Abstract
Patients with metastatic prostate cancer (PCa) have a poorer prognosis than patients with organ-confined tumors. We strove to uncover the proteome signature of primary PCa and associated lymph node metastases (LNMs) in order to identify proteins that may indicate or potentially promote metastases formation. We performed a proteomic comparative profiling of PCa tissue from radical prostatectomy (RPE) of patients without nodal metastases or relapse at the time of surgical resection (n = 5) to PCa tissue from RPE of patients who suffered from nodal relapse (n = 5). For the latter group, we also included patient-matched tissue of the nodal metastases. All samples were formalin fixed and paraffin embedded. We identified and quantified more than 1200 proteins by liquid chromatography tandem mass spectrometry with subsequent label-free quantification. An increase of ribosomal or proteasomal proteins in LNM (compared to corresponding PCa) became apparent, while extracellular matrix components rather decreased. Immunohistochemistry (IHC) corroborated accumulation of poly-(ADP-ribose)-polymerase 1 and N-myc-downstream-regulated-gene 3, alpha/beta hydrolase domain-containing protein 11, and protein phosphatase slingshot homolog 3 in LNM. These findings strengthen the present interest in examining PARP inhibitors for the treatment of aggressive PCa. IHC also corroborated increased abundance of retinol dehydrogenase 11 in metastasized primary PCa compared to organ-confined PCa. Generally, metastasizing primary tumors were characterized by an enrichment of proteins involved in cellular lipid metabolic processes with concomitant decrease of cell adhesion proteins. This study highlights the usefulness of a combined proteomic-IHC approach to explore novel aspects in tumor biology. Our initial results open novel opportunities for follow-up studies.
Abbreviations: PCa, prostate cancer; LNM, lymph node metastasis; TU, primary tumor; RPE, radical prostatectomy; LND, lymph node dissection; FFPE, formalin-fixed, paraffin-embedded; MS, mass-spectrometry; IHC, immunohistochemistry; FC, fold change
Introduction
The American Cancer Society estimates 160,000 new cases of prostate cancer (PCa) in 2017 and expects 26,000 patients to die from PCa [1]. PCa prognosis is often favorable, especially when the disease is organ-confined as 5-year survival rates of over 95% show. The total removal of the prostate, called radical prostatectomy (RPE), if necessary in combination with the radical removal of pelvic lymph nodes, offers curative treatment. Apart from surgery, external beam radiation or the implantation of radioactive metal seeds into the prostate, called brachytherapy, is a further treatment possibility [2]. However, PCa returns in 15% to 30% of all cases as local relapse or in terms of metastases [3]. Especially in metastatic state, the survival rates drop to around 28% [4]. PCa metastasizes predominantly via the lymphatic drainage ways to local lymph nodes in the small pelvis, e.g., to those alongside the iliacal vessels, in the fossa obturatoria, or presacral. In advanced stages or in case of relapse, PCa often affects also aortal or caval lymph nodes [5], which are classified as distant metastases.
The nodal relapse of PCa, meaning PCa-positive lymph nodes after primary therapy, can be treated with metastases-directed therapy, such as salvage lymph node dissection (salvage-LND) or targeted radiation therapy [2], [5], [6].
For PCa risk and prognosis assessment, several well-established prognostic schemes exist, e.g., the Gleason Grading/ISUP grade groups [7], TNM classification, surgical margin status, or the height of prostate-specific antigen (PSA) value at primary diagnosis. However, these tools cannot predict the biological course and the metastatic potential of primary PCa as precisely and reliable as desired.
In cancer research, mass spectrometry (MS)–based proteomic approaches are predestined methods for the initial identification of potential new prognostic, diagnostic, or therapeutic markers, hence opening novel opportunities for follow-up studies. The ability to perform proteomic researches on formalin-fixed, paraffin-embedded (FFPE) tissue [8], [9], [10] provided the possibility for retrospective proteomic investigations on sample cohorts with a long-term follow up.
Many studies on the proteome of PCa, e.g., from Aiello et al. [11], Davalieva et al. [12], or Iglesias-Gato et al. [13], compare malignant to benign tissue. We considered it especially interesting to investigate whether the proteome composition of malignant tissue differs from other malignant tissue. Therefore, we compare tissue of primary PCa from RPE of relapse-free patients (n = 5) to tissue of primary PCa from RPE with nodal relapse in the course of median 32 months (n = 5). Additionally, patient-matched primary tumor tissue was compared to the tissue of recurrent lymph node metastases (LNMs) (n = 5). Our focus laid on patients with LNM relapse only, as there is increasing evidence for superior overall survival rates compared to patients with metastases confined to other organs [14], [15].
On FFPE tissue of the above-described sample cohorts, we conducted a retrospective MS-based proteomic analysis with label-free quantification with twofold purpose: a) We strove to investigate whether metastasized primary PCa tissue, nonmetastasized primary PCa tissue, and tissue of corresponding nodal metastases exhibit measurable differences in their proteomic profile. b) We aimed to identify individual proteins, which are differentially expressed in metastasized versus nonmetastasized primary PCa tissue and furthermore with significant divergent abundance between metastasized primary PCa tissue of RPEs and corresponding tumor tissue of nodal relapse. We hypothesize that these proteins could on the one hand be predictors of LNM formation and/or indicate its promotion or suppression. Potential candidates identified in this initial study might also serve as starting point for biomarker research on larger and randomized sample cohorts.
Materials and Methods
Ethics Statement
The study was approved by the Ethics Committee of the University Medical Center Freiburg (no. 562/15). Before study inclusion, all patient data were pseudonymized. Patients gave written informed consent for the use of their tissues for research purposes.
Patient Cohort
This study includes 10 male patients with primary diagnosis of PCa between 2003 and 2011. All patients underwent RPE. Five patients (nos. 1-5) had LNM, either already at the time of RPE or LND or they developed biochemical recurrence over time with local (rpN1) or distant (rpM1 (LYM)) nodal relapse exclusively. In case of relapse, they underwent a salvage-LND with therapeutic approach. Before salvage-LND, patients received temporary androgen deprivation therapy with bicalutamide (no. 1), radiation of the prostatic loge (no. 2-4), or no additional therapy (no. 5). The remaining five patients (nos. 6-10) were free of biochemical relapse (postoperative PSA< 0.2 ng/ml). The median relapse-free interval was 10 years, with the latest follow-up examination performed in 2017. Patient characteristics including TNM classification at the time of RPE and salvage-LND, Gleason Score/ISUP grade group, and the localization of the recurrent LNM used for this study are specified in Table 1.
Table 1.
Clinical Characteristics of the Patient Cohort
| Patient No. | TNM Classification at RPE | Gleason Pattern | ISUP Grade Group | Relapse-Free-Interval [Years] | TNM Classification at Relapse | Localization of LNM Chosen for This Study | ||
|---|---|---|---|---|---|---|---|---|
| TU with LNM | #1 | pT3b pN0 cM0 | 4 + 4 (+5) | 4 | 4 | rpN1 rpM1 (LYM) | Iliaca ex. | recurrent LNM |
| #2 | pT3a pN1 cM0 | 4 + 5 | 5 | 3 | rpN1 rpM1 (LYM) | Iliaca ex. | ||
| #3 | pT2c pN0 cM0 | 4 + 5 | 5 | 2 | rpN1 rpM1 (LYM) | A. obt. | ||
| #4 | pT3b pN1 cM0 | 4 + 3 (+5) | 3 | 2 | rpN1 rpM1 (LYM) | Iliaca com. | ||
| #5 | pT3a pN0 cM0 | 4 + 5 | 5 | 2 | rpN1 rpM0 (LYM) | Iliaca ex. | ||
| TU without LNM | #6 | pT2c pN0 cM0 | 3 + 3 | 1 | 13 | |||
| #7 | pT2c pN0 cM0 | 3 + 3 | 1 | 9 | ||||
| #8 | pT3b pN0 cM0 | 3 + 4 | 2 | 7 | ||||
| #9 | pT3b pN0 cM0 | 3 + 5 | 4 | 10 | ||||
| #10 | pT3a pN0 cM0 | 3 + 4 | 2 | 10 |
This study comprised 10 patients with histologically confirmed PCa in radical prostatectomy. Five patients had lymph node metastases, either already at primary diagnosis or in kind of nodal relapse during the following 2 to 4 years. From these patients, specimens of the primary tumors and lymph node metastases were investigated. Five further patients remained relapse-free, with a median relapse-free interval since the radical prostatectomy of 10 years. Here, specimens of the primary tumors were investigated.
