Abstract
Glucosinolates (GLS) are a group of plant secondary metabolites mainly found in Cruciferous plants, share a core structure consisting of a β-thioglucose moiety and a sulfonated oxime, but differ by a variable side chain derived from one of the several amino acids. These compounds are hydrolyzed upon cell damage by thioglucosidase (myrosinase), and the resulting degradation products are toxic to many pathogens and herbivores. Human beings use these compounds as flavor compounds, anti-carcinogens, and bio-pesticides. GLS metabolism is complexly linked to auxin homeostasis. Indole GLS contributes to auxin biosynthesis via metabolic intermediates indole-3-acetaldoxime (IAOx) and indole-3-acetonitrile (IAN). IAOx is proposed to be a metabolic branch point for biosynthesis of indole GLS, IAA, and camalexin. Interruption of metabolic channeling of IAOx into indole GLS leads to high-auxin production in GLS mutants. IAN is also produced as a hydrolyzed product of indole GLS and metabolized to IAA by nitrilases. In this review, we will discuss current knowledge on involvement of GLS in auxin homeostasis.
Keywords: glucosinolate, auxin, metabolism, development, Arabidopsis
Introduction
Glucosinolates (GLS) are a group of secondary metabolites found almost exclusively in Brassicaceae (Agerbirk and Olsen, 2012). GLS are nitrogen and sulfur rich compounds, forming a two-component defense system (mustard oil bomb) with myrosinases against herbivores and microorganisms (Bones and Rossiter, 1996; Rask et al., 2000). Upon tissue damage by herbivores, the mustard oil bomb comes into action, where myrosinases hydrolyze GLS into different degradation products that are toxic to the enemy (Rask et al., 2000; Chen and Andreasson, 2001). Apart from plant defense, some of the GLS degradation products are physiologically significant in plant nutrition and growth regulation (Hull et al., 2000; Rask et al., 2000; Kutz et al., 2002; Grubb et al., 2004; Katz et al., 2015). GLS breakdown products are part of human consumption and health. For instance, some of the GLS metabolites give characteristic flavors to Brassica vegetables (cabbage, cauliflower, broccoli, etc.) and condiments (mustard, horseradish, wasabi, etc.); and some others such as sulforaphane, indole-3-carbinol and phenethyl isothiocyanate act as cancer-preventive agents (Zhang et al., 1994; Hecht, 2000; Nakajima et al., 2001; Keck and Finley, 2004; Hayes et al., 2008). Moreover, Brassica crops are used for crop rotation and/or biofumigation as certain GLS metabolites exhibit natural biopesticide properties (Gimsing and Kirkegaard, 2009).
Glucosinolates are evolutionarily younger and evolved from cyanogenic glucosides. Cyanogenic glucosides are widespread in planta, whereas GLS are restricted to the order Capparales and the genus Drypetes of Euphorbiaceae (Johnson et al., 2009). Cyanogenic glucosides and GLS do not coexist in plants, except for one species, Carica papaya that produces both phenylalanine-derived cyanogenic glucosides and GLS (Bennett et al., 1997). Evolutionary link between cyanogenic glucosides and GLS is supported by having similarities in their biosynthesis such as amino acids as precursors and CYTOCHROME P450s (CYPs) as aldoxime metabolizing enzymes (Bak et al., 1998, 2001; Hansen et al., 2001a; Naur et al., 2003).
Glucosinolates (GLS) are characterized by having a thioglucose moiety, a sulfonated oxime, and a side-chain derived from aliphatic, aromatic, or indole amino acids (Figure 1). Currently, more than 130 different GLS structures have been identified (Agerbirk and Olsen, 2015). GLS are biosynthesized from amino acids and stored in the vacuoles of specific laticifer-like sulfur-rich cells called S-cells localized in the phloem cap along the vasculature and the leaf margins (Koroleva et al., 2010). GLS hydrolyzing myrosinases are localized in myrosin cells and are spatially separated from GLS (Thangstad et al., 1991; Xue et al., 1993; Andreasson et al., 2001; Husebye et al., 2002). Biosynthesis and long-distance transport of GLS are critical for spatio-temporal distribution of the GLS in plants (Nour-Eldin et al., 2012; Andersen and Halkier, 2014; Jørgensen et al., 2015). In Arabidopsis, transport of GLS compounds is mediated by transporter proteins GTR1/NPF2.10 and GTR2/NPF2.11 (Nour-Eldin et al., 2012).
FIGURE 1.
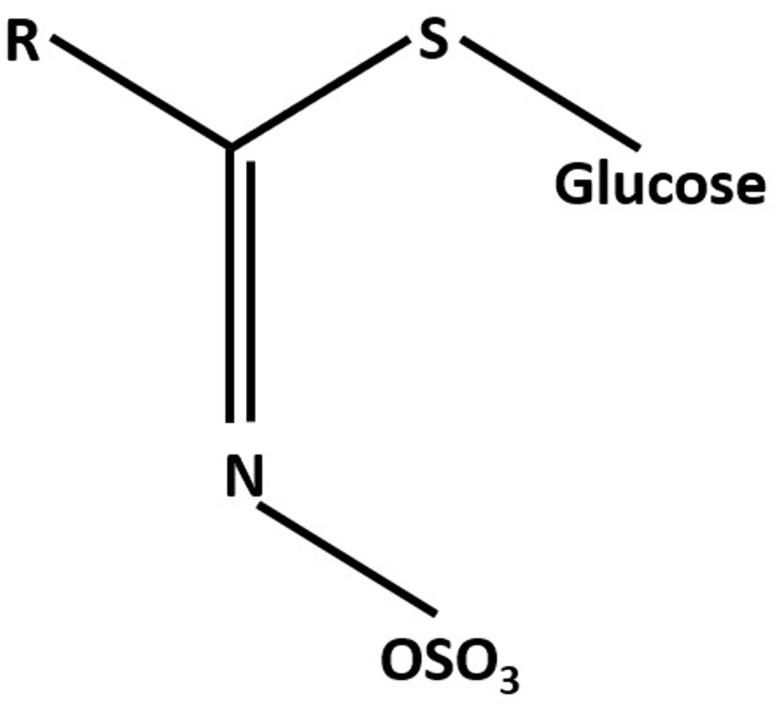
General structure of glucosinolates (GLS).
Several lines of evidence suggest that indole GLS are metabolically linked to auxin homeostasis. Interruption of GLS metabolism leads to severe defects in plant growth and development similar to high-auxin phenotypes (Boerjan et al., 1995; Mikkelsen et al., 2000, 2004; Bak and Feyereisen, 2001; Hansen et al., 2001b; Reintanz et al., 2001; Chen et al., 2003; Tantikanjana et al., 2004; Skirycz et al., 2006; Ueda et al., 2006). The high-auxin phenotypes of GLS mutants were an effect of blocking the indole GLS pathway downstream of indole-3-acetaldoxime (IAOx), which resulted in overflow of IAOx to indole-3-acetic acid (IAA) (Halkier and Gershenzon, 2006; Nafisi et al., 2006). In this review, we discuss current knowledge on potential involvement of GLS especially indole GLS metabolism in auxin homeostasis.
Structure and Classification of GLS
The typical chemical structure of GLS consists of β-D-glucopyranose residue linked via a sulfur atom to a (Z)-N-hydroximinosulfate ester plus a precursor amino acid derived R group (Figure 1). Based on the precursor amino acid and the types of modification to the variable R group, GLS can be classified into aromatic (phenylalanine or tyrosine), aliphatic (alanine, leucine, isoleucine, methionine, or valine), and indole GLS [tryptophan (TRP)] (Fahey et al., 2001; Agerbirk and Olsen, 2012). A list and structures of GLS can be found in an excellent review (Clarke, 2010).
An Overview of Biosynthesis of GLS and Camalexin
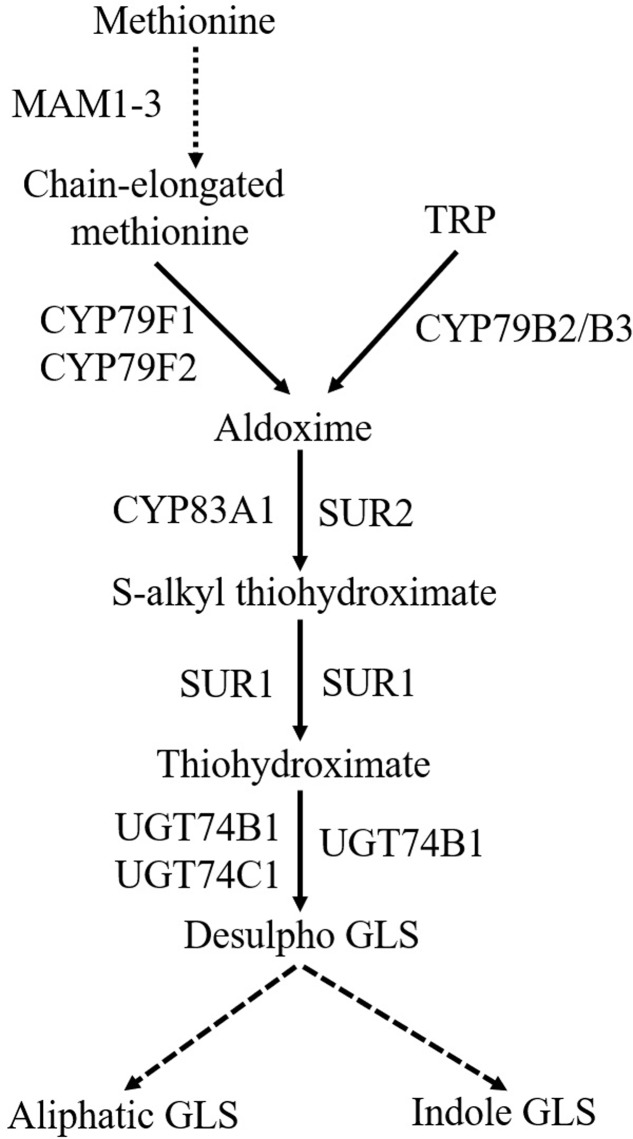
Glucosinolates biosynthetic pathway comprises three steps (Figure 2): amino acid chain-elongation, core structure formation, and secondary modifications (Chen and Andreasson, 2001; Halkier and Gershenzon, 2006). In the amino acid chain-elongation step, certain aliphatic and aromatic amino acids are elongated by insertion of methylene groups into their side chains. These reactions are mediated by the METHYLTHIOALKYLMALATE SYNTHASE (MAM) 1-3 and MAM-like (MAML) genes (Kroymann et al., 2001; Textor et al., 2007). The amino acid moiety of either chain elongated or not, is converted to a core GLS structure in a series of reactions. During this process, amino acids are converted to their corresponding aldoximes by CYP79s (Figure 2). CYP79A2 catalyzes phenylalanine (Wittstock and Halkier, 2000), CYP79F1 and CYP79F2 metabolize chain-elongated methionine (Hansen et al., 2001b; Chen et al., 2003), whereas CYP79B2/B3 convert TRP (Hull et al., 2000) to their aldoximes. The enzyme catalyzing homophenylalanine is unknown. Aldoximes are metabolized to S-alkylthiohydroximates by CYP83s. Methionine-derived aldoximes are catalyzed by CYP83A1, whereas aromatic- and indole-acetaldoximes are catalyzed by CYP83B1/SUPERROOT2 (SUR2) (Bak and Feyereisen, 2001; Hemm et al., 2003). S-alkylthiohydroximates are cleaved into thiohydroximates by a C-S lyase/SUPERROOT1 (SUR1) (Mikkelsen et al., 2004), followed by S-GLUCOSYLTRANSFERASE (UGT) mediated glycosylation (Petersen et al., 2001). Finally, sulfonation of the desulfo-GLS is carried out by sulfotransferases (Piotrowski et al., 2004). Later, GLS core structure undergoes several secondary modifications at the side chain and glucose moiety (Hopkins et al., 2009). Side chain of aliphatic GLS is modified by oxygenation, hydroxylation, alkenylation, and benzoylation, whereas side chain of indole GLS is modified by hydroxylation and methoxylation (Sønderby et al., 2010b).
