Abstract
The Hippo-YAP pathway is essential for controlling organ size and tumorigenesis. Previous studies have demonstrated that the primary outcome of YAP signaling in the nucleus is achieved by interaction with the transcription factor TEAD1. The YAP/TEAD1 complex binds to DNA element and regulates the expression of genes involved in cell growth. However, constitutive knockout of TEAD1 leads to early embryonic lethality in mice. Thus, generation of a floxed TEAD1 mouse becomes crucial for further understanding mid- to late-gestation and post-natal role of TEAD1. Herein, we created and characterized a mouse model that allows for conditional disruption of TEAD1. Embryonic fibroblasts derived from the floxed TEAD1 mice enabled the Cre-mediated deletion of TEAD1 in vitro using virally delivered Cre recombinase. Furthermore, crossing the floxed TEAD1 mouse with a ubiquitously expressing Cre mouse resulted in efficient ablation of the floxed allele in vivo, and the animals recapitulated early embryonic lethality defects. In conclusion, our data demonstrate an important role of TEAD1 in early development in mice, and the floxed TEAD1 mouse model will be a valuable genetic tool to determine the temporal and tissue-specific functions of TEAD1.
Keywords: Hippo-YAP pathway, TEAD1, floxed allele mouse, gene targeting, conditional knockout, Cre, embryonic lethality, transcription factor
Introduction
The Hippo pathway is evolutionarily conserved and is crucial for controlling organ size, development and tumorigenesis (Pan, 2010; Zhao et al., 2010; Zhou, 2014). In mammals, the core components of the Hippo pathway include cytosolic kinase proteins Mst1/2 (mammalian Ste20-like kinase 1 or 2) and Lats1/2 (large tumor suppressor 1 or 2), the major downstream effector YAP (yes-associated protein) that can shuttle between the cytoplasm and nucleus, and the TEAD (TEA domain transcription factor, also known as transcriptional enhancer factor) family transcription factors that bind with YAP to induce transcription. The Hippo pathway is activated by a spectrum of extracellular stimuli including cell-cell contact and a subset of G-protein-coupled receptors (Plouffe et al., 2015). Upon stimulation, Mst1/2 kinases phosphorylate downstream kinases Lats1/2, which subsequently phosphorylate YAP, resulting in retention of YAP in the cytoplasm. Conversely, when upstream kinases are inactivated, unphosphorylated YAP translocates into the nucleus where it binds to proteins of the TEAD family, to induce genes that involve in growth and differentiation (Zhao et al., 2008a).
We previously reported that the Hippo signaling effector, YAP not only plays a critical role in neointimal formation after arterial injury but also is critical for cardiac/smooth muscle proliferation during cardiovascular development in mice (Wang et al., 2012; Wang et al., 2014; Xu et al., 2015). Studies have suggested that the YAP activity is mediated primarily via TEAD transcription factors, through which YAP binds to a consensus DNA sequence 5′-CATTCC-3′, named the MCAT (muscle CAT) element (Stein et al., 2015; Vassilev et al., 2001; Zanconato et al., 2015; Zhao et al., 2008b). In contrast to this widespread model, recently Galli et al reported that YAP DNA occupancy is associated only with a very small subset of TEAD binding sites (Galli et al., 2015), suggesting that YAP and TEAD may play independent roles in different cell/tissue contexts at different time points. The TEAD family of proteins consist of four members, TEAD1 (TEF-1, NTEF-1), TEAD2 (ETF, TEF-4), TEAD3 (DTEF-1, TEF-5, ETFR-1) and TEAD4 (TEF-3, RTEF-1) (Yoshida, 2008). Among these TEAD family proteins, we found TEAD1 is the most abundant family member in vascular smooth muscle cells and plays a critical role in smooth muscle dedifferentiation (Liu et al., 2014). Previous studies showed that the TEAD1 global knockout (KO) in mice results in early embryonic lethality (Chen et al., 1994; Sawada et al., 2008). Thus we sought to develop a genetic approach by which TEAD1 is targeted via conditional allele disruption.
In this study we generated TEAD1 flox (F) mice to provide a tool to circumvent the global deletion of TEAD1-induced embryonic lethality. We demonstrated an effective Cre-mediated ablation of the TEAD1 floxed allele in vitro and in vivo. We anticipate this TEAD1 conditional allele mouse line will provide a valuable tool to dissect the temporal and tissue-specific roles of TEAD1 in physiological and pathological settings.