Tissue Collection, Fixation, and Macrodissection
Tissue specimens were harvested at the time of radical prostatectomy with pelvic lymphadenectomy or at salvage-LND. Formalin fixation was started latest within 20 minutes after surgical removal. Tissue fixation in formalin and paraffin embedding was conducted according to routine protocols. After processing, samples were immediately anonymized.
Hematoxylin-eosin (HE)–stained sections of all samples were inspected under a light microscope by experienced pathologists to confirm diagnosis and to mark eligible tumor areas of invasive PCa or LNM (0.5 cm2-3.0 cm2) for macrodissection of consecutive sections. These sections were obtained by cutting 10-μm–thick slices from the paraffin block on a microtome, mounted on a glass slide, and dried overnight at 37°C. Deparaffinization in xylene and decreasing concentrations of ethanol was performed as described previously [16] and in an automated manner using the Medite Tissue Stainer COT 20. Tumor areas corresponding to the adjacent HE-stained tumor template were macrodissected of the deparaffinized tissue sections with a scalpel or hollow needle. The tumor tissue was transferred into 1.5-ml microreaction tubes.
Sample Preparation for LC-MS/MS Analysis and Data Acquisition
For further tissue preparation for MS analysis, 100 μl of lysis buffer (0.09 mM HEPES pH 8.0, 0.02 mM DTT, 0.1% Rapigest, in water) was added per mm3 tissue. Samples were incubated at 95°C, shaking at 750 rpm, for 1 hour. When cooled down to 25°C, pH was adjusted to 7 to 8. Two micrograms of trypsin (sequencing grade, Worthington, Lakewood, NJ)/mm3 tissue was added to the samples, and they were incubated at 37°C for 18 hours. The samples were centrifuged at 19,000g for 15 minutes to remove cellular detritus, and the supernatant was transferred to a fresh microreaction tube. The peptide concentration was measured with a bicinchoninic acid assay [17]. Sodium dodecyl sulfate polyacrylamide gel electrophoresis with silver nitrate gel staining was performed to visualize tryptic digestion. In case of incomplete digestion, tryptic digestion was repeated. Rapigest was degraded by addition of final concentrations of 3 M guanidinium hydrochloride and 0.2 M hydrochloric acid, incubation for 30 minutes at 37°C, followed by centrifugation at 1900g for 10 minutes. The supernatant was desalted using self-packed C18 Stage tips [18]. Acetonitrile was removed from the samples by centrifugal vacuum evaporation. A total of 0.5 μg of each sample was analyzed on a Q-Exactive plus (Thermo Scientific) mass spectrometer coupled to an Easy nanoLC 1000 (Thermo Scientific) with a flow rate of 300 nl/min. Buffer A was 0.5% formic acid, and buffer B was 0.5% formic acid in acetonitrile (water and acetonitrile were at least high-performance liquid chromatography gradient grade quality). A gradient of increasing organic proportion was used for peptide separation (main ramp 5%-40% acetonitrile in 80 minutes). The analytical column was an Acclaim PepMap column (Thermo Scientific), 2 μm particle size, 100 Å pore size, length 150 mm, and inner diameter 50 μm. The mass spectrometer operated in data-dependent mode with a top 10 method at a mass-to-charge ratio of 300 to 2000.
Immunohistochemistry (IHC)
We performed immunohistochemical staining of five antibodies (AbCam, Cambridge, MA): poly (ADP-ribose) polymerase-1 (PARP1; mouse monoclonal anti-human, clone A6.4.12, [ab110915]), N-Myc downstream-regulated gene 3 protein (NDRG3; rabbit monoclonal anti-human, clone EPR9011(B) [ab133715]), retinol dehydrogenase 11 (RDH11; rabbit polyclonal anti-human, [ab85849]), ABHD11 (NBP2-33574, Novus), and protein phosphatase slingshot homolog 3 (SSH3) (NBP-100-0674). First, tissue sections (1 tissue section per paraffin sample block) of 2 μm were deparaffinized and subjected to heat-induced epitope retrieval [19]. The staining protocol included the following steps: incubation with H2O2 (5 minutes), with primary antibodies (60 minutes), with mouse/rabbit linker (15 minutes), and with horseradish peroxidase and secondary antibody (20 minutes) and finally incubation with 3,3′-diaminobenzidine (10 minutes). Samples were counterstained manually with hematoxylin; xylene was used as permanent mounting medium. Positive controls were performed on control tissue as suggested by the antibody manufacturer (human placenta and skin for PARP1 and human kidney for NDRG3, RDH11 ABHD11, and SSH3) and on PCa as well as LNM tissue. For negative controls, antibody-diluent solution instead of the primary antibody was added to the samples; all other steps were idem.
LC-MS/MS Data Analysis
MS data were analyzed by MaxQuant version 1.5.28 [20] with the Uniprot human database downloaded on November 26, 2013, containing 20,271 reviewed canonical sequences without isoforms [21]. The analysis included an initial search with a precursor mass tolerance of 20 ppm for mass recalibration and a main search with precursor mass and fragment mass tolerances of 6 ppm and 20 ppm, respectively. The search included a fixed modification of carbamidomethyl cysteine and no variable modifications. Tryptic cleavage specificity with up to two missed cleavages was used with a minimal peptide length of seven amino acids. The false discovery rate was set to 0.01 for peptide and protein identifications in individual analyses. Relative protein quantification was done label-free using MaxLFQ [22]. Proteins were only further considered if they were identified and quantified in at least four of the five patient cases for both comparisons. Files obtained by MaxQuant were further processed using RStudio v.0.99.446 (R Foundation for Statistical Computing, Vienna, Austria) as previously described [23]. Decoy sequences and potential contaminant entries were removed. Ratios were log2 transformed, and a linear model was fitted using the limma package [24]. Proteins with a 50% increased or decreased ratio (log2 delta LFQ >/< +/−0.58) and a limma-moderated P value of <.01 (for selection of individual proteins for immunohistochemical analysis) or <.05 [for gene ontology (GO) enrichment analysis and alike] were classified as having an altered abundance (Figure 1E). STRING [25], [26] and DAVID [27] were used for the biological interpretation of significantly affected proteins.
