Abstract
The population dynamics of the Pleistocene woolly mammoth (Mammuthus primigenius) has been the subject of intensive palaeogenetic research. Although a large number of mitochondrial genomes across Eurasia have been reconstructed, the available data remains geographically sparse and mostly focused on eastern Eurasia. Thus, population dynamics in other regions have not been extensively investigated. Here, we use a multi-method approach utilising proteomic, stable isotope and genetic techniques to identify and generate twenty woolly mammoth mitochondrial genomes, and associated dietary stable isotopic data, from highly fragmentary Late Pleistocene material from central Europe. We begin to address region-specific questions regarding central European woolly mammoth populations, highlighting parallels with a previous replacement event in eastern Eurasia ten thousand years earlier. A high number of shared derived mutations between woolly mammoth mitochondrial clades are identified, questioning previous phylogenetic analysis and thus emphasizing the need for nuclear DNA studies to explicate the increasingly complex genetic history of the woolly mammoth.
Introduction
Ancient DNA (aDNA) has uncovered complex intra-species population dynamics of the woolly mammoth (Mammuthus primigenius). Previously unknown coexistence and extinction events of different maternal lineages have been revealed, changing our understanding of the evolution and extinction of this emblematic megafaunal species. With previous research focusing on short mitochondrial DNA (mtDNA) fragments of the hypervariable region (HVR), two mitochondrial clades across Beringia, clade I (primarily in North America) and clade II (in Siberia), were originally identified1. Through similar markers, a third maternal population (clade III) has been recently identified in Europe; a lineage suggested to have been lost around 34 ka cal BP, and replaced through migration of individuals from eastern Eurasia carrying clade I mtDNAs that reached Europe by 32 ka cal BP2. Chang et al 3. reconstructed mitochondrial genomes of clade III individuals from Europe showing that the lineage existed until at least 24 ka cal BP (Dresden, Germany), and possibly until around 12 ka BP in western Eurasia; thus overlapping in time with the arrival of clade I. This is unlike the complete population replacement of mtDNA as suggested by Palkopoulou et al.2. Furthermore, Enk et al 4. identified a distinct deeply-diverged sister-lineage of clade III in eastern Eurasia present around 40 cal ka BP (3/B1 in Chang et al.3). Understanding the nature of replacement events in mammoth mitochondrial lineages has further importance due to previous debate of drift versus selection with the earlier loss of clade II after a period of coexistence with clade I around 44 ka cal BP1,5,6. Despite presentation of new dates showing a later survival of clade III in Europe, Chang et al 3. do not discuss the replacement event in context of the scenario suggested by Palkopoulou et al.2. Although sampling outside of permafrost environments for aDNA studies has increased, the total number of identified clade I and III individuals in Europe remains limited (n = 23 mitochondrial genomes, hypervariable mitochondrial region n = 17). Thus, as yet, the turnover event of western Eurasian woolly mammoths have not been extensively explored.
Gaining a greater understanding of mammoth population dynamics across Late Pleistocene Europe also has relevance in smaller geographic regions, due to the unusual interactions of the mammoth with other species at particular archaeological sites. Drucker et al 7. showed through 13C and 15N isotopic analysis of bones from the southern German site of Geißenklösterle that the normally distinctive carbon and nitrogen dietary position of the woolly mammoth8,9 was atypically being “shared” by horses during both Aurignacian and Gravettian cultural periods (42 ka–~26 ka cal BP10); a pattern not found further south-west at Abri Pataud (France). It was hypothesised by the authors7 that these mammoths may have belonged to clade III, and thus the stable isotopic pattern of the site may be related to the turnover proposed by Palkopoulou et al.2.
Here, we aimed to refine understanding of the European late Pleistocene woolly mammoth replacement event in a region that has been sparsely sampled. Furthermore, we do this in the context of unusual isotopic characteristics found at sites in the area. Utilising biomolecular methods (ZooMS, stable isotopes and/or ancient DNA), we screen for mammoth specimens in 48 fragmentary or previously analysed faunal material or collagen samples7,11–13, spanning ~38 ka–15 ka cal BP from across central Europe (Supplementary Data S1). Using in-solution enrichment methods on the bone samples, we nearly double the number of dated mammoth mitochondrial genomes from Europe, and in combination with new and previously published stable isotopic and mtDNA data we begin to improve our understanding of late Pleistocene woolly mammoth population turnover in western Eurasia.
Results
Collagen Preservation and Stable Isotopes
Collagen of sixteen previously unanalysed specimens from Geißenklösterle and Hohle Fels (Germany) produced here C:Ncoll values of between 2.9–3.614 and had percentages of Ccoll above 8% and Ncoll above 5%15,16, representing well preserved and reliable collagen values (Methods and Supplementary Data S2).
The corpus of previously published isotopic data from Geißenklösterle and Hohle Fels from the Late Pleistocene was augmented with the new 13C and 15N values of mammoth (n = 9), rhino (n = 6) and horse (n = 1) (Supplementary Data S2). Species identification was isotopically inferred through Ward’s minimum variance clustering alongside previously identified specimens (See Methods and Supplementary Information).
The new isotopic values on mammoth were consistent with those measured previously7,13, with typical ca. 3–5‰ higher 15N values than those of reindeer (Figure S1). The unusual overlap in 15N values between horse and mammoth were already described at Geißenklösterle from the same chronological context7,13. This unusual pattern at Geißenklösterle is emphasized by the different pattern at Hohle Fels whereby, based on current sampling, horses and mammoths seem to have maintained the ‘isotopic separation’ expected of the classic mammoth steppe during the Aurignacian cultural period (~37 ka cal BP). A single horse found in the Aurignacian-Gravettian transitional layer at Hohle Fels (IIe) suggests that the same pattern may have also occurred at Hohle Fels later - but more stable isotope data of all species from the secure Gravettian layers are required to verify this.
ZooMS
The osteological remains of woolly mammoth in central Europe, particularly at Geißenklösterle and Hohle Fels, are often highly fragmentary making morphological identification difficult17, limiting classification to “mammoth/rhino sized”-like categories18. This also often results in variability in DNA preservation. Additionally, previous molecular protocols required large amount of bone material, resulting in large deformations of specimens. In response, curators now often restrict sampling of well preserved morphologically-secure specimens, making access to this material difficult. As ZooMS uses collagen (a biomolecule that generally preserves better than aDNA), this represents a cheap and fast method of confirming species identification when other methods fail19.