Results
Validation of TEAD1 targeted allele
A targeted TEAD1 allele mouse line with gene-trap knockout, lacZ tagged insertion and conditional potential (TEAD1tm1a(komp)Wtsi, herein referred to as TEAD1 neo) has been generated by the Knockout Mouse Project (KOMP) consortium (figure 1A). A “conditional” (flox) allele can be achieved by crossing with flippase (FLP) recombinase expressing mice to remove the En2 SA/LacZ/neo cassette flanked by two FRT sites. Subsequently cre recombinase mediated deletion of TEAD1 floxed exon (E) 3 leads to a TEAD1 null allele because removal of E3 will cause a reading frameshift of the chimeric transcript of E2 and E4, thereby introducing an immediate formation of a stop codon (figure 1A, B). Breeding of heterozygous TEAD1 neo mice with C57BL/6 mice produced wildtype (WT) and TEAD1+/neo animals with expected Mendelian distribution. However, TEAD1+/neo intercross failed to obtain TEAD1 neo/neo mice after birth at postnatal (P) 21 days (n=0 of 105 mice). This data suggested the insertion of EN2 SA cassette led to prematurely terminating transcription, thereby leading to a possible embryonic lethality. Analysis of embryos from the timed mating TEAD1+/neo intercrossed female mice revealed TEAD1neo/neo embryos died by E10.5. The genotypes of the neo/neo or +/neo embryos at E10.5 confirmed the targeted allele by PCR of the yolk sac DNA with the primers as described in figure 1A (figure 1C). The yolk sac of mutant embryos at E10.5 exhibited a defect of vasculature (figure 1D). Furthermore, the E10.5 mutant embryos were smaller in mass, exhibited dilated the fourth ventricles of brain and had enlargement of the pericardial cavity and underdeveloped heart (figure 1E), similar to previous observations using retrovirus-mediated gene trap knock out strategy (Chen et al., 1994). To confirm TEAD1 gene deletion by the gene trap at protein level, western blotting was performed using total lysates harvested from E10.5 WT, neo/+ and neo/neo embryos. 90% of TEAD1 protein expression was ablated in TEAD neo/neo embryos, while TEAD1 neo/+ embryos showed a dosage effect, expressing about 50% TEAD protein level compared to WT littermates (figure 1F). The incomplete deletion of TEAD1 in TEAD1neo/neo embryos suggested that the gene-trap cassette did not completely abolish TEAD1 expression, as such a minor fraction of wild-type transcripts can still be produced from this allele by splicing around the insertion of SA/LacZ/neo cassettes. To test whether the lacZ can express β-galactosidase (β-gal) from the gene trap cassette or not, we performed western blot and X-gal staining using the E10.5 embryos from TEAD1 neo/+ intercrossing. Data from these assays showed undetectable β-gal expression by western blot (figure 1F) and very weak X-gal staining in the hearts of E10.5 neo/+ and neo/neo embryos (figure 1G), respectively. We speculate that the chimeric mRNA of TEAD1 E2 and LacZ resulted from the inserted trap cassette in TEAD1 gene locus is unstable or/and unable to efficiently translate to a fusion protein. Taken together, these data demonstrated that the targeted TEAD1 allele mouse line represents a knockout first allele for TEAD1 although it does not function as a complete null allele.
Figure 1. Gene targeting scheme for generating conditional TEAD1 allele in mice.
