19:1179–1185. doi: 10.1038/mp.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Carey V, Gentleman R, Dudoit S, Irizarry R, Huber W, editors. Vol. 1. Springer; 2005. pp. 397–420. [Google Scholar]
- 125.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. doi:gb-2004-5-10-r80 [pii] 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen LS, Storey JD. Eigen-R2 for dissecting variation in high-dimensional studies. Bioinformatics. 2008;24:2260–2262. doi: 10.1093/bioinformatics/btn411. doi:btn411 [pii] 10.1093/bioinformatics/btn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 129.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 130.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/jneurosci.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