To improve confidence of uncertain morphological, stable isotopically inferred, and/or DNA-based species assignment (see below) at Geißenklösterle and Hohle Fels (Methods and Supplementary Information), ZooMS was performed on sub-sampled collagen extracted from eleven previously or newly analysed collagen samples for stable isotope analysis (above). Spectral quality was good for all eleven collagen samples analysed, providing direct molecular species identification of the stable isotope results that nine should be identified as Elephantidae and two as Rhinocerotidae (See Supplementary Data S3). For both mammalian families, ZooMS is not able to differentiate between possible Late Pleistocene genera in western Eurasia20. For Elephantidae, these specimens probably represent Mammuthus (as opposed to Palaeoloxodon), and for Rhinocerotidae an attribution to Coelodonta seems likely (opposed to Stephanorhinus), as per zooarchaeological assessment of the sites21,22. ZooMS identifications are in agreement with previous morphological observations as well as dietary isotopic signatures (see Table 1, Supplementary Data S2 and S3).
Table 1.
Details of late Pleistocene Central European woolly mammoth specimens successfully generating more than three fold coverage and more than 66% complete mitochondrial genomes after insolution enrichment.
| Sample | Location | 14C Date (cal.) | ZooArch Ident. | δ13C(‰) | δ15N (‰) | ZooMS Ident. | % Endogenous | mtDNA (fold) | mtDNA % | Frag. Length | mtClade |
|---|---|---|---|---|---|---|---|---|---|---|---|
| JK2760 | Hohle Fels, DE | 31812 | Mammoth/Rhino Size | −21.0 | 8.5 | — | 4.296 | 86.54 | 98 | 80.95 | III |
| JK2761 | Hohle Fels, DE | — | Mammoth/Rhino Size | −21.3 | 8.3 | — | 0.222 | 5.29 | 72 | 79.02 | III |
| JK2762 | Geißenklösterle, DE | 37904 | Mammoth/Rhino Size | −21.0 | 9.0 | Elephantidae | 0.215 | 22.23 | 93 | 79.04 | III |
| JK2764 | Geißenklösterle, DE | 38031 | Mammoth | −21.1* | 8.0* | Elephantidae | 0.606 | 243.18 | 98 | 73.35 | III |
| JK2765 | Hohle Fels, DE | 32226* | Mammoth/Rhino Size | −21.1 | 8.5 | Elephantidae | 0.902 | 109.99 | 97 | 70.40 | III |
| JK2766 | Hohle Fels, DE | 35127* | Mammoth | −21.1* | 8.0* | — | 1.019 | 79.49 | 97 | 79.00 | III |
| JK2768 | Hohle Fels, DE | 31666 | Mammoth/Rhino Size | −20.8 | 8.5 | Elephantidae | 1.72 | 60.40 | 98 | 87.13 | III |
| JK2769 | Geißenklösterle, DE | 37360 | Mammoth | −21.1* | 8.9* | Elephantidae | 0.342 | 27.95 | 93 | 74.68 | III |
| JK2770 | Hohle Fels, DE | 31683 | Mammoth/RhinoSize | −20.8 | 8.4 | — | 2.728 | 164.57 | 98 | 82.25 | III |
| JK2771 | Geißenklösterle, DE | — | Mammoth | −21.2* | 8.8* | — | 0.126 | 8.89 | 82 | 82.26 | III |
| JK2772 | Hohle Fels, DE | 38336 | Mammoth/Rhino Size | −21.1 | 9.0 | — | 6.018 | 230.59 | 98 | 73.93 | III |
| JK2773 | Hohle Fels, DE | 34924 | Mammoth/Rhino Size | −21.1 | 9.0 | Elephantidae | 0.429 | 29.41 | 95 | 79.81 | III |
| JK2774 | Geißenklösterle, DE | — | Mammoth | −21.1* | 9.1* | — | 0.256 | 5.68 | 74 | 74.37 | III |
| JK2779 | Geißenklösterle, DE | — | Mammoth | −20.9* | 8.3* | — | 0.038 | 11.63 | 86 | 78.58 | III |
| JK2780 | Geißenklösterle, DE | 31762* | Mammoth | −20.9* | 8.7* | Elephantidae | 0.048 | 7.27 | 77 | 78.24 | III |
| JK2782 | Kesslerloch, CH | 15409* | Mammoth | −20.5* | 6.4* | — | 22.694 | 108.30 | 99 | 90.14 | I |
| JK2790 | Kraków Spadzista, PL | 26966* | Mammoth | −20.3* | 8.7* | — | 0.765 | 216.37 | 98 | 72.02 | I |
| JK2796 | Kraków Spadzista, PL | 22961* | Mammoth | −20.3* | 9.0* | — | 0.453 | 6.72 | 78 | 67.91 | I |
| JK2802 | Kraków Spadzista, PL | — | Mammoth† | −20.8* | 9.5* | — | 15.563 | 522.97 | 99 | 77.40 | I |
| JK2803 | Kraków Spadzista, PL | 26947* | Mammoth | −20.2* | 9.0* | — | 0.091 | 11.86 | 85 | 68.59 | I |
Radiocarbon (14C) dates were calibrated with OxCal 4.262 to the Intcal 13 curve63. Endogenous DNA calculation is derived from shotgun sequencing of the woolly mammoth NGS libraries. Fragment lengths are in base pairs (bp). Asterixis (*) indicate previously published data. Dagger (†) represents individuals with tooth furrows. More detailed summaries can be found in Supplementary Data S1–S6.
Ancient DNA
For ancient DNA analysis, 44 bone and teeth samples were selected from Kesslerloch (Switzerland), Kraków Spadzista (Poland), Předmostί (Czech Republic), and additional individuals from Geißenklösterle and Hohle Fels to those analysed above. A summary of analysis performed per sample can be found in Supplementary Data S1.
In-solution capture of mammoth mitochondrial genomes was successfully carried out with capture baits generated by long-range PCR using three pairs of newly designed primers (Supplementary Data S4) on DNA extracted from elephant blood (see Methods and Supplementary Information). Of the forty-four samples genetically tested, after capture twenty produced mitochondrial genomes meeting the ‘relaxed’ criteria of Chang et al 3. (see Table 1 and methods), resulting in 5.3 to 522.97-fold coverage with 72%–99% of the mtDNA genome covered (Table 1). Additionally, of these, 13 met the ‘strict’ Chang et al 3. criteria of >10-fold average coverage with 90% nucleotide support (range x 22.2–x522.97) and >80% (range 81%–98%) of the mitochondrial genome covered. Capture efficiency (endogenous DNA percentage after/before capture) of these 20 mtDNAs ranged from 0.8%–78.9% (Supplementary Data S5). All mitochondrial genomes had C to T damage patterns indicative of authentic aDNA23 (Supplementary Figure S3). The number of samples for each site producing mitochondrial genomes is as follows: Kesslerloch (n = 1), Geißenklösterle (n = 7), Hohle Fels (n = 8), Kraków Spadzista (n = 4). Shotgun data from the Předmostί specimens all presented low values for endogenous DNA (0.005%–0.024%, Supplementary Data S5), suggesting bad DNA preservation at the site. Both Kraków Spadzista and Předmostί are open air rather than the cave environments of the other sites, the former type making specimens more susceptible to weathering, temperature fluctuation, pH and precipitation changes, which could lead to variable levels of DNA preservation24.
