Abstract
Rhodococcus genome sequence analysis has revealed a surprisingly large (and unexplored) potential for the production of secondary metabolites. Also, putative γ-butyrolactone gene clusters have been identified in some Rhodococci. These signalling molecules are known to regulate secondary metabolism in Streptomyces. This work provides evidence for synthesis of a γ-butyrolactone(-like) molecule by Rhodococci (RJB), the first report in the Rhodococcus genus. The Rhodococcus jostii RHA1 RJB molecule was detected by a reporter system based on the γ-butyrolactone receptor protein (ScbR) of Streptomyces coelicolor. This RJB is structurally identical to 6-dehydro SCB2, the predicted precursor of the S. coelicolor γ-butyrolactone SCB2. The R. jostii RHA1 key RJB biosynthesis gene was identified (gblA): Deletion of gblA resulted in complete loss of RJB synthesis whereas higher RJB levels were detected when gblA was overexpressed. Interaction of the RJB molecule with ScbR indicates that communication may occur between these two Actinomycete genera in their natural habitat. Furthermore, RJB may provide a highly relevant tool for awakening cryptic secondary metabolic gene clusters in Rhodococci. This study provides preliminary evidence that R. jostii RHA1 indeed synthesizes diffusible molecules with antimicrobial activity, but a possible role for RJB in this remains to be established.
Introduction
Rhodococcus is a genus of aerobic, acid resistant, non-sporulating, Gram-positive soil bacteria (family Nocardiaceae, order Actinomycetales), which contain mycolic acids in their cell walls1. This genus is well-known for its catabolic versatility2–5, but little is known about its secondary metabolism. Computational analysis has shown that this genus has a great potential for synthesis of secondary metabolites4,6–10. Analysis of several Actinomycete genomes, including 4 strains of the genus Rhodococcus, R. jostii RHA1, R. equi 103 S, R. opacus B4 and R. erythropolis PR4, uncovered a relatively high percentage of genes encoding non-ribosomal peptide synthetases (NRPS) in Rhodococci. Also, conserved γ-butyrolactone biosynthesis gene clusters were identified in these Rhodococci7. The physiological roles of γ-butyrolactones have been extensively studied in members of the genus Streptomyces only, although their putative biosynthetic genes also appear to be present in other Actinomycete genera11. These signalling molecules are known to participate in the regulation of secondary metabolism and to induce a range of physiological responses12–15. γ-Butyrolactones bind to one or more receptor proteins, which belong to the TetR family of transcriptional regulators. Several of these receptor proteins have been described in the genus Streptomyces to be involved in controlling expression of secondary metabolite gene clusters15–19. The γ-butyrolactone TetR receptors bind to DNA, thus blocking the expression of the target genes. Binding of γ-butyrolactone ligands to the TetR regulators induces a change in the receptor protein conformation. The receptor therefore, cannot bind to the DNA anymore influencing the expression of the targeted genes. For example, in Streptomyces coelicolor the γ-butyrolactones receptor protein (ScbR) directly regulates the coelimycin gene cluster. Lack of γ-butyrolactones inhibits the production of coelimycin20. In Streptomyces griseus γ-butyrolactones are known to regulate streptomycin production and morphological differentiation15.
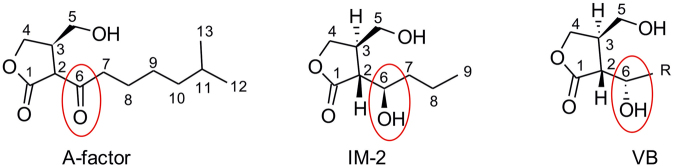
Three main groups of γ-butyrolactones have been described to date, classified according to their structures: A-factor type, which contains a keto group in the carbon 6 of the molecule; IM-2, with a hydroxyl group in the same carbon in the R configuration; and VB type, also with a hydroxyl group in the same carbon but in an S configuration21–25 (Fig. 1). Within each group there is further diversity depending on the structure of the acyl chain connected to carbon 6.
Figure 1.
Illustration of the three γ-butyrolactone structural types described to date, differing in group and conformation at the C6 position. A-factor was first described in Streptomyces griseus 15; IM-2 molecules were first identified in Streptomyces lavendulae 24; VB molecules were first reported for Streptomyces venezuelae 22. Adapted from Martin-Sanchez et al.27.
The enzymes involved in the biosynthesis of γ-butyrolactones have been described and partially characterized in several Streptomyces strains. They have been named according to the species that employ them or the compound that is regulated by these molecules15–17,19,26,27. For unification purposes, we have renamed these enzymes so that they can be applied to any butanolide system in any strain by employing the nomenclature Gbl (gamma-butyrolactone) (Table 1).
Table 1.
Proposed unified nomenclature of described GBL biosynthesis enzymes.
GblA (gamma-butyrolactone biosynthesis A) catalyses the first step of the biosynthesis by condensing a glycerol derivative with a fatty acid derivative (Compound 1, Fig. 2). This enzyme was first described in S. griseus where it was named AfsA (Fig. 2)15. After this step two different pathways have been predicted15. Reactions in Pathway A are believed to be catalysed by non-specific enzymes. Pathway B includes a reductase GblC (gamma-butyrolactone biosynthesis C), named BprA in S. griseus (Fig. 2)15 and a phosphatase that is thought to be a non-pathway-specific phosphatase. GblC is predicted to reduce the double bond in carbons 3 and 2 in the lactone ring (conversion of compound 4 into 5). In some species, there is also a short chain dehydrogenase (GblD, gamma-butyrolactone biosynthesis Dehydrogenase) that reduces the keto group in carbon 6 of 6-dehydro-γ-butyrolactone forms (compound 6 to compound 7 in Fig. 2).
Figure 2.
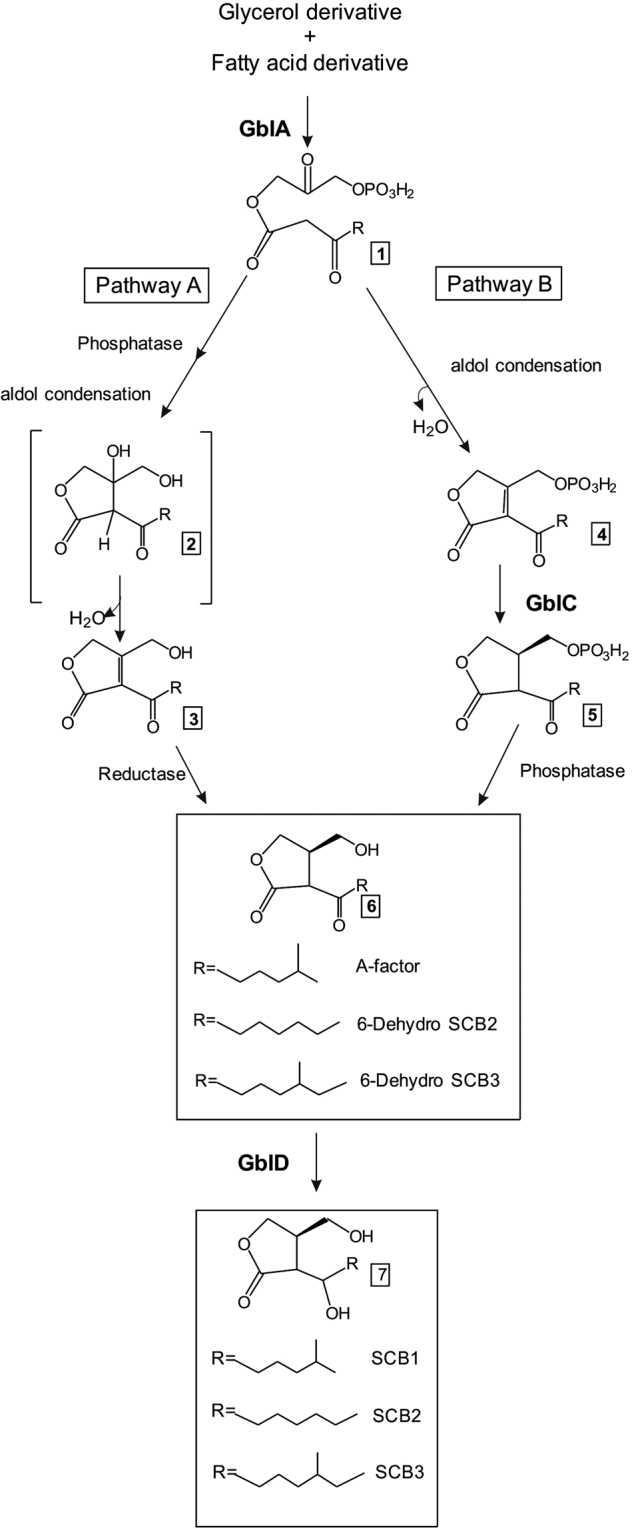
Predicted biosynthetic pathway of γ-butyrolactones in Streptomyces species (adapted from15,27,48) The generic names given to the enzymes in this work are shown at the (putative) steps that they catalyse. A-factor is the γ-butyrolactone from S. griseus. SCBs (Streptomyces Coelicolor Butyrolactones), γ-butyrolactones described in Streptomyces coelicolor.
