Abstract
Neuregulins, a four-member family of epidermal growth factor-like signaling molecules, have been studied for over two decades. They were first implicated in schizophrenia in 2002 with the detection of linkage and association at the NRG1 locus followed after a few years by NRG3. However the associations with disease have not been very consistently observed. In contrast, association of NGR3 variants with disease presentation, specifically the presence of delusions, has been more consistent. This appears to be mediated by quantitative changes in the alternative splicing of the gene, which has also been consistently observed. Additional diseases and phenotypes, psychiatric or not, have also been connected with NRG3. These results demonstrate two important aspects of behavioral genetics research. The first is that if we only consider simple risk and fail to examine the details of each patient’s individual phenotype, we will miss important insights on the disease biology. This is an important aspect of the goals of precision medicine. The second is that the functional consequences of variants are often more complex than simple alterations in levels of transcription of a particular gene, including, among others, regulation of alternative splicing. To accurately model and understand the biological consequences of phenotype - associated genetic variants, we need to study the biological consequences of each specific variant. Simply studying the consequences of a null allele of the orthologous gene in a model system, runs the risk of missing the many nuances of hypomorphic and/or gain of function variants in the genome of interest.
Keywords: schizophrenia, delusions, psychosis, neuregulin, NRG3, gene, association, linkage, gene regulation, gene expression, alternative splicing, precision medicine
INTRODUCTION
Neuregulin 3 (NRG3) has received growing attention over the last 10 years for its possible role in schizophrenia. While the function and processing of NRG3 has not been as well studied as its homolog, NRG1, their similarity and functional overlaps allow us to draw many parallels. I will start this review by summarizing our current knowledge of the neuregulin family, sometimes drawing from knowledge on NRG1, as a way to better understand NRG3 and its possible roles in disease. I will continue with specific information on the NRG3 gene and protein and summarize the connections that have been made with psychotic phenotypes and, in particular, schizophrenia.
THE NEUREGULIN GENE FAMILY
Neuregulins (NRGs) are a multi-member family of Epidermal Growth Factor (EGF)-like signaling molecules involved in cell-cell communication [Britsch, 2007]. Historically, depending on specific functions that have been attributed to them, members of the NRG family have been referred to as Neu Differentiation Factor (NDF) [Wen et al., 1992], Heregulins [Holmes et al., 1992], Glial Growth Gactor (GGF) [Marchionni et al., 1993], Acetylcholine Receptor-Inducing Activity (ARIA) [Falls et al., 1993] and Sensory and Motor Neuron-derived Factor (SMDF) [Ho et al., 1995]. This diverse array of names and corresponding functions is indicative of the versatility and importance of neuregulins in the human brain. There are 4 human neuregulin paralogs NRG1–4. Of these, NRG1 is the most extensively studied while NRG4 is the least well known. All 4 genes produce multiple isoforms through alternative splicing [Falls, 2003; Ponomareva et al., 2005; Carteron et al., 2006; Hayes et al., 2007] and are expressed in multiple tissue types. NRG1,2 and 3, are known to have significant roles in the development and function of the central nervous system [Mei and Xiong, 2008; Ponomareva et al., 2005; Zhang et al., 1997; Mei and Nave, 2014].
All neuregulins have N-terminal EGF-like domains and signal through tyrosine kinase receptors of the ErbB family including EGF-R, ErbB1, ErbB2, ErbB3, and ErbB4 [Britsch, 2007]. ErbB receptors function as either a homo- or hetero- dimer, however ERBB2 and ERBB3 have lost their capabilities for ligand interaction and kinase activity requiring heterodimerization with other ERBB receptors to generate signals [Citri and Yarden, 2006]. This multitude of neuregulin genes, splice forms and receptors create tremendous diversity, which coupled with variable receptor specificities [Sweeney et al., 2000], would better be understood and studied at the level of systems biology [Citri and Yarden, 2006], something that has not yet been accomplished.
An appreciation of the complexity generated through alternative splicing can be gained from studies on NRG1 [Falls, 2003; Steinthorsdottir et al., 2004]. Dozens of NRG1 transcripts have been described and grouped into 3 major types I, II and III [Falls, 2003]. All have the N-terminal EGF-like domain, while types I and II also contain an Immunoglobulin (Ig) - like domain but differ in other type-specific N-terminal sequences. Further, type I functions in paracrine signaling (membrane bound but cleaved from the cell surface) or as a cytosolic signaling molecule. Similarly, the N-terminus of type II can be cleaved from the cell surface or secreted, while type III remains attached to the cell membrane, signaling only to immediately neighboring cells [Falls, 2003]. Steinthorsdottir et al [Steinthorsdottir et al., 2004] expanded this classification describing NRG1 transcript types IV, V and VI. Liu et al [Liu et al., 2011] showed that the expression of different NRG1 transcripts varies during development and according to brain activity. Alternative splicing of NRG3 has not been characterized in as much detail, however multiple studies from others [Carteron et al., 2006; Kao et al., 2010; Paterson et al., 2016] and us [Zeledon et al., 2015] have confirmed the existence of multiple alternative splice forms. A neural-specific splice variant of NRG3 was first described by Carteron et al [Carteron et al., 2006] while Kao et al later described multiple NRG3 transcripts encoding structurally distinct protein isoforms. The levels of these splice variants are predicted, in part, by the genotype of a DNA variant (rs10748842) [Kao et al., 2010] that our group associated with delusion severity in schizophrenia [Chen et al., 2009]. Most recently, Paterson et all [Paterson et al., 2016] confirmed the effect of rs10748842 genotype, as well as the existence of multiple isoforms and their variability across development and in psychiatric disease. Subsequently, we also described allelic correlations with specific NRG3 splice forms, further highlighting their importance [Zeledon et al., 2015].
