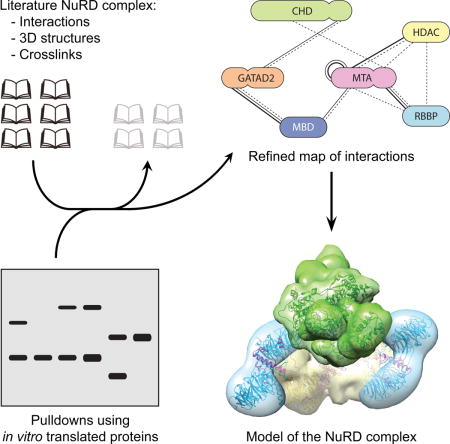
Abstract
The nucleosome remodelling and deacetylase (NuRD) complex is essential for the development of complex animals. NuRD has roles in regulating gene expression and repairing damaged DNA. The complex comprises at least six proteins with two or more paralogues of each protein routinely identified when the complex is purified from cell extracts. To understand the structure and function of NuRD, a map of direct subunit interactions is needed. Dozens of published studies have attempted to define direct inter-subunit connectivities. We propose that conclusions reported in many such studies are in fact ambiguous for one of several reasons. First, the expression of many NuRD subunits in bacteria is unlikely to lead to folded, active protein. Second, interaction studies carried out in cells that contain endogenous NuRD complex can lead to false positives through bridging of target proteins by endogenous components. Combining existing information on NuRD structure with a protocol designed to minimize false positives, we report a conservative and robust interaction map for the NuRD complex. We also suggest a 3D model of the complex that brings together the existing data on the complex. The issues and strategies discussed herein are also applicable to the analysis of a wide range of multi-subunit complexes.
Keywords: Nucleosome Remodelling and Deacetylase (NuRD) complex, Chromatin remodeling, Protein-protein interactions, Protein structure, Co-immunoprecipitations
Enzymes used: Micrococcal nuclease (MNase), EC 3.1.31.1; histone deacetylase (HDAC), EC 3.5.1.98
Graphical abstract
The NuRD complex is essential for gene regulation in complex animals. To understand its structure, a map of direct subunit interactions is needed. We assessed the reliability of published data on this area and experimentally mapped interactions to develop a high-confidence interaction map of the NuRD complex.
Introduction
Large multi-subunit protein complexes with ATP-dependent DNA translocase activity remodel and repair chromatin. There are four major classes of such complexes in mammals: (i) SWI/SNF, (ii) INO80, (iii) ISWI and (iv) CHD families (reviewed in [1–3]). Each class is categorized by sequence similarity in their translocase domain. Complexes from each class appear to have multiple functions in controlling gene transcription and repairing damaged DNA. Disrupting their activity results in significant impairment of growth, development and homeostasis.
The sets of proteins that comprise each class of complex are quite well defined. However, the arrangement of proteins within complexes and the mechanistic details that underpin their function is largely unknown. Low-resolution structures of complete or partial complexes have been derived from negative-stain electron microscopy [4–11] but these have not been sufficient to derive a clear understanding of the remodelling process. It is also likely that these complexes are very dynamic, which hinders analysis by either crystallization or electron microscopy. Innovative approaches will be required to surmount this issue. In the absence of a high-resolution structure, a map of direct contacts between subunits, combined with knowledge of their structure and biochemistry, could provide a snapshot of the architecture of the complex. These data would enable the creation of models to propose and test functional and mechanistic hypotheses.
Inter-subunit connections have been defined by a number of methods; we estimate co-immunoprecipitations (coIPs), in vitro pulldown experiments and yeast two-hybrid analysis to be the most common. Covalent crosslinking combined with mass spectrometry identification (XL-MS) is increasingly popular. Biophysical analysis of purified proteins (e.g., by surface plasmon resonance, NMR or titration calorimetry) is used mostly to confirm interactions defined by other methods. For all approaches, accurate interpretation of observations requires careful consideration of the experimental strengths and weaknesses. The known structure and biochemistry of the proteins concerned must also be taken into account.
The nucleosome remodelling and deacetylase (NuRD) complex is a CHD-class complex that is conserved among complex animals. It is important for development, stem cell biology and DNA repair [12–14]. The complex has six core subunits, each with several paralogues: the DNA translocase CHD4 (or -3 or -5), HDAC1 (or -2), MTA1 (or -2 or -3), GATAD2A (or -B), RBBP4 (or -7) and MBD2 (or -3) (Figure 1). Structural data on subcomplexes of NuRD is becoming available [15–19], but no structure of the intact complex exists. Since 1998, a considerable body of subunit interaction data has been built up that relies on the methods described above. However, we propose that there are issues in these experimental approaches that lead to either false negatives or, more often, false positives. We surveyed the literature to catalogue reported pairwise interactions between NuRD subunits, and assessed the reliability of these methods through our own subunit interaction studies. As a result, herein we present a set of high-confidence direct inter-subunit interactions. With this set we propose a model for the overall architecture of the NuRD complex. We also highlight inherent flaws in methods that are widely considered to be reliable for defining protein-protein interactions.
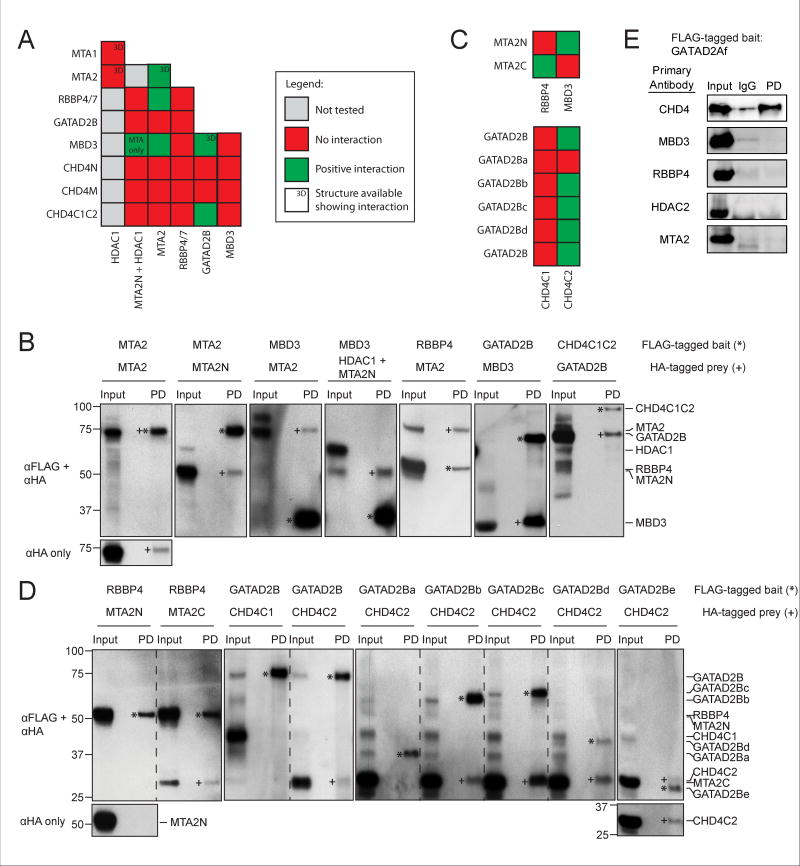
Figure 1. Topology of the NuRD components.
Only one paralogue of each protein is shown. Domains known or predicted to be ordered are shown in colours, and regions predicted to be disordered are shown in grey. Colouring is preserved in other figures. R indicates RBBP-binding motifs. CC indicates coiled coils. Numbering is for the human proteins (UniProt IDs are CHD4: Q14839; MTA2: O94776; GATAD2B: Q8WXI9; HDAC1: Q13547; RBBP4: Q09028; MBD3: O95983). Shorter constructs used in pulldown experiments are indicated with black lines and names in italics. MBD3cc is a construct that fuses MBD3 with the N-terminal coiled-coil domain of GATAD2A. This latter domain forms a dimer with the MBD3 coiled-coil, stabilizing MBD3 [34].
Results
Assessing reported NuRD subunit interactions: choice of expression system
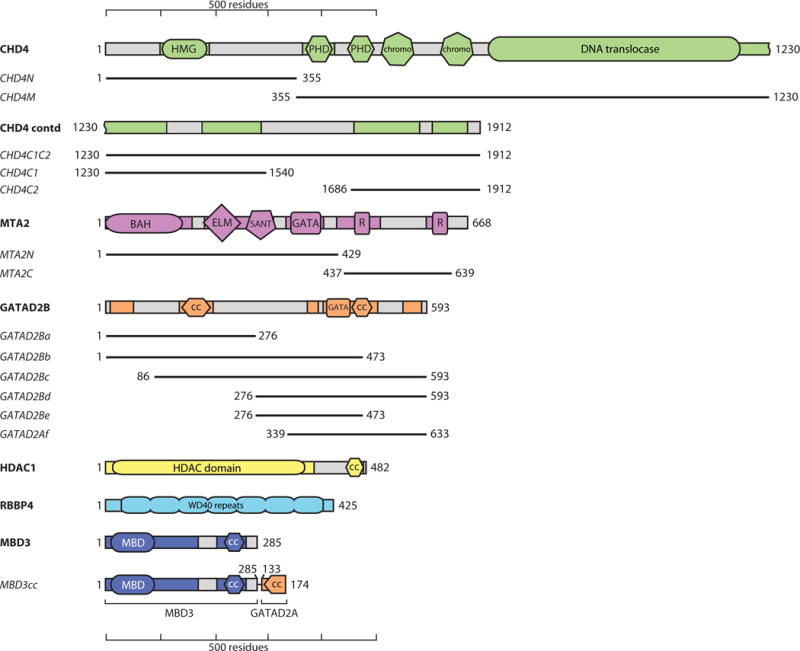
Our survey of the literature found 45 papers describing experiments that define interactions between core NuRD subunits. Table 1, Table 2 and Figure 2 summarise the data from these studies. In several papers (e.g., [20, 21]) proteins were expressed in bacteria, as is common for biochemical and structural analysis of proteins and protein complexes. However, it is often overlooked that the presence of soluble protein from bacterial expression does not guarantee correct folding. Soluble, partially-folded aggregates often form and can bind non-specifically to other proteins [22]. Affinity tags can also impact folding, leading to non-specific interactions [23]. Recently a ‘cloning scar’ in a commercially available ORF library was shown to lead to aberrant co-purification of endogenous proteins [24].