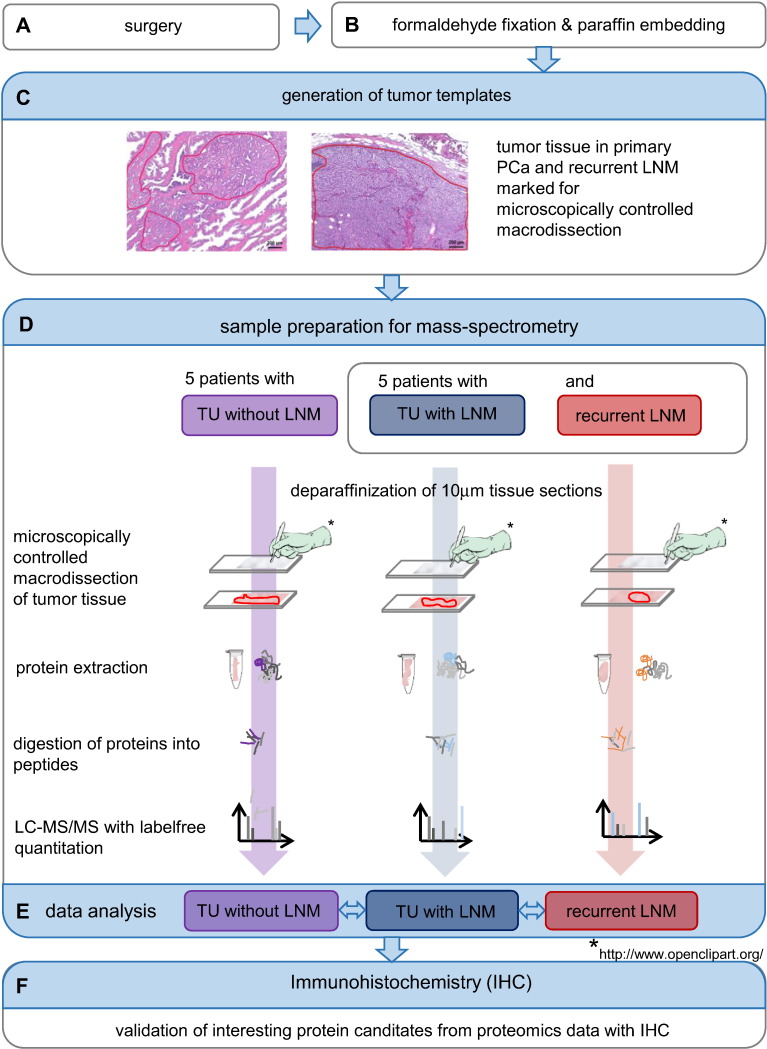
Figure 1.
Workflow of the study. After surgery (A), PCa and LNM tissues were formalin-fixed and paraffin-embedded (B). On HE-stained sections, experienced pathologists marked tissue areas for proteomic analysis. These were used as templates for the macrodissection of tumor tissue on the deparaffinized sections to analyze (C). Proteins were extracted from the tissue pieces, digested into peptides, and analyzed with LC-MS/MS (D). MaxQuant was used for peptide/protein identification and MaxLFQ for label-free quantification. During data analysis, we compared the sample groups TU with LNM, TU without LNM, and recurrent LNM against each other (E). IHC was performed to corroborate interesting protein candidates from proteomics data (F).
Positive controls stained positive; negative controls remained negative in all staining cycles. We evaluated the three immunohistochemical markers according to a well-established and widely used pathological scoring system. Thereby, the percentage of positively stained tumor tissue out of all tumor tissue and the staining intensity, graduated with 0 = negative, 1 = weak, 2 = moderate, or 3 = strong, are assessed [28], [29]. For both, we considered only those tumor areas which corresponded to the HE-stained tumor templates. Thus, we ensured to evaluate IHC on the same tumor areas that underwent proteomic analysis.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium [30] via the PRIDE partner repository with the dataset identifier PXD005833 (reviewer account details: username: reviewer29110@ebi.ac.uk, password: 2YrVi3YT).
Results
General Approach
We aimed for a comprehensive proteomic investigation of PCa, including five cases of primary tumors (TU) that formed neither primary nor secondary metastases, five cases of TUs that gave rise to recurrent LNMs, together with the corresponding, patient- matched, recurrent LNMs. FFPE specimen represents the standard tissue for histopathological diagnosis and evaluation of human malignancies. Formalin-induced crosslinks reduce tissue degradation to a minimum and ensure the maintenance of tissue architecture and correct appraisal of, e.g., the tumor infiltration in adjacent structures in lymph or vein vessels. FFPE specimen allows for a long-term follow-up as well as straightforward and easy sample preparation for proteomic investigations.
Protein extraction was performed with a “direct trypsinization” protocol that minimizes sample consumption and yields robust results in quantitative proteomics. For relative protein quantification, we employed a label-free approach. The entire workflow is summarized in Figure 1.
Protein Identification from FFPE Samples
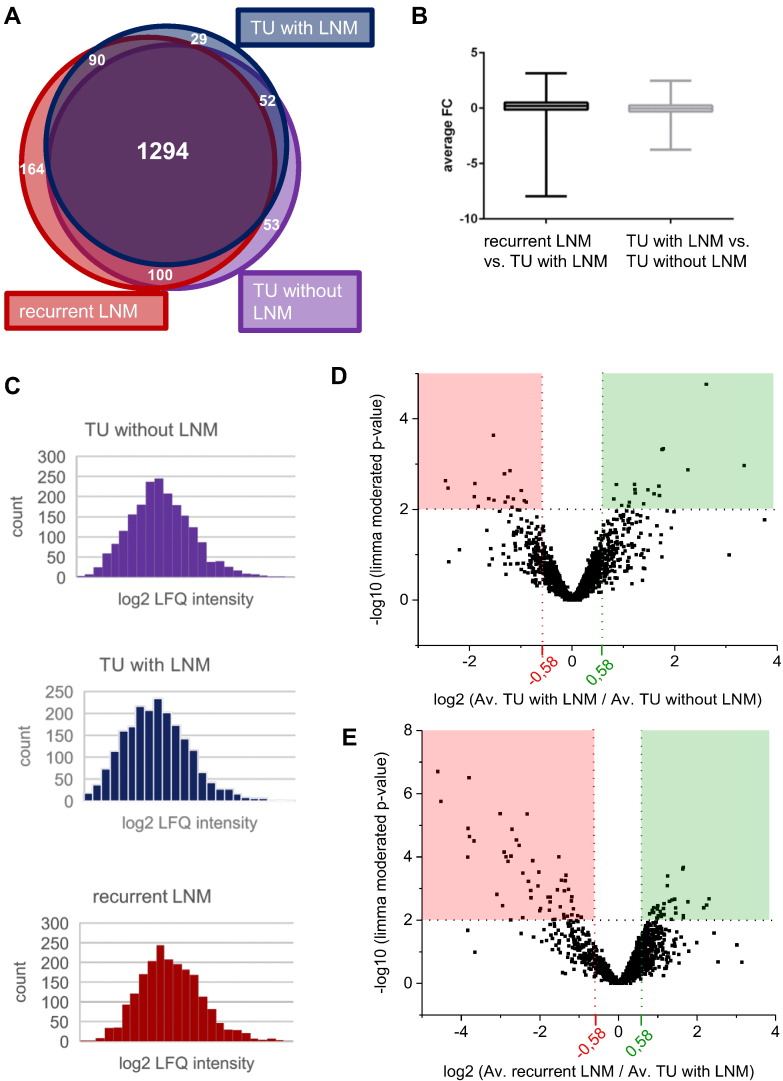
In each patient sample, up to 1867 proteins were identified and quantified at a false discovery rate <1% (Supplementary Table 1, Supplementary Table 2). However, incomplete overlap of proteome coverage is an intrinsic feature of MS-based proteomics [31], and some proteins may only be expressed in some cases. We focused on proteins that were identified and quantified in at least four cases of each subcohort (primary PCa without metastases and without relapse, primary PCa with secondary nodal metastases, recurrent lymph node metastases). With these conditions, a Venn representation of the proteome coverage shows that 1294 proteins were commonly identified in all three subcohorts (Figure 2A, Supplementary Table 3).
Figure 2.
Overview of proteomic results. (A) The Venn diagram shows that 1294 proteins were shared among all three groups. This number and the other numbers given refer to proteins which were present in at least four of five replicates per group. (B) The average FCs were visualized as boxplots. (C) Histograms showing the average log2-transformed LFQ protein intensities of the three sample groups. (D and E) Volcano plots showing average FC values and limma-moderated P values. Comparing recurrent LNM to TU with LNM, 87 proteins met significance criteria (P value < .01). Comparing TU with LNM to TU without LNM, 35 proteins displayed a P value < .01.
Differentially Regulated Proteins
Abundance differences were log2-transformed and visualized with boxplots (Figure 2B). As highlighted in Figure 2C, the log-transformed LFQ values display a prototypical profile [32].