The butanolide system has only been studied as a quorum-sensing system in Streptomyces but it has not been explored whether it can have a role in interspecies communication; there is evidence that other genera also have this system11,28. In this work, we identified a γ-butyrolactone(-like) molecule in R. jostii RHA1 (RJB), the first time that such a molecule has been identified in the genus Rhodococcus. A R. jostii RHA1 gblA deletion mutant did not induce the growth of a γ-butyrolactone reporter strain25, indicating that the gblA gene is essential for synthesis. Moreover, an overproduction of RJB was observed when gblA was overexpressed in R. jostii RHA1. LC-MS analysis of extracts from the R. jostii RHA1 WT strain and the derived gblA deletion and the gblA overexpression strains indicated that the RJB molecule synthesized has the same structure as 1) 6-dehydro SCB2, a stereoisomer of A-factor, the known γ-butyrolactone from S. griseus and 2) a predicted precursor of SCB2 (Streptomyces Coelicolor Butyrolactone 2) (Figs 1, 2), a known γ-butyrolactone from S. coelicolor 27.
Materials and Methods
Strains and growth conditions
All strains used in this study are described in Table 2 and the media used in Supplementary Table S1. Rhodococcus was grown at 30 °C. Luria-Bertani agar from Sigma-Aldrich was used as a standard medium. Appropriate antibiotics were used at the following concentrations: apramycin 50 μg/ml, kanamycin (Km) 200 μg/ml.
Table 2.
Microbial strains used in this work.
| Strain | Details | Reference |
|---|---|---|
| Rhodococcus jostii RHA1 | Wild type | 4 |
| RHA1-OE | R. jostii RHA1 + pRM4-gblA | This work |
| RHA1-ΔgblA | R. jostii RHA1ΔgblA | This work |
| RHA1-C | R. jostii RHA1ΔgblA + pRM4-gblA | This work |
| S. coelicolor LW16/pTE134 | γ-Butyrolactone reporter strain. S. coelicolor M145 ΔscbAΔscbR (LW16) containing the construct pTE134 (scbR and Km resistance gene under the control of a γ-butyrolactone inducible promoter (cpkOp) | 33 |
| S. coelicolor M145 | Wild type strain of S. coelicolor | 49 |
| E. coli DH5αTM | Cloning strain. F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK−, mK + ) phoA supE44 λ– thi-1 gyrA96 relA1 | Invitrogen |
| Aspergillus niger N402 | Bioactivity reporter strain. Wild type. | 50 |
| Micrococcus luteus ATCC9341/ Kocuria rhizophila | Bioactivity reporter strain. Wild type. | |
| Bacillus subtilis ATCC6633 | Bioactivity reporter strain. Wild type. | |
| E. coli JM101 | Bioactivity reporter strain F′ traD36 proA + B + lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV thi | NEB |
| Mycobacterium smegmatis MC2 155 | Bioactivity reporter strain. Wild type. |
Phenotypic characterization of the different strains
R. jostii RHA1 WT, RHA1-ΔgblA and RHA1-OE were grown on different solid media to check for phenotypic differences. Growth, pigmentation and antibiotic activity against Aspergillus niger, Mycobacterium smegmatis, Escherichia coli, Bacillus subtilis and Micrococcus luteus on solid media were monitored by visual observation. M. smegmatis and M. luteus were grown at 30 °C and E. coli and B. subtilis were grown at 37 °C before inoculating the media for the bioassay. Afterwards plates were kept at 30 °C. Cell shapes were studied under a Zeiss Axioskop 2 phase contrast microscope. The media used for antibiotic production tests were Trypton Soya Agar (TSA), Difco Nutrient Agar (DNA), Luria Broth Agar (LBA), Starch Casein Agar (SCA), Minimum Salt Medium (MSM) nitrogen deficient and complimented with casamino acids, Supplemented Minimum Medium Solid (SMMS) and low pH SMMS: in this case pH was not adjusted after mixing components (Supplementary Table S1). R. jostii RHA1 transformants were grown for 4 days before plating the bioassay test strains (Table 2) next to the Rhodococcus colonies. Growth of the antimicrobial bioassay test strains was followed over time and scored after 4 days.
Bioinformatic analysis
The following complete genome sequences of Rhodococci were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/): Rhodococcus jostii RHA1, Rhodococcus aetherivorans, Rhodococcus equi 103 S, Rhodococcus equi ATCC 33707, Rhodococcus erythropolis CCM2595, Rhodococcus erythropolis PR4, Rhodococcus erythropolis R138, Rhodococcus opacus B4, Rhodococcus opacus PD630, Rhodococcus opacus R7, Rhodococcus pyridinivorans SB3094, Rhodococcus sp. AD45, Rhodococcus sp. B7740, Rhodococcus sp. BCP1. AntiSMASH (http://antismash.secondarymetabolites.org/) was used for the detection of γ-butyrolactone biosynthetic gene clusters in these genomes using the ClusterFinder algorithm.
Deletion mutagenesis
Unmarked gene deletion mutagenesis29 was used to delete gblA. Primers afsA-del-GTG-FW-XbaI and afsA-del-GTG-Rv-EcoRI and afsA-del-UAG-Fw-PstI and afsA-del-UAG-Rv-XbaI (Table 3) were designed to amplify fragments of 1.5 Kb upstream and downstream of gblA, including the first and last ~200 bp from gblA to ensure that the surrounding genes were not affected by the deletion. Genomic DNA was used as template. Both fragments were cloned in pK18mobSacB (ATCC® 87097™) between EcoRI and PstI, producing the deletion construct pK18mobSacB-ΔgblA. The deletion construct was transformed into R. jostii RHA1 by electrotransformation. First cross-over colonies were selected for Km resistance and sucrose sensitivity. Second recombination was selected by Km sensitivity and sucrose resistance. Deletion mutants were checked by PCR using genomic DNA with primers outside the 1.5 Kb homologous regions and by sequencing of the resulting product using primers afsA-check-Fw-2 and afsA-check-Rv-1 and afsA-check-Fw-2 and afsA-check-Rv-2 (see Fig. S1 and Table 3).
Table 3.