Table 1. Selection of published interactions between NuRD subunits.
The interactions were classified as ‘high confidence’ (high resolution structural data and/or biophysical characterization), ‘low confidence, folding uncertainty’ (when one or both of the interacting proteins are probably not properly folded), or ‘low confidence, bridging uncertainty’ (when the observed interaction is likely to be indirect and is observed because of bridging by endogenous components). In the case of immunoprecipitations and pulldowns, ‘Protein 1’ is the bait, and ‘Protein 2’ the prey. 35S: SDS-PAGE and detection of 35S-labelled proteins; WB: western blot; Dm: Drosophila melanogaster.
| Protein 1 | Protein 2 | Expression System | Method/Detection | Reference |
|---|---|---|---|---|
| High confidence | ||||
|
| ||||
| MBD2/3 | GATAD2A | E. coli | NMR/ITC | [16, 42] |
| HDAC1 | MTA1 | HEK293 cells | X-ray | [15] |
| RBBP4 | MTA1 | Sf9 cells/HEK293 cells | X-ray | [17, 19] |
|
| ||||
| Low confidence, folding uncertainty | ||||
|
| ||||
| MBD3 | MTA2 | E. coli/IVT | GST Pulldown/35S | [37] |
| MBD3 | HDAC1 | E. coli/Sf9 cells | GST Pulldown/WB | [37] |
| MBD3 | RBBP4/7 | E. coli | GST Pulldown/WB | [37] |
| MBD3 | GATAD2B | E. coli/IVT | GST Pulldown/35S | [43] |
| HDAC1 | HDAC2 | E. coli/IVT | GST Pulldown/35S | [30] |
| HDAC1 | CHD4 | Sf9 cells/IVT | FLAG Pulldown/35S | [44] |
| MBD2/3 | GATAD2A/B | HEK293 cells/E. coli/IVT | GST Pulldown/WB/35S | [45] |
| MBD3 | MTA2 | E. coli | GST Pulldown/WB | [21] |
| MBD3 | HDAC1 | E. coli | GST Pulldown/WB | [21] |
| MBD3 | MBD2 | E. coli | GST Pulldown/WB | [20] |
| Dm. MBD2/3 | Dm. p55 (RBBP-like) | E. coli/IVT | GST Pulldown/35S | [46] |
| Dm. MBD2/3 | Dm. MI-2 (CHD4-like) | E. coli/IVT | GST Pulldown/35S | [46] |
|
| ||||
| Low confidence, bridging uncertainty | ||||
|
| ||||
| MBD2 | RBBP7 | HEK293 cells | FLAG Pulldown/WB | [34] |
| MBD2 | MTA2 | HEK293 cells | FLAG Pulldown/WB | [34] |
| MBD2 | HDAC2 | HEK293 cells | FLAG Pulldown/WB | [34] |
| MBD3 | GATAD2A/B | HEK293 cells | GST Pulldown/WB | [29] |
| HDAC2 | CHD4 | HeLa cells | Co-IP/WB | [47] |
| HDAC1 | RBBP4 | SAOS cells | Co-IP/WB | [48] |
| HDAC1 | GATAD2A/B | HEK293 cells | GST Pulldown/WB | [29] |
| RBBP4/7 | GATAD2A/B | HEK293 cells | GST Pulldown/WB | [29] |
| MTA1 | CHD4 | HeLa cells | Co-IP/WB | [49] |
| MTA1 | HDAC2 | HeLa cells | Co-IP/WB | [49] |
| MTA1 | MBD3 | HeLa cells | Co-IP/WB | [49] |
| Dm. p66-like | Dm. MI-2 (CHD4-like) | Dm. embryos | Co-IP/WB | [50] |
Table 2.
Published inter-subunit crosslinks derived from XL-MS data.
| Protein 1 | Protein 2 | Expression System | # crosslinks | Reference |
|---|---|---|---|---|
| RBBP4 | MTA1 | HEK293/HeLa cells | 26 | [19, 40] |
| HDAC1 | MTA1 | HEK293 cells | 19 | [19] |
| CHD3/4 | GATAD2A/B | HeLa cells | 6 | [40] |
| HDAC1 | RBBP4 | HEK293/HeLa cells | 5 | [19, 40] |
| MBD3 | GATAD2A/B | HeLa cells | 5 | [40] |
| MTA1/2 | CHD3/4 | HeLa cells | 4 | [40] |
| MBD3 | MTA2 | HeLa cells | 3 | [40] |
| MTA2 | GATAD2B | HeLa cells | 1 | [40] |
Figure 2. Summary of published interactions and crosslinks between NuRD subunits.
(a) Interactions are classified as “high confidence” (solid line), or “low confidence” (dotted line), according to the criteria in Table 1. (b) Representation of the XL-MS data presented in Table 2, taken from either [40] (red lines) or [19] (blue lines). Proteins are coloured in a similar way to Figure 1.
For a well-studied protein, an absence of biochemical or biophysical data in the literature for bacterially-produced protein is a likely indicator that the protein is not amenable to bacterial expression. For example, several structures of HDAC-containing complexes have been determined recently [15, 19, 25], and two groups have reported structures of RBBP4 and RBBP7 [17, 26, 27]; all have used eukaryotically expressed proteins. Similarly, no structures have been reported for domains in MTA1 other than the HEK293-expressed complex with HDAC1 [15]. Finally, despite extensive trials, our laboratories have never successfully expressed folded HDACs, RBBPs or MTAs (either domains or full-length proteins) in bacteria (JPM and DCW, unpublished data).
A number of published studies have examined inter-subunit interactions in NuRD using bacterially expressed subunits. With the above caveats in mind, it is our view that such observed interactions are in general not reliable. We have graded reported subunit interactions in Table 1 to account for the uncertainty introduced by the use of bacterially expressed proteins and, as discussed in the next section, by bridging interactions.
Assessing reported NuRD subunit interactions: the issue of endogenous components
The delineation of direct interactions between overexpressed components of a multi-protein complex can be compromised by the presence of either the endogenous complex or other interacting proteins in the chosen expression system; for eukaryotic proteins, this issue can be particularly significant for expression in insect or mammalian cells. Endogenous proteins could bridge the two subunits of interest, giving the false impression of a direct interaction. In the same vein, co-immunoprecipitations (coIPs) are often regarded as the ‘gold standard’ for detecting the presence of a physiologically relevant interaction [28]; they are convenient and use small quantities of cells. However, western blots are almost always used for detection. These can be misleading because host-derived proteins (or other bridging macromolecules) that might be present will not be detected.
This issue, which we believe affects many studies describing protein-protein interactions, is illustrated in Figure 3. As part of an effort to probe NuRD complex architecture, we asked whether direct RBBP4-RBBP4 interactions can take place. We co-expressed FLAG tagged RBBP4 (FLAG-RBBP4) and HA-tagged RBBP4 (HA-RBBP4) in HEK293 cells, and anti-FLAG beads were used to capture FLAG-RBBP4. Western blot analysis of the eluate with anti-HA antibody showed that HA-RBBP4 co-purified with FLAG-RBBP4 (Figure 3a), suggesting that RBBP4 forms homo-oligomers. However, when the gel was stained with SYPRO Ruby, numerous other proteins were observed (Figure 3b), even under more stringent conditions (500 mM NaCl, Figure 3c, d). The molecular weights of several of these additional proteins correlate with other NuRD subunits, and their identity was verified with western blots (Figure 3a, c). These data indicate that endogenous NuRD components can associate with overexpressed FLAG/HA-RBBP4, illustrating how a NuRD-type complex can assemble from endogenous and overexpressed components. Therefore, from this set of experiments it cannot be concluded that two RBBPs directly interact. In fact, we and others have demonstrated that MTA1 can simultaneously bind two molecules of RBBP4/7 [18, 19], indicating that the apparent RBBP-RBBP interaction is likely to be indirect – mediated by MTA1/2/3. We have observed a similar phenomenon when probing for other interactions between pairs of subunits, including CHD4 and RBBP7 (Figure 3e).
Figure 3. Endogenous proteins can bridge interactions between expressed proteins.