FIGURE 2.
General scheme of biosynthesis of aliphatic and indole GLS. Dashed arrow denotes the process with multiple steps. MAM1-3, METHYLTHIOALKYLMALATE SYNTHASE; TRP, tryptophan; CYP79F1, CYTOCHROME P450 MONOOXYGENASE 79F1; CYP79F2, CYTOCHROME P450 MONOOXYGENASE 79F2; CYP79B2/B3, CYTOCHROME P450 MONOOXYGENASE 79B2/CYTOCHROME P450 MONOOXYGENASE 79B3; SUR2, SUPERROOT2; SUR1, SUPERROOT1; UGT74B1, S-GLUCOSYLTRANSFERASE 74B1; UGT74C1, S-GLUCOSYLTRANSFERASE 74C1; GLS, glucosinolates.
Camalexin is a major phytoalexin found in specific group of Cruciferous plants including Arabidopsis (Glawischnig, 2007; Rauhut and Glawischnig, 2009; Bednarek et al., 2011). Camalexins are synthesized in response to fungal pathogens and play positive role in their resistance (Pedras et al., 2011). In camalexin biosynthetic pathway, CYP71A13/12 convert IAOx into indole-3-acetonitrile (IAN) (Nafisi et al., 2007; Müller et al., 2015) which is then oxidized and conjugated to glutathione by glutathione-S-transferase GSTF6 to produce GSH (IAN) (Su et al., 2011). This GSH (IAN) is metabolized to Cys(IAN) by γ-glutamyl peptidases GGP1 and GGP3 (Geu-Flores et al., 2011) which is metabolized to camalexin by CYP71B15/PAD3 (Zhou et al., 1999; Schuhegger et al., 2006; Böttcher et al., 2009).
An Overview of Auxin Biosynthesis
IAA is proposed to be biosynthesized from two pathways, TRP-independent and TRP-dependent pathway (Woodward and Bartel, 2005; Chandler, 2009; Normanly, 2010). TRP-dependent IAA biosynthesis is considered as the main route of IAA biosynthesis in plants (Figure 3).
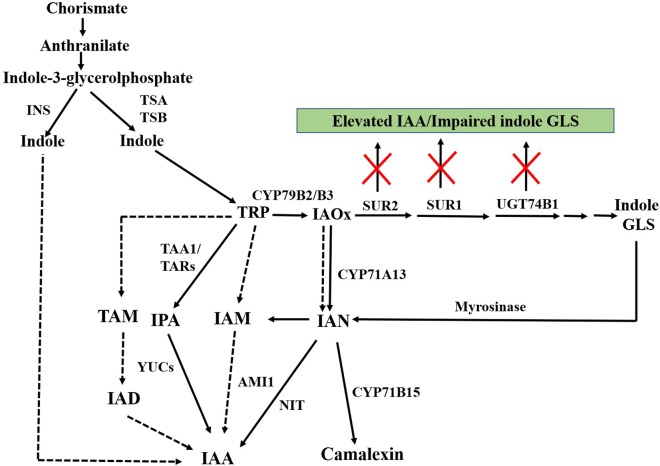
FIGURE 3.
Schematic representation of TRP-independent and TRP-dependent auxin biosynthesis. IAOx metabolized from TRP acts as key metabolic branching point for biosynthesis of indole GLS, camalexin, and IAA. Inactivation of genes of post-acetaldoxime GLS biosynthesis leads to elevated IAA along with impaired indole GLS. Dashed arrows indicate pathways that are not well defined. INS, INDOLE SYNTHASE; TSA, TRP SYNTHASE A; TSB, TRP SYNTHASE B; TRP, tryptophan; CYP79B2/B3, CYTOCHROME P450 MONOOXYGENASE 79B2/CYTOCHROME P450 MONOOXYGENASE 79B3; SUR2, SUPERROOT2; SUR1, SUPERROOT1; UGT74B1, S-GLUCOSYLTRANSFERASE 74B1; GLS, glucosinolates; IAOx, indole-3-acetaldoxime; TAA1, TRP AMINOTRANSFERASE OF ARABIDOPSIS; IPA, indole-3-pyruvic acid; YUC, YUCCA; TAM, tryptamine; IAD, indole-3-acetaldehyde; IAM, indole-3-acetamide; AMI1, AMIDASE1; IAN, indole-3-acetonitrile; NIT, NITRILASE; CYP71B15, CYTOCHROME P450 MONOOXYGENASE 71B15; IAA, indole-3-acetic acid.
TRP-Independent Pathway
In the TRP-independent pathway, indole-3-glycerol phosphate or indole is the likely precursor, while the genes and enzymes involved in this pathway are still largely unknown (Ouyang et al., 2000; Zhang et al., 2008; Wang et al., 2015). Studies on TRP auxotrophic mutants that were unable to synthesize TRP revealed the existence of TRP-independent pathway (Baldi et al., 1991; Wright et al., 1991; Normanly et al., 1993; Ouyang et al., 2000; Tivendale et al., 2014). When TRP auxotrophic mutants in maize and Arabidopsis were fed with isotope-labeled anthranilate and TRP, IAA was more enriched than TRP, and the incorporation of the label into IAA from TRP was low, indicating occurrence of TRP-independent IAA biosynthesis (Wright et al., 1991; Normanly et al., 1993). The TRP biosynthetic mutants trp3 and trp2, defective in TRP SYNTHASE A and TRP SYNTHASE B subunits, respectively, accumulated higher levels of IAA than the wild-type despite containing lower TRP levels (Last et al., 1991; Normanly et al., 1993; Radwanski et al., 1996; Ouyang et al., 2000). Recently, INDOLE SYNTHASE was suggested to catalyze indole-3-glyceralphosphate in TRP-independent pathway and play essential role in embryo development (Zhang et al., 2008; Wang et al., 2015).
TRP-Dependent Pathway
In the TRP-dependent pathway, IAA is synthesized via indole-3-pyruvic acid (IPA), indole-3-acetamide (IAM), tryptamine (TAM) and/or IAOx as intermediates (Woodward and Bartel, 2005; Chandler, 2009; Normanly, 2010; Figure 3). The IPA pathway is considered as a predominant auxin biosynthesis pathway in plants (Mashiguchi et al., 2011; Zhao, 2014).
The IAM Pathway
The IAM pathway is well established in bacteria where TRP MONOOXYGENASE (iaaM) converts TRP to IAM, which is further converted to IAA by IAM HYDROLASE (iaaH) (Patten and Glick, 1996). AMIDASE 1 (AMI1) was identified as a homolog of iaaH in Arabidopsis and Nicotiana (Pollmann et al., 2002, 2006; Nemoto et al., 2009). However, no iaaM homologs have been found in plants. IAM was identified as an endogenous compound in many plant species including Arabidopsis, maize, rice, and tobacco (Sugawara et al., 2009; Mano and Nemoto, 2012; Novak et al., 2012). Additionally, AMI1 enzyme activity was detected in various plants (Kawaguchi et al., 1991, 1993; Arai et al., 2004; Pollmann et al., 2009; Sánchez-Parra et al., 2014), suggesting existence of this pathway in the plants.
The IPA Pathway
In the IPA pathway, the TRP aminotransferase TAA1/TAR (TRP AMINOTRANSFERASE OF ARABIDOPSIS) gene family converts TRP to IPA (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009; Zhou et al., 2011; Tivendale et al., 2012), which is subsequently converted to IAA by YUCCA (YUC) flavin monooxygenase (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). Homologs of TAA1 were identified in maize (Chourey et al., 2010; Phillips et al., 2011), pea (Tivendale et al., 2012), and several other species (Chourey et al., 2010; Liu et al., 2012). There are 11 YUCs in Arabidopsis, overexpression of YUCs results in auxin-overproduction phenotypes in Arabidopsis (Zhao et al., 2001; Woodward et al., 2005; Cheng et al., 2006; Kim et al., 2007; Hentrich et al., 2013) and various other plants (Zhao, 2014). Conversely, loss-of-function yuc mutants displayed low auxin synthesis with developmental defects (Cheng et al., 2006, 2007; Zhao, 2014), which can be rescued by adding auxin to growth media (Chen et al., 2014) or by expressing bacterial auxin biosynthetic gene iaaM under the control of a YUC promoter (Cheng et al., 2006). Overexpression of TAAs does not cause any obvious developmental phenotypes. However, low auxin phenotypes of sav3 and wei8 caused by mutations in TAA1 were rescued by overexpressing iaaM or by using the synthetic auxin picloram (Stepanova et al., 2008; Tao et al., 2008).
The TAM Pathway
In the TAM pathway, TRP DECARBOXYLASE (TDC) converts TRP to TAM. Moreover, TDCs were functionally characterized to participate in indole alkaloid and serotonin biosynthesis. For instance, transgenic tobacco plants overexpressing the TDC gene of Catharanthus roseus accumulated very high levels of TAM, whereas IAA levels were unaffected (Songstad et al., 1990). Hence, the TAM pathway is not considered as a major player of auxin biosynthesis.