Primers used in this work.
| Primer | Sequence | Amplicon target |
|---|---|---|
| ScbA-jostii-NdeI-F | GCGATACATATGGCGCAAATTTCCCGGCCGAT | R. jostii gblA gene |
| ScbA-jostii-BamHI-R | CGCTATGGATCCCTAGCGAGCGCATGCGCTCA | R. jostii gblA gene |
| afsA-del-GTG-FW-XbaI | GATTATCTAGAGAAGACCTCGGCCACGGATTG | Upstream region of R. jostii gblA gene for deletion |
| afsA-del-GTG-Rv-EcoRI | TACTTGAATTCGGGCTTTCGTGAACGACCTC | Upstream region of R. jostii gblA gene for deletion |
| afsA-del-UAG-Rv-XbaI | GATTATCTAGAGACGAGCGAGCCACGATCC | Downstream region of R. jostii gblA gene for deletion |
| afsA-del-UAG-Fw-PstI | GTCAACTGCAGGCCGGGCGAGATCGTTCAC | Downstream region of R. jostii gblA gene for deletion |
| afsA-del-check-1-Fw | CGACGCCGACTAGCGAGC | Primer annealing outside the 1.5 KB homologous region used for the double recombination to delete afsA |
| afsA-del-check-1-Rv | TGGTCGCGGTTACTGGACAC | Primer annealing outside the 1.5 KB homologous region used for the double recombination to delete afsA |
| afsA-del-Check-2-FW | TGGCAGGCGTGGAACACGTC | Primer annealing outside the 1.5 KB homologous region used for the double recombination to delete afsA |
| afsA-del-check-2-Rv | GTCGTTCGAGCGGACCGTTC | Primer annealing outside the 1.5 KB homologous region used for the double recombination to delete afsA |
| pSET152-CS-Fw | TACCGCATCAGGCGCCATTC | Primer annealing at one side of the insertion in PMR4 |
| pSET152-CS-Rv | TTATGCTTCCGGCTCGTATG | Primer annealing at one side of the insertion in PMR4 |
gblA complementation and overexpression
Primers ScbA-jostii-NdeI-F and ScbA-jostii-BamHI-R (Table 3) were used to amplify the R. jostii RHA1 gblA gene (969 bp) and clone it into pRM430 under the control of the strong constitutive promoter ermE*(pRM4-gblA). This construct was introduced into R. jostii RHA1 wild type strain and RHA1-ΔgblA by electrotransformation obtaining the transformants RHA1-OE and RHA1-C, respectively. Clones were selected by apramycin resistance and checked by PCR with primers pSET152-CS-Fw and pSET152-CS-Rv (Table 3) annealing in pRM4 at both sides of the insert and by sequencing of the resulting products.
Transformation of Rhodococcus
A modified electrotransformation protocol from Arenskotter et al.31 was used to introduce the different constructs described in this work into R. jostii RHA1. Strains were grown in 50 ml LB containing 1% w/v of glycine in a 250 ml Erlenmeyer flask at 30 °C and 220 rpm to an OD600 of 0.8–1. Cells were washed twice with 15 ml of chilled deionized water and concentrated to 2.5 ml in 10% glycerol and aliquoted in 400 μl. Subsequently, 100 ng to 1 μg of DNA was added to each 400 μl and the sample was kept on ice for at least 10 min. Cells were pulsed with a Biorad Xcell gene pulser at 1.75 kV, 50 μF and 200 Ω (field strength of 8.75 kV cm−1). Ice cold LB was added immediately after the pulse and the cell samples were allowed to recover for 4 h at 30 °C and 220 rpm. Subsequently the cells were plated on selective media.
γ-Butyrolactone extraction
Rhodococcus strains were grown in modified SMM solid (SMMS) medium32 (Supplementary Table S1) at 30 °C. Extraction of γ-butyrolactones was performed following the procedure described in Hsiao et al.33. R. jostii RHA1 WT and transformant strains derived were grown on modified SMMS32 (Table S1). Per strain, 40 standard (90 mm diameter) petri dishes were used. After 4 days of growth at 30 °C, when the strains turned orange due to carotene production, which is an indication of an active secondary metabolism, the agar of each plate was cut into pieces and extracted with ethyl acetate as described in Hsiao et al.33. Extracts were dried at 30 °C in a rotary evaporator, and then resuspended in 160 μl of methanol per 40 petri dishes.
GBL specific reporter assay (Kanamycin (Km) assay)
The GBL-specific reporter assay performed in this study were done following the protocol from Hsiao et al.33. From the extract of each strain, 60 μl were concentrated to 6 μl by evaporation in a Savant DNA SpeedVac and spotted onto a DNA plate (Supplementary Table S1) containing 4.5 µg ml−1 of Km and plates were uniformly spread with S. coelicolor strain LW16/pTE134. Streptomyces was plated before adding the extracts. As positive control, 6 μl of a stock solution of 1.5 mg ml−1 of chemically synthesized 6-dehydro SCB2 was used. Dried ethyl acetate resuspended in methanol was used as negative control. Results were reproducible with 2–3 biological replicates for each strain.
Liquid chromatography-Mass spectrometry analysis
For identification of R. jostii RHA1 γ-butyrolactone-like molecules, HPLC-MS analysis was performed using an Accella1250™ HPLC system coupled with the benchtop ESI-MS Orbitrap Exactive™ (Thermo Fisher Scientific, San Jose, CA). A Reversed Phase C18 (Shim Pack Shimadzu XR-ODS 3 × 75 mm) column was used and a gradient from 2% to 95% of acetonitrile:water (0.1% Formic Acid) as follows: 2 min 2% acetonitrile, 2–10 min gradient to 95% acetonitrile, 1 min 95% acetonitrile. To separate further the peaks from A-factor and 6-dehydro SCB2, a gradient from 2% to 80% acetonitrile was applied to the separation: 2 min 2% acetonitrile, 2–25 min in 2–80% acetonitrile, 1 min 80% acetonitrile. Data was analysed using Xcalibur software from Thermo Scientific. LC-MS analysis was performed with 2–4 biological replicates per strain.
Synthesis of γ-butyrolactone standards
Synthetic SCB1, SCB2, A-factor and 2-dehydro SCB2 γ-butyrolactones used in this study were chemically synthesized as described by Martin-Sanchez27.
Analysis of the interactions between Streptomyces coelicolor and R. jostii RHA1
R. jostii RHA1 wild type, strains RHA1-OE and RHA1-ΔgblA were plated onto modified minimum salt media (MSM) containing casamino acids instead of NH4NO3 (see Supplementary Table S1). After 4 days, S. coelicolor M145 was plated next to the patches of the R. jostii strains. Following a further incubation for 18 h, the S. coelicolor growth stage and production of coloured antibiotics20 was checked every 2 h; after 24 h of incubation these parameters were checked once a day for a week. Experiments were performed in 5 independent replicas.
Data availability statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Results
γ-Butyrolactone gene clusters in Rhodococci
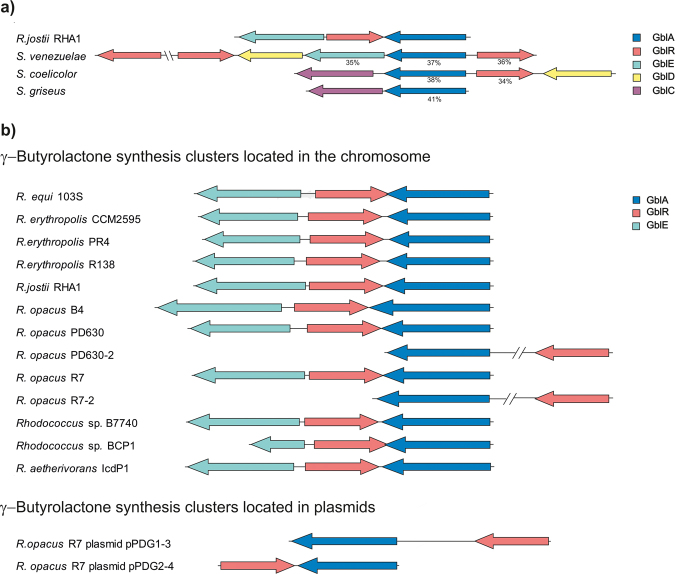
Analysis of the predicted γ-butyrolactone gene cluster of R. jostii RHA17,11 revealed the presence of a gblA gene (RHA1_RS22510) (Fig. 3a), encoding for GblA, the putative first enzyme in the biosynthetic pathway of γ-butyrolactones. It contains two AfsA repeats (Pfam03756 domain)15, the predicted active sites of GblA enzymes (see Fig. S2). A BLAST search with gblA in the R. jostii RHA1 genome sequence showed that this strain shares only a single copy. GblAjostii has 37–41% amino acid (AA) identity with (partially) characterized homologues of S. venezuelae (JadW1), S. coelicolor (ScbA) and S. griseus (AfsA) (Fig. 3a). The homologues of these three Streptomyces species have 43–65% AA identity between each other. The Gamma-butyrolactone Receptor protein (GblR) from R. jostii RHA1, annotated as a TetR regulator, has 34–36% AA identity with the corresponding proteins in the three Streptomyces strains (Fig. 3a); the homologues of the three Streptomyces species have 37–56% AA identity between each other. The γ-butyrolactone gene cluster of R. jostii RHA1 also includes a gene encoding a GblE enzyme (Gamma-butyrolactone biosynthesis Epimerase) with a NAD-epimerase/dehydratase predicted function. A (partially) characterized homologue is JadW2 of S. venezuelae (35% AA identity), shown to be essential for the synthesis of γ-butyrolactones34. However, it is not clear in which step of the biosynthesis pathway this enzyme acts. BLAST searches were also performed with GblD of Streptomyces coelicolor, a short chain dehydrogenase known to contribute to synthesis of γ-butyrolactones in some Streptomyces species, e.g. in S. coelicolor 27 and S. venezuelae 17. This yielded a large number of homologues with 30–40% AA identity to the query, encoded throughout the R. jostii RHA1 genome, and with a few of them located in the proximity of the gbl gene cluster. Also, two homologues of GblC were found encoded in the R. jostii RHA1 genome, with ~35% AA identity to the S. coelicolor GblC. These genes are not located in close proximity to the gbl gene cluster. Genes flanking the GBL gene cluster are not conserved between species.