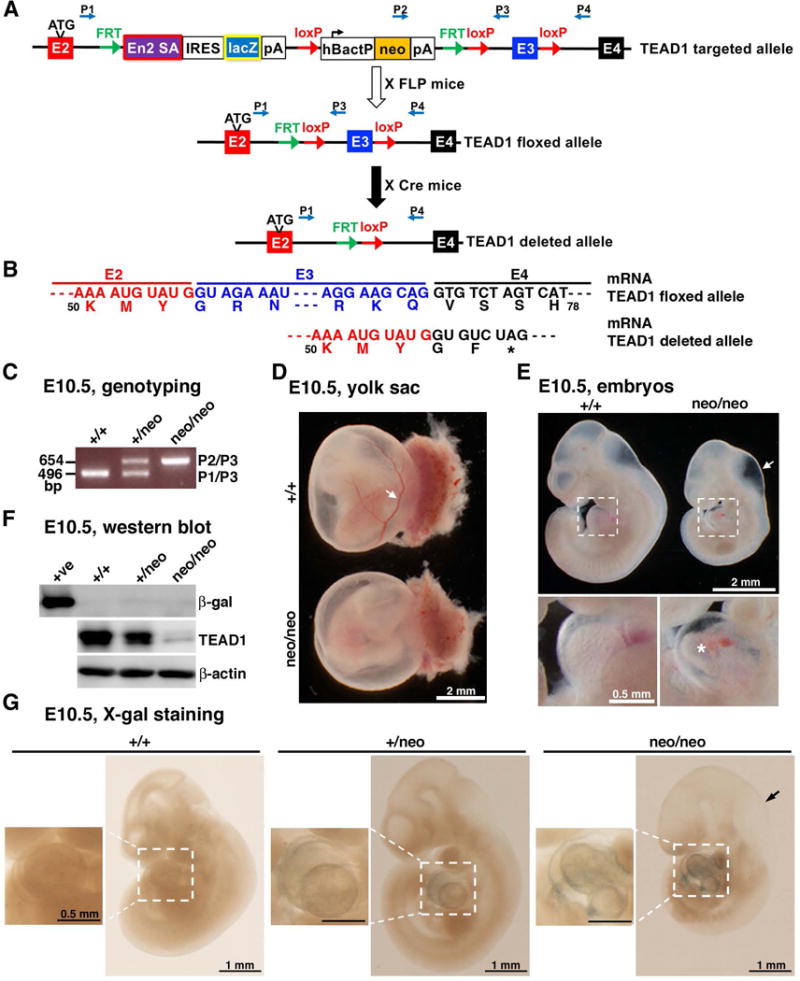
A. Schematic diagram demonstrates that TEAD1 targeted allele in which a FRT (green arrows)-flanked knockout-first (En2 SA/lacZ/neo) cassette was inserted into the intron (line) between TEAD1 gene exon (E) 2 and 3 (red and blue boxes, respectively, not to scale) and loxP sites (red arrows) were placed flanking E3. A floxed allele was produced after removal of En2 SA/lacZ/neo cassette by crossing with Flippase (FLP) mice. Subsequently a TEAD1 null allele was generated after deletion of loxP sites flanking E3 by crossing with Cre recombinase expressing mice. Alternative splicing of TEAD1 E1-2 to En2 SA in the trapping cassette was designed to disrupt the TEAD1 transcript. Primers (P) used for genotyping are shown as blue arrows. E2, E3 and E4, exons of TEAD1; En2 SA, mouse En2 splicing acceptor; IRES, internal ribosome entry site; lacZ, bacterial beta-galactosidase gene; pA, poly A; hBactP, human beta actin promoter; neo, promoter-driven neomycin resistant gene; FRT, flippase recognition target; loxP, locus of crossover in P1. B. mRNA transcripts from TEAD1 floxed allele or deleted allele are shown to demonstrate that removal of E3 causes a reading frameshift in the chimeric transcript of E2 and E4, thereby introducing an immediate formation of a stop codon (*). The predicted amino acids are shown and numbered below the mRNA encoded by its respective exons which are labeled in colors. C. Representative PCR genotyping results by using wild type (+/+), heterozygous (+/neo), and homozygous (neo/neo) embryos at E10.5. Note primer sets of 1, 2 and 3 can produce a 654 bp band in targeted allele (P2/P3) and a 496 bp band in wild type allele (P1/P3) of TEAD1 gene, respectively. D. Gross picture of E10.5 TEAD1 gene trapping (neo/neo) embryo shows paleness of the yolk sac with defects of vascular development compared to its littermate control (arrow). E. E10.5 TEAD1neo/neo embryos displayed severe retarded growth, dilation of 4th ventricle of brain (arrow), enlargement of pericardial cavity (boxed) and underdeveloped heart (*). F. E10.5 embryos from +/neo heterozygous mouse intercrossing were collected for western blot to validate the disruption of TEAD1 expression and β-gal expression due to En2 SA mediated trapping. Lysate using MEFs infected with virus encoding LacZ served as a positive control (far left). G. E10.5 embryos from +/neo heterozygous mouse intercrossing were collected at E10.5 for X-gal staining. The heart areas were boxed to show X-gal staining positive. Arrow points to the dilated 4th ventricle of brain.