To confirm isotopic observations and ZooMS taxonomic identification, all shotgun libraries were mapped to a white rhinoceros assembly (Ceratotherium simum simum; NCBI Accession: GCA_000283155.1). A ten-fold increase in DNA mapping to the rhinoceros genome sequence versus a hybrid elephant/mammoth genome sequence (see Methods) identified the morphologically identified “mammoth/rhino size” Hohle Fels rib fragment sample HF_67565_89_1632_Va as rhinoceros (Supplementary Data S5), in agreement with isotopic and ZooMS observations. When sufficient DNA preservation was present, all other “mammoth/rhino size” samples from Geißenklösterle and Hohle Fels taxonomically identified through isotopes or ZooMS were confirmed as mammoth.
The mitochondrial genomes from Geißenklösterle, Hohle Fels, Kraków Spadzista and Kesslerloch were aligned to those analysed in Chang et al 3. with Elephas maximus (NCBI Accession: EF588275) as outgroup, for a total of one hundred and sixty-four mitochondrial sequences spanning Eurasia and North America. The addition of the mtDNAs reconstructed here increases the number of European mitochondrial genomes (n = 22 in Chang et al 3. to n = 42). Due to inconsistent gaps in the alignment caused by different reconstruction methods across previous publications, the D-Loop (positions 15,422–16,770 on the ‘Krause’ reference sequence) was removed for downstream analysis.
Of the twenty samples preserving enough DNA for complete mitochondrial genome reconstruction, the Geißenklösterle and Hohle Fels specimens consisted of only clade III, whereas Kraków Spadzista and Kesslerloch only contained clade I individuals - representing the earliest mitochondrial genomes of the migrating clade I population in central Europe.
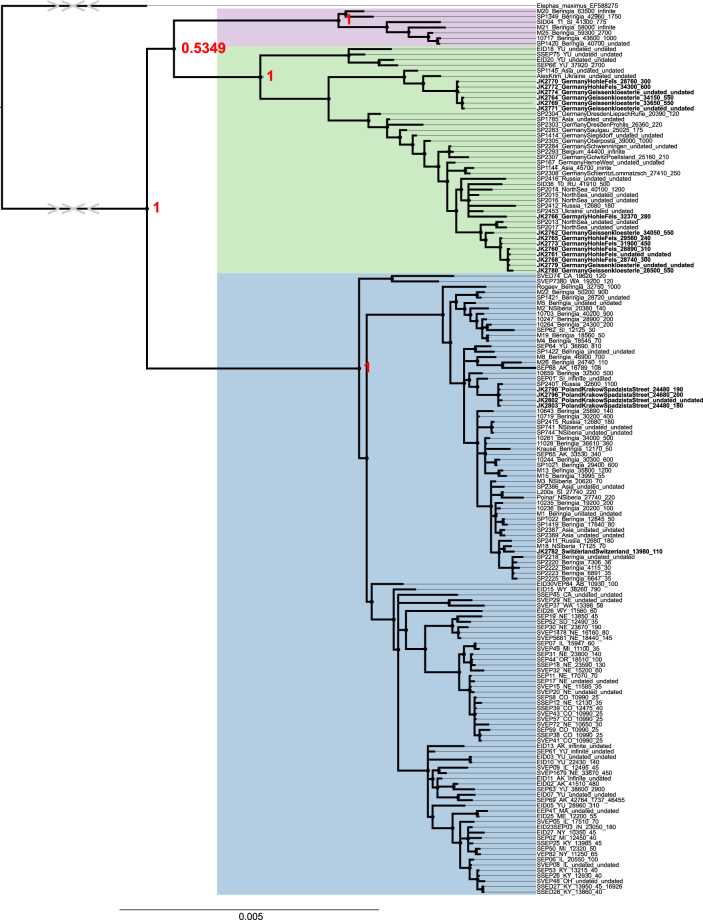
Clustering of the samples into three clades agrees with previous mammoth mitochondrial analyses1–5,25. However it was observed that here, as well as in all studies presenting Bayesian or Maximum Likelihood (ML) phylogenetic trees including all three mtDNA clades, the node connecting clade II and III in the majority of trees had low posterior probability or bootstrap values (Fig. 1) - regardless of phylogenetic method, evolutionary model and sequence completeness. While the BEAST coalescence tree from Enk et al.4 showed stronger support on this node (in contrast to ML), the Chang et al.3 tree that includes many more clade III mitochondrial sequences again shows low support. This low support could be considered to represent a trifurcation in the relationship between the three clades as explicitly presented in the ML tree presented by Enk et al.4, thus making these relationships difficult to resolve. This was further explored with our dataset by generating Neighbour-Joining (NJ), Maximum Parsimony (MP), ML, and Bayesian trees (see Methods and Supplementary Information) which again showed a low statistical support on the node connecting clade II and III (Fig. 1, Supplementary Figures S3–S6). As this suggested that there are potential difficulties in the ability to resolve the relationships between the three clades, an additional test to assess sequence relationships between each possible pairing of clades was performed by comparing the number of ‘shared’ derived SNPs between each clade (see Supplementary Information). This identified a rather high number of ‘homoplasic’ sites - the same mutations occurring independently on each clade, rather than following the expected phylogenetic relationships - suggesting that a typical ‘strict’ tree-like evolution in woolly mammoth mitochondrial lineages may not have occurred. The MP tree identified 65 derived positions for clade I, 70 for clade II, and 50 for clade III. In contrast, the average number of ‘shared’ derived SNPs sites are as follows: between clade II and clade I 24, between clade II and III 27, between clade I and III 17; out of approximately 15.4 thousand base pairs. The majority of the protein-coding gene related SNPs were found to be synonymous (n = 25), and the non-synonymous positions (n = 4) were distributed across the entire genome with no clustering on particular genes (Supplementary Data S6, Supplementary Information).