Figure 3.
Predicted γ-butyrolactone gene clusters in different strains. (a) Organization of the predicted γ-butyrolactone gene cluster of Rhodococcus jostii RHA1 compared to that of the known clusters in different Streptomyces strains. AA identity of the R. jostii RHA1 enzymes to the corresponding enzymes encoded by the genes in each strain is stated below each gene. (b) Comparison of the organization of the different γ-butyrolactone gene clusters of Rhodococci. A cluster is present in virtually all studied Rhodococci, with a highly similar organization. Only R. opacus R7 contains γ-butyrolactone clusters on its plasmids. GblA: γ-butyrolactone first biosynthetic enzyme. GblR: γ-butyrolactone receptor protein. GblE: γ-butyrolactone biosynthetic enzyme E, predicted to be a NAD-epimerase/dehydratase. GblD: γ-butyrolactone biosynthetic enzyme D, short chain dehydrogenase. GblC: γ-butyrolactone biosynthetic enzyme C, reductase.
All studied Rhodococcus strains possess a single predicted γ-butyrolactone gene cluster, except for R. pyridinovorans. R. opacus PD630 and R. opacus R7, which contain multiple γ-butyrolactone gene clusters in their genomes (Fig. 3b). R. opacus R7 contains γ-butyrolactone clusters on 2 different plasmids while in all other cases the clusters are located on the chromosome only. Most γ-butyrolactone gene clusters have a similar organization, with the gblR gene flanked by gblA and gblE but divergently oriented. The gblE gene however is not always present (Fig. 3b). We carefully checked whether other genes surrounding the predicted γ-butyrolactone gene clusters (Fig. 3) in the different Rhodococcus strains are conserved, but this was not the case.
γ-Butyrolactones from R. jostii RHA1
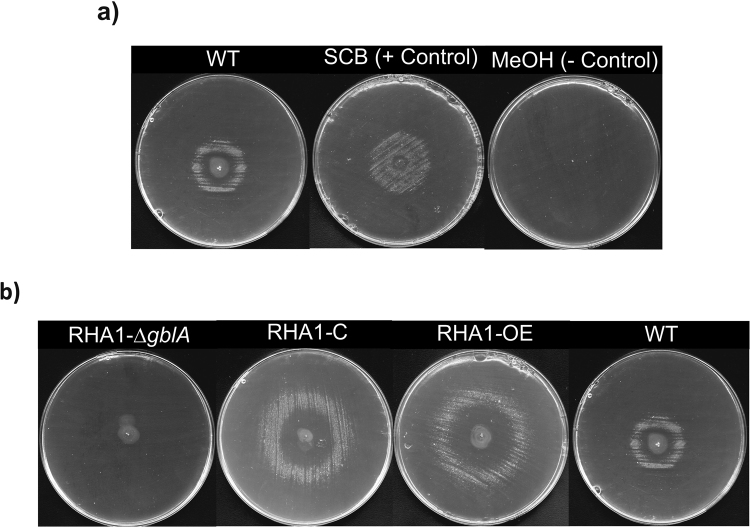
To analyse whether R. jostii RHA1 is producing any γ-butyrolactone(-like) molecules, a GBL-specific reporter assay developed for S. coelicolor (Km assay) was performed with ethyl acetate extracts of R. jostii RHA1 agar plates33. This test is based on release of the repression by the γ-butyrolactone receptor ScbR of transcription of a Km resistance gene in the S. coelicolor LW16/pTE134 indicator strain25. When plating this reporter strain on solid media with Km, it will only be able to grow if γ-butyrolactone molecules are present that are able to bind to the ScbR receptor protein and thereby allow transcription of the Km resistance gene. The R. jostii RHA1 extracts obtained, as described in the methods section, indeed induced the growth of the LW16/pTE134 reporter strain (Fig. 4a). R. jostii RHA1 thus indeed synthesizes γ-butyrolactone(-like) molecules (Rhodococcus Jostii Butyrolactone, RJB). This Km bioassay is very specific, since changes in the γ-butyrolactone aliphatic side chain are known to significantly affect the affinity of ScbR for these molecules25. Generic γ-butyrolactones also are not able to trigger this system35. R. jostii RHA1 apparently synthesizes one or more RJB molecules that are able to bind to the S. coelicolor γ-butyrolactone receptor protein ScbR, enabling growth of the reporter strain. In view of the high specificity of this assay it is likely that these R. jostii RHA1 RJB molecules are structurally most similar to SCBs, the γ-butyrolactone molecules of S. coelicolor 23,25.
Figure 4.
Detection of γ-butyrolactone(-like) molecules in R. jostii RHA1 ethyl acetate agar culture extracts by a GBL-specific reporter assay. When γ-butyrolactones are present in the sample the reporter strain S. coelicolor LW16/pTE134 forms a halo of growth around the sample application area in the centre of Km agar plates. Size of the halo of growth is indicative for the concentration of the diffusible γ-butyrolactones in the sample33. An excess of γ-butyrolactones inhibits the growth of the strain and a too low concentration cannot induce the expression of the Km resistance gene. (a) R. jostii RHA1 ethyl acetate extract (left), positive control with chemically synthesized 6-dehydro SCB2 (middle), negative control (solvent) (right). (b) R. jostii-ΔgblA (left), complemented strain (RHA1-C), gblA overexpression strain (RHA1-OE) and wild type strain (right). WT: wild type; MeOH: methanol
To further analyse the γ-butyrolactone synthesis of R. jostii RHA1 the putative γ-butyrolactone biosynthesis gene gblA was deleted from the genome using unmarked gene deletion mutagenesis as described in Van der Geize et al.29. Ethyl acetate extracts of the deletion strain RHA1-ΔgblA were made and tested for RJB synthesis. In contrast to the wild type R. jostii RHA1, extracts of RHA1-ΔgblA did not induce the growth of the LW16/pTE134 reporter strain in the GBL-specific reporter assay, indicating the absence of molecules structurally similar to γ-butyrolactones capable of binding to ScbR (Fig. 4b). No phenotypical differences in growth rate, colony shape, pigment or antibiotic production, or cell shape under the microscope, were observed for this strain compared to the WT strain. In order to verify that the observed phenotype was caused only by the deletion of gblA, the RHA1-ΔgblA deletion strain was transformed with the vector pRM4-gblA containing gblA jostii under the control of a constitutive strong promoter (ermE*) and the Cφ31 phage integrase. This strain showed production of γ-butyrolactones as detected by a GBL-specific reporter assay (Fig. 4b). pRM reintroduction of the wild type gblA gene resulted in a bigger halo of growth for the complemented strain than for the wild type strain indicating a higher production of γ-butyrolactones, probably due to the strong promoter used for the complementation (see Fig. 4b). In order to study the role of GblA in RJB production an overexpression strain (RHA1-OE) was constructed introducing pRM-gblA into wild type R. jostii RHA1. Next, RJB production by the RHA1-OE strain was analysed. Also in this case the Km bioassay showed a bigger halo of growth of the reporter strain compared to wild type R. jostii RHA1 extracts, which indicates a higher RJB concentration in the RHA1-OE sample due to a further diffusion from the application point. Thus, these experiments indicate that the overexpression of gblA results in an enhanced RJB production (Fig. 4b).