NRG3 GENE STRUCTURE
Neuregulin 3 was identified by Zhang et al in 1997 [Zhang et al., 1997] as an ERBB4 ligand enriched in neural tissue. In a study of ERBB receptor binding affinities of EGF domains, Jones et al [Jones et al., 1999] used ERBB receptor extracellular domains fused to immunoglobulins (IgGs) and confirmed that NRG3 only binds to ERBB4 - and ERBB2/4 IgGs, is in contrast with NRG1 whose isoforms bind to ERBB3 and ERBB4. NRG3 has a much higher affinity for ERBB4 than any of the other neuregulins [Jones et al., 1999]. The NRG3 gene maps to10q22-q23 [Gizatullin et al., 2000] at coordinates chr10:81,875,314–82,985,645 on the hg38 built of the human genome (Figure 1). It is a large gene spanning 1.11 Mb which based on UCSC genome browser data puts it on the 0.5 percentile of the genes in the genome in terms of size, similar in size to NRG1 but ~5 and 15 fold larger that NRG2 and NRG4, respectively. Unlike other neuregulin genes, NRG3 is flanked on either side by gene deserts greater than 1Mb. Gene deserts are also present around NRG1 but are only 400–600 Kb. The structure of NRG3 as reported in the UCSC Genome Browser is shown on Figure 1, including some of the observed alternative transcripts and an observed anti-sense transcript. Single nucleotide polymorphisms (SNPs) discussed in this paper are also shown. Of the observed or likely promoters only the most 5′ is at a CpG island. At the protein level, NRG3 isoform 1 (NCBI accession # NP_001010848) is 696 amino acids (aa), isoform 2 is 695 aa (NP_001159444) and isoform 3 is 499 aa (NP_001159445) with a significantly shorted N-terminus.
Figure 1.
The NRG3 gene as it appears on the UCSC genome browser (build hg38). Six alternative transcripts with their ENSEMBLE IDs and an antisense transcript are shown in the GENCODE v.24 track. SNPs discussed in this review are shown: schizophrenia associations in red, delusion factor associations in blue and others in green.
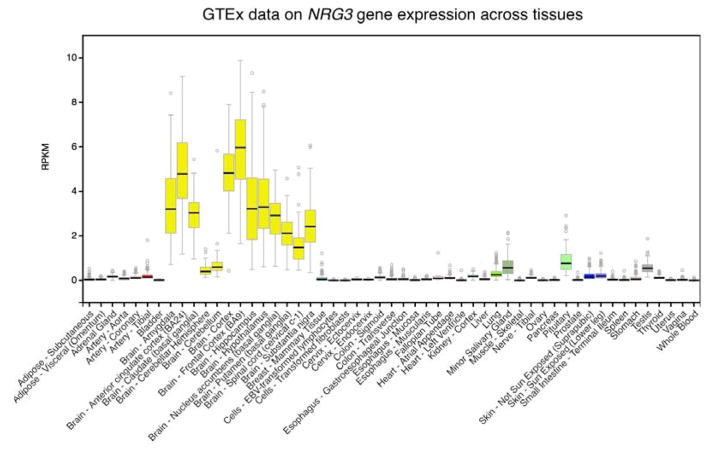
The original paper identifying NRG3 [Zhang et al., 1997] showed exclusive expression in the brain (excluding testis) by Northern blot analysis of RNA from 16 tissues not including mammary gland. In the Genotype-Tissue Expression (GTEx) project data (www.gtexportal.org and Figure 2) NRG3 is almost exclusively expressed in the nervous system with highest expression in the anterior cingulate and frontal cortex. The list of tissue types and developmental stages in GTEx is of course not complete. Some NRG3 expression in the developing breast and in breast cancer has also been reported [Howard, 2008; Dunn et al., 2004]. GTEx data show significant brain expression for all neuregulins but brain specificity is strongest for NRG3. Paterson et al using a PCR based approach have also described isoform specificity including isoforms expressed outside the CNS [Paterson et al., 2016].
Figure 2.
Data from GTEx on the expression of NRG3 across tissues
NRG3 PROTEIN DOMAINS AND DOWNSTREAM INTERACTING PARTNERS
When Zhang et al [Zhang et al., 1997] first identified NRG3 they also characterized a major transcript of the gene. The corresponding protein domains they identified based are shown in Figure 3. This transcript encoded a NRG3 protein that showed high similarity to specific NRG1 isoforms, particularly the SMDF type which lacks an Ig-like domain [Ho et al., 1995] and appears to correspond to type III NRG1 according to Falls [Falls, 2003]. There are two hydrophobic domains in the N-terminal segment of the protein, of which the most N-terminal likely serves as an internal signal sequence and the more C-terminal as a transmembrane domain [Zhang et al., 1997]. The extracellular domain contains the EGF domain, which is distinct from that of NRG1 and specifically binds ERBB4 [Zhang et al., 1997]. While it has been shown that NRG1 undergoes proteolytic cleavage in the transmembrane domain [Fleck et al., 2012; Willem, 2016] mediated by beta and gamma-secretases [Savonenko et al., 2008; Dejaegere et al., 2008] with the release of the EGF domain, this post-translational processing has not yet been explored for NRG3. Moreover, it is not known if NRG3 participates in “back-signaling” by the C-terminal domain, as has been shown for NRG1 [Bao et al., 2003].
Figure 3.
Domains of NRG3 as described by Zhang et. Amino acid positions are shown above. H/Ph: N-terminal hydrophobic segment, S/T rich: serine/threonine-rich domain, TM: predicted transmembrane domain.
Figure 4 summarizes the better studied downstream signaling of NRG3, after activation of ERBB4 by binding the EGF domain. The pathway information used for Figure 4 is from the NIH Cancer Genome Anatomy Project, BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta/h_erbB4pathway) and the cited literature. Binding of NRG3 (or other EGF domain proteins) leads to ERBB4 autophosphorylation as well as tyrosine phosphorylation of other cellular proteins [Zhou and Carpenter, 2002]. This activates PKC and ectodomain cleavage of ERBB4 by the metalloprotease ADAM17 (also known as TNF alpha converting enzyme; TACE) followed by an intramembranous cleavage by gamma-secretase [Ni et al., 2001] (a component of which, PSEN1, is shown here). This second cleavage releases the ERBB4 intracellular domain from the membrane into the cytoplasm, which is followed by its translocation to the nucleus and downstream transcription regulation [Ni et al., 2001].
Figure 4.