Generation of TEAD1 floxed allele mice and validation by Cre-mediated deletion of TEAD1 in vitro
To obtain TEAD1 floxed allele in mice, we crossed female TEAD1 targeted allele mice (neo/+) with the FLP recombinase germline deleter male mice (Farley et al., 2000) to remove the entire FRT-flanked En2 SA/LacZ/neo cassettes (figure 1A). PCR using tail genomic DNA from 2-week old offspring confirmed the removal of En2 SA/LacZ/neo cassettes (figure 2A). Sequencing results confirmed the deletion of En2 SA/LacZ/neo cassettes and the TEAD1 floxed allele in which loxP sites encompass the 3rd exon of TEAD1 (supplemental figure 1 and 2). Subsequently, TEAD1+/F mice were intercrossed to obtain TEAD1 flox/flox (TEAD1F/F) mice. Genotyping of the 2-week old progeny from the intercross revealed production of homozygous TEAD1F/F mice at the expected Mendelian ratio. Homozygous TEAD1F/F mice exhibited a normal life span and fertility without affecting endogenous TEAD1 expression in lung, skeletal muscle and heart, liver tissues (data not shown) compared to the age-matched WT control mice (figure 2B). To test if the TEAD1 floxed allele could be targeted and deleted by Cre recombinase in vitro, mouse embryonic fibroblasts (MEFs) harvested from E14.5 TEAD1F/F embryos were infected either with control GFP or Cre expressing adenovirus. Genomic PCR analysis and sequencing results demonstrated a complete conversion of flox allele to deleted allele of TEAD1 (figure 2C and supplement figure 3). qRT-PCR and western blot further confirmed the complete loss of TEAD1 expression at both mRNA level and protein level, respectively, without affecting TEAD1’s cofactor YAP expression (figure 2D and E). Taken together, these data demonstrate that the TEAD1 floxed allele mouse line was successfully generated and MEFs derived from the mice harboring TEAD1 floxed allele were fully capable of producing conditional Cre-mediated deletion of TEAD1 gene in vitro.
Figure 2. Knock out TEAD1 gene in vitro using the MEFs derived from floxed TEAD1 embryos.
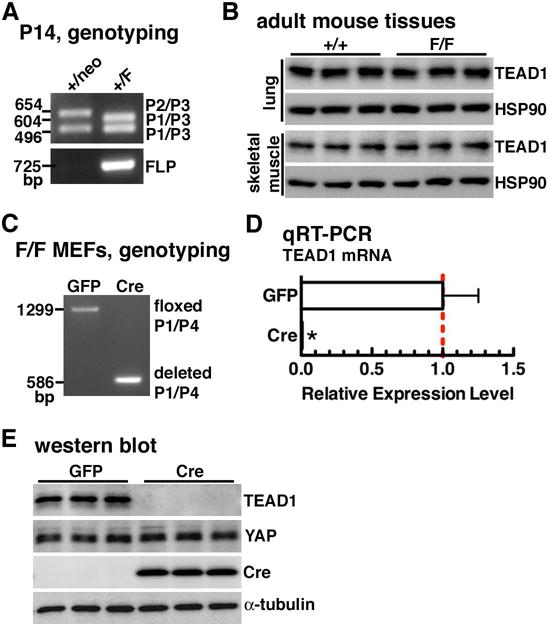
A. Representative PCR genotyping results by using the tail biopsy from postnatal (P) 14 days old offspring mice that were produced by mating +/neo female mice with FLP deleter male mice. PCR using primers 1, 2 and 3 produced a 654 bp band in targeted allele (P2/P3), a 604 bp band in floxed allele (P1/P3) and a 496 bp band in wild type allele (P1/P3) of TEAD1 gene, respectively (top panel). PCR result using primers specific for detecting FLP was shown at bottom panel. The floxed TEAD1 allele after FLP-mediated removal of En2 SA/lacZ/neo was detected only in FLP positive mouse as expected. B. Western blot demonstrated no difference of TEAD1 expression between adult WT and TEAD1F/F mouse tissues including lung and skeletal muscle. N=3 each group. C. DNA was extracted from adenovirus expressing GFP or Cre infected TEAD1F/F MEFs and PCR was performed to demonstrate Cre-mediated deletion of floxed E3 in TEAD1 gene locus. PCR using primers 1 and 4 will produce 1299 bp and 586 bp bands in TEAD1 flox and deleted allele, respectively. D. RNA was extracted from GFP or Cre adenovirus-infected TEAD1F/F MEFs for qRT-PCR to demonstrate a complete deletion TEAD1 expression at mRNA level after Cre excision. E. Western blot was performed to show an undetectable level of TEAD1 protein in Cre-infected TEAD1F/F MEFs without affecting TEAD’s cofactor YAP expression.
