Abstract
Usp9x has emerged as a potential therapeutic target in some hematologic malignancies and a broad range of solid tumors including brain, breast, and prostate. To examine Usp9x tumorigenicity and consequence of Usp9x inhibition in human pancreatic tumor models, we carried out gain- and loss-of-function studies using established human pancreatic tumor cell lines (PANC1 and MIAPACA2) and four spontaneously immortalized human pancreatic patient-derived tumor (PDX) cell lines. The effect of Usp9x activity inhibition by small molecule deubiquitinase inhibitor G9 was assessed in 2D and 3D culture, and its efficacy was tested in human tumor xenografts. Overexpression of Usp9x increased 3D growth and invasion in PANC1 cells and up-regulated the expression of known Usp9x substrates Mcl-1 and ITCH. Usp9x inhibition by shRNA-knockdown or by G9 treatment reduced 3D colony formation in PANC1 and PDX cell lines, induced rapid apoptosis in MIAPACA2 cells, and associated with reduced Mcl-1 and ITCH protein levels. Although G9 treatment reduced human MIAPACA2 tumor burden in vivo, in mouse pancreatic cancer cell lines established from constitutive (8041) and doxycycline-inducible (4668) KrasG12D/Tp53R172H mouse pancreatic tumors, Usp9x inhibition increased and sustained the 3D colony growth and showed no significant effect on tumor growth in 8041-xenografts. Thus, Usp9x inhibition may be therapeutically active in human PDAC, but this activity was not predicted from studies of genetically engineered mouse pancreatic tumor models.
Abbreviations: PDA, pancreatic ductal adenocarcinomas; PanIN, pancreatic epithelial lesions; GFR, growth factor reduced; DOX, doxycycline; 3D, three-dimensional; Usp9x-OV, Usp9x overexpression; DUB, deubiquitinase
Introduction
Pancreatic cancer is comprised of both exocrine and endocrine tumors of the pancreas with more than 90% of the pancreatic tumors classified as pancreatic ductal adenocarcinoma (PDA). PDA is relatively rare (11th most common) with 45,000 new cases per year but with a very poor prognosis [1]. Poor outcomes can be associated with the asymptomatic nature of early-stage disease, typically late clinical diagnosis, and high metastatic activity of the primary tumor resulting in a 5-year survival rate of only 6% [2], [3]. Even in cases where surgical intervention of localized disease is possible, 5-year survival (20%) is not markedly improved, highlighting the high propensity of PDA to seed micrometastatic disease and underscoring the need to fully interrogate initiating and promoting lesions or pathways that may provide new approaches in detection and therapy [4]. Toward that goal, animal models built on common genetic lesions detected in PDA patients have been informative and have led to the reassessment of the role of specific gene mutations, such as Kras, in tumorigenicity [5]. The Kras oncogene is frequently (90%) mutated in pancreatic cancer, with G12D being the most common mutation with capacity to constitutively activate Kras GTPase [6], [7]. Kras mutations occur in precursor lesions called the pancreatic epithelial lesions (PanIN), indicating an early role in the progression toward pancreatic cancer [6], [8]. In vivo mouse models have established the role of oncogenic Kras in the initiation of pancreatic cancer in mice [9], [10], while recent reports outline the importance of mutated Kras in pancreatic cancer maintenance [11]. Using a mouse model which allows for inducible, pancreas-specific, and reversible expression of oncogenic KrasG12D, with or without one allele of the tumor suppressor p53, Collins et al. showed that KrasG12D drives pancreatic tumorigenesis and is required for tumor maintenance [11]. However, KrasG12D induction alone causes only limited onset of tumorigenesis, which may reflect clinical observations which estimate that a single point mutation can occur 10 to 15 years prior to establishment of invasive disease and metastatic lesions [12]. Thus, complementation of Kras tumorigenicity with additional PDA-associated mutations reduces the latency of tumor development and provides useful PDA mouse models of human disease [12]. However, these models do not allow an unbiased assessment of other genes and epigenetic changes that may play a role in the emergence of invasive PDA [12]. This deficiency was recently addressed using insertional gene disruption technology provided by the “Sleeping Beauty” transposon [13], [14]. Using this transposon to interrogate gene disruption associated with shortened latency in a KrasG12D pancreatic tumor model, Perez-Mancera et al. described several cooperative genes that were previously described in PDA patients [13]. In addition, Usp9x, a DUB previously associated with tumor-permissive pathway control, was mapped as the most common insertionally disrupted gene in the KrasG12D background that cooperated in promoting KrasG12D tumorigenesis.