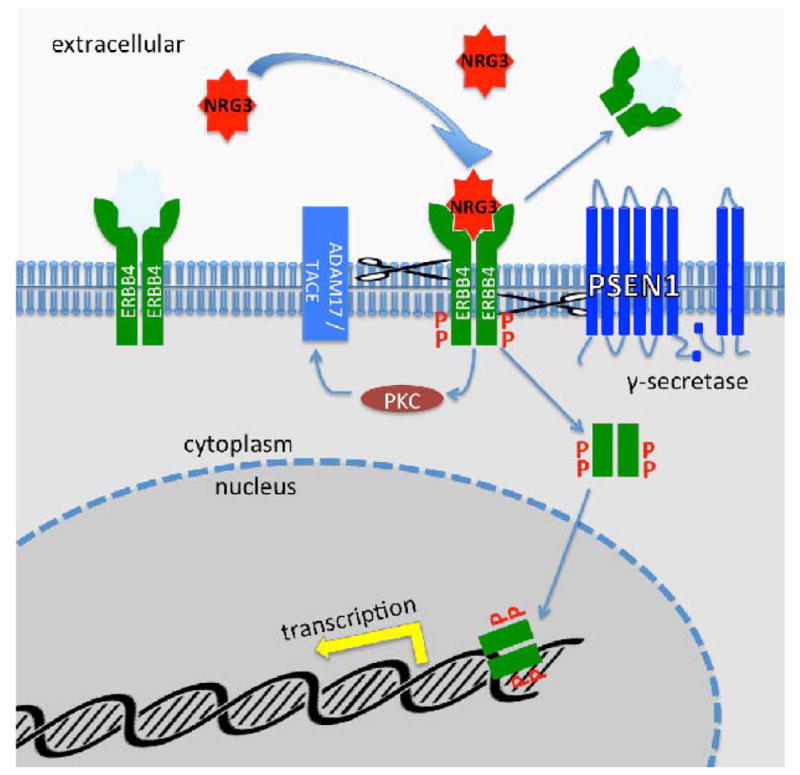
Downstream processing after binding of NRG3 to ERBB4 receptors. After binding ERBB4 is activated by autophosphorylation and phosphorylates other targets resulting in activation of ADAM17 which sheds its extracellular domain and Gamma-secretase which releases its intracellular domain. The latter translocates to the nucleus where it participates in transcriptional regulation.
The NRG3 structure outlined above and in Figure 3 corresponds to the main transcript described by Zhang et al [Zhang et al., 1997]. Other splice forms will differ in presence and spacing or these domains in the protein product including EGF, as well as their processing and function. This provides possible mechanisms for associations between regulatory variation and phenotype. However, until further research clarifies these relationships, this model remains only a hypothesis.
NEUREGULINS AND PSYCHIATRIC DISEASE
Identified in 1992 [Holmes et al., 1992] and studied extensively for its roles in the nervous system, the first neuregulin to draw the attention of psychiatric genetics research was NRG1, after a large study on an Icelandic population revealed linkage and association with schizophrenia at the NRG1 locus [Stefansson et al., 2002]. Many studies since then have shown mixed results. A meta analysis [Munafò et al., 2006] supported the association but only at the level of an NRG1 haplotype and the authors warned on the need for very large samples to dissect this result. The most recent and largest genome wide association study for schizophrenia did not implicate the NRG1 locus [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014], indicating either an absence of association with the disease risk per se or a pattern more complex than the additive effect model investigated by the GWAS. Additional studies from us and others have suggested that there may be interactions or variants involving more than one genes in the NRG/ERBB pathway that increase disease risk [Norton et al., 2006; Benzel et al., 2007; Walsh et al., 2008; Hatzimanolis et al., 2013]. Other studies have also found differences in the expression of NRG in certain brain regions of schizophrenic individuals, but this observation has not been uniform and not always in the same direction [Hashimoto et al., 2004; Law et al., 2006; Parlapani et al., 2010; Weickert et al., 2012; Nicodemus et al., 2009]. Despite these inconsistencies, NRG1 remains an important gene for nervous system development and function and likely to play roles in schizophrenia. For excellent reviews of the connections between NRG1 and schizophrenia see [Mostaid et al., 2016; Mei and Xiong, 2008; Mei and Nave, 2014].
NEUREGULIN 3 ASSOCIATIONS WITH SCHIZOPHRENIA
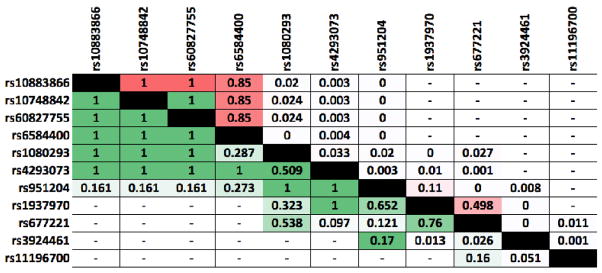
The NRG3 locus on 10q22.3 was first linked to schizophrenia by a study on pedigrees of Ashkenazi Jewish decent in 2003 [Fallin et al., 2003]. Two years after that, in a study of 64 candidate genes including NRG3, we reported association of SNP rs1080293 within the gene with schizophrenia [Fallin et al., 2005]. Later, Benzel et al in addition to a classic association study, investigated statistical interactions between SNP genotypes across components of the ERBB-NRG signaling network [Benzel et al., 2007]. They reported disease-associated SNPs in ERBB4, NRG1, NRG2, NRG3 and EGFR, as well as significant evidence of interaction between NRG3 and NRG1. The NRG3 associated SNPs, rs4293073 and rs3924461, were ~60 and 500 Kb away from the one we previously reported (see Figure 1). The methodology of their interaction analysis does not allow a clear assessment of overlaps with other variants we describe. In a later study we examined neuregulins 1 and 3 as part of a close-knit pathway, including ERBB4 and the genes producing beta and gamma secretases known to cleave NRGs and ERBB4 [Hatzimanolis et al., 2013]. In this rare-variant, sequencing-based study we found that damaging variants in genes - members of the pathway were co-occurring more often than expected in certain patients. This suggested a within-pathway burden of disease and identified a high-burden subgroup that also showed distinct phenotypes. NRG3 was included in the pathway, however the results did not support it as a participant to this phenomenon. The association of NRG3 variants with schizophrenia was also reported in a Chinese cohort [Wang et al., 2008] involving two SNPs, rs1937970 and rs677221 (r2 = 0.68 in Chinese/Japanese, source: http://archive.broadinstitute.org/mpg/snap) in intron two of isoforms 1 & 2. These SNPs however were not in LD with any other reported SNPs in terms of r2 (see Figure 5 for pairwise LD information in Caucasians on these and all other SNPs mentioned in this review and shown in Figure 1). Two more positive studies reported an association between rs10883866 and/or correlated SNPs and schizophrenia, [Kao et al., 2010] and [Morar et al., 2011], There have also been three negative studies [Zhang et al., 2013; Pasaje et al., 2011; Chen et al., 2009] and the genomic region does not show significant signals in the biggest genome wide association study for schizophrenia to date of 150,00 samples [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014]. Therefore, the involvement of these variants with risk for schizophrenia remains in question.