Validation of Cre-mediated deletion of TEAD1 in vivo
To validate Cre-mediated deletion of the TEAD1 conditional allele in vivo, we crossed the TEAD1+/F mice with CMV-Cre mice (figure 3A), which express Cre during early embryonic development resulting in deletion of loxP-flanked DNA sequences in all of tissues including developing germ line (Schwenk et al., 1995). The resultant heterozygous TEAD1+/− mice were viable and fertile, and appeared normal without any gross physical or behavioral abnormalities. We then intercrossed heterozygous TEAD1+/− mice to obtain TEAD1−/− mice (figure 3A). In the viable progeny from TEAD1+/− heterozygous intercrosses, no TEAD1−/− mice were collected after birth (n=0 of 120 mice), suggesting the TEAD−/− mutant embryos died prenatally. Indeed, isolation of embryos in utero revealed the global deletion of TEAD1 resulted in embryonic lethality before E9.5 (figure 3B). The E9.5 TEAD1 global KO embryos were much smaller than WT’s and displayed vascular defect within the yolk sac (figure 3C), as observed in TEAD1neo/neo embryos. qRT-PCR and western blot assays confirmed that there was no TEAD1 expression in TEAD1−/− mutant embryos, while approximately 50% mRNA and protein expression in heterozygous, respectively (figure 3D and E). Of note, complete deletion of TEAD1 led to mortality at E9.5, earlier than the death time observed in neo/neo embryos (figure 3C) and retrovirus-mediated gene trap embryos (Chen et al., 1994), most likely due to the difference of TEAD1 knockout efficiency in these animals.
Figure 3. Knock out TEAD1 gene in vivo using global promoter-driven Cre mouse.
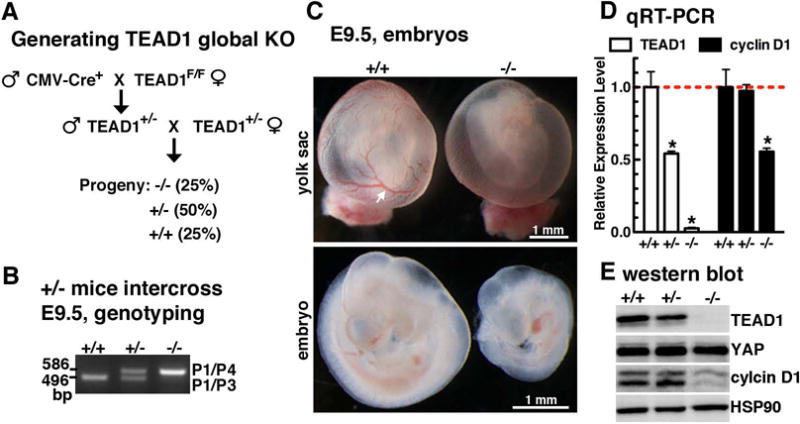
A. Schematic diagram to generate global TEAD1 KO mouse by using TEAD1 flox mice. After crossing CMV-Cre male mice with TEAD1F/F female mice, the resultant heterozygous mice were intercrossed to generate global TEAD1 KO mouse. This breeding strategy is expected to yield 25% progeny with the homozygous KO (−/−), 50% heterozygotes (+/−) and 25% wild-type (+/+) littermates. B. Genotyping of E9.5 embryos from TEAD1 heterozygous mouse intercrossing. The 496 bp band for WT allele (P1/P3) was converted to a 586 bp band of knockout allele (P1/P4). C. Gross pictures of E9.5 wild-type (+/+) and TEAD1 KO (−/−) embryos show vascular defect in KO yolk sac (top panel, arrow) and growth retardation of the KO embryos (bottom panel). D. E9.5 embryos collected from TEAD1 heterozygous intercrossed female mice were used for qRT-PCR or western blot (E) to examine TEAD1 and cylcin D1 expression as indicated. TEAD1 deletion did not change YAP but decreased cyclin D1 expression at both mRNA and protein levels.