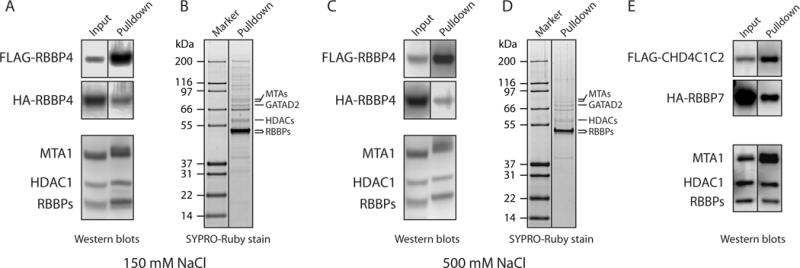
(a) Western blots showing investigation of a possible interaction between FLAG-RBBP4 and HA-RBBP4. The two proteins were co-expressed in HEK293 cells and purified on anti-FLAG beads with 150 mM NaCl washes. The blots were developed with anti-FLAG (top panel), anti-HA (middle panel) or a mixture of anti-MTA1, anti-HDAC1 and anti-RBBP4/7 (lower panel). Proteins detected in each band are indicated. In the middle panel, a band for HA-RBBP4 in the pulled down fraction suggests a direct RBBP4-RBBP4 interaction. However, endogenous NuRD components are detected in the lower blot that was run with a sample from the same pulldown. (b) SYPRO-Ruby stained gel of the purified protein fraction used in (a), showing that a number of endogenous proteins co-purify with FLAG-RBBP4. (c, d) Same as in (a, b), but anti-FLAG beads were washed with 500 mM NaCl during purification. Even under these higher stringency conditions, endogenous NuRD components were observed. (e) CoIP assessing a possible interaction between CHD4C1C2 (residues 1230–1912) and RBBP7. FLAG-CHD4C1C2 and HA-RBBP7 were co-expressed in HEK293 cells and FLAG-CHD4C1C2 was purified with anti-FLAG beads. Detection with anti-FLAG or anti-HA antibodies (top two panels) suggests a direct CHD4-RBBP7 interaction. However, endogenous NuRD components, which could be bridging the interaction, were also detected in the lower blot, which was run with a sample from the same pulldown and developed as in the lower panel in (a).
Many published studies report ‘direct’ interactions between NuRD complex subunits using proteins coexpressed in mammalian cells (Table 1). For example, Gong et al. showed GATAD2A/B interact with GST-tagged RBBP4/7, HDAC1 or MBD3 [29]. Their data consisted of pairwise HEK293 co-transfections and GST affinity pulldowns with western blot analysis. Similarly, Taplick et al. detected endogenous HDAC1 by western blot when purifying recombinant HDAC1 from 3T3 cell lysates [30]. They concluded that HDAC1 self-associates. However, the HDAC1-MTA1 crystal structure (see below, [15]) reveals two molecules of HDAC1 that are bridged by an MTA1 dimer; direct HDAC1-HDAC1 interactions do not occur.
An improved NuRD subunit interaction map derived from in vitro translated proteins
To address the artefacts caused by incorrect folding and endogenous complex components, we expressed NuRD subunits in vitro using rabbit reticulocyte lysate (RRL) with coupled transcription-translation. We reasoned that RRL is mammalian and more likely to produce correctly-folded NuRD components than E. coli. In addition, reticulocytes lack a nucleus and should have reduced quantities of NuRD components. Consistent with this hypothesis, we could not detect HDAC1 or MTA1 in the lysate by western blot, and only negligible quantities of RBBP4/7 (Figure 4a). In contrast, NuRD subunits could be readily detected in HEK293 cell lysates and also in a pulldown from HEK293 cells using FOG1(1–45) (which selectively enriches the NuRD complex from cell extracts [31–33]) as bait.
Figure 4. More reliable NuRD subunit interactions can be derived from in vitro rabbit reticulocyte lysate expression.
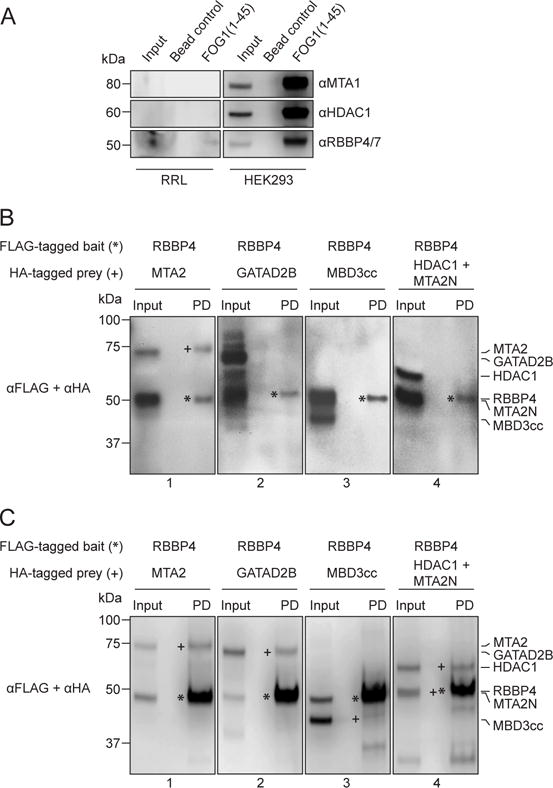
(a) Western blotting with NuRD-specific antibodies (indicated) shows negligible quantities of several NuRD subunits in rabbit reticulocyte lysates (RRL). As a positive control, all tested NuRD subunits could be readily detected in HEK293 cells. (b) Western blots of pulldowns of proteins co-expressed in RRL, using FLAG-RBBP4 as bait and various HA-tagged prey NuRD subunits as prey. The pulldowns show that RBBP4 interacts with MTA2 (Panel 1) but not GATAD2B (Panel 2), MBD3cc (Panel 3) or HDAC1+MTA2N (Panel 4). Bait proteins have been denoted with *, while the prey proteins, if observed in the pulldown lane (PD), are denoted with +. (c) The same experiment as (b), but with proteins co-expressed in HEK293 cells. In this case, RBBP4 appeared to interact with MTA2 (Panel 1), GATAD2B (Panel 2), MBD3cc (Panel 3) and HDAC1+MTA2N (Panel 4).
Using RRL, we conducted a pairwise interaction screen of all NuRD subunits: CHD4, HDAC1, MTA1/2, RBBP4/7, GATAD2B and MBD3. Our experimental design took into account the HDAC1-MTA1(162–335) structure [15]. The extensive HDAC1-MTA1(162–335) interface suggests that HDAC1 expressed in the absence of MTA1(162–335) might fold incorrectly or spuriously bind other proteins. The absence of an ‘HDAC1-alone’ crystal structure also hints at difficulties expressing recombinant HDAC1 (even in eukaryotic hosts). Thus, as a precaution we only assessed HDAC1 interactions with MTA2N (residues 1–429) present.
Figure 4b shows pulldowns carried out with FLAG-RBBP4 as bait and one of MTA2, GATAD2B, MBD3cc (a fusion of MBD3 and the coiled-coil region of GATAD2A, see methods and ref. [34]) or HDAC1+MTA2N as the prey. For comparison, Figure 4c shows pulldowns using the same proteins co-expressed in HEK293 cells. Using the RRL system, RBBP4 interacts with MTA2 (consistent with [17–19]) but not with GATAD2B, MBD3cc or the HDAC1+MTA2N complex. In contrast, and in line with the previous section, RBBP4 appeared to interact with every subunit tested in the HEK293 pulldowns.
Figure 5a summarizes the results of the RRL subunit interaction screen. Due to the large size of CHD4 (~1900 residues), we were unable to express the full-length protein in RRL. Therefore, the protein was divided into three polypeptides (shown in Figure 1): CHD4N (residues 1–355), CHD4M (residues 355–1230) and CHD4C1C2 (residues 1230–1912). The cut sites avoided crossing known domains or predicted secondary structures. CHD4N contains the recently reported HMG-box-like domain [35], CHD4M contains the PHD, chromo- and helicase domains and CHD4C1C2 contains two predicted helical domains with unknown folds or function. Figure 5b shows the western blots for which positive interactions were observed, while the rest of the pulldowns performed are shown in Figures 4b and 6. We observed five pairwise interactions: MTA2 and MTA2, MBD3 and MTA2, RBBP4 and MTA2, GATAD2B and MBD3, and CHD4C1C2 and GATAD2B.
Figure 5. The rabbit reticulocyte lysate subunit interaction screen identifies new interactions.