Characterization of the γ-butyrolactone(-like) molecules synthesized by R. jostii RHA1
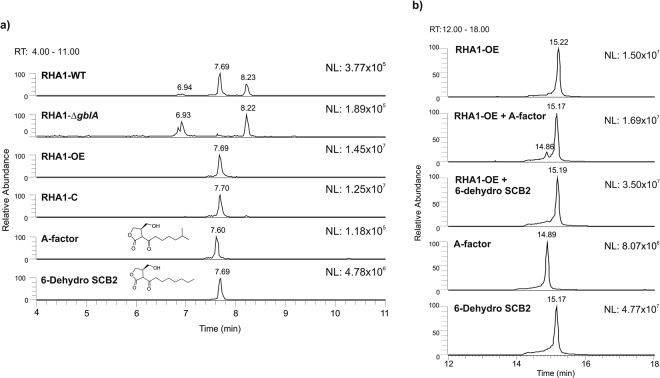
The extracts of wild type R. jostii RHA1, RHA1-ΔgblA, RHA1-OE and RHA1-C were analysed for RJBs by liquid chromatography coupled to a mass spectrometer (LC-MS). The structures of RJBs are apparently rather similar to those of the S. coelicolor γ-butyrolactones since they bind to ScbR, as evident from the Km bioassay (see above). Therefore, the LC-MS data was analysed searching for metabolites with masses similar to those of described for S. coelicolor γ-butyrolactones23,25,36. A peak eluting at 7.70 min with a mass of m/z 241.1441 amu [M-H]− was detected in the extracts from R. jostii RHA1 wild type strain, RHA1-OE and RHA1-C, but it was missing in RHA1-ΔgblA. The gblA gene thus is essential for its synthesis, indicating that this peak indeed represents a R. jostii RHA1 RJB molecule (Fig. 5a). The mass of the detected R. jostii RHA1 RJB molecule corresponds to the A-factor signalling molecule of S. griseus and also to the intermediate compound 6-dehydro SCB2 of S. coelicolor 27 (Figs 2, 5a). Synthetic standards of A-factor and 6-dehydro SCB2 were also analysed on LC-MS, which eluted at 7.60 and 7.69 min, respectively (Fig. 5a) and with an exact mass of 241.1444 amu [M-H]−. The extracts of the different Rhodococcus strains yielded peaks with the same retention time as 6-dehydro SCB2. The RHA1-OE and RHA1-C peaks had a higher intensity than in the R. jostii RHA1 wild type strain, corresponding to the Km bioassay results, showing a bigger halo than seen with the wild type strain (Fig. 4b). To analyse whether the molecule detected in the R. jostii RHA1 extracts is similar to 6-dehydro SCB2 or to A-factor, the extract from RHA1-OE was spiked with the synthetic standards of these compounds at 50 ng/µl and run in the LC-MS with a longer gradient (Fig. 5b). As a control, a mixture of both standards (A-factor and 6-dehydro SCB2) was also run in the same conditions. The mixture of both standards and the extract spiked with the standard of A-factor showed two different peaks at 14.86 min and 15.17 min, corresponding to A-factor and 6-dehydro SCB2, respectively. When the extract was spiked with 6-dehydro SCB2 the peaks completely overlapped, confirming that the single RJB detected is structurally identical to 6-dehydro SCB2 (Fig. 5b). The R. jostii RHA1 samples were also compared to the available chemically synthesized standards of S. coelicolor γ-butyrolactones27, but we were not able to find any other known γ-butyrolactone(-like) molecules in R. jostii RHA1 extracts.
Figure 5.
LC-MS analysis of ethyl acetate extracts of the various R. jostii RHA1 strains grown for 4 days on SMMS. (a) A peak eluting at 7.70 min with a mass of mass m/z 241.1441 amu [M-H]− was detected in all samples except in RHA1-ΔgblA. RHA1-C, the complemented strain, and RHA1-OE, the gblA overexpression strain, both showed a higher intensity of this peak. This mass corresponds to the described γ-butyrolactone from S. griseus (A-factor) and the stereoisomer 6-dehydro SCB2, known to be an intermediate in the synthesis of the γ-butyrolactone SCB2 in S. coelicolor. The standard of A-factor showed a peak eluting at 7.60 min while the standard of 6-dehydro SCB2 eluted at 7.69 min. (b) Extracts of RHA1-OE spiked with standards of A-factor or 6-dehydro SCB2 at 50 ng/µl using a longer gradient to separate both peaks further. The spiked extract of R. jostii RHA1 with both standards confirmed that the molecule synthesized by R. jostii RHA1 has the same retention time and mass as 6-dehydro SCB2. (NL: Normalization Level; RT: Retention Time).
When the samples were screened for masses between 187 and 350, a mass range that includes all described γ-butyrolactones, two peaks had a higher intensity in the RHA1-OE and RHA1-C strains than in the R. jostii RHA1 wild type; these two peaks were not visible in the RHA1-ΔgblA deletion strain (see Fig. S3). One peak eluted at 7.08 min and showed three different masses, m/z 211.0972 amu [M-H]−, m/z 279.1369 amu [M-H]− and m/z 289.1658 amu [M-H]−. Another peak eluted at 7.88 min that corresponds to a mass of m/z 255.1236 amu [M-H]−. None of these masses correspond to known γ-butyrolactones, including the recently described ones in Sidda et al.36 or Xu et al.37.
The absence of a halo in the GBL-specific reporter assay in the gblA deletion mutant, together with LC-MS analysis of the extracts of the gblA deletion mutant, are clear evidence that (no detectable amount of) GBL is not produced anymore by the deletion mutant and confirm the role of GblA in biosynthesis of this molecule.
Phenotypical characterization of constructed R. jostii gblA strains
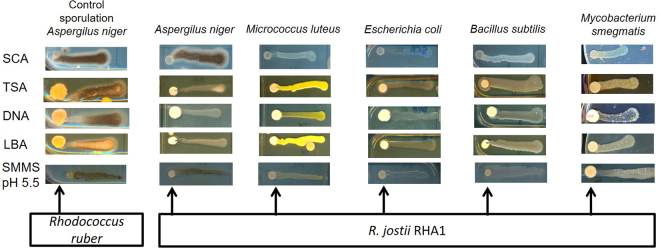
The γ-butyrolactone system is known to regulate secondary metabolite synthesis, morphogenesis or both, in Streptomycetes15,38. R. jostii RHA1 has almost 120 putative secondary metabolite biosynthetic gene clusters in its genome and most of them are uncharacterized4. Wild type R. jostii RHA1 was screened for secondary metabolite production during growth on different agar media. Production of bioactive compounds was tested with various indicator strains, two Gram-positive strains (Micrococcus luteus and Bacillus subtilis), one acid-resistant Gram-positive strain (Mycobacterium smegmatis), a Gram-negative strain (Escherichia coli) and a fungal species (Aspergillus niger). These antimicrobial bioassay strains were plated next to the Rhodococcus colonies. R. jostii RHA1 WT exerted clear inhibition of growth towards M. luteus on SCA medium and M. smegmatis in low pH SMMS (Fig. 6). Inhibition of sporulation of A. niger was observed on LBA, TSA and DNA agar media (Fig. 6).
Figure 6.
Bioactivity tests with R. jostii RHA1 spotted directly from glycerol stocks onto SCA, TSA, DNA, LBA and SMMS (pH 5.5) agar plates using A. niger, M. luteus, E. coli, B. subtilis and M. smegmatis as antimicrobial bioassay strains. After 4 days of incubation, the antimicrobial bioassay strains were applied on a horizontal line towards the R. jostii RHA1 patch. Inhibition of A. niger sporulation was observed on TSA, DNA and LBA (note loss of black pigment from the conidia compared to the control showing A. niger sporulation when plated next to Rhodococcus ruber). Growth inhibition of M. luteus and M. smegmatis was observed on SCA and SMMS, respectively (especially visible close to the R. jostii RHA1 patch at the left).