The IAOx Pathway
Indole-3-acetaldoxime is biosynthesized from TRP by CYP79B2/B3 (Hull et al., 2000). IAOx was first isolated from Brassica oleracea (Kindl, 1968), later conclusively identified from extracts of Brassica campestris (Ludwig-Müller and Hilgenberg, 1988). Earlier studies suggested that a cytochrome P450-like activity or plasma membrane-associated peroxidases might mediate conversion of TRP to IAOx (Kindl, 1968). Arabidopsis CYP9B2/B3 were identified in a yeast screen for cDNAs conferring resistance to 5-fluoroindole (the precursor of a toxic TRP derivative) and demonstrated their ability to specifically convert TRP to IAOx in vitro (Hull et al., 2000; Mikkelsen et al., 2000). YUC monooxygenase was assumed to take part in IAOx synthesis, however, endogenous IAOx levels were not significantly changed in yuc quadruple mutants demonstrating GLS metabolism as the main contributor of IAOx synthesis (Sugawara et al., 2009; Zhao, 2014). Endogenous IAOx has not been detected in non-GLS plants such as tomato, pea, rice, maize, or tobacco (Cooney and Nonhebel, 1991; Quittenden et al., 2009; Sugawara et al., 2009), correspondingly, CYP79B2/B3 have only been identified in GLS plants (Bak et al., 1998; Sugawara et al., 2009), suggesting IAOx synthesis is specific to GLS plants.
It has been proposed that IAOx is channeled into biosynthesis of IAA via IAN (Hull et al., 2000; Nafisi et al., 2006; Sugawara et al., 2009). It is well demonstrated that CYP71A13 catalyzes IAOx to IAN, but this gene is mainly induced in response to pathogen infection to produce camalexin (Glawischnig, 2007). IAN levels were increased in CYP79B2 overexpressing plants, and cyp79B2/B3 double mutants showed reduced IAN content (Zhao et al., 2002). Metabolite feeding studies showed that IAM and IAN are likely produced from IAOx (Sugawara et al., 2009). When the IAOx-deficient cyp79b2/b3 double mutants supplied with 13C6-labeled IAOx, 13C6 atoms were efficiently incorporated into IAM, IAN, and IAA, indicating that IAA can be produced from IAOx via IAM and IAN. In consistent with this, wild-type plants supplied with IAM and IAN showed auxin-overproduction phenotypes (Sugawara et al., 2009). When Arabidopsis CYP79B2 or CYP79B3 genes was ectopically expressed in tobacco, IAOx and IAN were identified as endogenous compounds in the transgenic plants together with elevated levels of IAA compared to their controls (Nonhebel et al., 2011).
In addition to being metabolized from IAOx, IAN is also produced from indole GLS by myrosinases (Halkier and Gershenzon, 2006). A tendency of IAN accumulation in accordance with turnover of glucobrassicin was observed in Arabidopsis (Müller and Weiler, 2000; Reintanz et al., 2001; Zhao et al., 2002). It has been reported that low concentration of IAN can induce a high-auxin phenotype in the Arabidopsis seedlings (Normanly et al., 1997). In maize, IAN was identified as an endogenous compound at lower magnitude than Arabidopsis, exogenously supplied IAN inhibited the root growth (Thimann, 1953; Park et al., 2003; Kriechbaumer et al., 2007). The altered auxin response in the root tips of Arabidopsis myrosinase double mutants tgg4 tgg5 was likely due to no IAN production from indole GLS under myrosinase disruption (Fu et al., 2016). In vivo conversion of IAN to IAA was observed in root tissue of Arabidopsis (Müller et al., 1998).
Nitrilases (NITs) are proposed to catalyze IAN to IAA (Bartling et al., 1992; Bartel and Fink, 1994; Normanly et al., 1997; Pollmann et al., 2002). Early studies assayed NIT enzyme activities from members of plant families including Cruciferae, Gramineae, and Musaceae (Mahadevan and Thimann, 1964; Thimann and Mahadevan, 1964). Arabidopsis genome contains four NITs named NIT1–NIT4, categorized into NIT4 and NIT1-subfamilies. The members of NIT1-subfamily, NIT1-3, were suggested to be emerged from phylogenetically older NIT4-subfamily by gene duplication events and subsequent neo-functionalization (Piotrowski, 2008). Transgenic plants overexpressing each of NIT1-3 were more sensitive to exogenously supplied IAN (Schmidt et al., 1996; Dohmoto et al., 2000a,b), whereas nit mutants were tolerant (Normanly et al., 1997). Increased NIT activity was appeared to alter levels of IAN and free IAA in NIT1 overexpressing transgenic Arabidopsis plants (Lehmann et al., 2017). In maize, loss-of-function of ZmNIT2, a homolog of AtNIT4, caused the mutants less sensitive to IAN with significantly lower amounts of total IAA in kernels and roots of young seedlings compared to wild-type plants (Kriechbaumer et al., 2007). NIT3 was proposed to catalyze IAN to IAA in sulfur deprived Arabidopsis roots (Kutz et al., 2002). In Brassica plants, development of root galls caused by Plasmodiophora brassicae infection appeared to be mediated by IAN-derived IAA (Grsic et al., 1999; Grsic-Rausch et al., 2000; Ishikawa et al., 2007).
Apart from several lines of supporting evidence, the role of NITs in IAA biosynthesis is still arguable (Piotrowski, 2008). It was shown that these enzymes have lesser substrate preference to IAN than the compounds including phenylpropionitrile, allylcyanide, phenylthio acetonitrile, and methylthio acetonitriles raising doubt on the role of these enzymes in IAA biosynthesis (Vorwerk et al., 2001). The substrate preference of NITs would be different if the enzymes were challenged with IAN as a predominant substrate (Pollmann et al., 2002). For instance, in the roots of sulfur-deprived plants, expression of NIT3 was strongly induced in response to intensified turnover of IAN precursor glucobrassicin and was suggested to metabolize IAA (Kutz et al., 2002). Additionally, in planta NITs were shown to much more efficient than the recombinant ones. For example, Arabidopsis NIT2 ectopically overexpressed in tobacco was able to convert IAN supplied at micromolar concentrations, 15-fold below the enzyme’s apparent in vitro Km (Schmidt et al., 1996). In the absence of exogenous IAN, neither the nit mutants nor NIT overexpressors showed severe auxin phenotypes, and endogenous IAN and IAA levels were not clearly distinguishable (Normanly et al., 1997). NIT1 overexpressors displayed strong reduction in their primary root length with clearly elevated levels of free IAA and IAN, while nit1-3 mutant displayed wild-type like roots with reduced total IAA levels (Lehmann et al., 2017). The cyp79b2/b3 mutants that are deficient in glucobrassicin have strongly reduced levels of IAN (Zhao et al., 2002), however, are not affected in infection rates of P. brassicae and consequent root gall development (Siemens et al., 2008). The role of NITs in the development of clubroot was also questioned, as cyp79b2/b3 mutants with low levels of IAN showed normal clubroot symptoms (Siemens et al., 2008). The involvement of indole GLS and NITs in the development of clubroot disease is seemingly more complicated. For instance, the transcripts of BnCYP83B1 and BnNit4 were up-regulated in the infected root of Brassica napus (Xu et al., 2016), whereas CYP83B1 and other GLS biosynthesis genes CYP79B2/B3 and CYP83A1 were differentially downregulated during different stages of infection in Brassica macrocarpa (Zhang et al., 2016). Hence, further studies are needed to understand this phenomenon. The role of IAN as a direct metabolite of IAOx and the involvement of NITs in IAA biosynthesis was argued, because the high-auxin phenotype of sur2 is not rescued in the nit1 genetic background (Bak et al., 2001). The mutant impaired with all NITs would exclude the possible redundancy of NIT activity and gives conclusive idea on the contribution of NITs in IAA biosynthesis.
Regulation of GLS Biosynthesis
Transcriptional Control of GLS Biosynthesis
Various Transcription Factors (TFs) are known to regulate GLS biosynthesis at transcriptional level. Of which, some TFs have been shown to control GLS production at global level. For instance, IQD1 positively affected production of both aliphatic and indole GLS. The gain- and loss-of-function of IQD1 led to increased and decreased accumulation of GLS, respectively (Levy et al., 2005). SLIM1, an EIN3-like TF, involved in sulfur deficiency response, was shown to repress the expression of GLS biosynthetic genes (Maruyama-Nakashita et al., 2006; Frerigmann and Gigolashvili, 2014). AtDOF1.1 was reported to promote GLS production (Skirycz et al., 2006). Finally, TFL2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, was shown to affect GLS biosynthesis (Kim et al., 2004; Bennett et al., 2005).
R2R3-MYBs constitute the largest MYB gene family in plants, characterized by possessing two repeats of DNA binding domains named R2 and R3 at the N-terminal end, and an activation or repression domain usually located at the C-terminus (Stracke et al., 2001). These TFs involve in various processes including development, metabolism and stress response (Chezem and Clay, 2016). Members of sub-group 12 R2R3-MYB are specific regulators of GLS biosynthesis: MYB34, MYB51, and MYB122 control indole GLS production, whereas MYB28, MYB29, and MYB76 regulate aliphatic GLS biosynthesis (Gigolashvili et al., 2009).
MYB28 is characterized as a dominant regulator of aliphatic GLS, whereas MYB29 and MYB76 are suggested to play additional accessory role. Overexpression of these MYBs has been shown to induce aliphatic GLS biosynthetic genes and aliphatic GLS content (Gigolashvili et al., 2007b, 2008; Hirai et al., 2007; Sønderby et al., 2007). Consistently, loss-of-function of MYB28 affected production of both short- and long-chain derived aliphatic GLS. However, myb29 and myb76 were defective in accumulation of only short-chain derived aliphatic GLS albeit to a lesser extent in myb76 (Hirai et al., 2007; Sønderby et al., 2007; Beekwilder et al., 2008; Gigolashvili et al., 2008), indicating dominance of MYB28 over other two MYBs in controlling aliphatic GLS production. In line with this, expression of aliphatic GLS biosynthetic genes was greatly affected in myb28 than in myb29 (Gigolashvili et al., 2007b, 2008; Hirai et al., 2007; Sønderby et al., 2007). Additionally, disruption of both MYB28 and MYB29 showed complete reduction of aliphatic GLS levels in myb28 myb29 double mutants (Sønderby et al., 2007; Beekwilder et al., 2008). This shows that MYB28 controls both short- and long-chain derived aliphatic GLS, while MYB29 and MYB76 regulate only the short-chain derived aliphatic GLS. However, Sønderby et al. (2010a) showed that MYB29 and MYB76 were able to regulate the production of long-chain derived aliphatic GLS in the genetic backgrounds of myb28 myb76 and myb28 myb29, respectively, suggesting interplay of MYBs in controlling GLS biosynthesis. Aliphatic biosynthetic genes were differentially transactivated by these MYBs, though MYB28 showed highest transactivation potential over the two MYBs (Gigolashvili et al., 2007b, 2008). For instance, MAML was strongly transactivated by MYB28 than MYB29; CYP79F2 was greatly transactivated by MYB28 and MYB29 but to a less extent by MYB76 (Gigolashvili et al., 2008). Additionally, the transcript levels of aliphatic biosynthetic genes were uncoupled from the levels of GLS metabolites in the myb28, myb29, and myb76 knockouts (Sønderby et al., 2010a). These reports suggest that a complex network of MYB28, MYB29, and MYB76 controls the aliphatic GLS biosynthesis specifically and coordinately.