To identify proteins that are significantly enriched or depleted in the aforementioned comparisons, we employed limma statistics [33], which is particularly powerful with regard to multiple testing correction and prevention of false-positive discoveries in the analysis of omics-style data. We chose the following criteria to distinguish significantly regulated proteins: 1) limma-moderated P value < .01 (P < .05 for enrichment analysis) and 2) average increase or decrease in abundance by more than 50% (log2 > 0.58 for increase; log2 < −0.58 for decrease). These criteria highlighted 35 differentially regulated proteins between nonmetastasizing, relapse-free primary PCa and metastasizing primary PCa with nodal relapse as well as 87 differentially regulated proteins between nodal metastasizing primary tumors and their corresponding secondary LNMs. The corresponding volcano plots are visualized in Figure 2D, E; P values and average log2 ratios are listed in Supplementary Table 1, Supplementary Table 2.
Among differentially regulated proteins between nonmetastasizing, relapse-free primary PCa and primary PCa with nodal relapse was the cell-surface protein prostate-specific membrane antigen (PSMA). Elevated levels of PSMA have been reported before, e.g., by Ross et al. in an IHC study on RPE specimen [34], and overexpression of PSMA correlates with advanced pathological state and predicts biochemical recurrence. In addition to elevated levels, Lapidus et al. showed the enzyme's activity to be increased in PCa compared to BPH or normal prostate tissue [35]. Therefore, PSMA is discussed as a promising target for immunotherapy [36] or for diagnosis of PCa [37]. In a recently published phase 1 clinical study, PSMA-based PET/CT scans were shown to be superior in the detection of bone metastases and equal in the detection rates of soft tissue metastases compared to conventional imaging techniques [38]. Our proteomic findings on PSMA are in line with the bulk of published studies: in comparison to nonmetastasizing and relapse-free PCa, we measured a >4.75-fold increase of PSMA in primary tumor tissue of relapsing and nodal metastasizing PCa (average log2 = 2.25; Plimma< .0013). This consistency validates the quality of our experimental proteomic approach.
Biological Processes and Motifs Distinguishing Metastasizing and Nonmetastasizing Primary Tumors
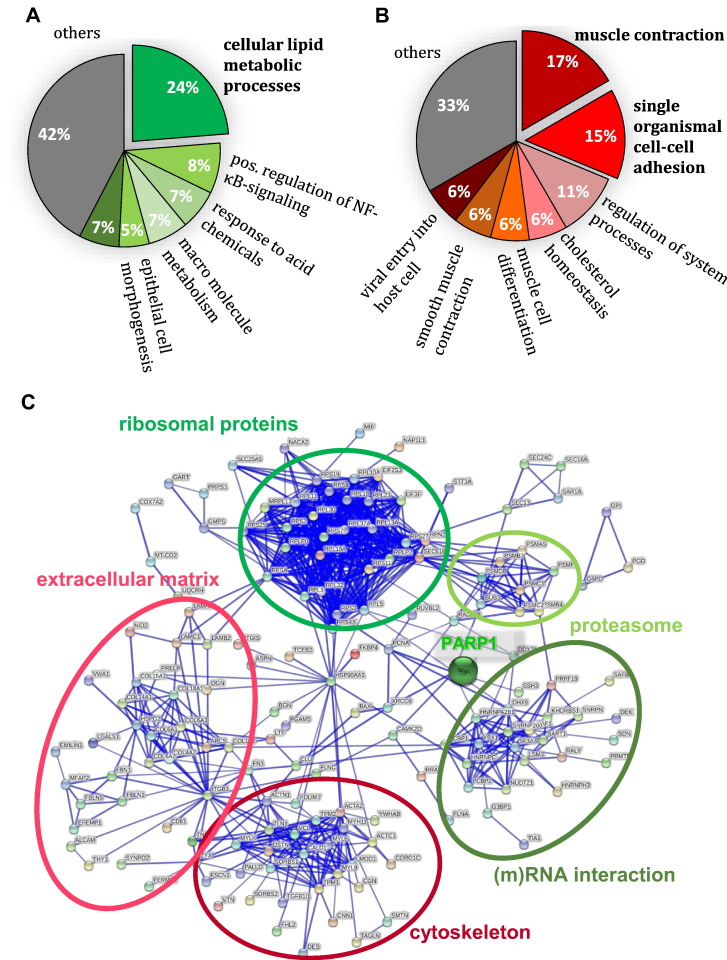
In order to functionally classify the proteins, which we found differentially regulated in metastasizing versus nonmetastasizing PCa tissue, we employed GO-term enrichment. For this, we focused on those proteins that were significantly more abundant in metastasizing PCa as compared to nonmetastasizing PCa. Twenty-four percent of these proteins map to cellular lipid metabolic process (Figure 3A); among contributing proteins were, e.g., acetyl-coa-carboxylase 1 (Uniprot accession Q13085), carnitine-O-acetyltransferase (Uniprot accession P43155), or beta-hexosaminidase (Uniprot accession P07686). This result accords with literature, as alterations in the lipid metabolism are described for many tumors, including PCa [39], which, e.g., overexpresses key enzymes of lipid de novo synthesis like the fatty acid synthetase [40]. In a recent review, it was pointed out that lipid accumulation plays a key role in cancer cell survival, protection against stress, and proliferation [41].
Figure 3.
Functional annotation of differentially regulated proteins.
(A/B) GO term enrichment in TU with LNM compared to TU without LNM. Predominantly enriched biological process (BP) was “cellular metabolic process,” while “muscle contraction” was reported the most depleted BP. (C) STRING functional annotation clustering revealed several clusters, which are marked in green if clustering proteins (moderated P value < .05; limma) were enriched or in red if they were decreased in recurrent LNM compared to TU with LNM. PARP1 laid at the interface of several clusters.
“Muscle contraction” and “single organism cell-cell adhesion” were found as the most depleted GO terms (Figure 3B). Proteins contributing to “muscle contraction” included, e.g., gelsolin (Uniprot accession P06396) or synemin (Uniprot accession O15061), proteins involved in the actin filament or intermediate filament organization. The cluster “single organismal cell-cell adhesion” included, e.g., integrin-linked protein kinase 1 (Uniprot accession Q13418), membrane primary amine oxidase (Uniprot accession Q16853), or tenascin (Uniprot accession P24821). DAVID bioinformatics analysis produced comparable results. Affected proteins did not cluster noteworthily in STRING functional annotation clustering (data not shown).
Altered Biological Processes and Motifs in Lymph Node Metastases Compared to Corresponding Primary Tumors
We further employed STRING clustering to functionally classify the differentially regulated proteins that distinguish lymph node metastases from metastasizing primary PCa tissue. We noticed several clusters (Figure 3C), e.g., increased abundance of ribosomal and proteasomal proteins in recurrent LNM. This points towards increased protein turnover in LNM as compared to the corresponding primary tumors. On the contrary, proteins with functions in focal adhesion, cytoskeleton organization, muscle contraction, or components of the extracellular matrix were depleted in recurrent LNM in comparison to tissue of the metastasizing primary TU. This could indicate less cohesive tumor cells in LNM compared to primary tumors. Other algorithms, like DAVID functional annotation clustering and GO term enrichment, yielded comparable results.
Immunohistochemical Investigation of Differential Levels of PARP1, NDRG3 RDH11, ABHD11, and SSH3
From the set of differentially regulated proteins, we selected five candidates for further investigation by IHC. These include PARP1, which participates in DNA repair [42] and for which we found elevated levels in LNMs as compared to metastasized and relapsing primary tumors. Furthermore, we selected NDRG3, for which a tumor-promoting role in PCa is suspected [43] and for which we also found elevated levels in LNMs as compared to metastasized and relapsing primary tumors. Lastly, we selected RDH11 for immunohistochemical investigation. This enzyme is highly expressed in the prostate and physiologically involved in the metabolism of short-chain aldehydes [44]. RDH11 was significantly increased in metastasizing primary tumors as compared to nonmetastasizing, relapse-free primary tumors. It caught our interest as a possible association or function in the context of PCa is rather unknown. In Table 2, proteins selected for IHC follow-up are listed with their respective statistical metrics.