The RHA1-ΔgblA deletion strain and the RHA1-OE overexpression strain were also analysed for changes in antimicrobial production in different growth media. No difference in bioactivity was observed between R. jostii RHA1 wild type and derived strains in any medium or with any antimicrobial bioassay strain tested. The growth on plate and colony shapes of all constructed strains were also analysed on all tested solid media, and their cell shapes in liquid LB medium, however, no differences were observed compared to the wild type strain (data not shown).
Interaction between R. jostii RHA1 and S. coelicolor M145
The GBL-specific reporter assay performed with extracts from R. jostii RHA1 indicate that the R. jostii RHA1 RJB interacts with the γ-butyrolactone receptor protein ScbR of S. coelicolor (see above). These different genera thus may be capable of interspecies communication. To study a possible interaction between R. jostii RHA1 and S. coelicolor M145, both strains were inoculated next to each other on agar plates. γ-Butyrolactones diffuse into the agar and therefore an exchange of signalling molecules between species is possible. R. jostii RHA1 was allowed to grow for 4 days, using carotene production (orange pigmentation) as indication that secondary metabolism was active. Subsequently, S. coelicolor M145 was plated next to it. As a control, S. coelicolor M145 and R. jostii RHA1 also were plated separately on the same agar media. After a further 24 h, S. coelicolor M145 developed aerial mycelium with its characteristic white pigmentation when grown next to the RHA1-OE overexpression strain, but not when growing next to the RHA1-WT and the RHA1-ΔgblA strains (Fig. 7).
Figure 7.
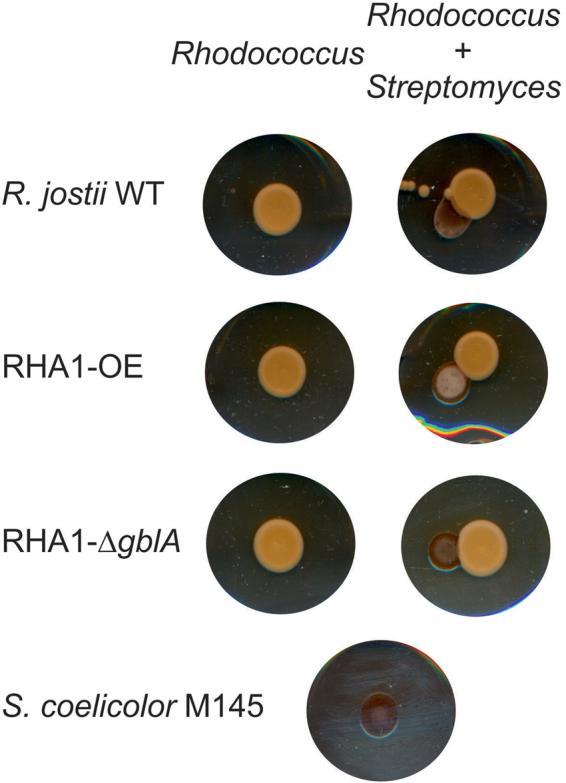
Interactions observed between R. jostii RHA1 WT, RHA1-OE and RHA1-ΔgblA with S. coelicolor M145 on MSM agar containing casamino acids. Rhodococcus strains were spotted directly from glycerol stocks and grown for 4 days before spores of S. coelicolor M145 were plated next to the patches of the Rhodococcus strains. On the left, the different strains from R. jostii RHA1 were grown separately. On the right, S. coelicolor M145 was grown next to the different R. jostii RHA1 strains. At the bottom, S. coelicolor M145 grown on its own.
Deletion of S. coelicolor gblA (scbA) did not affect the morphological development12. Effects of γ-butyrolactones on development of S. coelicolor was suggested in Kawabuchi et al.39, but they were unable to prove that this effect was due to γ-butyrolactones. This difference in sporulation (Fig. 7) may be due to the R. jostii RHA1 RJB alone, or caused by unknown RHA1-OE compounds accumulating in response to the enhanced synthesis of RJB.
Discussion
Rhodococci are Gram-positive soil bacteria known for the great variety of catabolic pathways which are encoded in their relatively large chromosomes, 9.6 Mb in case of R. jostii RHA14. Genome analyses showed that Rhodococci also contain a large number of uncharacterized putative secondary metabolite gene clusters4. Here we show that γ-butyrolactone gene clusters are not only present in the genomes of R. jostii RHA1, R. equi 103S4,7,10 , R. opacus B4 and R. erythropolis PR47, but occur even more widespread in Rhodococci (Fig. 3). Analysis of the γ-butyrolactone gene cluster in R. jostii RHA1 predicted the presence of genes encoding various homologues of enzymes known to be involved in the γ-butyrolactone biosynthesis in Streptomyces 15, namely GblA, GblE and GblR. R. jostii RHA1 lacks the GblD enzyme, which is also not present in S. griseus (Fig. 2), suggesting that R. jostii RHA1 may employ a similar biosynthetic pathway as S. griseus. However, the GblE enzyme encoded in R. jostii RHA1 is not present in S. griseus. Instead, this GblE enzyme is homologous to the γ-butyrolactone enzyme JadW2 from S. venezuelae, which is known to be essential for the production of γ-butyrolactones in this strain34 but it has an unknown biosynthetic role. In order to analyse whether there are more homologues to the γ-butyrolactone biosynthesis enzymes described in Streptomyces BLAST searches with gblD and gblC of S. coelicolor were performed. A large number of dehydrogenases with 30%-40% AA identity to GblD, were found spread throughout the R. jostii RHA1 genome. Also, two homologues of GblC were found encoded in the R. jostii RHA1 genome, with ~ 35% AA identity to the S. coelicolor GblC. Homologues of these enzymes with higher identity than the ones found in R. jostii RHA1 are also present in the genomes of different Streptomyces strains. These enzymes have never been reported to be part of the γ-butyrolactone biosynthesis pathways in these streptomycetes. Clearly, simple sequence analysis is not sufficient to predict the involvement of R. jostii RHA1 genes encoding homologous enzymes in the synthesis of RJB. Deletion mutagenesis of these putative gblC, gblD and gblE genes in R. jostii RHA1 followed by LC-MS analysis of cell extracts of these transformants strains, searching for intermediates accumulating, may serve to elucidate the biosynthetic pathway in this strain. Since this pathway has not been completely elucidated in Streptomyces, other not yet identified pathway specific enzymes may also be involved in the synthesis of these signalling molecules, but we were unable to identify them.
Our data provide the first evidence of γ-butyrolactone synthesis in the genus Rhodococcus. These molecules were proven to be able to bind to the γ-butyrolactone receptor protein from S. coelicolor. The binding of exogenous molecules to γ-butyrolactone receptor proteins from Streptomyces previously has been observed for extracts of the cultures of other non-Streptomyces species38,40. The latter study also suggested that Amycolatopsis mediterranei and Micromonospora echinospora produce an IM-2 type molecule and Actinoplanes teichomyceticus a VB type of γ-butyrolactone40. This conclusion was based on the efficiency by which these molecules bind to the S. virginiae and S. lavendulae γ-butyrolactone receptor proteins respectively, but no structural analysis has been performed. We used LC-MS analysis to compare the compounds in R. jostii RHA1 ethyl acetate extracts from solid media with different chemically synthesized standards of known γ-butyrolactones. We were unable to extract g-butyrolactones in high enough concentrations from liquid media for their detection, not even after 40 h of growth (data not shown). It thus appears that the system is not induced at all, or at least not strong enough, in liquid growth media. The R. jostii RHA1 RJB was identified as 6-dehydro SCB2 (Fig. 5). 6-dehydro SCB2 is an isomer of the γ-butyrolactone described in S. griseus (A-factor) and is a predicted precursor of one of the described γ-butyrolactones in S. coelicolor (SCB2)27. In S. coelicolor, a GblD enzyme reduces the keto group in carbon 6 to a hydroxyl group (Fig. 2). Many genes encoding enzymes with low similarity to GblD of S. coelicolor were found spread throughout the genome of R. jostii RHA1. However, we did not find a gblD homologue in the R. jostii RHA1 gene cluster (Fig. 3a), which corresponds to the observation that it is producing a 6-dehydro form of the molecule. When the samples were screened by LC-MS for a mass range that includes all known γ-butyrolactones, two peaks were observed in RHA1-OE and RHA1-C that were not present in the deletion strain and were in a lower intensity in the WT strain. The masses corresponding to these peaks did not match to those of any γ-butyrolactone described to date. These molecules could be γ-butyrolactones with novel structures, or totally different compounds, e.g. products of a biosynthesis pathway regulated by RJB. The detected mass of m/z 255.1236 amu [M-H]− differs in m/z 14 from 6-dehydro SCB2. In further work we will attempt the isolation of sufficient amounts of these molecules for NMR analysis to elucidate their structures.