Figure 1.
Bayesian phylogenetic tree of all previously published woolly mammoth mammoths mitochondrial genomes3–5,25–27 and 20 mitochondrial genomes presented here (in bold). Node values represent posterior probability values from 100,000,000 steps for the bayesian tree. For visibility reasons, only support values related to the relationship of the three clades are shown. Clades are indicated by the following colours: Blue - I; Purple - II; Green - III. Radiocarbon dates in sample names are uncalibrated. The full tree as well as Neighbour-Joining, Maximum Parsimony and Likelihood trees can be found in Supplementary Figures S4–S7. Trees were generated using Figtree (tree.bio.ed.ac.uk/software/figtree/) and modified in Inkscape (inkscape.org).
Discussion
A variety of methods have been used for the reconstruction of woolly mammoth mitochondrial genomes, including multiplex PCR3,5,26,27, shotgun sequencing5,25,28, array capture3, and in-solution capture with in silico designed baits4. Here, the in-solution capture method of Maricic et al 29. was to our knowledge for the first time successfully applied to mammoth specimens, utilising DNA baits amplified from elephant blood using only three pairs of primers. Baits generated in-house from DNA of related species is likely to be more cost-effective than commercial kits (as used in4) as well as expressing less reference bias for individual mtDNA lineages while producing similarly high mtDNA average coverages. We have nearly doubled the number of radiocarbon dated European clade III genomes (>3-fold coverage, n = 12 and n = 11 respectively). Five out of six tested “mammoth/rhino” sized bones actually belong to mammoths, which increases the proportion of securely identified mammoth remains found in the archaeological assemblages at Geißenklösterle and Hohle Fels. The identification of rhinoceros specimens as a byproduct of our main analysis highlights the benefits of screening otherwise species-of-interest ‘negative’ aDNA libraries or collagen extracts from ‘undetermined’ bone. Screening aDNA reads of off-target samples by mapping to different genomes (as above) requires no extra laboratory work, and collagen fingerprinting methods such as ZooMS can be cheaply applied to leftovers of collagen extraction for isotopic analysis or radiocarbon dating30,31. For example, ZooMS was still able to confirm the zooarchaeological identification for the Geißenklösterle sample JK2778 (GK_11499_58_279_IIb) when genetic identification failed.
Analysis of woolly mammoth mitochondrial genomes performed here has shown that the low statistical support for the relationship between clade I, II and III can be potentially attributed to a similarly high number of ‘shared’ derived SNPs between each pair of clades; complicating the resolving of the maternal phylogenetic tree. These derived SNPs comprise of around one third or more of the derived positions present on each branch between the MRCA of all clades and MRCA of each clade. To ensure these positions are real, the mapping strategy used in this study removes potential NUMTs fragments (nuclear-mitochondrial sequences) that could cause false SNP calls (despite the low likelihood of this happening25) by removing any fragment that mapped to both nuclear and mitochondrial genomes. We also find these homoplasic positions remain even when comparing mitochondrial genomes produced by different methods (multiplex PCR, array capture, in-solution capture and shotgun sequencing), suggesting this is not a protocol artefact. Additionally, the consensus genomes generated here were called with a penalising filter for damage, making typical aDNA damage, increased frequency of C to T miscoding lesions at the end of fragments, also unlikely to be the cause of these ‘homoplasies’. This was confirmed by visual inspection of read alignments. Biologically, although not the only possibilities, this pattern is consistent with processes such as recombination or convergent evolution. This would be unusual as mitochondria are typically considered unlikely to undergo recombination as having more than one mitochondrial haplotype after fertilisation is rare32,33, and convergent evolution is expected to generally lead to a higher number of non-synonymous than synonymous mutations33. Although not an issue for the aims of this study, this pattern should be further scrutinised as this could suggest current interpretations of the maternal evolutionary history of the mammoth may be more complicated than previously thought. As the statistical support and number of ‘shared’ derived SNPs seem to show that the data does not follow a tree-like pattern, it may be that it is not possible to apply dating or classical tree-based phylogenetic methods on mammoth mitochondrial data34. This could possibly call into question the reliability of the Bayesian dating analyses performed previously2–5.
New radiocarbon dates of the identified mammoth samples presented here (Table 1, Supplementary Data S1) confirm the continuation of the existence of clade III mtDNA3 after 34 ka cal BP in Europe. Whilst there is no mixture of individuals from the two clades at the sites analysed in this study, these data do not yet reject the possibility of a coexistence being found in eastern Europe between clade III and clade I. Indeed, the Kraków Spadzista mitochondrial genomes indicate that clade I was already established in central Europe by 27 ka cal BP, much further west and earlier than previously detected2,3 (Fig. 2). Thus, the increased sampling of Europe (Chang et al.3 this study) is indicating that our understanding of this replacement event may be less clear than ‘just’ a extirpation of clade III and subsequent arrival of clade I with no ‘temporal overlap’ as proposed previously2. Indeed, a synthesis of all mammoth dates in Europe by Lorenzen et al 35. and Puzachenko et al 36. has shown mammoth remains being present in Europe throughout this period.
Figure 2.