Figure 5.
Linkage Disequilibrium between NRG3 SNPs mentioned in this review and shown in Figure 1. R-squared is above the diagonal and D-prime below, and higher values are highlighted red and green respectively. Dashes indicate no data due to SNP distances >500 Kb
In 2009 we published a follow up study on the previously reported 10q22 linkage [Chen et al., 2009] with results that showed a modified story regarding the effects of NRG3 variation that seems to hold up more consistently than the disease associations. We covered the entire linkage region of 12.5 Mb with 1,414 SNPs in an Askenazi Jewish cohort. In addition to the binary phenotype of schizophrenia (affected/unaffected) we expanded our study to use as outcomes the factor scores from a principal components factor analysis we had recently completed [McGrath et al., 2009]. The results for the binary schizophrenia phenotype were not compelling although one of the best signals (rs1080293 at p=10−3) was located in intron 1 of NRG3. This is the same SNP as reported by Fallin et al [Fallin et al., 2005] yet note that this is a highly overlapping dataset. When we looked for associations across the 9 heritable factors derived from 73 items from our diagnostic ratings and assessment interviews we saw a remarkable signal in intron 1 for the “delusion” factor involving 3 SNPs (rs10883866, rs10748842, and rs6584400, all pairwise r2 >0.85 in Caucasians) located within 13 kb and reaching a p-value of 2.30 × 10−7. This suggested that the genotype at NRG3 had a strong influence on the phenotypic features of the disease, despite lack of significant influence on the risk to become affected. Weaker signals in NRG3 were observed for two other factors, disorganized (rs951204) and scholastic (rs11196700), while another signal for hallucinations was located in the 3′ gene desert. This was, to our knowledge, one of the first studies to explore and identify a strong association with the phenotypic presentation rather than the presence of a psychiatric disease. Importantly, replications of these results followed. First, Kao et al [Kao et al., 2010] found significant association of these same SNPs with patient delusions and positive symptom severity as well as an association with disease risk. They also showed that one of these variants, rs10748842 is in a conserved region and predicts the brain expression of developmentally regulated NRG3 transcripts differing in their 5′ exon, suggesting a mechanism for the association. Somewhat later Morar et al [Morar et al., 2011] replicated once more the association of these SNPs, this time with Schneiderian first-rank symptoms, very similar to our ‘delusion factor’. Additionally, they found these variants were associated with the degraded-stimulus continuous performance task (DS-CPT) and speculated that NRG3 genotypes may be modulating early attentional processes for perceptual sensitivity and vigilance. Interestingly this effect in opposite directions between cases and controls. Following these results, Meier et al [Meier et al., 2013] tried to replicate and extend them to bipolar disorder. They evaluated genotype at rs6584400 for associations with psychotic symptoms using the Operational Criteria Checklist for Psychotic Illness (OPCRIT) [McGuffin et al., 1991] and attention performance using the Trail Making Test (TMT) [Periáñez et al., 2007]. They found an association with OPCRIT psychotic symptoms in schizophrenia and with TMT score in both schizophrenia and bipolar disorder thus replicating and extending the previous findings.
NEUREGULIN 3 FUNCTIONAL VARIATION
The interesting genetic association results in NRG3, its perceived important functions in brain development and its extensive alternative splicing, prompted investigators to explore regulation of NRG3 expression, particularly as it related to the phenotype-associated SNPs. Kao et al [Kao et al., 2010] reported that NRG3 expression is increased in schizophrenia brains and that the variants identified earlier by us are expression Quantitative Trait Loci (eQTLs), regulating specific alternative transcripts. We also followed up on our initial finding looking for the functional variants underlying the association. We performed an extensive characterization of the 162 kb linkage disequilibrium block including the delusion associated SNPs in 47 AJ schizophrenia patients at the extremes of the distribution of the delusion factor score. We found multiple variants including the originally identified rs10883866 and rs60827755 (r-squared=1), which showed allele-specific differences in driving reporter gene expression in vitro. We also found that carriers of the risk allele (C) at rs10748842 have a decreased expression of transcripts containing downstream alternative exon 1 in both the DLPFC (N=94 human postmortem samples) and the superior temporal gyrus (N=190). This is the exon used for transcripts type II and III as described by Kao et al and the observed effect is in the same direction.
More recently, Paterson et al [Paterson et al., 2016], using postmortem human dorsolateral prefrontal cortex, quantified isoform classes I-IV of NRG3 for 286 controls individuals from gestational week 14 to 85 years old, 34 bipolar patients and 69 major depression patients. They found individually specific expression trajectories of the 4 classes of transcripts across development and through life. In addition to disease specific differences they also found that alternative transcripts producing isoforms II and III varied according to the genotype at rs10748842 and were brain specific, which is a third replication of the previous results. Therefore, the evidence that the expression of NRG3 alternative transcripts is important in the presentation of psychosis in schizophrenia is significant.