MYB34/ATR1 was initially identified as a regulator of TRP biosynthesis as it controls the expression of TRP biosynthetic gene ASA1 (Bender and Fink, 1998). The expression of indole GLS biosynthetic genes CYP79B2/B3 and CYP83B1/SUR2 were altered in myb34 mutants, and transcript levels of MYB34 were reduced in atr4/cyp83B1/sur2, indicating its involvement in GLS biosynthesis (Smolen and Bender, 2002). The plants constitutively overexpressing MYB34 accumulated 10-fold higher indole GLS compared to their control plants. Conversely, myb34 knockouts displayed low indole GLS with reduced transcript levels of indole GLS biosynthetic genes (Celenza et al., 2005). Similar to MYB34, both MYB51 and MYB122 positively regulated indole GLS production. Metabolite analysis showed increase of indole GLS levels in the plants overexpressing MYB51 and MYB122, and decrease in myb51 and myb122 knockouts (Gigolashvili et al., 2007a). Thus, MYB34, MYB51, and MYB122 positively regulate indole GLS production.
Because CYP79B2/B3 can convert TRP to IAOx, and the expression of CYP79B2/B3 is regulated by MYB34, MYB51, and MYB122, it is possible that production of other IAOx derived metabolites including IAA, camalexin and indole-3-carboxylic acids may also be regulated by these MYB genes. Indeed, it was reported that MYB34, MYB51, and MYB122 could show a conditional and probably indirect impact on the biosynthesis of camalexin and indole-3-carbolic acids (Frerigmann et al., 2015, 2016). Moreover, elevated IAA levels were found in MYB34, and MYB122 overexpression lines (Celenza et al., 2005; Gigolashvili et al., 2007a), suggesting a potential role of these TFs in auxin homeostasis.
Recently, it has been shown that bHLH04, bHLH05, and bHLH06 genes of sub-group IIIe of bHLH TF family take part in GLS biosynthesis together with R2R3-MYBs (Schweizer et al., 2013; Frerigmann et al., 2014). bHLH06/MYC2 was shown to negatively regulate indole GLS biosynthesis, as levels of indole GLS were increased in bhlh06/myc2 mutants (Dombrecht et al., 2007). However, it was later shown that bHLH06, bHLH04, and bHLH05 positively regulate indole GLS biosynthesis, as the triple mutants had reduced levels of indole GLS (Schweizer et al., 2013; Frerigmann et al., 2014). bHLH06 bound directly to the G-box motif in the promoters of GLS biosynthetic genes (Schweizer et al., 2013). Moreover, MYB-bHLH interactions can play essential role in controlling GLS biosynthesis. For instance, the reduction of indole GLS levels was more pronounced in myb51 bhlh05 plants than bhlh05 single mutants. In line with this, double gain-of-function mutants myb34-1D bhlh05D94 had 20-fold more indole GLSs than their single mutants and wild-type plants (Frerigmann et al., 2014). These findings suggest that MYB and bHLH TFs play critical roles in regulating indole GLS biosynthesis.
Hormonal Control of GLS Biosynthesis
Glucosinolates biosynthesis is regulated by various phytohormones, including jasmonic acid (JA), ethylene, salicylic acid (SA), and brassinosteroids (BRs) (Mikkelsen et al., 2003; Guo et al., 2013). JA is a well characterized stress signaling molecule known to integrate plant response to various environmental cues, and is involved in a wide variety of plant processes (Turner et al., 2002; Koo and Howe, 2009). JA and its precursors and derivatives collectively called as jasmonates (JAs). They are shown to induce various TFs that are involved in secondary metabolite production (Memelink et al., 2001). JAs signaling involves perception of JAs by F-box protein CORONATINE INSENSITIVE1 (COI1) protein of Skp–Cullin–F-box protein complex (SCFCOI1) that facilitates ubiquitin-26S proteasome mediated degradation of transcriptional repressors called JAZ (Jasmonate ZIM domain). JAZ proteins interact with and repress a variety of TFs; this repression is released upon JAZ degradation via JAs signaling. As JAZ proteins are known to repress sub-group IIIe of bHLH TFs, to inhibit interaction between MYB-bHLH (Chini et al., 2007), these proteins play critical role in controlling GLS biosynthesis in a JAs dependent manner. It was found that the interaction strength between bHLH-MYB proteins could affect the interaction between bHLH-JAZ proteins. An amino acid substitution in bHLH05 weakens its interaction with JAZ protein (Frerigmann et al., 2014) allowing the bHLH05 to induce indole GLS biosynthesis. Exogenous JAs treatment induced GLS biosynthetic genes and GLS content in various plant species (Brader et al., 2001; Gigolashvili et al., 2008; De Geyter et al., 2012). Therefore, MYB-bHLH interactions may play a crucial role in JAs responsive GLS biosynthesis.
Salicylic acid differentially regulates GLS biosynthesis. SA treatment has been shown to induce nearly all types of GLS, of which, 2-phenylethyl GLS showed highest accumulation in oilseed rape (Kiddle et al., 1994). In Arabidopsis, 4-methoxy-glucobrassicin was reported to be induced by SA, while the contents of glucobrassicin and neoglucobrassicin were decreased (Mikkelsen et al., 2003). Increased SA production in mpk4 and cpr1 induced 50% more GLS accumulation in the mutants compared to wild-type plants (Mikkelsen et al., 2003).
Glucosinolates production is negatively regulated by BR. Microarray analysis of BR-responsive genes showed that CYP79B2 was down-regulated by BR treatment in Arabidopsis (Goda et al., 2002). BR treatment reduced accumulation of both aliphatic and indole GLS. The inhibitory role of BR was confirmed by significantly higher accumulation of GLS content in BR-deficient mutant cpd, whereas transgenic plant overexpressing BR biosynthetic gene DWF4 showed dramatically reduced GLS levels (Guo et al., 2013). In another study, binding sites of BZR1 were identified in the promoters of MYB34 and MYB51 (Sun et al., 2010). Hence, it has been suggested that BR induced inhibition of GLS biosynthesis may be mediated by BR signaling TFs BZR1 and BES1 by binding directly to GLS biosynthetic genes or indirectly by interacting with MYB factors (Guo et al., 2013).
Further, ethylene has also been shown to induce the expression of GLS biosynthetic genes and their regulators (Mikkelsen et al., 2003; Frerigmann and Gigolashvili, 2014). Broccoli florets treated with ethylene were found to have higher quantities of specific indole GLS (Villarreal-Garcia et al., 2016). It was reported that abscisic acid (ABA) can also induce indole GLS biosynthesis (Frerigmann and Gigolashvili, 2014).
The distinct roles of indole GLS biosynthesis regulators MYB34, MYB51, and MYB122 in response to the phytohormones have been reported (Frerigmann and Gigolashvili, 2014). MYB34 was found to mediate ABA- and JA- induced indole GLS production. ABA and JA treatments strongly induced indole GLS biosynthesis in myb51 and myb122 but this tendency was not observed in myb34 mutants. Ethylene/SA induced accumulation of indole GLS was highly affected in myb51 than in myb34 and myb122, indicating a potential role of MYB51 in indole GLS synthesis in response to the treatment of these two hormones. MYB122 has been suggested to play a minor role in ethylene/JA induced GLS biosynthesis (Frerigmann and Gigolashvili, 2014; Frerigmann, 2016).
Hormonal cross-talks in controlling GLS biosynthesis were also suggested. For example, methyl-JA induced specific indole GLS accumulation was significantly decreased in SA-overproducing mutant cpr1 than in wild-type, indicating suppression of methyl-JA induced GLS biosynthesis by SA (Mikkelsen et al., 2003). In Brassica rapa, SA antagonistically affected methyl-JA induced GLS accumulation in the root regardless of the site of elicitation. Similar effect was found in the leaves when the roots were elicited, however, the effect was synergetic if the leaves were elicited (Zang et al., 2015).
GLS Metabolism Is A Modulator of Auxin Homeostasis
Some of the GLS mutants were isolated from the genetic screens aimed to identify genes involved in auxin homeostasis in Arabidopsis. For instance, sur1 was isolated in a screen designed to identify mutants with high-auxin phenotypes including small and epinastic cotyledons, an elongated hypocotyl, excess adventitious and lateral roots, and a reduced number of leaves (Boerjan et al., 1995). The different alleles of sur1, alf1, rty, and hsl3, which encodes a C-S lyase, identified in independent root morphology screens, also showed high-auxin related abnormal root morphology (Celenza et al., 1995; King et al., 1995; Lehman et al., 1996). Later, sur2 and rnt1, loss-of-function mutants of CYP83B1, were found to display the phenotypes similar to sur1 (Delarue et al., 1998; Winkler et al., 1998; Barlier et al., 2000; Bak et al., 2001). UGT74B1 encodes a UDP-glucose:thiohydroximate S-glycosyltransferase. Insertional mutations in UGT74B1 resulted in phenotypes reminiscent of auxin overproduction, such as epinastic cotyledons, elongated hypocotyls in light grown plants (Grubb et al., 2004).
The GLS mutants with high-auxin phenotypes were found to have altered levels of IAA along with impaired GLS content. In sur1/rty, free and conjugated IAA levels were over accumulated (Boerjan et al., 1995; King et al., 1995) with undetectable levels of all types of GLS (Mikkelsen et al., 2004). Similarly, the mutants of UGT74B1 having excess free and conjugated IAA were associated with reduction in all types of GLS compared to their control plants (Grubb et al., 2004). Indole GLS production was reduced to 50% in sur2 plants (Bak et al., 2001) compared to wild-type, whereas free IAA levels were increased at all developmental stages tested (Delarue et al., 1998). cyp79f1/bushy1/sps mutants showed extremely bushy phenotype (Reintanz et al., 2001; Tantikanjana et al., 2001). cyp79f1 mutant showed decreased levels of short-chain derived GLS but increased levels of indole-3-ylmethyl-GLS, IAA (Reintanz et al., 2001), and cytokinin (Tantikanjana et al., 2001). Because the cytokinin responsive reporter ARR5::uidA and auxin responsive reporter DR5::uidA in the cyp79f1 mutant showed that increased levels of cytokinin, but not auxin, correlate well with a root-specific expression pattern, the bushy phenotype might be caused by increased level of cytokinin (Tantikanjana et al., 2004). Both auxin and cytokinin can influence the hormone levels of each other. Increased cytokinin levels in cyp79f1 might induce accumulation of auxin. Alternatively, increased indole GLS production in cyp79f1 likely increased IAA biosynthesis (Mikkelsen et al., 2003). CYP79B2/B3 were up-regulated in stressed plants, resulting in increased indole GLS and IAA (Mikkelsen et al., 2003). Therefore, it is also possible that sps/cyp79f1 mutants were stressed because of the perturbation of cytokinin homeostasis, which in turn up-regulates CYP79B2/B3 genes (Tantikanjana et al., 2004).