Unmarked deletion mutagenesis of the gblA gene in R. jostii RHA1 abolished RJB synthesis. Various homologues of this gene in Streptomyces species are known to be essential for biosynthesis of γ-butyrolactone molecules, catalyzing the first step of the biosynthesis, the condensation of a glycerol derivative with a fatty acid derivative (Fig. 2). Both Km bioassays (Fig. 4) and LC-MS analysis of extracts of the various R. jostii RHA1 (mutant) strains (Fig. 5) confirmed that gblA is essen2yrolactones are known to regulate secondary metabolism and/or morphogenesis in the genus Streptomyces 15,38. R. jostii RHA1 contains a large number of putative secondary metabolite clusters that are mostly uncharacterized. RJB may be involved in control of the expression of one or more of these clusters. Although a few Rhodococcus antimicrobials are known41–44 this genus has remained largely unexplored for production of secondary metabolites. In this work, we detected bioactivity of R. jostii RHA1 against Micrococcus luteus, Aspergillus niger and Mycobacterium smegmatis. Lariatins, cyclic peptides that have bioactivity against Mycobacterium species, were found in R. jostii K01-B017142, but the enzymes involved in the synthesis of these compounds are not encoded in the genome from R. jostii RHA1. We have not been able to find a difference in antibiotic production, growth rate or colony shape between the R. jostii RHA1 WT, RHA1-ΔgblA and RHA1-OE strains, therefore further experiments are needed to analyse the role of γ-butyrolactone system in R. jostii RHA1. Mutagenesis analysis of GblR may help identify any R. jostii RHA1 genes that are regulated by its RJB. Various systems may be controlled by RJB in Rhodococci, analogous to the situation in the genus Streptomyces. In some species of Streptomyces γ-butyrolactones are known to be involved in morphogenesis and sporulation, as is the case in S. griseus. Deletion of afsA in S. griseus blocked its sporulation and streptomycin production38,45. The RJB in R. jostii RHA1 might be controlling the synthesis of one or more secondary metabolites that have remained unidentified, or it may be directly or indirectly influencing the primary metabolism in this strain.
The γ-butyrolactone system is known to be present in several Actinomycete genera21,25,33,40. γ-Butyrolactone molecules described in Streptomyces have differences in structure depending on the producing species. An exception is a γ-butyrolactone produced by S. venezuelae which was found to be identical to SCB3 from S. coelicolor. Actinomycetes are soil bacteria that live in a rich community of microorganisms. The γ-butyrolactone system may have developed as a way to communicate between different species19,46. To test whether such interspecies communication occurs between R. jostii RHA1 and S. coelicolor, we plated these strains next to each other. S. coelicolor sporulation clearly was accelerated when growing next to RHA1-OE compared to S. coelicolor growing alone or next to the RHA1 WT and RHA1-ΔgblA strains. These results indicate that compounds secreted by the R. jostii RHA1-OE strain affect morphological differentiation in S. coelicolor. This effect is known to be induced by γ-butyrolactone molecules in other Streptomyces species15,47 but has never been described before in S. coelicolor. The addition of 6-dehydro SCB2 to a confluent lawn of S. coelicolor however did not induce sporulation which indicated that the phenotypical difference observed is not a direct effect of this RJB. This phenomenon thus remains to be studied in more detail in future work.
This work reports on the synthesis of a γ-butyrolactone(-like) molecule by R. jostii RHA1. This RJB molecule appears to be structurally very similar to the γ-butyrolactones described in Streptomyces and interacts with the S. coelicolor butanolide system. In future work, we aim to elucidate the physiological roles of these signalling molecules in Rhodococcus metabolism, with specific interest in possible regulatory effects on representatives of the many secondary metabolite biosynthetic gene clusters in this genus. We have shown that R. jostii RHA1 produces compounds with antibiotic activity, with at least one of them active against M. smegmatis and therefore potentially also against the fast-emerging multidrug resistant Mycobacterium tuberculosis. Activation of cryptic secondary metabolite clusters in Rhodococci may potentially unlock the biosynthesis of novel compounds that are of interest to the pharmaceutical industry.
Electronic supplementary material
Acknowledgements
AC was financially supported by the University of Groningen, and by the Dutch Technology Foundation (STW), which is part or the Netherlands organization for Scientific Research (NWO) and partly funded by the Ministry of Economic Affairs (STW 10463). We thank Subramaniyan Mannathan and Adriaan J. Minnaard of the Bio-Organic Chemistry department of the University of Groningen for providing the chemically synthesised SCBs, and Lara Martin-Sanchez for fruitful discussions.
Author Contributions
A.C., M.P. and L.D. designed the study and planned the experiments. A.C. and M.P. performed the experimental work. A.C., M.P. and L.D. wrote the manuscript. All authors reviewed the manuscript and agree with its contents.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17853-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gürtler V, Mayall BC, Seviour R. Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol. Rev. 2004;28:377–403. doi: 10.1016/j.femsre.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Curragh H, et al. Haloalkane degradation and assimilation by Rhodococcus rhodochrous NCIMB 13064. Microbiology. 1994;140(Pt 6):1433–1442. doi: 10.1099/00221287-140-6-1433. [DOI] [PubMed] [Google Scholar]
- 3.van der Geize R, Dijkhuizen L. Harnessing the catabolic diversity of Rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 2004;7:255–261. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 4.McLeod MP, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor CR, et al. Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 2012;113:521–530. doi: 10.1111/j.1365-2672.2012.05352.x. [DOI] [PubMed] [Google Scholar]
- 6.Orro A, et al. Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: genetic determinants and metabolic abilities with environmental relevance. PLoS One. 2015;10:e0139467. doi: 10.1371/journal.pone.0139467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doroghazi, J. R. & Metcalf, W. W. Comparative genomics of Actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics14, 611-2164-14-611 (2013). [DOI] [PMC free article] [PubMed]
- 8.Hjerde E, et al. Draft genome sequence of the Actinomycete Rhodococcus sp. strain AW25M09, isolated from the Hadsel Fjord, Northern Norway. Genome Announc. 2013;1:e0005513–13. doi: 10.1128/genomeA.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letek M, et al. The genome of a pathogenic Rhodococcus: cooptive virulence underpinned by key gene acquisitions. PLoS Genet. 2010;6:e1001145. doi: 10.1371/journal.pgen.1001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceniceros, A., Dijkhuizen, L., Petrusma, M. & Medema, M. H. Genome-based exploration of the specialized metabolic capacities of the genus Rhodococcus. BMC Genomics18, 593-017-3966-1 (2017). [DOI] [PMC free article] [PubMed]
- 11.Polkade AV, Mantri SS, Patwekar UJ, Jangid K. Quorum sensing: an under-explored phenomenon in the phylum Actinobacteria. Front. Microbiol. 2016;7:131. doi: 10.3389/fmicb.2016.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2) Mol. Microbiol. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 13.Bibb MJ. Regulation of secondary metabolism in Streptomycetes. Curr. Opin. Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Li X, et al. ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci. Rep. 2015;5:14831. doi: 10.1038/srep14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato JY, Funa N, Watanabe H, Ohnishi Y, Horinouchi S. Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in. Streptomyces. Proc. Natl. Acad. Sci. USA. 2007;104:2378–2383. doi: 10.1073/pnas.0607472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Alia D, et al. Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3( Microb. Biotechnol. 2011;4(2):239–251. doi: 10.1111/j.1751-7915.2010.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shikura N, Yamamura J, Nihira T. barS1, a gene for biosynthesis of a gamma-butyrolactone autoregulator, a microbial signaling molecule eliciting antibiotic production in Streptomyces species. J. Bacteriol. 2002;184:5151–5157. doi: 10.1128/JB.184.18.5151-5157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takano E. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 2006;9:287–294. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Zou Z, et al. A gamma-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol. Microbiol. 2014;94:490–505. doi: 10.1111/mmi.12752. [DOI] [PubMed] [Google Scholar]
- 20.Gottelt M, Kol S, Gómez-Escribano JP, Bibb M, Takano E. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2) Microbiology. 2010;156:2343–2353. doi: 10.1099/mic.0.038281-0. [DOI] [PubMed] [Google Scholar]
- 21.Khokhlov, A. S. Problems of studies of specific cell autoregulators (on the example of substances produced by some Actinomycetes) In: Frontiers of Bioorganic Chemistry and Molecular Biology (ed Ananchenko, S. N.) 201–210 (Pergamon, 1980).