Table 2.
Immunohistochemical Investigation
| Uniprot Acc. | Protein Name | Gene Name | Ø FC | P Value | CI.L | CI.R | Ø Identified Peptides | Ø Sequence Coverage [%] | |
|---|---|---|---|---|---|---|---|---|---|
| Recurrent LNM vs TU with LNM | Q9UGV2 | Protein NDRG3 | NDRG3 | 1.43 | .00029 | 0.88 | 1.98 | 2.60 | 9.79 |
| P09874 | Poly-(ADP-Ribose)-Polymerase | PARP1 | 0.63 | .0361 | 0.05 | 1.22 | 6.20 | 8.42 | |
| Q8NFV4 | Alpha/beta hydrolase domain-containing protein 11 | ABHD11 | 2.30 | .0021 | 1.02 | 3.59 | 2.8 | 9.3 | |
| Q8TE77 | Protein phosphatase Slingshot homolog 3 | SSH3 | 1.77 | .0026 | 0.85 | 2.69 | 0.7 | 1.4 | |
| TU with LNM vs TU without LNM | Q8TC12 | Retinol dehydrogenase 11 | RDH11 | 1.23 | .00361 | 0.48 | 1.97 | 8.33 | 33.07 |
Proteins chosen for immunohistochemical corroboration are listed with their respective FC value, P value, confidence intervals, average number of identified peptides, as well as average sequence coverage among the five biological replicates per group. The FC value is the log2-transformed ratio of a protein's intensity in two samples, which is commonly calculated to show an increase (for positive values) or decrease (for negative values) of the respective proteins' abundance in one condition/sample.
IHC visualizes the presence of a protein with the help of a colorimetric reaction. We evaluated the staining intensity for PARP1, NDRG3, RDH11, ABHD11 and SSH3 with the above-described score to estimate the relative quantitation of a protein in the tissue.
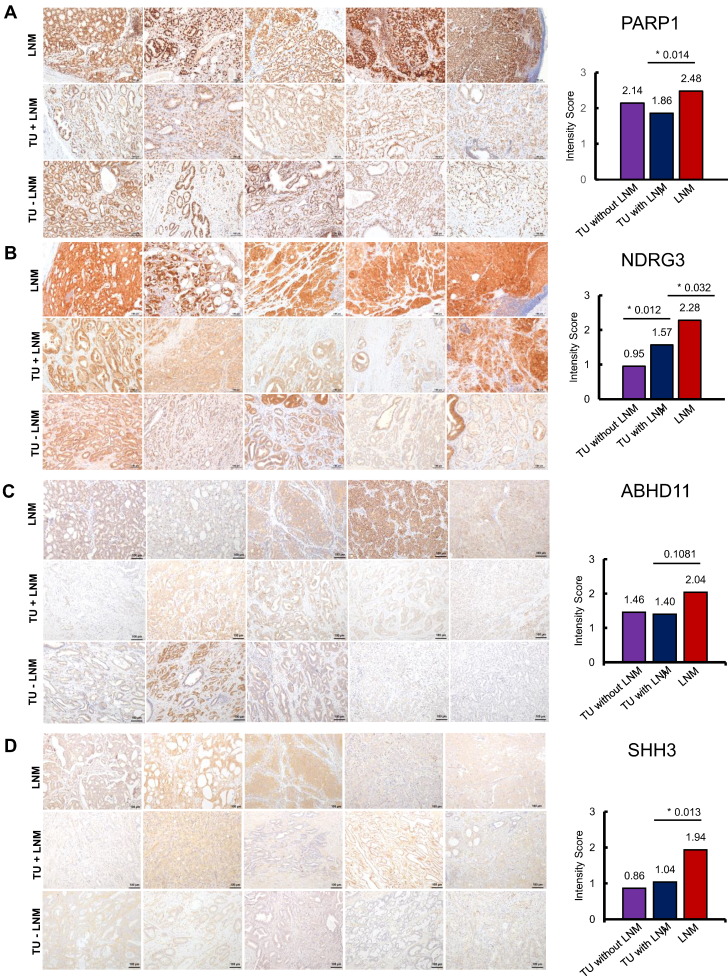
Regarding PARP1 IHC staining, all tumor areas in all three experimental subcohorts stained positive and displayed a nuclear staining pattern; representative images are shown in Figure 4A. PARP1 average staining intensity was highest in the LNM (medium intensity score 2.48), followed by nonmetastasizing primary TU (medium intensity score 2.14) and by metastasizing primary TU (medium intensity score 1.86), as displayed in the bar charts in Figure 4A. In line with our proteomic results, the stronger PARP1 staining intensity in LNMs, indicating a higher PARP1 abundance in the metastases as compared to the patient-matched primary TU, was significant (P = .014, Student's t test). There was no significant difference in PARP1 staining intensity in metastasizing and nonmetastasizing primary tumors.
Figure 4.
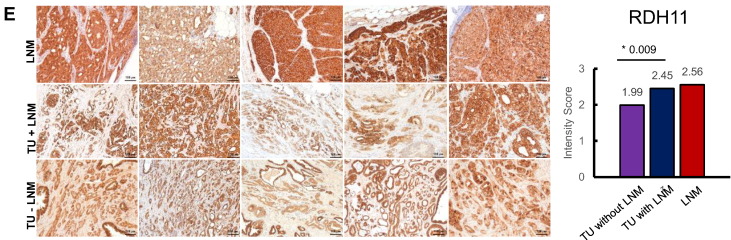
IHC stainings for (A) PARP1, (B) NDRG3 (C) ABHD11, (D) SSH3, and (E) RDH11 in recurrent LNM, TU with LNM, and TU without LNM, respectively. Staining intensity was evaluated manually and scored as follows: 0 = negative, 1 = weak, 2 = moderate, or 3 = strong. The average intensity scores for each group are plotted in the bar charts on the right side. Statistically significant differences were marked with an asterisk (*) and the respective P value (Student's t test).
NDRG3 is annotated as a cytoplasmic protein, and cytoplasmatic staining was detected in IHC on metastasizing and nonmetastasizing TU as well as on LNM (Figure 4B). NDRG3 staining intensity was lowest in nonmetastasizing PCa (medium intensity score 0.95) followed by metastasizing PCa (medium intensity score 1.55), and LNM displayed the strongest intensity with a medium score of 2.28. The stronger staining intensity in LNM compared to metastasizing TU was significant with a P value = .012, Student's t test.
Furthermore, we noticed significantly elevated NDRG3 staining in metastasizing primary tumors as compared to nonmetastasizing primary tumors (P value = .03, Student's t test).
ABHD11 showed a cytoplasmic staining pattern in IHC (Figure 4C), although some reports also point to a mitochondrial localization [45]. ABHD11 intensity displayed a trend for increased intensity in LNM (medium intensity score 2.04) as compared to patient-matched metastasizing TU (medium intensity score 1.46, P value = .1081, Student's t test). The ABHD11 staining intensity in metastasizing compared to nonmetastasizing TU was almost comparable (medium intensity score 1.40 and 1.46, P value = .8183, Student's t test).
SSH3 is expressed at a low level in normal prostate glandular cells [46] and also in the primary PCa of our staining cohort; the IHC intensity was rather weak (Figure 4D), with a medium intensity score of 0.86 in nonmetastasizing tumors and 1.04 in the metastasized group, with no statistically significant difference. The LNM displayed a stronger IHC intensity (medium IHC intensity score 1.94), which was significant in comparison to the metastasized primary tumors (P value = .013, Student's t test).