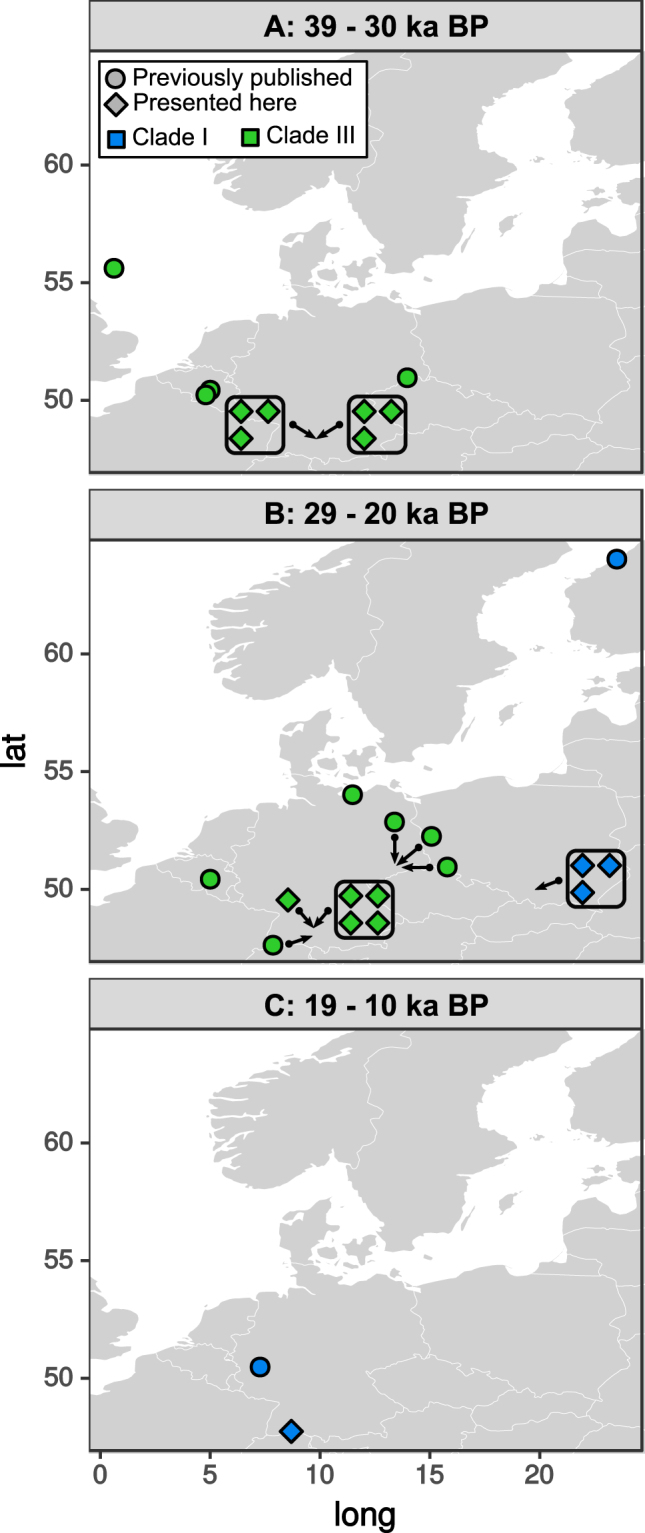
Time series of all genetically typed and radiocarbon dated European woolly mammoths from this study (diamonds), and dated HVR samples and full mitochondrial genomes analysed in2,3 (circles). Time slices are in uncalibrated BP. Clade III lineages (green) are present until 25 ky BP in Germany, but clade I (blue) is now shown to be present in central Europe by 24 ka BP (27 ka cal BP), as represented by the new Kraków Spadzista mitochondrial sequences. Maps were generated in R60 using the ggplot2 package61 and modified in Inkscape (inkscape.org).
The increasing number of mtDNA sequences from western Eurasia is uncovering a curiously similar pattern to one that occurred ten thousand years earlier in eastern Eurasia between clade I and clade II. Barnes et al 1. originally concluded that drift was the likely cause of the loss of clade II in a restricted population. In contrast, further Bayesian skyline analysis by Debruyne et al 6. did not detect a demographic bottleneck that would result from genetic drift in clade II, and thus put forward the possibility of some form of competitive replacement causing the rather sudden loss, when considered in the context of the timing of the arrival of clade I just prior to the loss of clade II. However, Gilbert et al 5. at the same time suggested that the mitochondria themselves did not show any functional differences that could provide some form of selective advantage. Both arguments could also potentially be applied to the situation occurring in Europe between clade III and I ten thousand years later. More recent BEAST analysis on whole Eurasian datasets by Palkopoulou et al 2. did not detect any noticeable reduction of the effective population size during this period. Nuclear DNA would potentially allow to test for functional differences between all three populations, defined by mtDNA clades, in particular why clade I seems to be more ‘dominant’ than the other two clades. This type of data would be particularly pertinent due to the ‘contradictions’ identified in our study in the homoplasies observed between the different clades along the mitochondrial genome, thus questioning our ability of current models to elucidate the intra-population dynamics of the woolly mammoth.
Generated from highly fragmentary remains, the 20 central European Late Pleistocene mitochondrial genomes produced here increases the number of mitochondrial genome sequences from this region. Unusual patterns and difficulties in resolving the relationship between the three clades due to sequences not seemingly following a strict ‘binary’ tree-like pattern have been raised here. This questions previous demographic and genetic dating analyses and thus requires further investigation and scrutiny of these ancient data. The new mitochondrial genomes represent the earliest European clade I individuals at 27 ka cal BP, and corroborate the existence of clade III in Europe past 34 ka cal BP. Increasing numbers of samples are reducing the ‘temporal gap’ between the respective occupations of the two clades in Europe. This pattern is starting to indicate parallels with a replacement event that occurred previously between clade I and clade II in Beringia, however the nature of both events remain unresolved and are becoming even less clear than previously considered. More genetic sampling from chronologically secured sites in Europe before and throughout the LGM is required to confirm a loss of clade III before the loss of clade I. Analysis of male lineage markers as well as autosomal chromosomes is required to help disentangle the increasingly complex relationship of different mammoth groups and may provide additional evidence for the local and global extinction of one of the largest members of the Pleistocene megafauna.
Methods
Sample Selection and Zooarchaeological Identification
Forty-four samples spanning ~34 ka to 13 ka BP (38 ka −15 ka cal BP) were obtained from archaeological collections or previous stable isotopic analysis7,11–13 from the Late Pleistocene sites: Kesslerloch (Switzerland, n = 1), Geißenklösterle (Germany, n = 12), Hohle Fels (Germany, n = 10), Předmostί (Czech Republic n = 6), and Kraków Spadzista (Poland, n = 15) (See Supplementary Data S1). Three further collagen samples from Geißenklösterle that were previously radiocarbon dated10 were analysed for stable isotopic and ZooMS analysis. An additional collagen sample from Hohle Fels from previous dating was also analysed for carbon and nitrogen stable isotopes.
For samples from Geißenklösterle and Hohle Fels, a zooarchaeological identification of woolly mammoth was not always possible due to high fragmentation of remains (especially of ribs, an important raw material for the production of tools at the site13). Although the megafaunal remains also include woolly rhino, identifiable remains from this species are not as abundant; therefore the megafaunal remains are considered to be dominated by woolly mammoth in the Aurignacian and Gravettian cultural layers22. In cases of unclear identification the size class of “mammoth/rhino” was assigned (Supplementary Data S1).
The samples from all sites were additionally chosen based on previously published direct 14C dates or secure stratigraphic layers prior the Last Glacial Maximum ranging from ~36 ka–~19 ka 14C (based on dates of archaeological layers) and a single individual after the Last Glacial Maximum dated to 13 ka BP (Kesslerloch). Overall, the samples selected here represent the date range of the estimated extinction of clade III and earliest European clade I woolly mammoths2,3.