The altered IAA levels found in the GLS mutants were proposed to be synthesized from IAOx (Hull et al., 2000), a common precursor for indole GLS, camalexin, and IAA (Figure 3). The CYP71 clade genes SUR2/CYP83B1 and CYP71A13/12 channel IAOx into biosynthesis of indole GLS and camalexin, respectively. SUR2 catalyzes IAOx into an indole-3-S-alkyl-thiohydroximate and is subsequently metabolized to indole GLS (Bak et al., 2001).
Indole-3-acetaldoxime channeling into production of either IAA, or secondary metabolites indole GLS or camalexin must be tightly controlled. In CYP79B2 overexpressing plants, elevated IAOx has been found to be channeled into biosynthesis of indole GLS (Mikkelsen et al., 2000) and IAA (Zhao et al., 2002). In response to the increased production of IAA in CYP79B2 overexpressors, transcripts of IAA-inducible genes including IAA/AUX, SAUR, and GH3s were induced (Zhao et al., 2002). Consistently, disruption of CYP79B2/B3 abolished production of indole GLS, and affected rate of IAA biosynthesis in the mutants (Zhao et al., 2002). CYP79B2 was shown to highly express in response to silver nitrate treatment that induces camalexin synthesis. Consistently, cyp79B2 single and cyp79B2/B3 double mutants were unable to synthesize camalexin under induced conditions (Glawischnig et al., 2004; Ljung et al., 2005). In the absence of pathogen attack, IAN levels were not altered in cyp71A13 knockout mutants (Sugawara et al., 2009), indicating fine-tuned regulation of IAOx metabolic channeling into the corresponding pathways. Loss-of-function mutations of SUR2 restricted IAOx flux into biosynthesis of indole GLS, resulting in decreased indole GLS production (Bak et al., 2001) and increased free IAA levels (Delarue et al., 1998). In agreement with the elevated free IAA levels, sur2 disruption induced transcription of early auxin responsive genes such as Aux/IAAs and GH3s (Mikkelsen et al., 2009; Morant et al., 2010). Conversely overexpression of SUR2 led to increased production of indole GLS (Bak et al., 2001). It was shown that TAM can competitively inhibit SUR2 that resulted in conversion of IAOx into IAA (Bak and Feyereisen, 2001). These studies indicate that IAOx plays important roles in plant development and defense responses as a branch point for biosynthesis of indole GLS, camalexin, and IAA.
Differential activation of IAOx pathway resulted in altered auxin homeostasis in post-acetaldoxime mutants or overexpressors of CYP79B2 (Nafisi et al., 2006). For instance, in sur2, increased endogenous IAA levels were associated with up-regulation of CYP79B2, IAA conjugation genes such as GH3s (Morant et al., 2010), and subsequent accumulation of IAA catabolites such as IAA-aspartate and oxindole-3-acetic acid (Barlier et al., 2000). Consistently, IAA-leucine resistant 1-like family of amidohydrolases ILL1 and ILL2 which release IAA from amide conjugates were down-regulated indicating that increased IAA levels were catalyzed to irreversible conjugates in sur2 plants (Morant et al., 2010). Similar to sur2 plants, overexpression of CYP79B2 induced expression of auxin responsive genes and increased accumulation of IAN (Zhao et al., 2002). It appears that impaired aliphatic GLS production indirectly affects production of IAOx derived indole GLS and IAA levels, as demonstrated by upregulation of CYP79B2/B3 genes in CYP79F1 co-suppressed plants (Hansen et al., 200lb), and increased accumulation of indole-3-ylmethyl-GLS, IAA in cyp79f1 mutant (Reintanz et al., 2001). Nevertheless, the increased IAA levels in cyp79f1 mutants may not be responsible for the bushy phenotype.
It was shown that cyp79b2/b3 double mutants displayed wild-type like IAA levels under normal growth conditions, but showed a modest decrease of free IAA levels under high temperature (Zhao et al., 2002; Sugawara et al., 2009). Overexpression of CYP79B2 significantly elevated levels of indole GLS and IAN but with normal IAA levels (Zhao et al., 2002). It was suggested that CYP79B2/B3 were primarily responsible for production of secondary metabolites GLS and camalexin (Glawischnig et al., 2004). These observations question whether IAOx pathway can contribute to basal IAA production, and its role in regulating plant growth and development.
Besides this, potential involvement of IAOx pathway in certain circumstances is well documented. It has been proposed that root growth under sulfur starvation is initiated by extra IAA produced from IAN (Kutz et al., 2002). IAA produced from IAOx and IPA-pathways has been shown to involve in PIF4 mediated hypocotyl elongation in response to high temperature (Franklin et al., 2011). The expression of TAA1 and CYP79B2 genes were induced in response to high temperature, however, their expression was greatly reduced in pif4-101 mutants (Franklin et al., 2011). Similarly, IAOx and IPA pathways were shown to be hyperactive during high temperature induced microsporogenesis as demonstrated by tremendous increase of transcripts of NIT2 and TAA1 (Rodríguez-Sanz et al., 2015). Recently, it has been demonstrated that miR10515 promotes IAA biosynthesis via IAOx pathway under high temperature by suppressing SUR1 (Kong et al., 2015). Overexpression of miR10515 partially phenocopied sur1 phenotype with repressed SUR1 expression and elevated IAA concentration. NIT3 expression was strongly induced or repressed in the miR10515 overexpressing and silenced plants, respectively (Kong et al., 2015). Low auxin phenotype of cyp79b2/b3 double mutants appeared only in high temperature grown plants (Zhao et al., 2002). These reports suggest that IAOx pathway may provide extra auxin in response to environmental stresses.
It was reported that upregulation of the IAOx pathway can compensate defects in the IPA pathway (Stepanova et al., 2008). The elevated IAA in sur2 plants had attenuated the meristem maintenance and lateral root formation defects of TAA1 and TAR2 double mutants wei8 tar2. Additionally, dwarf phenotypes of sur2/rty1 were alleviated in wei8 tar2 sur2 and wei8 tar2 rty1 genetic backgrounds suggesting an existence of a functional overlap between the IPA- and IAOx-dependent routes of auxin biosynthesis (Stepanova et al., 2008). Developmental defects in ASA1/WEI2 mutants were suppressed by excess IAA levels accumulated in sur1 and sur2 mutants, respectively (Stepanova et al., 2005). These findings indicate that IAOx pathway may be operative under normal growth conditions as well; however, further studies are needed to confirm this idea. Regardless of how significant this pathway is in controlling plant growth and development, endogenous IAOx and its metabolizing enzymes CYP79B2/B3 were found only in GLS plants. Thus, IAOx pathway is considered as a species-specific pathway (Sugawara et al., 2009).
Conclusion
Biosynthetic pathways of indole GLS, camalexin and IAA are metabolically connected by their common metabolic intermediate IAOx. Disruption of indole GLS production leads to altered auxin homeostasis via differential activation of IAOx pathway. Physiological role of NITs in IAA biosynthesis is so far not conclusive. Our current knowledge on IAOx pathway suggests that this pathway is likely operative under special circumstances. Identification of the enzyme responsible for conversion of IAOx to IAA, and its functional characterization under normal and induced conditions would help us to better understand the role of this pathway in plant development.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the projects from the National Basic Research Program of China (2014CB943400), National Natural Science Fund of China (31171389; 91217310; 91017008; 31270330) and One-hundred Talent Project, and the President’s International Fellowship Initiative of the Chinese Academy of Sciences.