(a) Summary of all the interaction crosses performed. The ‘MTA only’ label for the MBD3-MTA2N+HDAC1 cross indicates that MBD3 interacted only with MTA2N but not HDAC1. (b) Western blots of positive interactions observed in (a) are shown here. Five pairs of interactions were observed: MTA2-MTA2, MBD3-MTA2, RBBP4-MTA2, GATAD2B-MBD3 and CHD4C1C2-GATAD2B. Bait proteins have been denoted with * while the prey proteins, if observed in the pulldown lane (PD), have been denoted with +. For the MTA2-MTA2 interaction, the corresponding αHA-only blot has been provided to show that the HA-tagged MTA2 bait protein has been pulled down. (c) Summary of all the pulldowns performed with the fragments of MTA2, CHD4 and GATAD2B. (d) Corresponding western blots of all tested interactions as summarised in (c). RBBP4 was observed to interact with MTA2C and GATAD2B with CHD4C2. The interaction between GATAD2B and CHD4C2 was then further narrowed down to the residues 276–473 of GATAD2B (construct GATAD2Be). Bait proteins have been denoted with * while the prey proteins, if observed in the pulldown lane (PD), have been denoted with +. Where the bait and prey proteins are of similar molecular masses, the corresponding αHA-only blot is provided. (e) Immunoprecipitations using FLAG-GATAD2Af (339–633) as bait in transfected HEK293 cells. The western blots were incubated with the antibodies indicated on the left. GATAD2Af is able to pull down CHD4 but not other NuRD components. IgG: negative control using IgG for the immunoprecipitation. Note that in all cases, the relative intensities of bait and prey bands do not correlate with the concentrations of the relevant proteins because of the different activity of the anti-FLAG and anti-HA antibodies.
Figure 6. Western blots from the RRL subunit interaction screen.
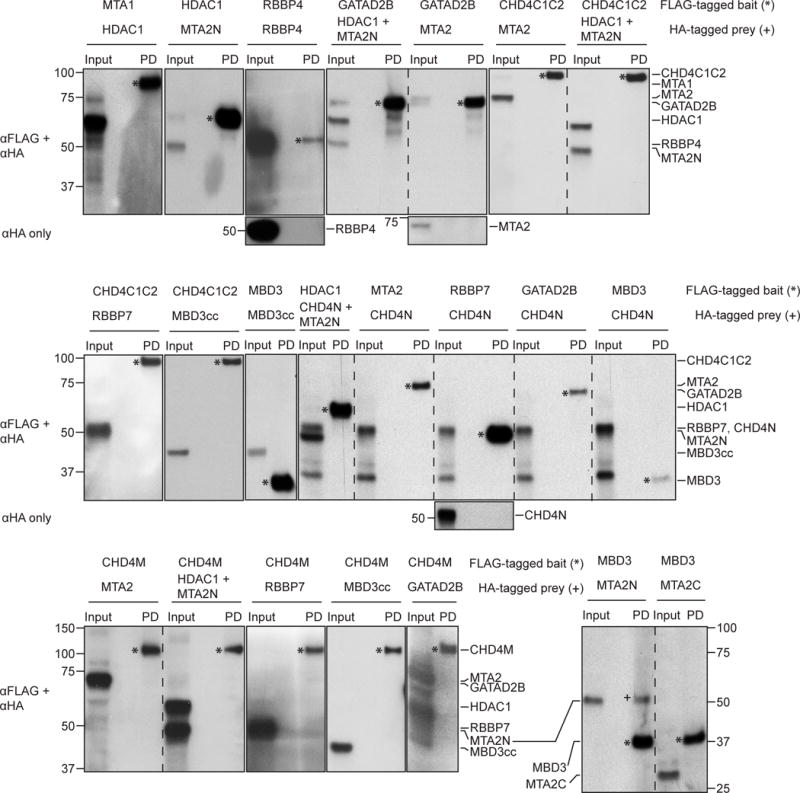
Rabbit reticulocyte lysate pulldowns performed but not shown in Figures 4 and 5 are shown here, including most of the negative interactions. Bait proteins are denoted with * while the prey proteins, if observed in the pulldown lane (PD), have been denoted with +. Where the bait and prey proteins are of similar molecular masses, the corresponding αHA-only blot is provided.
To further refine the set of observed interactions, we divided MTA2, CHD4C1C2 and GATAD2B into shorter fragments (shown in Figure 1): MTA2N (residues 1–429), MTA2C (residues 437–639), CHD4C1 (residues 1230–1540), CHD4C2 (residues 1686–1902), GATAD2Ba (residues 1–276), GATAD2Bb (residues 1–473), GATAD2Bc (residues 86–593), GATAD2Bd (residues 276–593) and GATAD2Be (residues 276–473). Figure 5c summarizes the results of pulldowns with these fragments and Figure 5d shows the corresponding western blots. RBBP4 interacted with MTA2C but not MTA2N, whereas CHD4C2 but not CHD4C1 interacted with GATAD2B. Furthermore, GATAD2Bb, GATAD2Bc, GATAD2Bd and GATAD2Be but not GATAD2Ba bound CHD4. In line with these results, we also observed an interaction between GATAD2Af (residues 339–633) expressed in mammalian cells and endogenous CHD4 (Figure 5e). No other bridging NuRD components were detected in this pulldown, consistent with the interaction being direct. This positive result with full length CHD4 also complements the RRL experiments, given that we were unable to express full length CHD4 in the RRL system.
Surprisingly, HDAC1 did not show an interaction with any NuRD components, including MTA1 or MTA2. To test if this was due to incorrect folding of RRL-produced HDAC1, we conducted a deacetylase assay (Figure 7). Our results showed that RRL-produced HDAC1, when in isolation, has negligible deacetylase activity. When co-purified with MTA2N, some activity is observed, but significantly higher activity is observed when HDAC1 is immunoprecipitated from mammalian cells. From these data, we conclude that a large proportion of RRL-prepared HDAC1 does not form active enzyme, consistent with our observation that HDAC1 did not show reliable interactions with any of the NuRD components, including its high-confidence interaction partner MTA1/2. We do note, however, that the increase in HDAC1 deacetylase activity following co-expression with MTA2N suggests that at least a small fraction of RRL-derived HDAC1 can interact with MTA2. This observation also underscores the high sensitivity of enzyme assays as a means to assess correct folding.
Figure 7. HDAC activity assays show low activity for RRL-produced HDAC1.
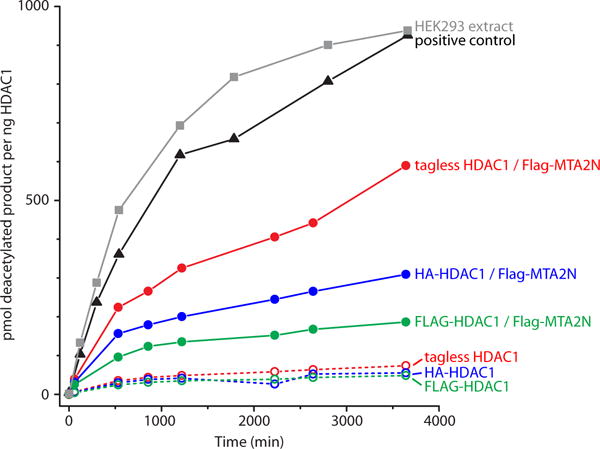
Deacetylase activity is shown at eight time points for samples of immunoprecipitated HDAC1 extracted from HEK293 cells (squares, grey), or produced with rabbit reticulocyte lysate (circles). The versions of HDAC1 tested were: tagless (red), HA-tagged (blue) or FLAG-tagged (green), with (filled circles, solid lines) or without (open circles, dashed lines) co-expression of FLAG-MTA2N. The amount of deacetylase activity observed for the HDAC positive control (supplied by the assay kit’s manufacturer) is also shown (triangles, black). Similar and consistent values were obtained for two to three independent experiments performed with separate samples of each protein, but only one is shown here for clarity.
The folding considerations of RRL-produced HDAC1 raise the question of whether the other NuRD subunits expressed by this method are correctly-folded and able to establish native interactions. However, it is significant that we only observed very few positive hits relative to the large number of negative hits (6 positive hits from 29 crosses, Figure 5a). This suggests some specificity in the observed interactions. In addition, of the six positive hits, three agree well with what we consider as high-confidence published data. The remaining three describe two new interactions (between MBD3 and MTA2, and between CHD4 and GATAD2B) which, as described above, were further analysed and confirmed using a number of different constructs (Figure 5c); the use of different constructs reduces the likelihood of false positive interactions due to misfolding of a particular construct. It is noteworthy that we also observed consistent negative hits in these follow up constructs. Taken together, all these point to the detected positive interactions to be bona fide interactions. If anything else, our setup makes it more likely to have more false negatives than false positive interactions, an example of which is our failure to detect the high confidence MTA-HDAC interaction, as discussed above.
Discussion
How reliable are reported NuRD subunit interactions?
False-positive protein-protein interactions arising from bridging or non-specific interactions can become embedded in the literature through subsequent citation and incorporation into interaction databases and can significantly skew interactome maps, complicating the process of delineating bona fide interactions.
Fundamentally, these problems arise when insufficient controls are conducted. For example, correct folding of proteins used in pulldowns/coIPs is rarely assessed. We suggest that many researchers associate aggregation with precipitation, therefore inferring that soluble proteins are correctly folded. One’s confidence in a reported protein-protein interaction should therefore take into account whether or not evidence of correct folding is given, especially for proteins that do not have structural or biophysical data available. This evidence could be biophysical (e.g., circular dichroism spectropolarimetry or one-dimensional 1H NMR spectroscopy) or functional (e.g., binding to other partner molecules or enzyme activity). Frequently, this might require proteins to be produced and purified in larger quantities, which might not be practical. Corroborating data from other labs showing functional proteins can be expressed and purified in the same system are also valuable.