Bone fragments from Geißenklösterle and Hohle Fels were sampled from ribs, vertebrae, a skull and a radius. From Kraków Spadzista, the dentine of the molars and the bone from a mandible were taken using a dremel circular saw. The remaining samples came from previous isotopic analysis in the form of bone or tooth powder, or collagen11–13.
Collagen Extraction and Isotopic Analysis
For collagen preservation screening, bone powder remaining from aDNA sampling (see below) from Geißenklösterle and Hohle Fels was analysed through elemental analysis37, performed on a Vario EL III elemental analyzer (Department of Geography, University of Tübingen). Collagen was extracted following a modified Longin38 protocol as by Bocherens et al.39. Collagen δ13C and δ15N values were then measured on a ThermoQuest Delta + XL mass spectrometer (Department of Geosciences, University of Tübingen). International standards for 13C (V-PDB) and 15N (atmospheric air) were used and measurements were normalised to δ13C values of USGS24 (δ13C = −16.00) and to δ15N values of IAEA 305 A (δ15 n = 39.80). Analytical error, based on within-run replicate measurements of laboratory standards (albumen, modern collagen, USGS 24, IAEA 305 A) was ±0.1 for δ13C and ±0.2 for δ15N.
Collagen from the isotopic analysis of specimens yielding mitochondrial genomes (see below) and that had not been previously dated was additionally sub-sampled for radiocarbon dating at the Oxford Radiocarbon Accelerator Unit (ORAU). 14C dates were converted to F 14C dates and then calibrated in OxCal 4.2 using the IntCal13 calibration curve. Calibrated 14C dates are indicated by ‘cal BP’ (Supplementary Data S1).
Cluster analysis was performed in JMP v11.1.1 (SAS) using Ward’s minimum variance.
ZooMS Identification
Collagen extracted from samples from Geißenklösterle and Hohle Fels during isotopic analysis was subsampled (<1 mg dry collagen) for collagen fingerprinting by Zooarchaeology by Mass Spectrometry (ZooMS40). Eleven samples either identified as “mammoth” or “rhino” were chosen to provide within-site spectral references when assessing samples assigned “mammoth/rhino size” from morphological and isotopic observations. These included seven samples from Geißenklösterle and four from Hohle Fels (Supplementary Data S2). ZooMS MALDI-TOF-MS analysis was conducted following Welker et al.41, (see Supplementary Information) and peptide markers were identified against the ZooMS sequence database presented by Welker et al.20. ZooMS relies on collagen type I (COL1) amino acid sequence variation to differentiate between closely related species. As collagen contains less substitutions compared to genetic sequences and ZooMS only uses a subset of the phylogenetic information present in COL142, ZooMS taxonomic identifications are commonly on the (sub)family level. Hence, for Elephantidae standard ZooMS screening is not able to differentiate between Mammuthus and Palaeoloxodon, the two Late Pleistocene Elephantid genera present in Eurasia43.
Sample Preparation, DNA Extraction and Sequencing
All aDNA work prior to amplification was performed in a dedicated aDNA laboratory at the University of Tübingen, following aDNA clean room protocols44. In brief, the positive pressure laboratory is UV irradiated daily and surfaces regularly cleaned with bleach, and separate dedicated hoods for reagent preparation, sampling and reactions are used. All laboratory consumables and reagents are UV irradiated when possible. Full clean room suits with hairnets, masks and double pairs of gloves are worn. Positive extraction controls in the form of an internal well-characterised cave bear bone were included, and one negative control during each extraction and two controls per library preparation session.
aDNA extraction was performed using a modified version of a silica-based protocol45. Extracts were converted to double indexed sequencing libraries46,47 and transferred to a modern DNA lab for amplification (Supplementary Data S4).
Enrichment of mitochondrial DNA was performed following the protocol of Maricic et al.29. Template material was provided by Wilhelma Zoo (Stuttgart, Germany) as a subsample of blood taken from two Asian elephants after routine veterinary care by the in-house Veterinary practitioner. Blood was drawn using standard venipuncture techniques in accordance with the relevant veterinary regulations and practises of the Zoo. No experimentation was performed on the elephants for the purpose of this study. The DNA extracted using a QIAamp DNA Blood Mini kit (Qiagen). Three new primer pairs (Supplementary Data S4) were manually designed to produce three ~5.5 k bp amplicons spanning the entire Elephas maximus mitochondrial genome (NCBI Accession: EF588275.2) through Long-Range PCR (LR-PCR). Amplicons sonicated to an length of <1000 bp were converted to biotinylated bait libraries and in-solution bead capture was performed29.
Pre- and post-enrichment libraries were then sequenced on an Illumina HiSeq 2500 (University of Tübingen). More information about methods from all laboratory protocols can be found in the Supplementary Information.
aDNA Pre-Processing, Consensus Calling and Phylogenetic Analysis
Endogenous DNA quantification, aDNA authentication and mitochondrial genome reconstruction were performed with an in-house version of the NGS data processing pipeline EAGER v1.9248 using a hybrid reference consisting of the African elephant nuclear genome and the woolly mammoth mitochondrial genome (NCBI Accession: GCA_000001905.1 and NC_007596.2 respectively).
The final consensus sequences were called by GenConS (Generate Consensus Sequence, v1), a new consensus caller tool designed here to reduce the risk of false positive SNP positions that may be erroneously called due to typical aDNA damaged molecules49,50. In brief, taking a SNP and optionally an InDel VCF file as input (both were used in this study), a position is analysed to assess whether an aDNA miscoding pattern49 is present prior to SNP calling. If a damage pattern is detected, the coverage of the miscoding base is artificially reduced in order to reduce the chance of a false-positive SNP call due to damage. Additional thresholds apply: a minimum coverage for a position to be called (5 in this study) and a minimal allele frequency for a call to made (75% in this study). When either are not met an ‘N’ is called. The resulting consensus sequence is written in FASTA format. Furthermore, a file in Consensus Call Format is generated giving detailed information of each consensus call (further information about GenConS can be found in the Supplementary Information). GenConS is available in the TOPAS package at www.github.com/subwaystation/TOPAS.
Previously published mitochondrial genomes3–5,25–27,51 were downloaded from the NCBI SRA database, and the consensus sequences generated here were aligned with these using ‘Consensus Align’ in Geneious vR852. Evolutionary models were estimated using modelgenerator v8553, jModelTest2 v2.1.1054 and IQ-Tree v1.4.255, and phylogenetic analysis performed using Geneious vR8 for Neighbour Joining, MEGA for Maximum Parsimony56, IQ-Tree v1.4.2 for Maximum Likelihood57,58, and MrBayes v3.2.6 for Bayesian59 methods (Supplementary Information, Supplementary Data S6).