References
- Agerbirk N., Olsen C. E. (2012). Glucosinolate structures in evolution. Phytochemistry 77 16–45. 10.1016/j.phytochem.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Agerbirk N., Olsen C. E. (2015). Glucosinolate hydrolysis products in the crucifer Barbarea vulgaris include a thiazolidine-2-one from a specific phenolic isomer as well as oxazolidine-2-thiones. Phytochemistry 115 143–151. 10.1016/j.phytochem.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Andersen T. G., Halkier B. A. (2014). Upon bolting the GTR1 and GTR2 transporters mediate transport of glucosinolates to the inflorescence rather than roots. Plant Signal. Behav. 9:e27740. 10.4161/psb.27740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E., Bolt Jorgensen L., Hoglund A. S., Rask L., Meijer J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127 1750–1763. 10.1104/pp.010334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y., Kawaguchi M., Syono K., Ikuta A. (2004). Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J. Plant Res. 117 191–198. 10.1007/s10265-004-0146-6 [DOI] [PubMed] [Google Scholar]
- Bak S., Feyereisen R. (2001). The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127 108–118. 10.1104/pp.127.1.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Kahn R. A., Nielsen H. L., Moller B. L., Halkier B. A. (1998). Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 36 393–405. 10.1023/A:1005915507497 [DOI] [PubMed] [Google Scholar]
- Bak S., Tax F. E., Feldmann K. A., Galbraith D. W., Feyereisen R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch paint in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 101–111. 10.1105/Tpc.13.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi B. G., Maher B. R., Slovin J. P., Cohen J. D. (1991). Stable isotope labeling, in vivo, of d- and l-tryptophan pools in Lemna gibba and the low incorporation of label into indole-3-acetic acid. Plant Physiol. 95 1203–1208. 10.1104/pp.95.4.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I., Kowalczyk M., Marchant A., Ljung K., Bhalerao R., Bennett M., et al. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. U.S.A. 97 14819–14824. 10.1073/pnas.260502697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B., Fink G. R. (1994). Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 91 6649–6653. 10.1073/pnas.91.14.6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling D., Seedorf M., Mithofer A., Weiler E. W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205 417–424. 10.1111/j.1432-1033.1992.tb16795.x [DOI] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Ver Loren van Themaat E., Maddula R. K., Svatos A., Schulze-Lefert P. (2011). Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192 713–726. 10.1111/j.1469-8137.2011.03824.x [DOI] [PubMed] [Google Scholar]
- Beekwilder J., van Leeuwen W., van Dam N. M., Bertossi M., Grandi V., Mizzi L., et al. (2008). The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLOS ONE 3:e2068. 10.1371/journal.pone.0002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J., Fink G. R. (1998). A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 5655–5660. 10.1073/pnas.95.10.5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. N., Kiddle G., Wallsgrove R. M. (1997). Involvement of cytochrome P450 in glucosinolate biosynthesis in white mustard (a biochemical anomaly). Plant Physiol. 114 1283–1291. 10.1104/pp.114.4.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. N., Wenke T., Freudenberg B., Mellon F. A., Ludwig-Müller J. (2005). The tu8 mutation of Arabidopsis thaliana encoding a heterochromatin protein 1 homolog causes defects in the induction of secondary metabolite biosynthesis. Plant Biol. 7 348–357. 10.1055/s-2005-837634 [DOI] [PubMed] [Google Scholar]
- Boerjan W., Cervera M. T., Delarue M., Beeckman T., Dewitte W., Bellini C., et al. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. 10.1105/tpc.7.9.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bones A. M., Rossiter J. T. (1996). The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol. Plant. 97 194–208. 10.1111/j.1399-3054.1996.tb00497.x [DOI] [Google Scholar]
- Böttcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21 1830–1845. 10.1105/tpc.109.066670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G., Tas E., Palva E. T. (2001). Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 126 849–860. 10.1104/pp.126.2.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Jr., Grisafi P. L., Fink G. R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. 10.1101/gad.9.17.2131 [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Quiel J. A., Smolen G. A., Merrikh H., Silvestro A. R., Normanly J., et al. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137 253–262. 10.1104/pp.104.054395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. W. (2009). Local auxin production: a small contribution to a big field. Bioessays 31 60–70. 10.1002/bies.080146 [DOI] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55 1072–1079. 10.1093/pcp/pcu039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Andreasson E. (2001). Update on glucosinolate metabolism and transport. Plant Physiol. Biochem. 39 743–758. 10.1016/S0981-9428(01)01301-8 24589761 [DOI] [Google Scholar]
- Chen S., Glawischnig E., Jorgensen K., Naur P., Jorgensen B., Olsen C. E., et al. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33 923–937. 10.1046/j.1365-313X.2003.01679.x [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20 1790–1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19 2430–2439. 10.1105/tpc.107.053009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem W. R., Clay N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131 26–43. 10.1016/j.phytochem.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernandez G., Adie B., Chico J. M., Lorenzo O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671. 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- Chourey P. S., Li Q. B., Kumar D. (2010). Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol. Plant 3 1026–1036. 10.1093/mp/ssq057 [DOI] [PubMed] [Google Scholar]
- Clarke D. B. (2010). Glucosinolates, structures and analysis in food. Anal. Methods 2 310–325. 10.1039/b9ay00280d [DOI] [Google Scholar]
- Cooney T. P., Nonhebel H. M. (1991). Biosynthesis of indole-3-acetic acid in tomato shoots: measurement, mass-spectral identification and incorporation of -2H from -2H2O into indole-3-acetic acid, D- and L-tryptophan, indole-3-pyruvate and tryptamine. Planta 184 368–376. 10.1007/BF00195339 [DOI] [PubMed] [Google Scholar]
- De Geyter N., Gholami A., Goormachtig S., Goossens A. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 17 349–359. 10.1016/j.tplants.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Delarue M., Prinsen E., Onckelen H. V., Caboche M., Bellini C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14 603–611. 10.1046/j.1365-313X.1998.00163.x [DOI] [PubMed] [Google Scholar]
- Dohmoto M., Sano J., Isaji G., Yamaguchi K. (2000a). Indole acetonitrile-sensitivity of transgenic tobacco containing Arabidopsis thaliana nitrilase genes. Plant Cell Rep. 19 1027–1032. 10.1007/s002990000218 [DOI] [PubMed] [Google Scholar]
- Dohmoto M., Tsunoda H., Isaji G., Chiba R., Yamaguchi K. (2000b). Genes encoding nitrilase-like proteins from tobacco. DNA Res. 7 283–289. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G. P., Sprague S. J., Kirkegaard J. A., Ross J. J., Reid J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 2225–2245. 10.1105/tpc.106.048017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey J. W., Zalcmann A. T., Talalay P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 5–51. 10.1016/S0031-9422(00)00316-2 [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Lee S. H., Patel D., Kumar S. V., Spartz A. K., Gu C., et al. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108 20231–20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H. (2016). Glucosinolate regulation in a complex relationship - MYC and MYB - no one can act without each other. Glucosinolates 80 57–97. 10.1016/bs.abr.2016.06.005 [DOI] [Google Scholar]
- Frerigmann H., Berger B., Gigolashvili T. (2014). bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 166 349–369. 10.1104/pp.114.240887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H., Gigolashvili T. (2014). Update on the role of R2R3-MYBs in the regulation of glucosinolates upon sulfur deficiency. Front. Plant Sci. 5:626. 10.3389/fpls.2014.00626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H., Glawischnig E., Gigolashvili T. (2015). The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 6:654. 10.3389/fpls.2015.00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H., Pislewska-Bednarek M., Sanchez-Vallet A., Molina A., Glawischnig E., Gigolashvili T., et al. (2016). Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 9 682–695. 10.1016/j.molp.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Fu L., Wang M., Han B., Tan D., Sun X., Zhang J. (2016). Arabidopsis myrosinase genes AtTGG4 and AtTGG5 are root-tip specific and contribute to auxin biosynthesis and root-growth regulation. Int. J. Mol. Sci. 17:E892. 10.3390/ijms17060892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F., Moldrup M. E., Böttche C., Olsen C. E., Scheel D., Halkier B. A. (2011). Cytosolic gamma-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23 2456–2469. 10.1105/tpc.111.083998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Berger B., Flugge U. I. (2009). Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem. Rev. 8 3–13. 10.1007/s11101-008-9112-6 [DOI] [Google Scholar]
- Gigolashvili T., Berger B., Mock H. P., Müller C., Weisshaar B., Flügge U. I. (2007a). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50 886–901. 10.1111/j.1365-313X.2007.03099.x [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Engqvist M., Yatusevich R., Müller C., Flugge U. I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177 627–642. 10.1111/j.1469-8137.2007.02295.x [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Berger B., Müller C., Flugge U. I. (2007b). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51 247–261. 10.1111/j.1365-313X.2007.03133.x [DOI] [PubMed] [Google Scholar]
- Gimsing A. L., Kirkegaard J. A. (2009). Glucosinolates and biofumigation: fate of glucosinolates and their hydrolysis products in soil. Phytochem. Rev. 8 299–310. 10.1007/s11101-008-9105-5 [DOI] [Google Scholar]
- Glawischnig E. (2007). Camalexin. Phytochemistry 68 401–406. 10.1016/j.phytochem.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Glawischnig E., Hansen B. G., Olsen C. E., Halkier B. A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 8245–8250. 10.1073/pnas.0305876101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., Shimada Y., Asami T., Fujioka S., Yoshida S. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130 1319–1334. 10.1104/pp.011254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grsic S., Kirchheim B., Pieper K., Fritsch M., Hilgenberg W., Ludwig-Müller J. (1999). Induction of auxin biosynthetic enzymes by jasmonic acid and in clubroot diseased Chinese cabbage plants. Physiol. Plant. 105 521–531. 10.1034/j.1399-3054.1999.105318.x [DOI] [Google Scholar]
- Grsic-Rausch S., Kobelt P., Siemens J. M., Bischoff M., Ludwig-Müller J. (2000). Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol. 122 369–378. 10.1104/Pp.122.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C. D., Zipp B. J., Ludwig-Müller J., Masuno M. N., Molinski T. F., Abel S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 893–908. 10.1111/j.1365-313X.2004.02261.x [DOI] [PubMed] [Google Scholar]
- Guo R., Qian H., Shen W., Liu L., Zhang M., Cai C., et al. (2013). BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 64 2401–2412. 10.1093/jxb/ert094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Hansen C. H., Du L., Naur P., Olsen C. E., Axelsen K. B., Hick A. J., et al. (2001a). CYP83b1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 276 24790–24796. 10.1074/jbc.M102637200 [DOI] [PubMed] [Google Scholar]
- Hansen C. H., Wittstock U., Olsen C. E., Hick A. J., Pickett J. A., Halkier B. A. (2001b). Cytochrome p450 CYP79F1 from arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276 11078–11085. 10.1074/jbc.M010123200 [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Kelleher M. O., Eggleston I. M. (2008). The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 47(Suppl. 2), 73–88. 10.1007/s00394-008-2009-8 [DOI] [PubMed] [Google Scholar]