Overall, we recommend that, when reporting protein-protein interactions by pulldown analysis, the question of whether proteins are correctly folded is addressed whenever possible. Additionally, gels stained for total protein would help detect possible bridging proteins. In our system, approximately one million HEK293 cells (0.25 mL of culture for suspension-adapted cells) produced good-quality SYPRO-stained gels for NuRD interactors. This is a reasonable quantity for most situations. Unfortunately, the quantities of protein typically produced in the RRL system are too small to readily permit biophysical analysis or SYPRO staining. In this study, for example, we were only able to directly assess the folding of HDAC1 using an enzymatic activity assay. However, existing structural and biochemical data served to increase our confidence in the conclusions that we drew about NuRD subunit interactions.
A refined subunit interaction map for the NuRD complex
RRL is a eukaryotic and cell-free system for the production of recombinant proteins. In our experience, balancing the possibility of incorrect folding with bridging issues, we find interactions detected by RRL to be a good compromise that yields higher overall confidence than those detected in cellular expression systems.
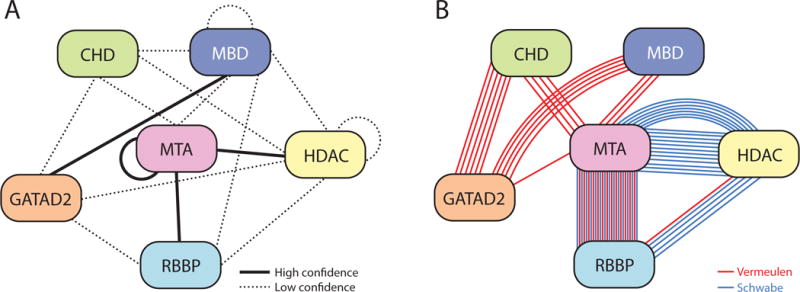
Our results suggest that, contrary to the picture that a survey of the literature paints, the NuRD complex is constructed from relatively few direct inter-subunit interactions. Encouragingly, our data here agree closely with existing (orthogonal) structural data. That is, interactions between RBBP4 and the C-terminal half of MTA1 have been observed in several studies (Table 1 and [17–19]), MBD3 and GATAD2B are known to interact through a short coiled-coil (analogous to the MBD2-GATAD2A interaction [16]) and the ELM-SANT domain region of MTA1/2 homodimerizes in the HDAC-MTA1 structure [15].
The other two observed pairwise interactions, MBD3-MTA2 and CHD4-GATAD2B, have not been structurally characterized. MBD3 was reported to bind MTA2 [21, 37], although one of the studies ([21]) used MTA2 expressed in E. coli, which was probably not well-folded (based on extensive work done on MTA1/2 in our labs; data not shown). CHD4 has not been observed previously to bind GATAD2B or GATAD2A in the mammalian NuRD complex. Here, we have localized this interaction to a predicted helical domain within the C-terminus of CHD4. Our data suggest that GATAD2B(276–473), which contains a GATA-type zinc finger and a predicted coiled coil, is the region interacting with CHD4. It is notable that the COILS server [38] predicts two coiled-coil motifs with moderate confidence within the CHD4C2 region.
To clarify the major contacts made in the NuRD complex, we combined our RRL data with existing high-confidence interactions observed in 3D structures. We also considered information from XL-MS (Table 2 and Figure 2b) [19, 39, 40]. In two studies using XL-MS on intact NuRD complex [39, 40], five interactions were identified by multiple independent crosslinks, namely MTA1/2/3-RBBP4/7 (>10 crosslinks), GATAD2A/B-MBD3 (5), MTA2–MBD3 (3), GATAD2A/B-CHD3/4 (6) and MTA1/2-CHD3/4 (4). The first two corroborate both our RRL data and existing 3D structures. The third and fourth corroborate interactions observed in our RRL experiments. Further, five of the six crosslinks between CHD4 and GATAD2A/B directly reflect our observed interaction between CHD4C2 and GATAD2Bb/c. In addition, XL-MS data for a recombinant HDAC1-MTA1-RBBP4 complex show multiple connections between HDAC1 and both MTA1 and RBBP4 [19]. Crosslinks observed within a multi-protein complex do not unambiguously imply two proteins directly interact. However, agreement between pulldown and XL-MS data from different studies increase one’s confidence that a direct interaction exists.
Figure 8 summarises NuRD subunit interactions from known structures, our RRL screen and reported XL-MS data. Each connection is corroborated by two orthogonal methods, except HDAC-RBBP and CHD-MTA (see below), giving rise to a highly self-consistent map of inter-subunit interactions within the NuRD complex.
Figure 8. Refined subunit interactions map for the NuRD complex.
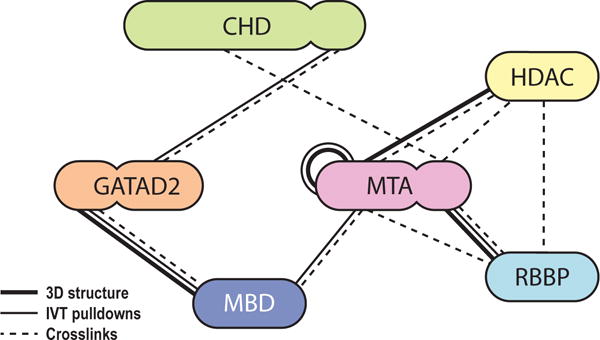
Thick lines: interactions based on 3D structures [15–17, 19]. Thin lines: interactions observed in our screen carried out using RRL-produced proteins. Dashed lines: interactions suggested by at least 3 crosslinks in XL-MS studies [19, 40]. Proteins are coloured in similar way to Figures 1 and 2.
A model of the NuRD complex
Several recent papers report structures of NuRD subcomplexes (Figure 9a, b). Gnanapragasam et al. determined the structure of the MBD2-GATAD2A coiled coil by NMR spectroscopy [16]. Millard et al. showed by X-ray crystallography that HDAC1 forms a dimer of dimers with the MTA1 ELM and SANT domains [MTA1(162–335)] [15]. Subsequently they showed by negative-stain single-particle EM that a longer MTA1 polypeptide [MTA1(162–546)] allows the HDAC1-MTA1 dimer to bind two RBBP4 subunits in a 2:2:2 fashion (HDAC:MTA:RBBP; Figure 9b) [19]. We showed through multiple approaches including single-particle EM that MTA1(449–715) binds two RBBP molecules simultaneously (Figure 9b) [18].
Figure 9. Model of the NuRD complex based on the interaction map and structures of NuRD subcomplexes.
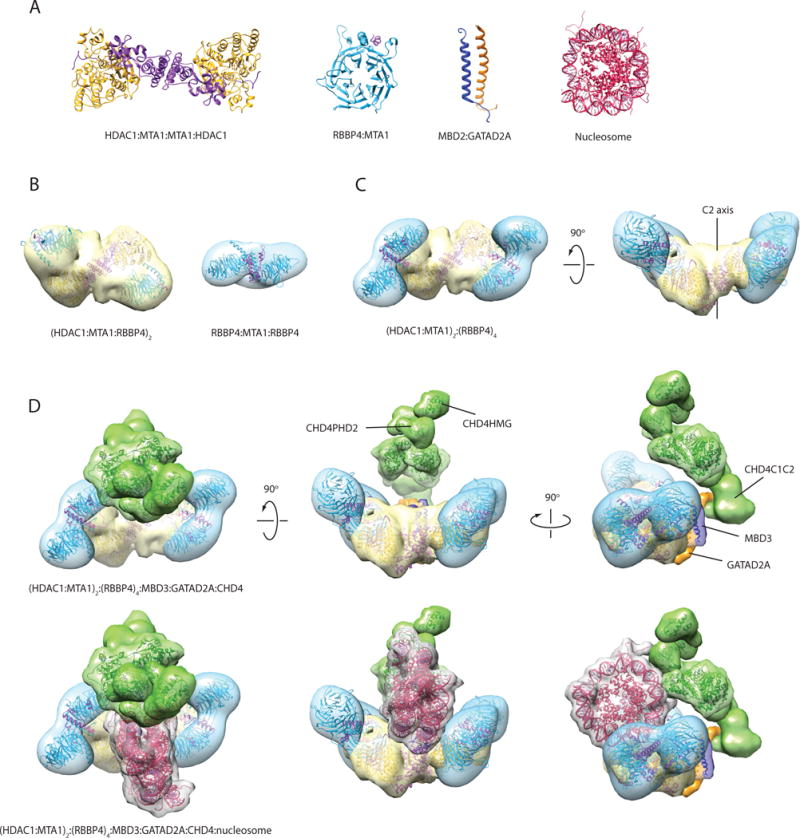
(a) X-ray structure of the HDAC1-MTA1(ELM-SANT) dimer (PDB ID: 4BKX) [15], X-ray structure of RBBP4-MTA1(674–686) (PDB ID: 4PBY) [17], NMR structure of MBD2-GATAD2A coiled-coil (PDB ID: 2L2L) [16], and X-ray structure of the nucleosome (PDB ID: 1EQZ). (b) Single-particle EM envelopes for the (HDAC1-MTA1-RBBP4)2 sub-complex (PDB ID: 5FXY and EMDB ID: EMD-3399) [19] and the MTA1-(RBBP4)2 subcomplex (EMDB ID: EMD-3431) [18]. The structures have been rendered as a reduced resolution envelope by applying a Gaussian filter in Chimera [36]. X-ray crystal structures of HDAC1-MTA1 and RBBP4 have been fitted into the maps using the ‘fit’ function in Chimera. (c) Model of an (HDAC1-MTA1)2-RBBP44 sub-complex generated using two copies of the MTA1-(RBBP4)2 model, and superimposing an RBBP4 subunit from each copy onto each RBBP4 in the (HDAC1-MTA1-RBBP4)2 model. (d) Model of the NuRD complex generated using the subcomplex in (c) and positioning MBD3, GATAD2A and CHD4 in agreement with the observed protein-protein interactions. MBD3 (dark blue) bridges MTA1 and GATAD2A (orange), while GATAD2A contacts the C-terminal third of CHD4 (green). MBD3 is represented by the MBD2:GATAD2A structure and the MBD domain of MBD3 (PDB ID: 2MB7). A density envelope derived from a disordered polypeptide chain was used to depict GATAD2A. CHD4 is represented by the structures of CHD4HMG (PDB ID: 2N5N), CHD4PHD1 (PDB ID: 2L5U), CHD4PHD2 (PDB ID: 1MM2), and the yeast Chd1 chromodomain-ATPase domains (PDB ID: 3MWY); a volume made from two copies of the DNA-binding domain of CHD1 (PDB ID: 4B4C) was used to represent the CHD4C1C2 region. The three bottom views are the same as the top views, with the addition of a manually placed nucleosome (PDB ID: 1AOI). Structures and modelled maps were generated using Chimera. Proteins are coloured as in Figures 1, 2 and 8. It is important to emphasise that, while (a) and (b) show published experimental structures, for (c) and (d) the envelopes shown are hypothetical and built as just described, but not based on novel experimental EM or SAXS data on the respective complexes.