Usp9x has been described as a critical mediator of cell survival. Increased expression of Usp9x is associated with hematologic malignancies including follicular lymphoma, diffuse large B cell lymphoma, multiple myeloma [15], chronic myelogenous leukemia [16], as well as solid tumors such as brain tumors [17], esophageal squamous cell carcinomas [18], prostate [19] and breast cancers [15], [20]. High expression levels of Usp9x associate with poor prognosis in multiple myeloma [15] and esophageal squamous cell carcinomas [18]. Some cancers, including primary breast cancer, demonstrate an association between Usp9x and Mcl-1, a prosurvival BCL2 family member that is essential for stem and progenitor cell survival and is known to confer chemo- and radioresistance in a variety of tumors including lymphoma, breast, renal, lung, bladder, and prostate cancer [18], [21], [22]. Inhibition of Usp9x has emerged as a therapeutic strategy in the treatment of hematologic malignancies, melanoma, and ERG-positive prostate tumors [15], [19], [23]. Usp9x inhibition is also shown to sensitize tumor cells to chemo- and radiotherapy by reducing Mcl-1 levels [21], [22], [24], [25].
In the present study, we examined the role of Usp9x in pancreatic tumors. We established a 3D culture model of genetically engineered mouse tumor derived cell lines, established human pancreatic cancer cell lines, and patient-derived pancreatic cancer cell lines. Using these models, we assessed the in vitro pancreatic phenotype resulting from Usp9x overexpression as well as the consequence of short hairpin RNA (shRNA)–mediated Usp9x knockdown and small molecule–mediated inhibition on that phenotype. We performed parallel assessments in murine pancreatic tumor–derived cell lines established from mice with constitutive or doxycycline-inducible expression of KrasG12D and Tp53R172H. The results suggest that Usp9x serves as a tumor suppressor in genetically engineered mouse pancreatic tumors, as previously demonstrated. However, in established human pancreatic tumor cells, Usp9x supports tumor cell survival and the malignant phenotype, illustrating wide distinctions in function in murine tumor cell models and bona fide human pancreatic cancer while also highlighting the potential for Usp9x inhibitors to be used in the treatment of human PDAC.
Material and Methods
Reagents
All cell culture reagents and culture media were purchased from Invitrogen (Grand Island, NY). Growth factor reduced (GFR) Matrigel (# 354230) was purchased from BD Biosciences (Bedford, MA). Tet-free fetal bovine serum was from Clontech (Mountain View, CA). HA-UbVS was from Boston Biochem (Cambridge, MA). Doxycycline (DOX), Puromycin, MTT, and other laboratory reagents were from Sigma (St. Louis, MO). EOAI3402143 (or G9) was synthesized and purified by Evotec Ltd. (Abingdon OX14 4RZ, United Kingdom).
Cell Culture and Establishment of Stable shRNA KD and Usp9x Overexpressing Cell Lines
All the established cell lines were procured from ATCC more than a year ago, and early passage cells were frozen in 10% DMSO complete media in liquid nitrogen following standard protocols for future use. Cell lines were authenticated by morphology and growth characteristics and were tested for mycoplasma. These cells were grown under standard culture conditions of 5% CO2 at 37°C following ATCC recommendations. Murine cell lines 8041, 4668, 65671, and 13342 were obtained from our collaborator Marina Pasca Di Magliano; have been well characterized for morphology, growth characteristics, and xenograft studies [26]; and were grown in 10% FBS-RPMI. The 4668 cell line was grown in the presence of 1 μg/ml of DOX unless DOX depletion was required to inhibit Kras expression/activity. To avoid induction of Kras from basal DOX levels in fetal bovine serum, DOX depletion experiments were carried out using tetracycline-free serum.
The human patient-derived cell lines UM-2, UM-6, UM-16, and UM-76 were established in the laboratory of Diane Simeone by xenotransplantation of the human pancreatic adenocarcinomas into mice. Briefly, after surgical removal, the tumor was digested using the Tissue Dissociation Kit (Miltenyi Biotec, #130-095-929) and were subjected to single cell isolation using Magnetic Cell Isolation following MACS technology (Miltenyi Biotec, Gladbach, Germany, # 130-090-753, #130-093-235, and #130-093-237) as instructed by the manufacturer. The cells were plated and subcloned in 10% RPMI medium. Cultures were purified of mouse stroma by labeling them with a biotin-labeled anti-H2Kd mouse antibody (Southern Biotech, Alabama, USA, #1911-08) and removing them using anti-biotin beads (Miltenyi Biotec Gladbach, Germany, #130-090-485) on a magnetic bead column (Miltenyi Biotec, Gladbach, Germany, #130-042-901).
The lentiviral expression system for shRNA against human Usp9x was kindly provided by Dr. Dzwokai Ma (University of California, Santa Barbara), and production of virus and infection were previously described [27]. Sequences targeting murine Usp9x 5′-CCCTTGCAAGATCTTGATAATA-3′ (KD1; HP_274673) and 5′-CGCAATCAAGTTCAATGATTAT-3′ (KD2; HP_13310) described in RNAi Codex (http://cancan.cshl.edu/cgi-bin/Codex/Codex.cgi) were cloned into the LMP retroviral shRNA system (Open Biosystems, Pittsburgh, PA) following the manufacturers’ protocol [28]. Oligo's and primers for the procedure were purchased from IDT (Coralville, IA). The sequence and viral packaging of LMP control shRNA targeting luciferase have been described previously [28].