RDH11 displays a cytoplasmatic staining pattern in IHC. RDH11 intensity was lowest in nonmetastasizing TU (medium intensity score 1.99); it was higher in the metastasized group (medium intensity score 2.45) and highest in the LNM (medium intensity score 2.56). The RDH11 intensity difference between nonmetastasizing TU and metastasizing TU was statistically significant (P value = .0093, Student's t test), and IHC thereby corroborated the proteomic analysis. The difference in RDH11 staining intensity between LNMs and corresponding primary tumors was not significant.
Discussion
In this study, we compared the proteome composition of different PCa tissues (nonmetastasizing primary TU, metastasizing primary TU, secondary LNM). Comparing malignant to malignant tissue, we aimed to carve out differences among those and thereby contribute to a better characterization of heterogenous-appearing PCa. Our study differs from the vast majority of PCa proteomic studies, which typically compare tumor tissue to “normal” prostate tissue or benign hyperplasia.
We identified and quantified over 1200 proteins in FFPE tissue of metastatic and nonmetastatic primary PCa and in recurrent LNMs and highlighted different proteome motifs. The total case number of our study is comparably small with five biological replicates per experimental group.
Our protocol included a macrodissection step to enrich for tumor tissue. Macrodissection has been shown to produce equally accurate protein identification numbers as laser capture dissection [47] in FFPE samples. Besides, we chose macrodissection because of the possibility to cover relatively large tumor areas and thereby avoid bias due to intratumorous heterogeneity, which is often distinct in PCa. To further strengthen our analysis, we focused on proteins that were shared among at least four replicates and among at least two experimental groups.
We measured an increased abundance of PARP1, NDRG3, ABHD11, and SSH3 in metastases and confirmed this by IHC. We showed that PARP1, NDRG3, ABHD11, and SSH3 are enriched in recurrent LNMs as compared to patient-matched primary tumors. Furthermore, we found that RDH11 is more abundant in metastasized compared to nonmetastasized primary PCa tumors.
PARP1 is involved in DNA damage sensation and induces its repair when the damage is within physiological limits and otherwise induces apoptosis of the cell [48]. PARP1 also contributes to the transcriptional regulation of protumorigenic processes like cell growth or epithelial-to-mesenchymal transition [49], [50], [51]. Monotherapy with specific PARP inhibitors (PARPi) is highly effective in patients harboring defects in BRCA 1/2 genes, most often ovarian or breast cancer patients [52]. These mutations are rare in PCa patients [53], but other genomic alterations have been proposed to account for susceptibility to PARPi. Important candidates in this context are, e.g., ETS gene rearrangements, as PARP has been shown to physically interact with the gene fusion product TMPRSS2:ERG and seems to be required for ERG-mediated gene transcription, cell invasion, and PCa progression [54], [55]. Several strategies to treat PCa with PARPi are currently investigated in clinical trials. First results show that PARPi are able to induce antitumoral effects, e.g., as measured by PSA response or a decrease in circulating tumor cells [56]. In our study, we found an increased PARP1 abundance in recurrent LNM, which was corroborated by IHC stainings. Suggesting an underlying functional correlate for this, we can herewith support the idea that PARP1 inhibition might be a promising therapeutic option in patients with metastatic disease.
Protein NDRG3 is a member of the NDRG family, a group of proteins highly conserved among species and expressed in various tissues, including a high expression in the normal prostate. The physiological function of NDRG3 is only partly known so far [57]. Lee et al. shed some light onto the functional mechanisms and pathways NDRG3 is involved in and describe a lactate-induced cellular response to hypoxia with NDRG3 playing a key role: In normoxia, NDRG3 undergoes proteasomal degradation; under prolonged hypoxia, however, lactate accumulates and stabilizes NDRG3. The stabilized NDRG3 protein then acts via the Raf-ERK signaling pathway and mediates cell growth and angiogenesis [58].
In hepatitis B virus–related hepatocellular carcinoma NDRG3 expression was overexpressed in clinical tumor specimen as well as in HBV expressing hepatocellular carcinoma cell lines. NDRG3 levels were inversely correlated with levels of the liver specific micro-RNA miRNA-122. Transfection of miRNA-122 into hepatocellular carcinoma cell lines repressed the transcription and expression of NDRG3. Furthermore, the viral replication rate and the cell division rates were significantly reduced when NDRG3 levels were low. Due to the described role in HBV-related hepatocellular carcinoma, the authors suggest NDRG3 (and miRNA-122) as a candidate molecular diagnostic biomarker or future therapeutic target [59].
Regarding PCa, several studies reported alterations in NDRG3 levels or NDRG3-mediated cancer-promoting effects. In PCa cell lines, NDRG3 expression is upregulated upon androgen treatment, increases cell growth and migration capacities, and promotes tumor growth in mouse models. Furthermore, NDRG3 overexpression led to an increase of several angiogenetic chemokines (CXCL1, CXCL3, CXCL5), which is in the line with the results of Lee et al. and suggest a potential role in angiogenesis [43].
An IHC study on 206 pairs of PCa and corresponding noncancerous tissue reported an overexpression of NDRG3 in tumor compared to normal tissue, which was positively correlated with advanced pathological state, presence of metastases, as well as shortened relapse-free and overall survival [60].
In our study, we found increased levels of NDRG3 in metastasizing and nodal relapsing primary PCa compared to nonmetastasizing, relapse-free primary PCa.
ABHD11 is expressed only to a low level in glandular prostate cells [61] and, to date, has not been associated with PCa. In other human malignancies, such as ovarian cancer, increased levels induced cancer cell proliferation, invasion, and migration and inhibited apoptosis in cell culture [62]. Regarding its physiological function, ABDH11 was suggested to be a regulator of lipid metabolism [63]. Increased ABDH11 enzymatic activity in human lung adenocarcinoma could be mapped to the development of distant metastases and aggressive cancer phenotypes, which is why the authors suggested ABDH11 as a potential biomarker for lung adenocarcinoma [64]. In our proteomic data, ABDH11 was among the top candidates enriched in LNM compared to metastasizing TU [ave. fold change (FC) = 2.8, P value = .0021]. We therefore aimed to confirm this result by IHC. A difference in staining intensity was clearly visible and measurable. Although this difference failed to reach statistical significance, we were able to show a rectified trend in line with proteomic data and suggest ABHD11 for further investigation in the context of PCa.
To the current state of knowledge, SSH3 is physiologically involved in actin filament dynamics [65]. This is interesting per se, as actin filaments and the cytoskeleton in general play a crucial role in cancer cells’ ability to migrate and form metastasis. Whether SSH3 has a role in the formation of LNM in PCa remains to be determined; nevertheless, its increased abundance in LNM of PCa compared to corresponding primary tumors, which we show in proteomic and IHC data, is a novel finding.
Apart from PARP1 and NDRG3, RDH11 was selected for IHC corroboration of proteomics data. RDH11 is an enzyme involved in the metabolism of short-chain aldehydes and is highly expressed in the prostate [21], [66]. In a gene expression study on profiling androgen-regulated transcripts, RDH11 expression was induced upon androgen stimulation in cell culture [44], which makes RDH11 a possible mediator of androgen effects. Furthermore, the authors describe a substantial structural homology of RDH11 to the short-chain dehydrogenase family and hypothesized that RDH11 might likewise be involved in steroid synthesis or degradation. In our study, a GO-term–based cluster named “cellular lipid metabolism” was predominantly enriched in nodal metastasized PCa compared to nonmetastasized, relapse-free PCa. Among the contributing proteins was RDH11. Our MS data revealed increased levels of RDH11 in primary PCa with nodal relapse in comparison to nonmetastasizing, relapse-free PCa, which could be confirmed by IHC. So far, a differential expression of RDH11 in metastasized and nonmetastasized PCa has not been described. However, RDH11 has been proposed previously to function as a marker for malignant prostate tissue, as reported in a gene expression study on the cancer cell line LNCaP, where RDH11 contributed to a panel of marker genes for malignant prostate tissue [67]. Taken together, the results of our and other studies point towards a noteworthy yet rather unknown precise function of RDH11 in the biology of PCa and provide a starting point for further research.