Together, these structural data suggest that a HDAC-MTA-RBBP subcomplex containing full-length MTA has a 2:2:4 stoichiometry. This stoichiometry was also observed recently for the homologous Drosophila melanogaster complex, using multiangle laser light scattering (MALLS) and label-free iBAQ quantitative MS [41]. Two papers from the Vermeulen laboratory also used iBAQ MS to estimate the subunit stoichiometry of the mammalian NuRD complex [39, 40]. These studies reported HDAC:MTA ratios of 1:3 [39] and ~2:2.5 [40], the second of which is more consistent with the ratio inferred above. Using these data, we constructed a simple model of the 2:2:4 HDAC-MTA-RBBP sub-complex that is suggested by the structural data (Figure 9c). This model, like the HDAC1-MTA1 structure, has a two-fold axis of rotational symmetry. It is also likely that the MTA-(RBBP)2 units (blue) exhibit some dynamics relative to the HDAC-MTA unit, perhaps allowing them to engage effectively with nucleosomal substrates during remodelling.
The observed interaction between the CHD4C2 domain and GATAD2 in our RRL screen and in published XL-MS data point to a model in which the MBD-GATAD2 dimer connects CHD4 to the HDAC-MTA-RBBP unit. A similar suggestion has been made based on work on Drosophila NuRD [41]. Further, the observation that immunoprecipitation of GATAD2Af from mammalian cells pulls down CHD4 but no other NuRD components (Figure 5e) suggests that the connection between CHD4 and the rest of the NuRD complex is predominantly through the GATAD2Af region, not via multiple interactions with other subunits.
In contrast, the stoichiometry of GATAD2 and MBD relative to HDAC-MTA-RBBP is less clear. A 1:1 ratio of GATAD2 and MBD is observed in the coiled-coil NMR structure. This ratio is corroborated in the label-free iBAQ quantitative MS of Kloet et al. [40]. The same paper also observes a 2:2 for HDAC:MBD, suggesting two MBD-GATAD2 units might exist in a single NuRD complex. In contrast, however, MALLS and iBAQ data on D. melanogaster NuRD argue for a 2:1 HDAC:MBD ratio and a sub-stoichiometric amount of GATAD2 [41]. A sub-stoichiometric GATAD2 is unlikely; we have previously shown, using blue-native-PAGE (BN-PAGE) and two-dimensional BN-SDS-PAGE that GATAD2 is associated with the NuRD complex in all its forms (including the CHD-less NuDe complex) [32]. This apparent discrepancy might be attributable to biological differences between the mammalian and fly NuRD.
On balance, we favour a HDAC:MBD:GATAD2 ratio of 2:1:1 for two main reasons: (i) the 1:1 MBD:GATAD2 ratio has been observed in a high-resolution NMR structure, a method that is in general prefarable for quantification purposes to MS-based techniques; (ii) despite the possible differences between fly and mammalian NuRD, the HDAC:MBD data obtained by Zhang et al. were based on two orthogonal methods, whereas Kloet et al. had used MBD as the purification ‘handle’ — this could have led to excess MBD that complicated quantification.
Together, these data suggest an overall stoichiometry for the NuRD complex of 2:2:4:1:1:1 (HDAC:MTA:RBBP:MBD:GATAD2:CHD). We have created an indicative model based on these data (Figure 9d) that docks the MBD-GATAD2 dimer with the N-terminal region of MTA, consistent with our RRL pulldown data. We also show a nucleosome in this image and it is easy to imagine how interactions made with the nucleosome by the histone- and DNA-binding sites of RBBP, MBD and MTA subunits could position the single CHD4 translocase enzyme to act on the DNA. It is also likely that the substrate preferences of entities such as the PHD domains of CHD4 and the RBBP subunits will play a role in determining the preferred remodelling targets of NuRD. A more complete model, and from it, a better understanding of the biochemical mechanism by which NuRD remodels chromatin, awaits detailed structural and biophysical analysis of the intact complex.
Materials and Methods
Plasmids
The pcDNA3.1 expression vector was used for suspension-adapted HEK Expi293F™ cell expression and in vitro rabbit reticulocyte lysate transcription-translation. Full-length genes for human HDAC1 (UniProt ID: Q13547) and human CHD4 (UniProt ID: Q14839) with an N-terminal FLAG-tag were kind gifts from Prof. Gerd Blobel. These constructs were sub-cloned to substitute the FLAG-tag for a HA-tag. Full-length genes for human RBBP4 (UniProt ID: Q09028), human RBBP7 (UniProt ID: Q16576), mouse MTA2 (UniProt ID: Q9R190), human MTA1 (UniProt ID: Q13330), mouse GATAD2B (UniProt ID: Q8VHR5), human GATAD2B (UniProt ID: Q8WXI9) and mouse MBD3 (UniProt ID: Q9Z2D8) were cloned with N-terminal FLAG- and HA-tags. We also generated a construct, MBD3cc, consisting of murine MBD3 fused to residues 133–174 of mouse GATA2DA (UniProt ID: Q8CHY6). This region of GATA2DA has been previously shown to form a coiled-coil interaction with MBD3 [42]. We hypothesised that fusing this region of GATA2DA to MBD3 could stabilise and improve expression of MBD3. Shorter constructs were also generated from these full-length genes: MTA2N (1–429), MTA2C (437–639), GATAD2Ba (1–276), GATAD2Bb (1–473), GATAD2Bc (86–593), GATAD2Bd (276–593), GATAD2Be (276–473), CHD4N (1–355), CHD4M (355–1230), CHD4C1C2 (1230–1912), CHD4C1 (1230–1540) and CHD4C2 (1686–1912). For expression in adherent HEK293T cells, the fragment GATAD2Af (339–633) was cloned into the pCMV-Tag2b vector, with an N-terminal FLAG-tag sequence.
HEK293 cell protein expression and lysate preparation
Suspension-adapted HEK Expi293F™ cells (Thermo Fisher Scientific, Waltham, MA, USA) were grown to a density of 2×106 cells/mL in Expi293™ Expression Medium (Thermo Fisher Scientific). Combinations of equimolar quantities of plasmids were co-transfected into cells using linear 25-kDa polyethylenimine (PEI) (Polysciences, Warrington, PA, USA). 3.8 μg of DNA mix was first diluted in 205 μL of PBS and vortexed briefly. 7.6 μg of PEI was then added and the mixture was vortexed again, incubated for 20 min at room temperature, and then added to 1.9 mL of HEK cell culture. The cells were incubated for 65 h at 37 °C with 5% CO2 and horizontal orbital shaking at 130 rpm. 1-mL aliquots of cells were then harvested, washed twice with PBS, centrifuged (300 g, 5 min), snap-frozen in liquid nitrogen and stored at −80 °C. Lysates were prepared by sonicating thawed cell pellets in 0.5 mL of lysis buffer (50 mM Tris-HCl, 150 or 500 mM NaCl, 1% (v/v) Triton X-100, 1× cOmplete® EDTA-free protease inhibitor (Roche, Basel, Switzerland), 0.2 mM DTT, pH 7.9), incubating on ice for 30 min to precipitate chromatin and then clarifying the lysate via centrifugation (≥16,000 g, 20 min, 4 °C). The cleared supernatant was used for FLAG-affinity pulldowns as described below.