- 22.Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J. Antibiot. (Tokyo) 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 23.Takano E, et al. Purification and structural determination of SCB1, a gamma-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2) J. Biol. Chem. 2000;275:11010–11016. doi: 10.1074/jbc.275.15.11010. [DOI] [PubMed] [Google Scholar]
- 24.Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J. Ferment. Bioeng. 1989;68:170–173. doi: 10.1016/0922-338X(89)90131-1. [DOI] [Google Scholar]
- 25.Hsiao NH, et al. Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem. Biol. 2009;16:951–960. doi: 10.1016/j.chembiol.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Aroonsri A, Kitani S, Choi SU, Nihira T. Isolation and characterization of bamA genes, homologues of the gamma-butyrolactone autoregulator-receptor gene in Amycolatopsis mediterranei, a rifamycin producer. Biotechnol. Lett. 2008;30:2019–2024. doi: 10.1007/s10529-008-9794-2. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Sanchez, L. et al. Identification and characterisation of enzymes involved in γ-butyrolactone biosynthesis in Streptomyces coelicolor. In: Quorum sensing in Streptomyces coelicolor: Regulation of the SCB signalling system that controls the synthesis of antibiotics. PhD thesis L. Martin-Sanchez, University of Groningenhttp://www.rug.nl/research/portal/files/26679124/Chapter_3.pdf, 91–138 (2016).
- 28.Zhang XM, Zhang DF, Li WJ, Lu CH. Pseudonocardides A–G, new γ-butyrolactones from marine-derived Pseudonocardia sp. YIM M13669. Helv. Chim. Acta. 2016;99:191–196. doi: 10.1002/hlca.201500109. [DOI] [Google Scholar]
- 29.van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 2001;205:197–202. doi: 10.1111/j.1574-6968.2001.tb10947.x. [DOI] [PubMed] [Google Scholar]
- 30.Menges R, Muth G, Wohlleben W, Stegmann E. The ABC transporter Tba of Amycolatopsis balhimycina is required for efficient export of the glycopeptide antibiotic balhimycin. Applied Microbiology and Biotechnology. 2007;77:125–134. doi: 10.1007/s00253-007-1139-x. [DOI] [PubMed] [Google Scholar]
- 31.Arenskötter M, Baumeister D, Kalscheuer R, Steinbüchel A. Identification and application of plasmids suitable for transfer of foreign DNA to members of the genus Gordonia. Appl. Environ. Microbiol. 2003;69:4971–4974. doi: 10.1128/AEM.69.8.4971-4974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. in Practical Streptomyces Genetics (John Innes Foundation, Norwich, 2000).
- 33.Hsiao NH, Gottelt M, Takano E. Chapter 6. Regulation of antibiotic production by bacterial hormones. Methods Enzymol. 2009;458:143–157. doi: 10.1016/S0076-6879(09)04806-X. [DOI] [PubMed] [Google Scholar]
- 34.Lee YJ, Kitani S, Kinoshita H, Nihira T. Identification by gene deletion analysis of barS2, a gene involved in the biosynthesis of gamma-butyrolactone autoregulator in Streptomyces virginiae. Arch. Microbiol. 2008;189:367–374. doi: 10.1007/s00203-007-0327-5. [DOI] [PubMed] [Google Scholar]
- 35.Bhukya H, Bhujbalrao R, Bitra A, Anand R. Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2) Nucleic Acids Res. 2014;42:10122–10133. doi: 10.1093/nar/gku587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidda JD, et al. Overproduction and identification of butyrolactones SCB1-8 in the antibiotic production superhost Streptomyces M1152. Org. Biomol. Chem. 2016;14:6390–6393. doi: 10.1039/C6OB00840B. [DOI] [PubMed] [Google Scholar]
- 37.Xu, J. H., Gu, K. B., Zhang, D. J., Li, Y. G. & Tian, L. Ghanamycins A and B, two novel gamma-butyrolactones from marine-derived Streptomyces ghanaensis TXC6-16. J. Antibiot. (Tokyo) (2017). [DOI] [PubMed]
- 38.Khokhlov, A. S. A-factor and analogous Actynomicete autoregulators of lactone nature In: Microbial Autoregulators (Harwood Academic Publishers, 1991).
- 39.Kawabuchi M, Hara Y, Nihira T, Yamada Y. Production of butyrolactone autoregulators by Streptomyces coelicolor A3(2) FEMS Microbiology Letters. 1997;157:81–85. doi: 10.1111/j.1574-6968.1997.tb12756.x. [DOI] [Google Scholar]
- 40.Choi SU, Lee CK, Hwang YI, Kinosita H, Nihira T. Gamma-butyrolactone autoregulators and receptor proteins in non-Streptomyces Actinomycetes producing commercially important secondary metabolites. Arch. Microbiol. 2003;180:303–307. doi: 10.1007/s00203-003-0591-y. [DOI] [PubMed] [Google Scholar]
- 41.Chiba H, Agematu H, Sakai K, Dobashi K, Yoshioka T. Rhodopeptins, novel cyclic tetrapeptides with antifungal activities from Rhodococcus sp. III. Synthetic study of rhodopeptins. J. Antibiot. (Tokyo) 1999;52:710–720. doi: 10.7164/antibiotics.52.710. [DOI] [PubMed] [Google Scholar]
- 42.Iwatsuki M, et al. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J. Am. Chem. Soc. 2006;128:7486–7491. doi: 10.1021/ja056780z. [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa W, Tamura T. A quinoline antibiotic from Rhodococcus erythropolis JCM 6824. J. Antibiot. (Tokyo) 2008;61:680–682. doi: 10.1038/ja.2008.96. [DOI] [PubMed] [Google Scholar]
- 44.Borisova, R. B. Isolation of a Rhodococcus soil bacterium that produces a strong antibacterial compound. Electronic Theses and Dissertations. East Tennessee State UniversityPaper 1388 (2011).
- 45.Horinouchi S, Beppu T. Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 2007;83:277–295. doi: 10.2183/pjab.83.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nodwell JR. Are you talking to me? A possible role for γ-butyrolactones in interspecies signalling. Mol. Microbiol. 2014;94:483–485. doi: 10.1111/mmi.12787. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Vining LC. Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 byjadW 1, a member of the afsA family of γ-butyrolactone regulatory genes. Microbiology. 2003;149:1991–2004. doi: 10.1099/mic.0.26209-0. [DOI] [PubMed] [Google Scholar]
- 48.Sakuda S, Higashi A, Tanaka S, Nihira T, Yamada Y. Biosynthesis of virginiae butanolide A, a butyrolactone autoregulator from Streptomyces. J. Chem. Soc. Perkin Trans. 1992;114:663–668. [Google Scholar]
- 49.Bentley SD, et al. Complete genome sequence of the model Actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 50.Bos CJ, et al. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 1988;14:437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).