In summary, we successfully investigated the proteome of PCa using FFPE specimens of primary tumors and of LNMs with LC-MS/MS and IHC. We showed that different PCa tissues exhibit measurable different biological motifs and found several proteins with possibly interesting function in the biology of PCa, such as RDH11, PARP, and NdRG3.
The following are the supplementary data related to this article.
Protein Identified and Quantified in Proteome Comparison “Recurrent LNM” Versus “TU with LNM”
Proteome Comparison “TU with LNM” Versus “TU without LNM,” Significant Proteins for Biological Enrichment Analysis: FC >0.58 or <−0.58 and P value < .05
Venn Representation
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
Acknowledgement
O. S. acknowledges support by Deutsche Forschungsgemeinschaft [SCHI 871/5, SCHI 871/6, SCHI 871/8, SCHI 871/9, GR 1748/6, INST 39/900-1, and INST 39/935-1 (SFB850-Project B8)], European Research Council (ERC-2011-StG 282111-ProteaSys), and the Excellence Initiative of the German Federal and State Governments (EXC 294, BIOSS).
A. K. M. was funded by the Integrated Research Training Group of the CRC 850 “Control of Cell Motility in Morphogenesis, Cancer Invasion and Metastasis.”
We acknowledge Alejandro Gomez-Aulì for providing us with a limma script for statistical analysis.
References
- 1.American Cancer Society . 2017. Cancer Facts & Figures 2017. [Google Scholar]
- 2.Wirth M, Weißbach L, Ackermann R, Alberti W, Albrecht C, Wolff J, Wörmann B. Interdisziplinäre Leitlinie der Qualität S3 zur Früherkennung, Diagnose und Therapie der verschiedenen Stadien des Prostatakarzinoms. Therapie. 2009:1–620. [Google Scholar]
- 3.Han M, Partin AW, Zahkurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169(2):517–523. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- 4.Hodson R. Small organ, big reach. Nature. 2015;528(7582):S118–S119. doi: 10.1038/528S118a. [DOI] [PubMed] [Google Scholar]
- 5.Jilg CA, Rischke HC, Reske SN, Henne K, Grosu AL, Weber W, Drendel V, Schwardt M, Jandausch A, Schultze-Seemann W. Salvage lymph node dissection with adjuvant radiotherapy for nodal recurrence of prostate cancer. J Urol. 2012;188(6):2190–2197. doi: 10.1016/j.juro.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 6.Rischke HC, Schultze-Seemann W, Wieser G, Krönig M, Drendel V, Stegmaier P, Krauss T, Henne K, Volegova-Neher N, Schlager D. Adjuvant radiotherapy after salvage lymph node dissection because of nodal relapse of prostate cancer versus salvage lymph node dissection only. Strahlenther Onkol. 2015;191(4):310–320. doi: 10.1007/s00066-014-0763-5. [DOI] [PubMed] [Google Scholar]
- 7.Kristiansen G, Egevad L, Amin M, Delahunt B, Srigley JR, Humphrey PA, Epstein JI. Konsenskonferenz 2014 der ISUP zur Gleason-Graduierung des Prostatakarzinoms. Pathologe. 2016;37(1):17–26. doi: 10.1007/s00292-015-0136-6. [DOI] [PubMed] [Google Scholar]
- 8.Gustafsson OJR, Arentz G, Hoffmann P. Proteomic developments in the analysis of formalin-fixed tissue. Biochim Biophys Acta. 2014;1854(6):559–580. doi: 10.1016/j.bbapap.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4(11):1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Hood BL, Conrads TP, Veenstra TD. Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics. 2006;6(14):4106–4114. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- 11.Aiello D, Casadonte F, Terracciano R, Damiano R. Targeted proteomic approach in prostatic tissue: a panel of potential biomarkers for cancer detection. Oncoscience. 2016;3:220–241. doi: 10.18632/oncoscience.313. [no. July] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davalieva K, Kostovska IM, Kiprijanovska S, Markoska K, Kubelka-Sabit K, Filipovski V, Stavridis S, Stankov O, Komina S, Petrusevska G. Proteomics analysis of malignant and benign prostate tissue by 2D DIGE/MS reveals new insights into proteins involved in prostate cancer. Prostate. 2015;75:1586–1600. doi: 10.1002/pros.23034. [DOI] [PubMed] [Google Scholar]
- 13.Iglesias-Gato D, Wikström P, Tyanova S, Lavallee C, Thysell E, Carlsson J, Hägglöf C, Cox J, Andrén O, Stattin P. The proteome of primary prostate cancer. Eur Urol. 2016;69(5):942–952. doi: 10.1016/j.eururo.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 14.Ost P, Decaestecker K, Lambert B, Fonteyne V, Delrue L, Lumen N, Ameye F, De Meerleer G. Prognostic factors influencing prostate cancer-specific survival in non-castrate patients with metastatic prostate cancer. Prostate. 2014;74(3):297–305. doi: 10.1002/pros.22750. [DOI] [PubMed] [Google Scholar]
- 15.Pond GR, Sonpavde G, de Wit R, Eisenberger MA, Tannock IF, Armstrong AJ. The prognostic importance of metastatic site in men with metastatic castration-resistant prostate cancer. Eur Urol. 2014;65(1):3–6. doi: 10.1016/j.eururo.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 16.Bronsert P, Weißer J, Biniossek ML, Kuehs M, Mayer B, Drendel V, Timme S, Shahinian H, Küsters S, Wellner UF. Impact of routinely employed procedures for tissue processing on the proteomic analysis of formalin-fixed paraffin-embedded tissue. Proteomics Clin Appl. 2014;8(9–10):796–804. doi: 10.1002/prca.201300082. [DOI] [PubMed] [Google Scholar]
- 17.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 18.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 19.Lillie RD, Fullmer HM. 4th ed. McGraw-Hill Incorporation; USA: 1976. Histopathologic Technic and Practical Histochemistry. [Google Scholar]
- 20.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 21.Bateman A, Martin MJ, O'Donovan C, Magrane M, Apweiler R, Alpi E, Antunes R, Arganiska J, Bely B, Bingley M. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(D1):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox J, Hein MY, Luber CA, Paron I. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Auli A, Hillebrand LE, Biniossek ML, Peters C, Reinheckel T, Schilling O. Impact of cathepsin B on the interstitial fluid proteome of murine breast cancers. Biochimie. 2016;122:88–98. doi: 10.1016/j.biochi.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):1–13. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447–452. doi: 10.1093/nar/gku1003. [no. database issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, Lempicki RA, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Van Diest PJ, Van Dam P, Henzen-logmans SC, Berns E, Van Der Burg MEL, Green J, Vergote I. A scoring syetem for immunohistochemical staining: consensus report of the task force for basic research of the EORTG-GCCG. J Clin Pathol. 1997:801–804. doi: 10.1136/jcp.50.10.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosman FT, de Goeij AF, Rousch M. Quality control in immunocytochemistry: experiences with the oestrogen receptor assay. J Clin Pathol. 1992;45(2):120–124. doi: 10.1136/jcp.45.2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vizcaíno J, Deutsch EEW, Wang R, Vizcaino JA, Deutsch EEW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen VK, Steinsland H, Wiker HG. Improving genome annotation of enterotoxigenic Escherichia coli TW10598 by a label-free quantitative MS/MS approach. Proteomics. 2015;15(22):3826–3834. doi: 10.1002/pmic.201500278. [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):1–26. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Ross JS, Sheehan CE, Fisher HAG, Kaufman RP, Kaur P, Gray K, Webb I, Gray GS, Mosher R, Kallakury BVS. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin Cancer Res. 2003;9(17):6357–6362. [PubMed] [Google Scholar]