Alternatively, adherent HEK293T cells were transfected with the plasmid pCMV-Tag2b-GATAD2Af. At 48 h post transfection, the cells were lysed in micrococcal nuclease (MNase) digestion buffer (25 mM HEPES-KOH pH 7.6, 100 mM NaCl, 5 mM MgCl2, 3 mM CaCl2, 10% (v/v) glycerol, 0.2% (v/v) NP40 and 1× EDTA-free protease inhibitor cocktail (Roche)) and then subjected to MNase digestion using 1500 U/mL of MNase (Worthington Biochemical, Lakewood, NJ, USA) for 2 h on ice. Ethidium bromide was then added to the lysate at 300 μg/mL followed by centrifugation at 10,000 g for 15 min at 4 °C.
In vitro rabbit reticulocyte lysate protein expression
Proteins were expressed in vitro using the TNT® Quick Coupled Transcription/Translation System (Promega, Fitchburg, WI, USA). Groups of one, two or three proteins were co-expressed in the same reaction mixture. 25 μL (one protein), 50 μL (two proteins) or 75 μL (three proteins) of lysate master mix (MM) was used for each reaction. 25 μL of MM contained: 20 U of Ribosafe RNase inhibitor (Bioline, London, UK), 0.7 μL of 1 mM methionine and 1.5 μL cOmplete® EDTA-free protease inhibitor (from 50× stock; Roche). 1.5 μg of each plasmid DNA was used per expressed protein. The reactions were incubated at 30 °C for 4 h. Prior to immunoprecipitation, lysis buffer was added to a final concentration of 50 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) Triton X-100, 1× cOmplete® EDTA-free protease inhibitor (Roche), 1 mM DTT, pH 7.5.
Immunoprecipitation of proteins produced in HEK293 cells
When Expi293F™ cells were used, 20 μL of anti-FLAG Sepharose 4B beads (Biotool, Houston, TX, USA; pre-equilibrated with 50 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) Triton X-100, 1× cOmplete® EDTA-free protease inhibitor (Roche), 0.2 mM DTT, pH 7.5) was added to 0.5 mL of cleared HEK cell lysate. The mixtures were incubated overnight at 4 °C with orbital rotation. Post-incubation, the beads were washed with 5×1 mL ‘wash’ buffer (50 mM Tris-HCl, 150 or 500 mM NaCl, 0.5% (v/v) IGEPAL® CA630, 0.2 mM DTT, pH 7.5). Bound proteins were eluted by 3×20 μL treatment with ‘elution’ buffer (10 mM HEPES, 150 mM NaCl, 300 μg/mL 3×FLAG peptide (MDYKDHDGDYKDHDIDYKDDDDK), pH 7.5) for 1 h at 4 °C. Elution fractions were pooled for downstream analyses.
When adherent HEK293T cells were used, the cleared cell lysate was subjected to immunoprecipitation with anti-flag M2 antibody (Sigma-Aldrich, St. Louis, MO, USA, F1804) and mouse IgG (Santa Cruz Biotechnology, Dallas, TX, USA) control according to the Sigma Flag-IPT kit protocol (Sigma-Aldrich).
Immunoprecipitation of proteins produced in rabbit reticulocyte lysates
20 μL of anti-FLAG Sepharose 4B beads (Biotool) (pre-equilibrated with 50 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) Triton X-100, 1× cOmplete® EDTA-free protease inhibitor (Roche), 1 mM DTT, pH 7.5) was added to 50–75 μL rabbit reticulocyte lysate reactions. The mixtures were incubated overnight at 4 °C with orbital rotation. Post-incubation, the beads were washed with 5×800 μL ‘wash’ buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% (v/v) IGEPAL® CA630, 1 mM DTT, pH 7.5). Bound proteins were eluted by 2×20 μL treatment with ‘elution’ buffer (50 mM Tris-HCl, 150 mM NaCl, 10% (v/v) glycerol, 1 mM DTT, 300 μg/mL 3×FLAG peptide (MDYKDHDGDYKDHDIDYKDDDDK), pH 7.5) for 30 min on ice. Elution fractions were pooled for downstream analyses.
SDS-PAGE and western blot analysis
Samples of Expi293F™ lysates/rabbit reticulocyte lysate reactions, anti-FLAG bead washes and protein elutions were subjected to protein gel electrophoresis using Bolt™ 4–12% Bis-Tris Plus gels (Thermo Fisher Scientific) run in MES buffer at 165 V for 45 min. Gels were washed, fixed and stained with SYPRO Ruby (Thermo Fisher Scientific) according to the manufacturer’s instructions. The gels were scanned with a Typhoon FLA-9000 laser scanner (473 nm excitation, ≥510 nm emission filter; GE Healthcare, Chicago, IL, USA). For western blot analysis, the gel-separated proteins were blotted onto PVDF membranes and probed with antibodies from Cell Signalling Technology, Danvers, MA, USA: anti-FLAG-HRP (2044S, 1:2,000), anti-HA-HRP (2999S, 1:10,000), anti-MTA1 (5647S, 1:1,000), anti-HDAC1 (5356S, 1:1,000), anti-RBBP4/7 (9067S, 1:1,000); from Santa Cruz Biotechnology: anti-mouse-HRP (sc-2096, 1:10,000) and anti-rabbit-HRP (sc-2030, 1:10,000); and from Leinco Technologies, St. Louis, MO, USA: anti-mouse-HRP (M114, 1:10,000). The dilution factors used for each antibody are indicated in parentheses. Whenever needed, the blots were probed with anti-HA-HRP first and imaged before re-incubating with both anti-HA-HRP and anti-FLAG-HRP and re-imaging.
For the experiment using adherent HEK293T cells, the precipitated proteins were analyzed for different components of the NuRD complex by western blot using antibodies against RBBP4 (Novus Biologicals, Littleton, CO, USA, NBP1–40622), HDAC2 (Millipore, Billerica, MA, USA, 05–814), MTA2 (Santa Cruz Biotechnology sc-28731), MBD3 (Thermo Fisher Scientific MA1–41126), CHD4 (Millipore 06–1306) and TrueBlot HRP-conjugated secondaries (Rockland, Limerick, PA, USA).
HDAC activity assays
The IVT-produced samples analysed in the HDAC activity assays were prepared as described above. For the HEK293 extract control, a cleared cell lysate was prepared from 1×106 non-transfected Expi293F™ cells (Thermo Fisher Scientific) as described above. In both cases, HDAC1 was enriched by an anti-HDAC1 immunoprecipitation before the assays as follows. For each sample, 20 μL of protein A/G magnetic beads (Biotool) were washed three times with 500 μL buffer A (50 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) Triton X-100, pH 7.5). Anti-HDAC1 (0.5 μL, Cell Signalling Technology 5356S) was diluted in 25 μL of buffer A, and the mixture added to the washed beads and rotated for 3 h at 4 °C. The beads were then washed twice with 500 μL buffer A, and incubated with the IVT reactions or HEK293 cleared lysates overnight at 4 °C. Post-incubation, the beads were washed with 5×800 μL ‘wash’ buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% (v/v) IGEPAL® CA630, 1 mM DTT, pH 7.5), and finally resuspended in 60 μL of 10 mM HEPES, 150 mM NaCl, 1 mM DTT, pH 7.5.
Deacetylase activity of two–three separate preparations of each sample was measured using a HDAC activity assay kit (Cayman Chemical Company, Ann Arbor, MI, USA) as per the manufacturer’s instructions, using 15 μL of each bead suspension per 170-μL reaction. Eight data points were taken per sample, ranging from 1 to 3,660 min. Fluorescence was quantified using an Infinite M1000 Pro plate reader (Tecan, Zürich, Switzerland). The amount of protein in each assay was quantified from SYPRO-Ruby stained SDS-PAGE gels, using a known amount of monomeric BSA as reference. Western blots (anti-HDAC1) were used to ensure a precise estimation of the amount of HDAC1 in each sample relative to each other. As a positive control, 4 ng of the soluble fraction of the manufacturer-supplied HDAC was used per reaction well. All samples were assayed twice as technical duplicates.
Acknowledgments
This work was supported by National Health and Medical Research Council Grants APP1012161, 1063301, and 571099, Australian Research Council Grant DE120102857, and National Institutes of Health Grant R01-GM098264. We thank Gerd Blobel for supplying plasmids for FLAG-HDAC1 and FLAG-CHD4.
Abbreviations
- SWI/SNF
SWItch/Sucrose Non-Fermentable
- INO80
DNA helicase INO80
- ISWI
Imitation SWI
- CHD
Chromodomain helicase DNA binding protein
- co-IP
co-immunoprecipitation
- NuRD
Nucleosome remodelling and deacetylase complex
- RBBP
Retinoblastoma binding protein
- MTA
Metastasis associated
- HEK
Human embryonic kidney
- FLAG
epitope DYKDDDDK
- HA
human Influenza haemagglutinin epitope YPYDVPDY
- HDAC
Histone deacetylase
- GATAD2
Transcriptional repressor p66
- MBD
Methy-CpG binding domain protein
- PBS
phosphate buffered saline
- PEI
Polyethyleneimine
- DTT
dithiothreitol
- HRP
horseradish peroxidase
- MES
2-(N-morpholino)ethanesulfonic acid
- RRL
Rabbit reticulocyte lysate
- EM
electron microscopy
- XL-MS
Crosslinking combined with mass spectrometry
- ELM
egl-27 and MTA1 homology
- SANT
SWI, ADA2, N-CoR, TFIIIB-B
- IVT
in vitro translation
- GST
glutathione S-transferase
- ITC
isothermal titration calorimetry
- Y2H
yeast two-hybrid system
Footnotes
Author Contributions
M. T. performed most of the mammalian cell pulldowns, performed the HDAC activity assays, analysed data and edited the manuscript.