Accession Codes
Sequencing libraries have been deposited in the European Nucleotide Archive (ENA) with the project accession PRJEB21582. Final mitochondrial sequences have been stored in the NCBI GenBank database with accession numbers MF579931-MF579950.
Electronic supplementary material
Acknowledgements
We thank Maria Malina for assistance with the Geißenklösterle and Hohle Fels collections. We also express our gratitude to Elephants Pama and Zella, as well as Annika Weigold and Tobias Knauf-Witzens at Wilhelma Zoo, Stuttgart for donation of elephant blood. We also acknowledge Maria Spyrou for help with primer design. Matthew J. Collins, Annabell Reiner and Luke Spindler are thanked for support in the contexts of the proteomics work. We thank Dan Chang and Beth Shapiro for providing sequences from their study. M.L-G. is funded by a grant from the Czech Science Foundation (GACR 15-06446S) entitled “The relationships Between humans and larges canids - the dogs and wolves of the Gravettian Pˇredmostί site (Moravia)”. This work was partly supported by a DAAD One Year Postgraduate Scholarship to J.A.F.Y. and the Max Planck Society.
Author Contributions
J.A.F.Y., D.G.D., S.C.M., H.B., A.H. and J.K. conceived and designed the research. D.G.D., S.C.M., P.W., M.L-G., N.J.C., and H.B. provided samples and archaeological context. D.G.D conducted collagen extraction, stable isotopic analysis and analysed data. F.W conducted ZooMS analysis and analysed data. J.A.F.Y. and E.R performed sampling, DNA extraction and library preparation of sequencing libraries. J.A.F.Y., S.H. and A.H. performed aDNA bioinformatic analysis. J.A.F.Y. wrote the manuscript with input from all authors. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17723-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
James A. Fellows Yates, Email: fellows@shh.mpg.de
Johannes Krause, Email: krause@shh.mpg.de.
References
- 1.Barnes I, et al. Genetic structure and extinction of the woolly mammoth. Mammuthus primigenius. Current Biology. 2007;17:1072–1075. doi: 10.1016/j.cub.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 2.Palkopoulou E, et al. Holarctic genetic structure and range dynamics in the woolly mammoth. Proceedings of the Royal Society B: Biological Sciences. 2013;280:20131910. doi: 10.1098/rspb.2013.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang D, et al. The evolutionary and phylogeographic history of woolly mammoths: a comprehensive mitogenomic analysis. Sci. Rep. 2017;7:44585. doi: 10.1038/srep44585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enk, J. et al. Mammuthus population dynamics in Late Pleistocene North America: Divergence, phylogeography, and introgression. Front. Ecol. Evol. 4 (2016).
- 5.Gilbert MTP, et al. Intraspecific phylogenetic analysis of Siberian woolly mammoths using complete mitochondrial genomes. Proc. Natl. Acad. Sci. USA. 2008;105:8327–8332. doi: 10.1073/pnas.0802315105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debruyne R, et al. Out of America: ancient DNA evidence for a new world origin of late quaternary woolly mammoths. Current Biology. 2008;18:1320–1326. doi: 10.1016/j.cub.2008.07.061. [DOI] [PubMed] [Google Scholar]
- 7.Drucker DG, et al. Tracking possible decline of woolly mammoth during the Gravettian in Dordogne (France) and the Ach valley (Germany) using multi-isotope tracking (13C, 14C, 15N, 34S, 18O) Quat. Int. 2015;359–360:304–317. doi: 10.1016/j.quaint.2014.11.028. [DOI] [Google Scholar]
- 8.Bocherens H. Isotopic biogeochemistry and the palaeoecology of the mammoth steppe fauna. Deinsea. 2003;9:57–76. [Google Scholar]
- 9.Schwartz-Narbonne R, Longstaffe FJ, Metcalfe JZ, Zazula G. Solving the woolly mammoth conundrum: amino acid 15N-enrichment suggests a distinct forage or habitat. Sci. Rep. 2015;5:9791. doi: 10.1038/srep09791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higham T, et al. Testing models for the beginnings of the Aurignacian and the advent of figurative art and music: the radiocarbon chronology of Geißenklösterle. J. Hum. Evol. 2012;62:664–676. doi: 10.1016/j.jhevol.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Drucker, D. G., Rivals, F., Münzel, S. C. & Bocherens, H. Stable isotope and microwear investigation on the mammoth (Mammuthus primigenius) of Kraków Spadzista: insights into diet and environment. In Wojtal, P., Wilczyński, J. & Haynes, G. (eds.) A Gravettian Site in Southern Poland: Kraków Spadzista, 189–202 (ISEA PAS, Kraków 2015).
- 12.Bocherens H, et al. Reconstruction of the Gravettian food-web at Pȓedmost I using multi-isotopic tracking (13c, 15N, 34S) of bone collagen. Quat. Int. 2015;359–360:211–228. doi: 10.1016/j.quaint.2014.09.044. [DOI] [Google Scholar]
- 13.Münzel SC, Wolf S, Drucker DG, Conard NJ. The exploitation of mammoth in the Swabian Jura (SW-Germany) during the aurignacian and gravettian period. Quat. Int. 2017;445:184–199. doi: 10.1016/j.quaint.2016.08.013. [DOI] [Google Scholar]
- 14.DeNiro MJ. Postmortem preservation and alteration of in vivo bone collagen isotope ratios in relation to palaeodietary reconstruction. Nature. 1985;317:806–809. doi: 10.1038/317806a0. [DOI] [Google Scholar]
- 15.DeNiro MJ, Weiner S. Chemical, enzymatic and spectroscopic characterization of “collagen” and other organic fractions from prehistoric bones. Geochim. Cosmochim. Acta. 1988;52:2197–2206. doi: 10.1016/0016-7037(88)90122-6. [DOI] [Google Scholar]
- 16.Ambrose SH. Preparation and characterization of bone and tooth collagen for isotopic analysis. J. Archaeol. Sci. 1990;17:431–451. doi: 10.1016/0305-4403(90)90007-R. [DOI] [Google Scholar]
- 17.Münzel, S. C. The production of Upper Palaeolithic mammoth bone artifacts from southwestern Germany. In Caverretta, G., Gioia, P., Muss, M. & Palombo, M. R. (eds) The World of Elephants - Proceedings Of The First International Congress, Rome2001, 448–454 (Consiglio Nazionale delle Ricerche - Roma, Rome, 2001).