Other studies have extended the evidence on the importance of variation in NRG3 for brain function. An interesting study on evolution and how the human genome has been shaped by varying environmental factors, highlighted NRG3 and other schizophrenia candidate genes as likely to have biological relevance in adapting to the multiple factors that vary across geographical latitude [Di Gaetano et al., 2013]. Another study highlighted the importance of NRG3 in signaling related to pro-neuronal survival and neuritic elongation, a function mediated by an Elk-1 binding site 1 Kb upstream of NRG3 which promotes its expression [Plani-Lam et al., 2015]. Tost et al reported a highly significant increase difference in ventrolateral prefrontal cortex activation between rs10748842 genotypes, with higher activation for the low delusion C allele [Tost et al., 2014]. Finally in a magnetoencephalography study of 210 healthy siblings Salmela et al found evidence for linkage at the NRG3 locus with variation of the human parieto-occipital 10-Hz rhythmic activity [Salmela et al., 2016]. Of note, this linkage region contains other important brain function related genes including GRID1 and ATAD1.
ANIMAL MODELS
Validation of NRG3 as a schizophrenia-relevant gene has more recently come from experiments in mice. Loos et al [Loos et al., 2014] used recombinant-inbred and congenic mice to map impulsive behavior to the mouse Nrg3 locus. They found that increased impulsivity correlated with increased Nrg3 gene expression in the medial prefrontal cortex. They also showed that mice overexpressing Nrg3 showed more impulsivity, while the opposite was observed in loss of function mutants. Paterson and Law [Paterson and Law, 2014] using a synthetic peptide with the NRG3 EGF domain that crosses the blood brain barrier showed that overexposure of mice to this NRG3-EGF peptide during early postnatal development (postnatal days 2–10) produced an anxiogenic-like phenotype coupled with impaired social behavior in adulthood. Finally we recently reported on a novel Nrg3 knockout mouse [Hayes et al., 2016] which exhibited multiple schizophrenia related phenotypes. The mouse was engineered to lack exon 2 leading to premature stop codons and we confirmed the lack of Nrg3 protein in homozygous animals. These displayed novelty-induced hyperactivity, impaired prepulse inhibition of the acoustic startle response, and deficient fear conditioning while they lacked gross cytoarchitectonic or layer abnormalities.
These NRG3 animal models share one thing in common, the manifestation of traits and behaviors that are related to human psychiatric phenotypes. The phenotypes differ in each case, but so do the models, and none of them recapitulates the effects of genetic variation observed in humans. Altogether, these models add to the evidence supporting that NRG3 is an important gene whose disruptions can be involved in psychiatric disease.
NEUREGULIN 3 BEYOND SCHIZOPHRENIA
Since GWAS have started to leverage large samples and provide robust results, it has become increasingly accepted that psychiatric/behavioral phenotype show significant genetic overlaps [Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013]. It is also clear that genes and variants sometimes show pleiotropic effects in that they are associated with multiple often unrelated phenotypes [Sivakumaran et al., 2011]. NRG3 is no exception to this pattern. We have already mentioned above associations with attention deficits in schizophrenia and bipolar disorder [Meier et al., 2013] and disease specific splice form expression trajectories in Affective Disorders [Paterson et al., 2016]. There is also convincing evidence including GWAS for involvement of NRG3 in smoking behavior [Loukola et al., 2014; Turner et al., 2014], Alzheimer’s disease [Wang et al., 2014] and body mass index [Lee et al., 2016]. Moving beyond links to behavior, NRG3 has been implicated in Hirschsprung’s disease (a disorder of the of the enteric nervous system) by three studies, one involving copy number variation [Tang et al., 2012], another based on exome sequencing [Yang et al., 2013] and a common variant association study [Wang et al., 2016]. It has further been implicated in prolongation of the electrocardiogram’s QT interval (time for depolarization and repolarization of the ventricles) during iloperidone treatment [Volpi et al., 2009] and through interactions with other genes in susceptibility to tuberculosis [Daya et al., 2014].
CONCLUSIONS
NRG3 followed NRG1 into the spotlight of schizophrenia research, but has its own unique history, showing us how genetic variation may influence the specifics of a phenotype - not only the risk per se for disease. The association of NRG3 with specific traits within the schizophrenia phenotype has been consistent, with three independent positive studies with strong results and no negative reports. This stresses how the accurate and detailed phenotyping of each patient will help us identify genes and variants that determine it and move us a step closer to precision medicine. Even if we do not cure or prevent the disease, modifying the symptoms that lead to disability would be a valuable service to the patients.
The links of the phenotype-associated variation to NRG3 alternative splicing have also been consistent. Combined with the extensive developmental regulation of NRG3 they emphasize the importance of accurately identifying the effects of DNA variation and modeling it exactly in our efforts to understand the resulting phenotypes. Knocking out or overexpressing NRG3 has already provided interesting results but is not directly relevant the increased delusions of the patients and the effects of rs10883866 which involve specific transcripts with likely diverse consequences across development.
On the other hand, the evidence for an association of NRG3 variants with disease risk is not conclusive. The associations are not always replicable, and the gene does not show up in the biggest genome wide association study for schizophrenia to date [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014]. Many studies have suggested interactions of variation in NRG3 and related genes and these need to be further explored. It is also important to remember that the lack of conclusive association of common variants in a gene with disease risk does not exclude the involvement of the gene in the disease. For example, PSEN1, an established Alzheimer’s disease gene with ~100% penetrance for certain alleles, does not show association in large GWAS despite its clear connection to the risk. Lack of common variation substantially increasing risk is not synonymous to lack of relevance. As our tools for genetic research become more powerful, particularly genome sequencing, functional variant characterization and genome editing, and as we expand our views on the consequences of genetic variation, we will find ourselves in a much better place for understanding psychiatric disease in the not so distant future.
The uncertainties rising from genetic epidemiology studies may be alleviated by recent advances in genome editing technologies that make it possible to study the consequences of a disease associated variant within its native genomic context [Pham et al., 2016] and even in combination with others. While, in the case of humans, one can not apply this in an intact organism, the advent of induced pluripotent stem cells and their ability to differentiate and to be cultured in 2 or 3 dimensions forming organoids, can be harnessed to help us understand the disease related consequences and identify ways to reverse these outcomes.