- 35.Lapidus RG, Tiffany CW, Isaacs JT, Slusher BS. Prostate-specific membrane antigen (PSMA) enzyme activity is elevated in prostate cancer cells. Prostate. 2000;45(4):350–354. doi: 10.1002/1097-0045(20001201)45:4<350::aid-pros10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 36.Tasch J, Gong M, Sadelain M, Heston WD. A unique folate hydrolase, prostate-specific membrane antigen (PSMA): a target for immunotherapy? Crit Rev Immunol. 2001;21(1–3):249–261. [PubMed] [Google Scholar]
- 37.Haberkorn U, Eder M, Kopka K, Babich JW, Eisenhut M. New strategies in prostate cancer: prostate-specific membrane antigen (PSMA) ligands for diagnosis and therapy. Clin Cancer Res. 2016;22(1):9–15. doi: 10.1158/1078-0432.CCR-15-0820. [DOI] [PubMed] [Google Scholar]
- 38.Pandit-Taskar N, O'Donoghue JA, Durack JC, Lyashchenko SK, Cheal SM, Beylergil V, Lefkowitz RA, Carrasquillo JA, Martinez DF, Fung AM. A phase I/II Study for analytic validation of 89Zr-J591 immunoPET as a molecular imaging agent for metastatic prostate Cancer. Clin Cancer Res. 2015;21(23):5277–5285. doi: 10.1158/1078-0432.CCR-15-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogene. 2016;5(1):1–10. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swinnen JV, Van Veldhoven PP, Esquenet M, Heyns W, Verhoeven G. Androgens markedly stimulate the accumulation of neutral lipids in the human prostatic Adenocarcinoma cell line LNCaP. Endocrinology. 1996;137(10):4468–4474. doi: 10.1210/endo.137.10.8828509. [DOI] [PubMed] [Google Scholar]
- 41.Deep G, Schlaepfer IR. Aberrant lipid metabolism promotes prostate cancer: role in cell survival under hypoxia and extracellular vesicles biogenesis. Int J Mol Sci. 2016;17(7):1061–1074. doi: 10.3390/ijms17071061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhanraj Deshmukh YQ. Role of PARP1 in prostate cancer. Am J Clin Exp Urol. 2015;3(1):1–12. [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Li Y, Li Y, Hong A, Wang J, Lin B, Li R. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int J Cancer. 2009;124(3):521–530. doi: 10.1002/ijc.23961. [DOI] [PubMed] [Google Scholar]
- 44.Lin B, White JT, Ferguson C, Wang S, Vessella R, Bumgarner R, True LD, Hood L, Nelson PS. Prostate short-chain dehydrogenase reductase I (PSDR1): a new member of the short-chain steroid dehydrogenase/reductase family highly expressed in normal and neoplastic prostate epithelium. Cancer Res. 2001;61(4):1611–1618. [PubMed] [Google Scholar]
- 45.UniProtKB - Q8NFV4 (ABHDB_HUMAN) http://www.uniprot.org/uniprot/Q8NFV4 Uniprot.org. [Online]. Available:
- 46.The Human Protein Atlas SSH3. http://www.proteinatlas.org/ENSG00000172830-SSH3/tissue/prostate [Online]. Available:
- 47.Ostasiewicz P, Zielinska DF, Mann M, Wisniewski JR. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 2010;9(7):3688–3700. doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 48.Burkle A. PARP-1: a regulator of genomic stability linked with mammalian longevity. Chembiochem. 2001;2(10):725–728. doi: 10.1002/1439-7633(20011001)2:10<725::AID-CBIC725>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 49.Pu H, Horbinski C, Hensley PJ, Matuszak EA, Atkinson T, Kyprianou N. PARP-1 regulates epithelial-mesenchymal transition (EMT) in prostate tumorigenesis. Carcinogenesis. 2014;35(11):2592–2601. doi: 10.1093/carcin/bgu183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiewer MJ, Knudsen KE. Transcriptional roles of PARP1 in cancer. Mol Cancer Res. 2014;12(8):1069–1080. doi: 10.1158/1541-7786.MCR-13-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiewer MJ, Goodwin JF, Han S, Chad Brenner J, Augello MA, Dean JL, Liu F, Planck JL, Ravindranathan P, Chinnaiyan AM. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnenblick A, de Azambuja E, Azim HAJ, Piccart M. An update on PARP inhibitors—moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12(1):27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- 53.Edwards SM, Kote-Jarai Z, Meitz J, Hamoudi R, Hope Q, Osin P, Jackson R, Southgate C, Singh R, Falconer A. Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet. 2003;72(1):1–12. doi: 10.1086/345310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng FY, Brenner JC, Hussain M, Chinnaiyan AM. Molecular pathways: targeting ETS gene fusions in cancer. Clin Cancer Res. 2014;20(22):4442–4448. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J. Poly (ADP-ribose) polymerase inhibitor: an evolving paradigm in the treatment of prostate cancer. Asian J Androl. 2014;16(3):401–406. doi: 10.4103/1008-682X.123684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mateo J. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;368(24):2255–2265. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y, He F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol Cell Biochem. 2002;229(1–2):35–44. doi: 10.1023/a:1017934810825. [DOI] [PubMed] [Google Scholar]
- 58.Lee DC, Ahm H. A Lactate-Induced Response to Hypoxia. Cell. 2015;161:595–609. doi: 10.1016/j.cell.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Fan CG, Wang CM, Tian C, Wang Y, Li L, Sun WS, Li RF, Liu YG. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. 2011;26(5):1281–1286. doi: 10.3892/or.2011.1375. [DOI] [PubMed] [Google Scholar]
- 60.Ren G, Tang L, Yang A, Jiang W, Huang Y. Prognostic impact of NDRG2 and NDRG3 in prostate cancer patients undergoing radical prostatectomy. Histol Histopathol. 2014;29(4):535–542. doi: 10.14670/HH-29.10.535. [DOI] [PubMed] [Google Scholar]
- 61.The Human Protein Atlas, “ABHD11” [Online]. Available: https://www.proteinatlas.org/search/ABHD11, 14 November 2017
- 62.Wu D, Chen X, Sun K, Wang L, Chen S, Zhao Y. 2017. Role of the lncRNA ABHD11-AS 1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC; pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arya M, Srinivasan M, Rajasekharan R. Biochemical and biophysical research communications human alpha beta hydrolase domain containing protein 11 and its yeast homolog are lipid hydrolases. Biochem Biophys Res Commun. 2017;487(4):875–880. doi: 10.1016/j.bbrc.2017.04.145. [DOI] [PubMed] [Google Scholar]
- 64.Wiedl T, Arni S, Roschitzki B, Grossmann J, Collaud S, Soltermann A, Hillinger S, Aebersold R, Weder W. Activity-based proteomics: identification of ABHD11 and ESD activities as potential biomarkers for human lung adenocarcinoma. J Proteomics. 2011;74(10):1884–1894. doi: 10.1016/j.jprot.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 65.UniProtKB - Q8TE77 (SSH3_HUMAN) http://www.uniprot.org/uniprot/Q8TE77 Uniprot.org. [Online]. Available:
- 66.UniProtKB - Q8TC12 (RDH11_HUMAN) http://www.uniprot.org/uniprot/Q8TC12 Uniprot.org. [Online]. Available:
- 67.Cooper C, Edwards S, Campbell C, Flohr P, Shipley J, Giddings I, Te-Poele R, Dodson A, Foster C, Clark J. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br J Cancer. 2005;92(2):376–381. doi: 10.1038/sj.bjc.6602261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein Identified and Quantified in Proteome Comparison “Recurrent LNM” Versus “TU with LNM”
Proteome Comparison “TU with LNM” Versus “TU without LNM,” Significant Proteins for Biological Enrichment Analysis: FC >0.58 or <−0.58 and P value < .05
Venn Representation
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium [30] via the PRIDE partner repository with the dataset identifier PXD005833 (reviewer account details: username: reviewer29110@ebi.ac.uk, password: 2YrVi3YT).