J. K. K. L. cloned the MTA2C fragment and most of the CHD4 fragment constructs, performed most of the rabbit reticulocyte experiments (and some of the mammalian cell pulldowns), analysed data and edited the manuscript.
A. P. G. S. performed some of the rabbit reticulocyte experiments, analysed data, modelled the NuRD structure and edited the manuscript.
J. W. S. analysed data.
M. S. cloned the GATAD2B fragment constructs and performed the HDAC1 IPs for the HDAC activity assays.
M. S. T. cloned the MBD3cc construct.
E. I. performed pulldown experiments and analysed data.
C. S. W. cloned the MTA2N fragment construct.
C. K. cloned several of the CHD4 fragment constructs.
M. J. B. performed the FOG1(1–45) affinity NuRD complex purification from HEK293 cells.
M. J. S. carried out immunoprecipitation experiments.
D. C. W. Jr. designed and interpreted immunoprecipitation experiments.
N. E. S. performed literature searches, compiled subunit interactions, analysed data and edited the manuscript.
J. P. M. designed experiments, analysed data and edited the manuscript.
References
- 1.Mueller-Planitz F, Klinker H, Becker PB. Nucleosome sliding mechanisms: new twists in a looped history. Nat Struct Mol Biol. 2013;20:1026–32. doi: 10.1038/nsmb.2648. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–96. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manelyte L, Längst G. Chromatin Remodelers and Their Way of Action. In: Radzioch D, editor. Chromatin Remodelling. 2013. pp. 2–28. [Google Scholar]
- 4.Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Structural analysis of the RSC chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2002;99:13477–80. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–7. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–21. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leschziner AE, Lemon B, Tjian R, Nogales E. Structural studies of the human PBAF chromatin-remodeling complex. Structure. 2005;13:267–75. doi: 10.1016/j.str.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci U S A. 2007;104:4913–8. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen VQ, Ranjan A, Stengel F, Wei D, Aebersold R, Wu C, Leschziner AE. Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell. 2013;154:1220–31. doi: 10.1016/j.cell.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–21. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–19. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 13.O’Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans. 2013;41:777–82. doi: 10.1042/BST20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basta J, Rauchman M. The Nucleosome Remodeling and Deacetylase (NuRD) Complex in Development and Disease. Transl Res. 2015;165:36–47. doi: 10.1016/j.trsl.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millard CJ, Watson PJ, Celardo I, Gordiyenko Y, Cowley SM, Robinson CV, Fairall L, Schwabe JW. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell. 2013;51:57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnanapragasam MN, Scarsdale JN, Amaya ML, Webb HD, Desai MA, Walavalkar NM, Wang SZ, Zu Zhu S, Ginder GD, Williams DC., Jr p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc Natl Acad Sci U S A. 2011;108:7487–92. doi: 10.1073/pnas.1015341108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alqarni SS, Murthy A, Zhang W, Przewloka MR, Silva AP, Watson AA, Lejon S, Pei XY, Smits AH, Kloet SL, Wang H, Shepherd NE, Stokes PH, Blobel GA, Vermeulen M, Glover DM, Mackay JP, Laue ED. Insight into the architecture of the NuRD complex: structure of the RbAp48-MTA1 subcomplex. J Biol Chem. 2014;289:21844–55. doi: 10.1074/jbc.M114.558940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidberger JW, Sharifi Tabar M, Torrado M, Silva AP, Landsberg MJ, Brillault L, AlQarni S, Zeng YC, Parker BL, Low JK, Mackay JP. The MTA1 subunit of the nucleosome remodeling and deacetylase complex can recruit two copies of RBBP4/7. Protein Sci. 2016;25:1472–82. doi: 10.1002/pro.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millard CJ, Varma N, Saleh A, Morris K, Watson PJ, Bottrill AR, Fairall L, Smith CJ, Schwabe JW. The structure of the core NuRD repression complex provides insights into its interaction with chromatin. eLife. 2016;5:e13941. doi: 10.7554/eLife.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. J Biol Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 21.Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–35439. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- 22.Wissmueller S, Font J, Liew CW, Cram E, Schroeder T, Turner J, Crossley M, Mackay JP, Matthews JM. Protein-protein interactions: analysis of a false positive GST pulldown result. Proteins. 2011;79:2365–2371. doi: 10.1002/prot.23068. [DOI] [PubMed] [Google Scholar]
- 23.Lee BM, Buck-Koehntop BA, Martinez-Yamout MA, Dyson HJ, Wright PE. Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed β-Sheet and Zinc Binuclear Cluster. J Mol Biol. 2007;371:1274–1289. doi: 10.1016/j.jmb.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banks CAS, Boanca G, Lee ZT, Florens L, Washburn MP. Proteins interacting with cloning scars: a source of false positive protein-protein interactions. Sci Rep. 2015;5:8530. doi: 10.1038/srep08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–40. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, Laue ED, Mackay JP. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48.FOG-1 complex. J Biol Chem. 2011;286:1196–1203. doi: 10.1074/jbc.M110.195842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–85. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey MC, Sokolowski BH. In vivo verification of protein interactions in the inner ear by coimmunoprecipitation. Methods Mol Biol. 2009;493:299–310. doi: 10.1007/978-1-59745-523-7_18. [DOI] [PubMed] [Google Scholar]
- 29.Gong Z, Brackertz M, Renkawitz R. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol Cell Biol. 2006;26:4519–28. doi: 10.1128/MCB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taplick J, Kurtev V, Kroboth K, Posch M, Lechner T, Seiser C. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J Mol Biol. 2001;308:27–38. doi: 10.1006/jmbi.2001.4569. [DOI] [PubMed] [Google Scholar]
- 31.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–78. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low JK, Webb SR, Silva AP, Saathoff H, Ryan DP, Torrado M, Brofelth M, Parker BL, Shepherd NE, Mackay JP. CHD4 Is a Peripheral Component of the Nucleosome Remodeling and Deacetylase Complex. J Biol Chem. 2016;291:15853–66. doi: 10.1074/jbc.M115.707018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saathoff H, Brofelth M, Trinh A, Parker BL, Ryan DP, Low JK, Webb SR, Silva AP, Mackay JP, Shepherd NE. A peptide affinity reagent for isolating an intact and catalytically active multi-protein complex from mammalian cells. Bioorg Med Chem. 2015;23:960–5. doi: 10.1016/j.bmc.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 34.Desai MA, Webb HD, Sinanan LM, Scarsdale JN, Walavalkar NM, Ginder GD, Williams DC., Jr An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res. 2015;43:3100–3113. doi: 10.1093/nar/gkv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva AP, Ryan DP, Galanty Y, Low JK, Vandevenne M, Jackson SP, Mackay JP. The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose) J Biol Chem. 2016;291:924–38. doi: 10.1074/jbc.M115.683227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: an Integrated Modeling System. J Struct Biol. 2012;179:269–78. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 39.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kloet SL, Baymaz HI, Makowski M, Groenewold V, Jansen PW, Berendsen M, Niazi H, Kops GJ, Vermeulen M. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 2014 doi: 10.1111/febs.12972. [DOI] [PubMed] [Google Scholar]
- 41.Zhang W, Aubert A, Gomez de Segura J, Karuppasamy M, Basu S, Murthy A, Diamante A, Drury T, Balmer J, Cramard J, Watson A, Lando D, Lee S, Palayret M, Kloet S, Smits A, Deery M, Vermeulen M, Hendrich B, Klenerman D, Schaffitzel C, Berger I, Laue E. The Nucleosome Remodeling and Deacetylase Complex NuRD Is Built from Preformed Catalytically Active Sub-modules. J Mol Biol. 2016;428:2931–42. doi: 10.1016/j.jmb.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walavalkar NM, Gordon N, Williams DC., Jr Unique features of the anti-parallel, heterodimeric coiled-coil interaction between methyl-cytosine binding domain 2 (MBD2) homologues and GATA zinc finger domain containing 2A (GATAD2A/p66alpha) J Biol Chem. 2013;288:3419–27. doi: 10.1074/jbc.M112.431346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Q, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Zhang Y. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol Cell Biol. 2002;22:536–46. doi: 10.1128/MCB.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 45.Brackertz M, Boeke J, Zhang R, Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J Biol Chem. 2002;277:40958–66. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- 46.Marhold J, Brehm A, Kramer K. The Drosophila methyl-DNA binding protein MBD2/3 interacts with the NuRD complex via p55 and MI-2. BMC Mol Biol. 2004;5:20. doi: 10.1186/1471-2199-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt DR, Schreiber SL. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 1999;38:14711–14717. doi: 10.1021/bi991614n. [DOI] [PubMed] [Google Scholar]
- 48.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 49.Nair SS, Li DQ, Kumar R. A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell. 2013;49:704–718. doi: 10.1016/j.molcel.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kon C, Cadigan KM, da Silva SL, Nusse R. Developmental roles of the Mi-2/NURD-associated protein p66 in Drosophila. Genetics. 2005;169:2087–100. doi: 10.1534/genetics.104.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]