Acknowledgments
The author would like to thank Dr. David Valle for reviewing the manuscript and providing important feedback for its improvement. The author is supported in part by P50 MH094268 (project 1 PI) and R01 MH106522 (PI: McCallion).
Footnotes
Conflict of Interest: The author has no conflicts of interest
References
- Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzel I, Bansal A, Browning BL, Galwey NW, Maycox PR, McGinnis R, Smart D, St Clair D, Yates P, Purvis I. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct BBF. 2007;3:31. doi: 10.1186/1744-9081-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britsch S. The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- Carteron C, Ferrer-Montiel A, Cabedo H. Characterization of a neural-specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J Cell Sci. 2006;119:898–909. doi: 10.1242/jcs.02799. [DOI] [PubMed] [Google Scholar]
- Chen P-L, Avramopoulos D, Lasseter VK, McGrath JA, Fallin MD, Liang K-Y, Nestadt G, Feng N, Steel G, Cutting AS, Wolyniec P, Pulver AE, Valle D. Fine mapping on chromosome 10q22-q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet. 2009;84:21–34. doi: 10.1016/j.ajhg.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya M, van der Merwe L, van Helden PD, Möller M, Hoal EG. Investigating the Role of Gene-Gene Interactions in TB Susceptibility. PloS One. 2014;10:e0123970. doi: 10.1371/journal.pone.0123970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejaegere T, Serneels L, Schäfer MK, Van Biervliet J, Horré K, Depboylu C, Alvarez-Fischer D, Herreman A, Willem M, Haass C, Höglinger GU, D’Hooge R, De Strooper B. Deficiency of Aph1B/C-gamma-secretase disturbs Nrg1 cleavage and sensorimotor gating that can be reversed with antipsychotic treatment. Proc Natl Acad Sci U S A. 2008;105:9775–9780. doi: 10.1073/pnas.0800507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gaetano C, Matullo G, Piazza A, Ursino M, Gasparini M. A proximity-based method to identify genomic regions correlated with a continuously varying environmental variable. Evol Bioinforma Online. 2013;9:29–42. doi: 10.4137/EBO.S10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M, Sinha P, Campbell R, Blackburn E, Levinson N, Rampaul R, Bates T, Humphreys S, Gullick WJ. Co-expression of neuregulins 1, 2, 3 and 4 in human breast cancer. J Pathol. 2004;203:672–680. doi: 10.1002/path.1561. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang K-Y, Huganir RL, Valle D, Pulver AE. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, Nestadt G, Valle D, Liang K-Y, Pulver AE. Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am J Hum Genet. 2003;73:601–611. doi: 10.1086/378158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- Falls DL, Rosen KM, Corfas G, Lane WS, Fischbach GD. ARIA, a protein that stimulates acetylcholine receptor synthesis, is a member of the neu ligand family. Cell. 1993;72:801–815. doi: 10.1016/0092-8674(93)90407-h. [DOI] [PubMed] [Google Scholar]
- Fleck D, Garratt AN, Haass C, Willem M. BACE1 dependent neuregulin processing: review. Curr Alzheimer Res. 2012;9:178–183. doi: 10.2174/156720512799361637. [DOI] [PubMed] [Google Scholar]
- Gizatullin RZ, Muravenko OV, Al-Amin AN, Wang F, Protopopov AI, Kashuba VI, Zelenin AV, Zabarovsky ER. Human NRG3 gene Map position 10q22-q23. Chromosome Res Int J Mol Supramol Evol Asp Chromosome Biol. 2000;8:560. doi: 10.1023/a:1009232025144. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- Hatzimanolis A, McGrath JA, Wang R, Li T, Wong PC, Nestadt G, Wolyniec PS, Valle D, Pulver AE, Avramopoulos D. Multiple variants aggregate in the neuregulin signaling pathway in a subset of schizophrenia patients. Transl Psychiatry. 2013;3:e264. doi: 10.1038/tp.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes LN, Shevelkin A, Zeledon M, Steel G, Chen P-L, Obie C, Pulver A, Avramopoulos D, Valle D, Sawa A, Pletnikov MV. Neuregulin 3 Knockout Mice Exhibit Behaviors Consistent with Psychotic Disorders. Mol Neuropsychiatry. 2016;2:79–87. doi: 10.1159/000445836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes NVL, Blackburn E, Smart LV, Boyle MM, Russell GA, Frost TM, Morgan BJT, Baines AJ, Gullick WJ. Identification and characterization of novel spliced variants of neuregulin 4 in prostate cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2007;13:3147–3155. doi: 10.1158/1078-0432.CCR-06-2237. [DOI] [PubMed] [Google Scholar]
- Ho WH, Armanini MP, Nuijens A, Phillips HS, Osheroff PL. Sensory and motor neuron-derived factor. A novel heregulin variant highly expressed in sensory and motor neurons. J Biol Chem. 1995;270:26722. [PubMed] [Google Scholar]
- Holmes WE, Sliwkowski MX, Akita RW, Henzel WJ, Lee J, Park JW, Yansura D, Abadi N, Raab H, Lewis GD. Identification of heregulin, a specific activator of p185erbB2. Science. 1992;256:1205–1210. doi: 10.1126/science.256.5060.1205. [DOI] [PubMed] [Google Scholar]
- Howard BA. The role of NRG3 in mammary development. J Mammary Gland Biol Neoplasia. 2008;13:195–203. doi: 10.1007/s10911-008-9082-8. [DOI] [PubMed] [Google Scholar]
- Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 1999;447:227–231. doi: 10.1016/s0014-5793(99)00283-5. [DOI] [PubMed] [Google Scholar]