- 18.Münzel, S. C. Mammoth remains in the Upper and Middle Paleolithic layers of Geißenklösterle cave (Ach valley, Swabian Jura, southwestern Germany): Hunting season, acquisition of raw material and tool production at Geißenklösterle cave. In Vialou, D., Renault-MIskovsky, J. & Patou-Mathis, M. (eds) Comportements des hommes du Paléolithique moyen et supérieur enEurope: territoires et milieux, 39–49 (ERAUL 111, Liege 2005).
- 19.Prendergast ME, et al. Reconstructing asian faunal introductions to eastern africa from multi-proxy biomolecular and archaeological datasets. PloS one. 2017;12:e0182565. doi: 10.1371/journal.pone.0182565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welker F, et al. Palaeoproteomic evidence identifies archaic hominins associated with the Châtelperronian at the Grotte du Renne. Proc. Natl. Acad. Sci. USA. 2016;113:11162–11167. doi: 10.1073/pnas.1605834113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Münzel, S. C. Subsistence patterns in the Gravettian of the Ach valley, a former tributary of the Danube in the Swabian Jura. In Svoboda, J. A. & Sedláčková, L. (eds) The Gravettian along the Danube, Proceedings of the Mikulov Conference, 20–21November 2002, 71–85 (Archeologicky ustav AV ČR, Brno, 2004).
- 22.Conard, N. J., Kitagawa, K., Krönneck, P., Böhme, M. & Münzel, S. C. The importance of fish, fowl and small mammals in the Paleolithic diet of the Swabian Jura, southwestern Germany. In Zooarchaeology and Modern Human Origins, Vertebrate Paleobiology and Paleoanthropology, 173–190 (Springer Netherlands 2013).
- 23.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bollongino R, Tresset A, Vigne J-D. Environment and excavation: Pre-lab impacts on ancient DNA analyses. C. R. Palevol. 2008;7:91–98. doi: 10.1016/j.crpv.2008.02.002. [DOI] [Google Scholar]
- 25.Gilbert MTP, et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
- 26.Krause J, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 27.Rogaev EI, et al. Complete mitochondrial genome and phylogeny of Pleistocene mammoth Mammuthus primigenius. PLoS Biol. 2006;4:e73. doi: 10.1371/journal.pbio.0040073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poinar HN, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 29.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talamo S, et al. Direct radiocarbon dating and genetic analyses on the purported neanderthal mandible from the Monti Lessini (Italy) Sci. Rep. 2016;6:29144. doi: 10.1038/srep29144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charlton, S. et al. Finding Britain’s last hunter-gatherers: A new biomolecular approach to ‘unidentifiable’bone fragments utilising bone collagen. J. Archaeol. Sci. (2016).
- 32.Piganeau G, Eyre-Walker A. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity. 2004;92:282–288. doi: 10.1038/sj.hdy.6800413. [DOI] [PubMed] [Google Scholar]
- 33.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294X.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 34.Rieux, A. & Balloux, F. Inferences from tip-calibrated phylogenies: a review and a practical guide. Mol. Ecol. (2016). [DOI] [PMC free article] [PubMed]
- 35.Lorenzen ED, et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 2011;479:359–364. doi: 10.1038/nature10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puzachenko, A. Y. et al. The Eurasian mammoth distribution during the second half of the Late Pleistocene and the Holocene: Regional aspects. Quat. Int. (2016). In Press.
- 37.Bocherens H, Drucker D, Billiou D, Moussa I. Une nouvelle approche pour évaluer l'état de conservation de l’os et du collagène pour les mesures isotopiques (datation au radiocarbone, isotopes stables du carbone et de l’azote) Anthropologie. 2005;109:557–567. doi: 10.1016/j.anthro.2005.06.005. [DOI] [Google Scholar]
- 38.Longin R. New method of collagen extraction for radiocarbon dating. Nature. 1971;230:241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 39.Bocherens H, et al. Paleobiological implications of the isotopic signatures (13C, 15N) of fossil mammal collagen in Scladina Cave (Sclayn, Belgium) Quat. Res. 1997;48:370–380. doi: 10.1006/qres.1997.1927. [DOI] [Google Scholar]
- 40.Buckley M, Collins M, Thomas-Oates J, Wilson JC. Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:3843–3854. doi: 10.1002/rcm.4316. [DOI] [PubMed] [Google Scholar]
- 41.Welker F, Soressi M, Rendu W, Hublin J-J, Collins MJ. Using ZooMS to identify fragmentary bone from the late Middle/Early Upper Palaeolithic sequence of Les Cottés, France. J. Archaeol. Sci. 2015;54:279–286. doi: 10.1016/j.jas.2014.12.010. [DOI] [Google Scholar]
- 42.Welker F, et al. Ancient proteins resolve the evolutionary history of Darwin’s South American ungulates. Nature. 2015;522:81–84. doi: 10.1038/nature14249. [DOI] [PubMed] [Google Scholar]
- 43.Buckley M, Larkin N, Collins M. Mammoth and mastodon collagen sequences; survival and utility. Geochim. Cosmochim. Acta. 2011;75:2007–2016. doi: 10.1016/j.gca.2011.01.022. [DOI] [Google Scholar]
- 44.Pääbo S, et al. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 45.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010:db.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 47.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peltzer A, et al. EAGER: efficient ancient genome reconstruction. Genome Biol. 2016;17:1–14. doi: 10.1186/s13059-016-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briggs AW, et al. Patterns of damage in genomic DNA sequences from a neandertal. Proc. Natl. Acad. Sci. USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parks M, Lambert D. Impacts of low coverage depths and post-mortem DNA damage on variant calling: a simulation study. BMC Genomics. 2015;16:19. doi: 10.1186/s12864-015-1219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Enk J, et al. Complete Columbian mammoth mitogenome suggests interbreeding with woolly mammoths. Genome Biol. 2011;12:R51. doi: 10.1186/gb-2011-12-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kearse M, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chernomor, O., von Haeseler, A. & Minh, B. Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. (2016). [DOI] [PMC free article] [PubMed]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronquist F, et al. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org.
- 61.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2009).
- 62.Ramsey CB. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51:337–360. doi: 10.1017/S0033822200033865. [DOI] [Google Scholar]
- 63.Reimer PJ, et al. IntCal13 and marine13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon. 2013;55:1869–1887. doi: 10.2458/azu_js_rc.55.16947. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.