- Hecht S. S. (2000). Inhibition of carcinogenesis by isothiocyanates. Drug Metab. Rev. 32 395–411. 10.1081/DMR-100102342 [DOI] [PubMed] [Google Scholar]
- Hemm M. R., Ruegger M. O., Chapple C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15 179–194. 10.1105/tpc.006544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M., Böttcher C., Düchting P., Cheng Y., Zhao Y., Berkowitz O., et al. (2013). The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 74 626–637. 10.1111/tpj.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104 6478–6483. 10.1073/pnas.0611629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. J., van Dam N. M., van Loon J. J. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54 57–83. 10.1146/annurev.ento.54.110807.090623 [DOI] [PubMed] [Google Scholar]
- Hull A. K., Vij R., Celenza J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97 2379–2384. 10.1073/pnas.040569997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H., Chadchawan S., Winge P., Thangstad O. P., Bones A. M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128 1180–1188. 10.1104/pp.010925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Okazaki K., Kuroda H., Itoh K., Mitsui T., Hori H. (2007). Molecular cloning of Brassica rapa nitrilases and their expression during clubroot development. Mol. Plant Pathol. 8 623–637. 10.1111/J.1364-3703.2007.00414.X [DOI] [PubMed] [Google Scholar]
- Johnson S. D., Griffiths M. E., Peter C. I., Lawes M. J. (2009). Pollinators, “mustard oil” volatiles, and fruit production in flowers of the dioecious tree Drypetes natalensis (Putranjivaceae). Am. J. Bot. 96 2080–2086. 10.3732/ajb.0800362 [DOI] [PubMed] [Google Scholar]
- Jørgensen M. E., Nour-Eldin H. H., Halkier B. A. (2015). Transport of defense compounds from source to sink: lessons learned from glucosinolates. Trends Plant Sci. 20 508–514. 10.1016/j.tplants.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Katz E., Nisani S., Yadav B. S., Woldemariam M. G., Shai B., Obolski U., et al. (2015). The glucosinolate breakdown product indole-3-carbinol acts as an auxin antagonist in roots of Arabidopsis thaliana. Plant J. 82 547–555. 10.1111/tpj.12824 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Fujioka S., Sakurai A., Yamaki Y. T., Syono K. (1993). Presence of a pathway for the biosynthesis of auxin via indole-3-acetamide in trifoliata orange. Plant Cell Physiol. 34 121–128. [Google Scholar]
- Kawaguchi M., Kobayashi M., Sakurai A., Syono K. (1991). The presence of an enzyme that converts indole-3-acetamide into IAA in wild and cultivated rice. Plant Cell Physiol. 32 143–149. 10.1093/oxfordjournals.pcp.a078058 [DOI] [Google Scholar]
- Keck A. S., Finley J. W. (2004). Cruciferous vegetables: cancer protective mechanisms of glucosinolate hydrolysis products and selenium. Integr. Cancer Ther. 3 5–12. 10.1177/1534735403261831 [DOI] [PubMed] [Google Scholar]
- Kiddle G. A., Doughty K. J., Wallsgrove R. M. (1994). Salicylic acid-induced accumulation of glucosinolates in oilseed rape (Brassica napus L) leaves. J. Exp. Bot. 45 1343–1346. 10.1093/jxb/45.9.1343 [DOI] [Google Scholar]
- Kim J. H., Durrett T. P., Last R. L., Jander G. (2004). Characterization of the Arabidopsis TU8 glucosinolate mutation, an allele of TERMINAL FLOWER2. Plant Mol. Biol. 54 671–682. 10.1023/B:PLAN.0000040897.49151.98 [DOI] [PubMed] [Google Scholar]
- Kim J. I., Sharkhuu A., Jin J. B., Li P., Jeong J. C., Baek D., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145 722–735. 10.1104/pp.107.104935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindl H. (1968). Oxidases and oxygenases in higher plants.1. Occurrence of Indolyl-(3)-acetaldehyde oxime and its formation from L-tryptophan. Hoppe Seylers Z. Physiol. Chem. 349 519–520. [PubMed] [Google Scholar]
- King J. J., Stimart D. P., Fisher R. H., Bleecker A. B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037. 10.1105/tpc.7.12.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Li Y., Zhang M., Jin F., Li J. (2015). A novel Arabidopsis microRNA promotes IAA biosynthesis via the indole-3-acetaldoxime pathway by suppressing SUPERROOT1. Plant Cell Physiol. 56 715–726. 10.1093/pcp/pcu216 [DOI] [PubMed] [Google Scholar]
- Koo A. J., Howe G. A. (2009). The wound hormone jasmonate. Phytochemistry 70 1571–1580. 10.1016/j.phytochem.2009.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva O. A., Gibson T. M., Cramer R., Stain C. (2010). Glucosinolate-accumulating S-cells in Arabidopsis leaves and flower stalks undergo programmed cell death at early stages of differentiation. Plant J. 64 456–469. 10.1111/j.1365-313X.2010.04339.x [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Park W. J., Piotrowski M., Meeley R. B., Gierl A., Glawischnig E. (2007). Maize nitrilases have a dual role in auxin homeostasis and beta-cyanoalanine hydrolysis. J. Exp. Bot. 58 4225–4233. 10.1093/jxb/erm279 [DOI] [PubMed] [Google Scholar]
- Kroymann J., Textor S., Tokuhisa J. G., Falk K. L., Bartram S., Gershenzon J., et al. (2001). A gene controlling variation in arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 127 1077–1088. 10.1104/Pp.010416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz A., Müller A., Hennig P., Kaiser W. M., Piotrowski M., Weiler E. W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30 95–106. 10.1046/j.1365-313X.2002.01271.x [DOI] [PubMed] [Google Scholar]
- Last R. L., Bissinger P. H., Mahoney D. J., Radwanski E. R., Fink G. R. (1991). Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase beta genes. Plant Cell 3 345–358. 10.1105/tpc.3.4.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A., Black R., Ecker J. R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194. 10.1016/S0092-8674(00)81095-8 [DOI] [PubMed] [Google Scholar]
- Lehmann T., Janowitz T., Sanchez-Parra B., Alonso M. P., Trompetter I., Piotrowski M., et al. (2017). Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front. Plant Sci. 8:36. 10.3389/fpls.2017.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Wang Q., Kaspi R., Parrella M. P., Abel S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43 79–96. 10.1111/j.1365-313X.2005.02435.x [DOI] [PubMed] [Google Scholar]
- Liu H., Ying Y. Y., Zhang L., Gao Q. H., Li J., Zhang Z., et al. (2012). Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria x ananassa Duch.). Plant Cell Rep. 31 1425–1435. 10.1007/s00299-012-1258-4 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A. K., Celenza J., Yamada M., Estelle M., Nonmanly J., et al. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. 10.1105/tpc.104.029272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J., Hilgenberg W. (1988). A plasma membrane-bound enzyme oxidizes l-tryptophan to indole-3-acetaldoxime. Physiol. Plant. 74 240–250. 10.1111/j.1399-3054.1988.tb00627.x [DOI] [Google Scholar]
- Mahadevan S., Thimann K. V. (1964). Nitrilase. Ii. Substrate specificity and possible mode of action. Arch. Biochem. Biophys. 107 62–68. 10.1016/0003-9861(64)90269-3 [DOI] [PubMed] [Google Scholar]
- Mano Y., Nemoto K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63 2853–2872. 10.1093/jxb/ers091 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251. 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J., Verpoorte R., Kijne J. W. (2001). ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci. 6 212–219. 10.1016/S1360-1385(01)01924-0 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Fuller V. L., Hansen B. G., Nafisi M., Olsen C. E., Nielsen H. B., et al. (2009). Controlled indole-3-acetaldoxime production through ethanol-induced expression of CYP79B2. Planta 229 1209–1217. 10.1007/s00425-009-0907-5 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Hansen C. H., Wittstock U., Halkier B. A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275 33712–33717. 10.1074/jbc.M001667200 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Naur P., Halkier B. A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37 770–777. 10.1111/j.1365-313X.2004.02002.x [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Petersen B. L., Glawischnig E., Jensen A. B., Andreasson E., Halkier B. A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131 298–308. 10.1104/pp.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., Ekstrøm C., Ulvskov P., Kristensen C., Rudemo M., Olsen C. E., et al. (2010). Metabolomic, transcriptional, hormonal, and signaling cross-talk in superroot2. Mol. Plant 3 192–211. 10.1093/mp/ssp098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Hillebrand H., Weiler E. W. (1998). Indole-3-acetic acid is synthesized from L-tryptophan in roots of Arabidopsis thaliana. Planta 206 362–369. 10.1007/s004250050411 [DOI] [PubMed] [Google Scholar]
- Müller A., Weiler E. W. (2000). Indolic constituents and indole-3-acetic acid biosynthesis in the wild-type and a tryptophan auxotroph mutant of Arabidopsis thaliana. Planta 211 855–863. 10.1007/s004250000353 [DOI] [PubMed] [Google Scholar]
- Müller T. M., Böttcher C., Morbitzer R., Gotz C. C., Lehmann J., Lahaye T., et al. (2015). TRANSCRIPTION ACTIVATOR-LIKE EFFECTOR NUCLEASE-mediated generation and metabolic analysis of camalexin-deficient cyp71a12 cyp71a13 double knockout lines. Plant Physiol. 168 849–858. 10.1104/pp.15.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C. J., Glawischnig E., Olsen C. E., Halkier B. A., et al. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19 2039–2052. 10.1105/tpc.107.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M., Sønderby I. E., Hansen B. G., Geu-Flores F., Nour-Eldin H. H., Nørholm M. H. H., et al. (2006). Cytochromes P450 in the biosynthesis of glucosinolates and indole alkaloids. Phytochem. Rev. 5 331–346. 10.1007/s11101-006-9004-6 22954587 [DOI] [Google Scholar]
- Nakajima M., Yoshida R., Shimada N., Yamazaki H., Yokoi T. (2001). Inhibition and inactivation of human cytochrome P450 isoforms by phenethyl isothiocyanate. Drug Metab. Dispos. 29 1110–1113. [PubMed] [Google Scholar]
- Naur P., Petersen B. L., Mikkelsen M. D., Bak S., Rasmussen H., Olsen C. E., et al. (2003). CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133 63–72. 10.1104/pp.102.019240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto K., Hara M., Suzuki M., Seki H., Muranaka T., Mano Y. (2009). The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 583 487–492. 10.1016/j.febslet.2008.12.049 [DOI] [PubMed] [Google Scholar]
- Nonhebel H., Yuan Y., Al-Amier H., Pieck M., Akor E., Ahamed A., et al. (2011). Redirection of tryptophan metabolism in tobacco by ectopic expression of an Arabidopsis indolic glucosinolate biosynthetic gene. Phytochemistry 72 37–48. 10.1016/j.phytochem.2010.10.018 [DOI] [PubMed] [Google Scholar]
- Normanly J. (2010). Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2:a001594. 10.1101/cshperspect.a001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Cohen J. D., Fink G. R. (1993). Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. U.S.A. 90 10355–10359. 10.1073/pnas.90.21.10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G. R., Bartel B. (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9 1781–1790. 10.1105/tpc.9.10.1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Andersen T. G., Burow M., Madsen S. R., Jorgensen M. E., Olsen C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488 531–534. 10.1038/nature11285 [DOI] [PubMed] [Google Scholar]
- Novak O., Henykova E., Sairanen I., Kowalczyk M., Pospisil T., Ljung K. (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72 523–536. 10.1111/j.1365-313X.2012.05085.x [DOI] [PubMed] [Google Scholar]
- Ouyang J., Shao X., Li J. (2000). Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 24 327–333. 10.1046/j.1365-313x.2000.00883.x [DOI] [PubMed] [Google Scholar]
- Park W. J., Kriechbaumer V., Moller A., Piotrowski M., Meeley R. B., Gierl A., et al. (2003). The Nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol. 133 794–802. 10.1104/pp.103.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten C. L., Glick B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42 207–220. 10.1139/m96-032 [DOI] [PubMed] [Google Scholar]
- Pedras M. S., Yaya E. E., Glawischnig E. (2011). The phytoalexins from cultivated and wild crucifers: chemistry and biology. Nat. Prod. Rep. 28 1381–1405. 10.1039/c1np00020a [DOI] [PubMed] [Google Scholar]
- Petersen B. L., Andreasson E., Bak S., Agerbirk N., Halkier B. A. (2001). Characterization of transgenic Arabidopsis thaliana with metabolically engineered high levels of p-hydroxybenzylglucosinolate. Planta 212 612–618. 10.1007/s004250000429 [DOI] [PubMed] [Google Scholar]