- Kao W-T, Wang Y, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, Law AJ. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci U S A. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kwon DY, Kim M-S, Choi CR, Park M-Y, Kim AJ. Genome-wide association study for the interaction between BMR and BMI in obese Korean women including overweight. Nutr Res Pract. 2016;10:115–124. doi: 10.4162/nrp.2016.10.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bates R, Yin D-M, Shen C, Wang F, Su N, Kirov SA, Luo Y, Wang J-Z, Xiong W-C, Mei L. Specific regulation of NRG1 isoform expression by neuronal activity. J Neurosci Off J Soc Neurosci. 2011;31:8491–8501. doi: 10.1523/JNEUROSCI.5317-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos M, Mueller T, Gouwenberg Y, Wijnands R, van der Loo RJ, Birchmeier C, Smit AB, Spijker S Neuro-BSIK Mouse Phenomics Consortium. Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatry. 2014;76:648–655. doi: 10.1016/j.biopsych.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Loukola A, Wedenoja J, Keskitalo-Vuokko K, Broms U, Korhonen T, Ripatti S, Sarin A-P, Pitkäniemi J, He L, Häppölä A, Heikkilä K, Chou Y-L, Pergadia ML, Heath AC, Montgomery GW, Martin NG, Madden PaF, Kaprio J. Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol Psychiatry. 2014;19:615–624. doi: 10.1038/mp.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchionni MA, Goodearl AD, Chen MS, Bermingham-McDonogh O, Kirk C, Hendricks M, Danehy F, Misumi D, Sudhalter J, Kobayashi K. Glial growth factors are alternatively spliced erbB2 ligands expressed in the nervous system. Nature. 1993;362:312–318. doi: 10.1038/362312a0. [DOI] [PubMed] [Google Scholar]
- McGrath JA, Avramopoulos D, Lasseter VK, Wolyniec PS, Fallin MD, Liang K-Y, Nestadt G, Thornquist MH, Luke JR, Chen P-L, Valle D, Pulver AE. Familiality of novel factorial dimensions of schizophrenia. Arch Gen Psychiatry. 2009;66:591–600. doi: 10.1001/archgenpsychiatry.2009.56. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–770. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- Mei L, Nave KA. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron. 2014;83:27–49. doi: 10.1016/j.neuron.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Strohmaier J, Breuer R, Mattheisen M, Degenhardt F, Mühleisen TW, Schulze TG, Nöthen MM, Cichon S, Rietschel M, Wüst S. Neuregulin 3 is associated with attention deficits in schizophrenia and bipolar disorder. Int J Neuropsychopharmacol. 2013;16:549–556. doi: 10.1017/S1461145712000697. [DOI] [PubMed] [Google Scholar]
- Morar B, Dragović M, Waters FaV, Chandler D, Kalaydjieva L, Jablensky A. Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol Psychiatry. 2011;16:860–866. doi: 10.1038/mp.2010.70. [DOI] [PubMed] [Google Scholar]
- Mostaid MS, Lloyd D, Liberg B, Sundram S, Pereira A, Pantelis C, Karl T, Weickert CS, Everall IP, Bousman CA. Neuregulin-1 and schizophrenia in the genome-wide association study era. Neurosci Biobehav Rev. 2016;68:387–409. doi: 10.1016/j.neubiorev.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Law AJ, Luna A, Vakkalanka R, Straub RE, Kleinman JE, Weinberger DR. A 5′ promoter region SNP in NRG1 is associated with schizophrenia risk and type III isoform expression. Mol Psychiatry. 2009;14:741–743. doi: 10.1038/mp.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Moskvina V, Morris DW, Bray NJ, Zammit S, Williams NM, Williams HJ, Preece AC, Dwyer S, Wilkinson JC, Spurlock G, Kirov G, Buckland P, Waddington JL, Gill M, Corvin AP, Owen MJ, O’Donovan MC. Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet. 2006;141B:96–101. doi: 10.1002/ajmg.b.30236. [DOI] [PubMed] [Google Scholar]
- Parlapani E, Schmitt A, Wirths O, Bauer M, Sommer C, Rueb U, Skowronek MH, Treutlein J, Petroianu GA, Rietschel M, Falkai P. Gene expression of neuregulin-1 isoforms in different brain regions of elderly schizophrenia patients. World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2010;11:243–250. doi: 10.3109/15622970802022376. [DOI] [PubMed] [Google Scholar]
- Pasaje C-F, Bae J-S, Park B-L, Cheong HS, Kim J-H, Park T-J, Lee J-S, Kim Y, Park C-S, Kim B-J, Cha B, Kim JW, Choi WH, Shin T-M, Choi I-G, Hwang J, Shin H-D, Woo SI. Neuregulin 3 does not confer risk for schizophrenia and smooth pursuit eye movement abnormality in a Korean population. Genes Brain Behav. 2011;10:828–833. doi: 10.1111/j.1601-183X.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- Paterson C, Law AJ. Transient overexposure of neuregulin 3 during early postnatal development impacts selective behaviors in adulthood. PloS One. 2014;9:e104172. doi: 10.1371/journal.pone.0104172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson C, Wang Y, Hyde TM, Weinberger DR, Kleinman JE, Law AJ. Temporal, Diagnostic, and Tissue-Specific Regulation of NRG3 Isoform Expression in Human Brain Development and Affective Disorders. Am J Psychiatry. 2016 doi: 10.1176/appi.ajp.2016.16060721. appiajp201616060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periáñez JA, Ríos-Lago M, Rodríguez-Sánchez JM, Adrover-Roig D, Sánchez-Cubillo I, Crespo-Facorro B, Quemada JI, Barceló F. Trail Making Test in traumatic brain injury, schizophrenia, and normal ageing: sample comparisons and normative data. Arch Clin Neuropsychol Off J Natl Acad Neuropsychol. 2007;22:433–447. doi: 10.1016/j.acn.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Pham X, Song G, Lao S, Goff L, Zhu H, Valle D, Avramopoulos D. The DPYSL2 gene connects mTOR and schizophrenia. Transl Psychiatry. 2016;6:e933. doi: 10.1038/tp.2016.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plani-Lam JH-C, Chow T-C, Siu K-L, Chau WH, Ng M-HJ, Bao S, Ng CT, Sham P, Shum DK-Y, Ingley E, Jin D-Y, Song YQ. PTPN21 exerts pro-neuronal survival and neuritic elongation via ErbB4/NRG3 signaling. Int J Biochem Cell Biol. 2015;61:53–62. doi: 10.1016/j.biocel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Ponomareva ON, Ma H, Dakour R, Raabe TD, Lai C, Rimer M. Stimulation of acetylcholine receptor transcription by neuregulin-2 requires an N-box response element and is regulated by alternative splicing. Neuroscience. 