Previous studies showed that TEAD1 mediated YAP function to promote cell proliferation by directly activating cell cycle genes including cycline D1 (Mizuno et al., 2012; Zhao et al., 2008b). Our qRT-PCR and western blot assays revealed approximately 50% reduction of cyclin D1 expression in TEAD1 null embryos, suggesting the down-regulated pro-proliferative gene cyclin D1 expression is, at least in part, attributable to the embryonic growth retardation. Collectively, the early embryonic lethality observed in TEAD1 global KO embryos provides the proof of principle evidence that Cre-mediated deletion of E3 not only disrupts TEAD1 gene expression but also abolishes the function of TEAD1 in vivo in development. During the preparation of the manuscript, we realized a floxed TEAD1 mouse line was generated using the similar strategy as we described in this study (Liu et al., 2017). Our current study independently validated this mouse line and extensively characterized the TEAD1 conditional allele in vitro and in vivo.
In summary, the TEAD1F/F mouse generated in this study provides us with a valuable genetic tool to decipher tissue-specific roles of TEAD1 in both embryonic and postnatal stages. Information gained from these studies will advance our understating in the function and underlying mechanisms of Hippo/YAP/TEAD signaling in variety of tissues under physiological and pathological settings.
Methods
Generation of the TEAD1 conditional allele in mouse
The Tead1 mouse strain Tead1tm1a(KOMP)Wtsi used for this study was created from ES cell clone EPD0207_2_C09 generated by the Wellcome Trust Sanger Institute and made into mice and provided to the KOMP Repository (WWW.KOMP.org) as part of the KOMP2 Project by the Baylor College of Medicine as part of the BaSH consortium (targeting project: CSD34183) (Skarnes et al., 2011). Briefly, the TEAD1 gene targeting vector PRPGS00097_A_H05 was constructed containing 5169 bp TEAD1 partial intron 2 sequence and the downstream 4103 bp sequences of TEAD1 exon 3, 4 and 5 as long 5′ and short 3′ homologous arms, respectively. This targeting vector also contains the L1L2_Bact_P cassettes including an FRT site followed by En2 SA/lacZ/neo cassettes in which the first loxP site is inserted between En2 SA/lacZ and neomycin (neo) cassettes under the control of the human beta-actin promoter. A second FRT site and a second loxP site were inserted at position 112838936 of mouse Chromosome 7 upstream of the TEAD1 E3 which encodes partial TEAD1 DNA binding domain (Xiao et al., 1991). A third loxP site is inserted downstream of the targeted E3 at position 112839770. Thus the TEAD1 E3 is flanked by 2 loxP sites. The targeted vector was transfected into C57BL/6N origin JM8.N4 ES cells by electroporation. Long range PCR was used to confirm positive ES cell clones with correct homologous recombination. The TEAD1 targeted ES cells (clone EPD0207_2_C09) were used by the KOMP to generate chimeras by injection into C57BL/6 host blastocysts. The resulted chimera mice were bred with wild-type C57BL/6 mice to obtain germline transmission neo/+ mice. A sperm resuscitation has been carried out at UC, Davis and the resultant TEAD1neo/+ mice were transferred to Augusta University. To generate TEAD1 floxed allele mice, TEAD1neo/+ female mice were crossed to male FLP deleter mice (Jackson lab, stock #: 009086) (Farley et al., 2000) to excise the EN2 SA/lacZ/neo cassettes. Resulting mice that transmitted the TEAD1 flox allele to their progeny were intercrossed and maintained as TEAD1F/F on a C57BL/6 background. The TEAD1 flox mice described in this study will be available to the research community at the KOMP. Embryonic day (E) 0.5 was defined as noon of the day when the vaginal plug was detected. The use of experimental animals is approved by the Institutional Animal Care and Use at Augusta University.
Generation of TEAD1 global knockout mouse
Female TEAD1 conditional allele mice were bred with male mice ubiquitously expressing Cre (CMV-Cre, Jackson lab, stock#: 006054) (Schwenk et al., 1995) that is under the transcriptional control of a human cytomegalovirus minimal promoter to delete loxP-flanked TEAD1 gene in all tissues, including germ cells. The resultant heterozygous mice were subsequently intercrossed to generate mice with global deletion of TEAD1.
DNA extraction, genotyping by PCR (Polymerase Chain Reaction) and Sanger sequencing
DNA was extracted from yolk sac tissue, tail biopsy or MEFs essentially following the manufacturer’s protocol (Viagen) and genotyping by PCR were performed using the primers listed below as we previously described (Wang et al., 2014). PCR products were then run on agarose gel and visualized under UV after stained with ethidium bromide. In some cases, PCR product band after separated in agarose gel was purified by an agarose gel extraction kit (Thermo Fisher Scientific) for Sanger sequencing. All sequencing was performed by Genewiz.