- Phillips K. A., Skirpan A. L., Liu X., Christensen A., Slewinski T. L., Hudson C., et al. (2011). vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23 550–566. 10.1105/tpc.110.075267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M. (2008). Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69 2655–2667. 10.1016/j.phytochem.2008.08.020 [DOI] [PubMed] [Google Scholar]
- Piotrowski M., Schemenewitz A., Lopukhina A., Müller A., Janowitz T., Weiler E. W., et al. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279 50717–50725. 10.1074/jbc.M407681200 [DOI] [PubMed] [Google Scholar]
- Pollmann S., Düchting P., Weiler E. W. (2009). Tryptophan-dependent indole-3-acetic acid biosynthesis by ‘IAA-synthase’ proceeds via indole-3-acetamide. Phytochemistry 70 523–531. 10.1016/j.phytochem.2009.01.021 [DOI] [PubMed] [Google Scholar]
- Pollmann S., Müller A., Piotrowski M., Weiler E. W. (2002). Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 216 155–161. 10.1007/s00425-002-0868-4 [DOI] [PubMed] [Google Scholar]
- Pollmann S., Neu D., Lehmann T., Berkowitz O., Schäfer T., Weiler E. W. (2006). Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 224 1241–1253. 10.1007/s00425-006-0304-2 [DOI] [PubMed] [Google Scholar]
- Quittenden L. J., Davies N. W., Smith J. A., Molesworth P. P., Tivendale N. D., Ross J. J. (2009). Auxin biosynthesis in Pea: characterization of the tryptamine pathway. Plant Physiol. 151 1130–1138. 10.1104/pp.109.141507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski E. R., Barczak A. J., Last R. L. (1996). Characterization of tryptophan synthase alpha subunit mutants of Arabidopsis thaliana. Mol. Gen. Genet. 253 353–361. 10.1007/PL00008602 [DOI] [PubMed] [Google Scholar]
- Rask L., Andreasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42 93–113. 10.1023/A:1006380021658 [DOI] [PubMed] [Google Scholar]
- Rauhut T., Glawischnig E. (2009). Evolution of camalexin and structurally related indolic compounds. Phytochemistry 70 1638–1644. 10.1016/j.phytochem.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., et al. (2001). Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13 351–367. 10.1105/tpc.13.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Sanz H., Solis M. T., Lopez M. F., Gomez-Cadenas A., Risueno M. C., Testillano P. S. (2015). Auxin biosynthesis, accumulation, action and transport are involved in stress-induced microspore embryogenesis initiation and progression in Brassica napus. Plant Cell Physiol. 56 1401–1417. 10.1093/pcp/pcv058 [DOI] [PubMed] [Google Scholar]
- Sánchez-Parra B., Frerigmann H., Alonso M. M., Loba V. C., Jost R., Hentrich M., et al. (2014). Characterization of four bifunctional plant IAM/PAM-amidohydrolases capable of contributing to auxin biosynthesis. Plants 3 324–347. 10.3390/plants3030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. C., Müller A., Hain R., Bartling D., Weiler E. W. (1996). Transgenic tobacco plants expressing the Arabidopsis thaliana nitrilase II enzyme. Plant J. 9 683–691. 10.1046/j.1365-313X.1996.9050683.x [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B. L., Olsen C. E., Svatos A., et al. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141 1248–1254. 10.1104/pp.106.082024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F., Fernandez-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25 3117–3132. 10.1105/tpc.113.115139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J., Glawischnig E., Ludwig-Müller J. (2008). Indole glucosinolates and camalexin do not influence the development of the clubroot disease in Arabidopsis thaliana. J. Phytopathol. 156 332–337. 10.1111/j.1439-0434.2007.01359.x [DOI] [Google Scholar]
- Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47 10–24. 10.1111/j.1365-313X.2006.02767.x [DOI] [PubMed] [Google Scholar]
- Smolen G., Bender J. (2002). Arabidopsis cytochrome p450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby I. E., Burow M., Rowe H. C., Kliebenstein D. J., Halkier B. A. (2010a). A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis1[C][W][OA]. Plant Physiol. 153 348–363. 10.1104/pp.109.149286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby I. E., Geu-Flores F., Halkier B. A. (2010b). Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 15 283–290. 10.1016/j.tplants.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Sønderby I. E., Hansen B. G., Bjarnholt N., Ticconi C., Halkier B. A., Kliebenstein D. J. (2007). A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLOS ONE 2:e1322. 10.1371/journal.pone.0001322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad D. D., Deluca V., Brisson N., Kurz W. G. W., Nessler C. L. (1990). High-levels of tryptamine accumulation in transgenic tobacco expressing tryptophan decarboxylase. Plant Physiol. 94 1410–1413. 10.1104/Pp.94.3.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Hoyt J. M., Hamilton A. A., Alonso J. M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242. 10.1105/tpc.105.033365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D. Y., Dolezal K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Yun J., Robles L. M., Novak O., He W., Guo H., et al. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23 3961–3973. 10.1105/tpc.111.088047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Su T., Xu J., Li Y., Lei L., Zhao L., Yang H., et al. (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23 364–380. 10.1105/tpc.110.079145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., et al. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 5430–5435. 10.1073/pnas.0811226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X. Y., Cao D. M., Tang W. Q., He K., Zhu J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19 765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T., Mikkelsen M. D., Hussain M., Halkier B. A., Sundaresan V. (2004). Functional analysis of the tandem-duplicated P450 genes SPS/BUS/CYP79F1 and CYP79F2 in glucosinolate biosynthesis and plant development by Ds transposition-generated double mutants. Plant Physiol. 135 840–848. 10.1104/pp.104.040113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T., Yong J. W., Letham D. S., Griffith M., Hussain M., Ljung K., et al. (2001). Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 15 1577–1588. 10.1101/gad.887301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Ferrer J. L., Ljung K., Pojer F., Hong F., Long J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 164–176. 10.1016/j.cell.2008.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S., de Kraker J. W., Hause B., Gershenzon J., Tokuhisa J. G. (2007). MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144 60–71. 10.1104/pp.106.091579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangstad O. P., Evjen K., Bones A. (1991). Immunogold-Em localization of myrosinase in Brassicaceae. Protoplasma 161 85–93. 10.1007/Bf01322721 [DOI] [Google Scholar]
- Thimann K. V. (1953). Hydrolysis of indoleacetonitrile in plants. Arch. Biochem. Biophys. 44 242–243. 10.1016/0003-9861(53)90030-7 [DOI] [PubMed] [Google Scholar]
- Thimann K. V., Mahadevan S. (1964). Nitrilase. I. Occurrence, preparation, and general properties of the enzyme. Arch. Biochem. Biophys. 105 133–141. 10.1016/0003-9861(64)90244-9 [DOI] [PubMed] [Google Scholar]
- Tivendale N. D., Davidson S. E., Davies N. W., Smith J. A., Dalmais M., Bendahmane A. I., et al. (2012). Biosynthesis of the halogenated auxin, 4-chloroindole-3-acetic acid. Plant Physiol. 159 1055–1063. 10.1104/pp.112.198457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale N. D., Ross J. J., Cohen J. D. (2014). The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 19 44–51. 10.1016/j.tplants.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Turner J. G., Ellis C., Devoto A. (2002). The jasmonate signal pathway. Plant Cell 14(Suppl.), S153–S164. 10.1105/tpc.000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Nishiyama C., Shimada T., Koumoto Y., Hayashi Y., Kondo M., et al. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47 164–175. 10.1093/pcp/pci232 [DOI] [PubMed] [Google Scholar]
- Villarreal-Garcia D., Nair V., Cisneros-Zevallos L., Jacobo-Velazquez D. A. (2016). Plants as biofactories: postharvest stress-induced accumulation of phenolic compounds and glucosinolates in broccoli subjected to wounding stress and exogenous phytohormones. Front. Plant Sci. 7:45. 10.3389/fpls.2016.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S., Biernacki S., Hillebrand H., Janzik I., Müller A., Weiler E. W., et al. (2001). Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212 508–516. 10.1007/s004250000420 [DOI] [PubMed] [Google Scholar]
- Wang B., Chu J., Yu T., Xu Q., Sun X., Yuan J., et al. (2015). Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112 4821–4826. 10.1073/pnas.1503998112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler R. G., Frank M. R., Galbraith D. W., Feyereisen R., Feldmann K. A. (1998). Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Isolation of mutations in the cytochrome p450 gene superfamily. Plant Physiol. 118 743–750. 10.1104/pp.118.3.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U., Halkier B. A. (2000). Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. Catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 275 14659–14666. 10.1074/jbc.275.19.14659 [DOI] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A. W., Bartel B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95 707–735. 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C., Bemis S. M., Hill E. J., Sawa S., Koshiba T., Torii K. U. (2005). Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 139 192–203. 10.1104/pp.105.063495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. D., Sampson M. B., Neuffer M. G., Michalczuk L., Slovin J. P., Cohen J. D. (1991). Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254 998–1000. 10.1126/science.254.5034.998 [DOI] [PubMed] [Google Scholar]
- Xu L., Ren L., Chen K., Liu F., Fang X. (2016). Putative role of IAA during the early response of Brassica napus L. to Plasmodiophora brassicae. Eur. J. Plant Pathol. 145 601–613. 10.1007/s10658-016-0877-y [DOI] [Google Scholar]
- Xue J. P., Pihlgren U., Rask L. (1993). Temporal, cell-specific, and tissue-preferential expression of myrosinase genes during embryo and seedling development in Sinapis alba. Planta 191 95–101. 10.1007/BF00240900 [DOI] [Google Scholar]
- Yamada M., Greenham K., Prigge M. J., Jensen P. J., Estelle M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151 168–179. 10.1104/pp.109.138859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y. X., Ge J. L., Huang L. H., Gao F., Lv X. S., Zheng W. W., et al. (2015). Leaf and root glucosinolate profiles of Chinese cabbage (Brassica rapa ssp. pekinensis) as a systemic response to methyl jasmonate and salicylic acid elicitation. J. Zhejiang Univ. Sci. B 16 696–708. 10.1631/jzus.B1400370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang B., Ouyang J., Li J., Wang Y. (2008). Arabidopsis indole synthase, a homolog of tryptophan synthase alpha, is an enzyme involved in the Trp-independent indole-containing metabolite biosynthesis. J. Integr. Plant Biol. 50 1070–1077. 10.1111/j.1744-7909.2008.00729.x [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu Y., Fang Z., Li Z., Yang L., Zhuang M., et al. (2016). Comparative transcriptome analysis between broccoli (Brassica oleracea var. italica) and wild cabbage (Brassica macrocarpa Guss.) in response to Plasmodiophora brassicae during different infection stages. Front. Plant Sci. 7:1929. 10.3389/fpls.2016.01929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kensler T. W., Cho C. G., Posner G. H., Talalay P. (1994). Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc. Natl. Acad. Sci. U.S.A. 91 3147–3150. 10.1073/pnas.91.8.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2014). Auxin biosynthesis. Arabidopsis Book 12:e0173. 10.1199/tab.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S. K., Fankhauser C., Cashman J. R., Cohen J. D., Weigel D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309. 10.1126/science.291.5502.306 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hull A. K., Gupta N. R., Goss K. A., Alonso J., Ecker J. R., et al. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16 3100–3112. 10.1101/gad.1035402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T. L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428. 10.1105/tpc.11.12.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. Y., Zhang C. G., Wu L., Zhang C. G., Chai J., Wang M., et al. (2011). Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J. 66 516–527. 10.1111/j.1365-313X.2011.04509.x [DOI] [PubMed] [Google Scholar]