2005;134:495–503. doi: 10.1016/j.neuroscience.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Salmela E, Renvall H, Kujala J, Hakosalo O, Illman M, Vihla M, Leinonen E, Salmelin R, Kere J. Evidence for genetic regulation of the human parieto-occipital 10-Hz rhythmic activity. Eur J Neurosci. 2016;44:1963–1971. doi: 10.1111/ejn.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savonenko AV, Melnikova T, Laird FM, Stewart K-A, Price DL, Wong PC. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci U S A. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T, Rudan I, McKeigue P, Wilson JF, Campbell H. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89:607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, Olafsson O, Stefansson K, Gulcher JR. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Sweeney C, Lai C, Riese DJ, Diamonti AJ, Cantley LC, Carraway KL. Ligand discrimination in signaling through an ErbB4 receptor homodimer. J Biol Chem. 2000;275:19803–19807. doi: 10.1074/jbc.C901015199. [DOI] [PubMed] [Google Scholar]
- Tang CS-M, Cheng G, So M-T, Yip BH-K, Miao X-P, Wong EH-M, Ngan ES-W, Lui VC-H, Song Y-Q, Chan D, Cheung K, Yuan Z-W, Lei L, Chung PH-Y, Liu X-L, Wong KK-Y, Marshall CR, Scherer SW, Scherer S, Cherny SS, Sham P-C, Tam PK-H, Garcia-Barceló MM. Genome-wide copy number analysis uncovers a new HSCR gene: NRG3. PLoS Genet. 2012;8:e1002687. doi: 10.1371/journal.pgen.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost H, Callicott JH, Rasetti R, Vakkalanka R, Mattay VS, Weinberger DR, Law AJ. Effects of neuregulin 3 genotype on human prefrontal cortex physiology. J Neurosci Off J Soc Neurosci. 2014;34:1051–1056. doi: 10.1523/JNEUROSCI.3496-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR, Ray R, Lee B, Everett L, Xiang J, Jepson C, Kaestner KH, Lerman C, Blendy JA. Evidence from mouse and man for a role of neuregulin 3 in nicotine dependence. Mol Psychiatry. 2014;19:801–810. doi: 10.1038/mp.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi S, Heaton C, Mack K, Hamilton JB, Lannan R, Wolfgang CD, Licamele L, Polymeropoulos MH, Lavedan C. Whole genome association study identifies polymorphisms associated with QT prolongation during iloperidone treatment of schizophrenia. Mol Psychiatry. 2009;14:1024–1031. doi: 10.1038/mp.2008.52. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King M-C, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang K-S, Xu N, Wang L, Aragon L, Ciubuc R, Arana TB, Mao C, Petty L, Briones D, Su BB, Luo X, Camarillo C, Escamilla MA, Xu C. NRG3 gene is associated with the risk and age at onset of Alzheimer disease. J Neural Transm Vienna Austria 1996. 2014;121:183–192. doi: 10.1007/s00702-013-1091-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang J, Zhou Y, Wei Z, Xiao Y, Zhou K, Wen J, Yan J, Cai W. Contribution of Common Variants in GABRG2, RELN and NRG3 and Interaction Networks to the Risk of Hirschsprung Disease. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2016;40:509–526. doi: 10.1159/000452565. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Chen J-Y, Chen M-L, Chen C-H, Lai I-C, Chen T-T, Hong C-J, Tsai S-J, Liou YJ. Neuregulin 3 genetic variations and susceptibility to schizophrenia in a Chinese population. Biol Psychiatry. 2008;64:1093–1096. doi: 10.1016/j.biopsych.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Tiwari Y, Schofield PR, Mowry BJ, Fullerton JM. Schizophrenia-associated HapICE haplotype is associated with increased NRG1 type III expression and high nucleotide diversity. Transl Psychiatry. 2012;2:e104. doi: 10.1038/tp.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Peles E, Cupples R, Suggs SV, Bacus SS, Luo Y, Trail G, Hu S, Silbiger SM, Levy RB. Neu differentiation factor: a transmembrane glycoprotein containing an EGF domain and an immunoglobulin homology unit. Cell. 1992;69:559–572. doi: 10.1016/0092-8674(92)90456-m. [DOI] [PubMed] [Google Scholar]
- Willem M. Proteolytic processing of Neuregulin-1. Brain Res Bull. 2016;126:178–182. doi: 10.1016/j.brainresbull.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Yang J, Duan S, Zhong R, Yin J, Pu J, Ke J, Lu X, Zou L, Zhang H, Zhu Z, Wang D, Xiao H, Guo A, Xia J, Miao X, Tang S, Wang G. Exome sequencing identified NRG3 as a novel susceptible gene of Hirschsprung’s disease in a Chinese population. Mol Neurobiol. 2013;47:957–966. doi: 10.1007/s12035-012-8392-4. [DOI] [PubMed] [Google Scholar]
- Zeledon M, Eckart N, Taub M, Vernon H, Szymanski M, Wang R, Chen P-L, Nestadt G, McGrath J, Sawa A, Pulver AE, Avramopoulos D, Valle D. Identification and Functional Studies of Regulatory Variants Responsible for the Association of NRG3 with a Delusion Phenotype in Schizophrenia. Mol Neuropsychiatry. 2015;1:36–46. doi: 10.1159/000371518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, Godowski PJ. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci U S A. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Du X-Y, Yu J, Xu N, Zheng Y-W, Zhao Y-L, Zhang H, Ma J. No genetic evidence for Neuregulin 3 conferring risk of schizophrenia in the Chinese population. Psychiatry Res. 2013;205:279–281. doi: 10.1016/j.psychres.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Zhou W, Carpenter G. ErbB-4: a receptor tyrosine kinase. Inflamm Res Off J Eur Histamine Res Soc Al. 2002;51:91–101. doi: 10.1007/BF02684009. [DOI] [PubMed] [Google Scholar]