Protein extraction and western blot
Tissues were harvested from 3-month old TEAD1F/F mice and their littermate WT C57BL/6 mice as we previously described (Wang et al., 2011). Mouse embryos were removed from uterus and ground with a glass homogenizer in 100 μl RIPA buffer (Fisher) with 1% proteinase inhibitor cocktail (Pierce) and 1% PMSF as previously described (Wang et al., 2012; Wang et al., 2014; Xu et al., 2015). After sonication and centrifugation of the cell lysate, proteins were quantified by BCA assay and then loaded in a 9% SDS-PAGE gel at 5–10 μg per lane. Antibodies used in this study are: α-tubulin (Cell Signaling, #2144, rabbit, 1:5000), β-actin (Sigma, A5316, mouse, 1:5000), β-galactosidase (Bio-Rad, AHP1292GA, rabbit, 1:1000), Cre (Novagen, 69050-3, rabbit, 1:5000), cyclin D1 (Cell signaling, #2978, rabbit, 1:2000), HSP90 (Cell Signaling, #4874, rabbit, 1:2000), TEAD1 (abcam, ab133533, rabbit, 1:2000), YAP (Sigma, WH0010413M1, mouse, 1:2000). Images were acquired and band intensity was quantified by ImageQuant LAS 4000 Imaging Station (GE).
Whole mount X-gal staining
Whole mount X-gal staining for embryos were carried out by a standard protocol. Briefly, E10.5 embryos were dissected out from uterus followed by fixing with 4% paraformaldehyde in PBS (phosphate buffered saline) for 30 minutes on ice. After rinsed the fixed embryos 3 time with PBS for 15 minutes at room temperature, embryos were then incubated with 1mg/ml X-gal staining buffer over-night at 37°C in the dark. Subsequently embryos were rinsed with PBS, post-fixed with 4% paraformaldehyde and taken photos under a dissecting microscope.
Isolation of mouse embryonic fibroblasts (MEFs) and adenoviral infection
Embryonic fibroblasts were prepared by a standard protocol. Briefly, E14.5 embryos were dissected from TEAD1F/F intercrossed pregnant female mice. After all inner organs, head and limbs were removed, the remaining tissues were minced by scissors in 0.25% trypsin digestion buffer. Subsequently the minced tissue was collected and incubated in 0.25% trypsin at 4°C over-night and then cells were plated on 15-cm plates in 10% FBS/DMEM medium. After three passages, fibroblast cells were split for infection with adenovirus expressing control GFP or cre as described previously (Chen and Herring, 2013; Liu et al., 2014; Xu et al., 2015).
Total RNA isolation and quantitative reverse transcription PCR (qRT-PCR)
Total RNA from embryos or MEFs was extracted by TRIzol reagent (Invitrogen). 0.5–1 μg of RNA was utilized as a template for RT with random hexamer primers using the High Capacity RNA-to-cDNA Kit (Invitrogen). qRT-PCR was performed with respective gene-specific primers as described below (Liu et al., 2014; Wang et al., 2012; Wang et al., 2010; Wang et al., 2014; Xu et al., 2015). All samples were amplified in duplicate and every experiment was repeated independently 2 times. Relative gene expression was converted using the 2−ΔΔct method against the internal control hypoxanthine phosphor ribosyl transferase 1 (HPRT) for mouse.
Oligonucleotides for genotyping
Primers (P) for TEAD1 genotyping are: P1, forward: 5′-TGCCATCATGCCAAGCTATACTGG-3′; P2, forward: 5′-GGGATCTCATGCTGGAGTTCTTCG-3′; P3, reverse: 5′-TGAACT CACATGGTGGTTTACAGCC-3′; P4, reverse: 5′-CCTGCTTTATAGTCACAGCAGAGGC-3′; Primers for flippase mouse genotyping are: forward: 5′-CACTGATATTGTAAGTAGTTTGC-3′; reverse: 5′-CTAGTGCGAAGTAGTGATCAGG-3′ and the PCR product size is 725 bp.
Oligonucleotides for qRT-PCR
Mouse TEAD1, forward: 5′-AGCCAGATACATCAAACTCAGGACG-3′, reverse: 5′-CTTA ATGGCGGCTTGAATTTCTCGAAC-3′; Mouse cyclin D1, forward: 5′-CATCAAGTGTGA CCCGGACTG-3′, reverse: 5′-AGACCAGCCTCTTCCTCCACTT-3′; Mouse HPRT, forward: 5′-TGGCCCTCTGTGTGCTCAA-3′, reverse: 5′-TGATCATTACAGTAGCTCTTCAGTCTGA-3′.
Statistical analysis
Data are expressed as means ± SE, and statistical analysis using unpaired t test was done with Prism software (Graphpad). Differences with p values < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Drs. Dan Rudic and David Fulton for a critical reading of the manuscript.
Grant Support: The project described was supported by grants from the National Heart, Lung, and Blood Institute, NIH and AHA to J. Z.; J. Z. is an Established Investigator of the American Heart Association. T. W. and M. W. were supported by High Level Construction Disciplines in the Cardiology of Jiangxi Province (to the Department of Cardiology, the First Affiliated Hospital of Nanchang University); Q. Y., L. Y, J. L., W. Z. and L. H. were supported by a fund from Chinese National Key Specialty Program of Clinical Medicine.
References
- Chen M, Herring BP. Regulation of microRNAs by Brahma-related gene 1 (Brg1) in smooth muscle cells. J Biol Chem. 2013;288:6397–6408. doi: 10.1074/jbc.M112.409474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, Calogero RA, Camargo FD. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell. 2015;60:328–337. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang X, Hu G, Wang Y, Zhou J. The transcription factor TEAD1 represses smooth muscle-specific gene expression by abolishing myocardin function. J Biol Chem. 2014;289:3308–3316. doi: 10.1074/jbc.M113.515817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lee J, Kim BS, Wang Q, Buxton SK, Balasubramanyam N, Kim JJ, Dong J, Zhang A, Li S, Gupte AA, Hamilton DJ, Martin JF, Rodney GG, Coarfa C, Wehrens XH, Yechoor VK, Moulik M. Tead1 is required for maintaining adult cardiomyocyte function, and its loss results in lethal dilated cardiomyopathy. JCI Insight. 2017;2 doi: 10.1172/jci.insight.93343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Murakami H, Fujii M, Ishiguro F, Tanaka I, Kondo Y, Akatsuka S, Toyokuni S, Yokoi K, Osada H, Sekido Y. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31:5117–5122. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe SW, Hong AW, Guan KL. Disease implications of the Hippo/YAP pathway. Trends Mol Med. 2015;21:212–222. doi: 10.1016/j.molmed.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schubeler D, Bauer A. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu G, Betts C, Harmon EY, Keller RS, Van De Water L, Zhou J. Transforming growth factor-beta1-induced transcript 1 protein, a novel marker for smooth muscle contractile phenotype, is regulated by serum response factor/myocardin protein. J Biol Chem. 2011;286:41589–41599. doi: 10.1074/jbc.M111.250878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu G, Gao X, Wang Y, Zhang W, Harmon EY, Zhi X, Xu Z, Lennartz MR, Barroso M, Trebak M, Chen C, Zhou J. The induction of yes-associated protein expression after arterial injury is crucial for smooth muscle phenotypic modulation and neointima formation. Arterioscler Thromb Vasc Biol. 2012;32:2662–2669. doi: 10.1161/ATVBAHA.112.254730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu G, Zhou J. Repression of versican expression by microRNA-143. J Biol Chem. 2010;285:23241–23250. doi: 10.1074/jbc.M109.084673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu G, Liu F, Wang X, Wu M, Schwarz JJ, Zhou J. Deletion of yes-associated protein (YAP) specifically in cardiac and vascular smooth muscle cells reveals a crucial role for YAP in mouse cardiovascular development. Circ Res. 2014;114:957–965. doi: 10.1161/CIRCRESAHA.114.303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Xu F, Ahmed AS, Kang X, Hu G, Liu F, Zhang W, Zhou J. MicroRNA-15b/16 Attenuates Vascular Neointima Formation by Promoting the Contractile Phenotype of Vascular Smooth Muscle Through Targeting YAP. Arterioscler Thromb Vasc Biol. 2015;35:2145–2152. doi: 10.1161/ATVBAHA.115.305748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol. 2008;28:8–17. doi: 10.1161/ATVBAHA.107.155788. [DOI] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008a;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008b;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. An emerging role for Hippo-YAP signaling in cardiovascular development. J Biomed Res. 2014;28:251–254. doi: 10.7555/JBR.28.20140020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