For overexpression of Usp9x, p3xFlag-USP9X was created by three-way cloning using PCR to amplify a 320-bp N-terminal fragment of USP9X with the StuI site in Usp9X. Forward: 5′-tgtacgaagcttacagccacgactcgtggctc-3′, reverse: 5′-ggaaccacccatcgaggcc-3′. The PCR product was cut with HindIII and StuI. pCDNA5-TAP-USP9X was cut with StuI and NotI. These fragments were ligated into p3XFlag-CMV10 (Sigma) linearized with HindIII/NotI. A PCR was performed with forward primer 5′-gctctagatctatggactacaaagacc-3′ and the same reverse primer described above. This product was cut with BglII and StuI, and ligated together with the StuI/BamHI fragment from p3XFlag-USP9X together with MIGR1 linearized with BglII. p3xFlag-Vector and p3xFlag-USP9X were transfected into 8041 or PANC1 cell lines using linear polyethylenimine (PEI). A total of 106 cells were plated in 10-cm dishes and were grown for 24 hours under normal culture conditions. Five micrograms of each p3xFlag-Vector or p3xFlag-Usp9X plasmid was diluted in 800 μl of 150 mM NaCl and mixed with 40 μl of PEI, followed by 30-minutes incubation at RT. The mixture was added to the cells containing fresh complete media. The cells were given a fresh media change after 24 hours and assessed for phenotypic changes, 3D growth, and invasion at day 6 posttransfection.
MTT Assay
Cells were seeded in a 96-well plate at 5000 per well in the presence of the indicated concentration of G9 for 3 days in a CO2 incubator at 37°C. Twenty microliters of 5 g/l MTT solution were added to each well for 2 hours at 37°C. The cells were then lysed in 10% SDS buffer, and absorbance at 570 nm relative to a reference wavelength of 630 nm was determined with a microplate reader. To examine proliferation using the MTT assay, cells were plated in triplicates, and the samples were processed for MTT assay as described above at day 0, 1, 2, 3, and 4.
Three-Dimensional Cultures, Immunoblotting, and DUB Activity Assay
Cells were grown on top of GFR Matrigel for 8 days following a published protocol [29]. Phase contrast images were acquired at 5× or 10× resolution on a Leica inverted microscope. For treatment studies, the cells were plated with the desired dose of small molecule inhibitors, and the inhibitors were replenished every 3 days in fresh complete media. To quantify the number of colonies, the total number of colonies from two to three wells of an eight-well chamber slide was counted using phase contrast images acquired at 5× resolution.
For spheroid culture, 106 cells were plated in complete media on 100-mm dishes coated with 1% agarose. The cells were allowed to grow for 5 to 6 days with fresh media supplemented every third day. The spheroids were collected, pelleted, and lysed in RIPA lysis buffer and subjected to immunoblot analysis.
For monolayer studies, control and Usp9x KD cells were grown in 100-mm dishes for 24 hours and lysed in RIPA lysis buffer followed by immunoblot analysis with antibodies against Usp9x (#A301-350A, 1:2000, Bethyl Laboratories, Montgomery, TX), PARP1 (#9542, 1:2000, Cell Signaling, Danvers, MA), phospho-ERK1/2 (#9101, 1:1000, Cell Signaling), anti-HA (#11867423001, 1:5000, Roche Applied Science, Indianapolis), anti-Mcl1 (#J2312, 1:2000, Santa Cruz Biotechnology, Santa Cruz, CA), anti-ITCH (#611198, 1:2000, BD Biosciences, San Jose, CA), anti-KRas (#OP24, 1:1000, Calbiochem, Billerica, MA), and β-actin (#69879, 1:2000, Santa Cruz Biotechnology, Santa Cruz, CA,).
For DUB activity assays, monolayer adherent cells grown on 100-mm dishes for 24 hours were scraped in conditioned media using a cell scraper. The cells were collected, pelleted, and washed two times with ice-cold PBS. The cell pellets were resuspended in 100 μl of DUB assay buffer [containing 50 mmol/L Tris-HCl (pH 7.5), 250 mmol/L sucrose, 5 mmol/L MgCl2, and 1 mmol/L phenylmethylsulfonyl fluoride] and lysed by sonication. The cell debris was pelleted by centrifugation at 12,000g for 15 minutes. Total protein of the lysates was estimated by Bradford reagent. Twenty micrograms of each lysate was incubated with 200 nM of HA-labeled ubiquitin vinyl sulfone (HA-UbVS) for 45 minutes at 37°C. The HA-UbVS-labeled lysates were boiled with SDS Laemmli buffer and run on a 6-15% SDS-PAGE gel and subjected to immunoblot analysis with anti-HA (as a measure of DUB activity) or anti-Usp9x (#A301-351A, Bethyl, 1:2000).
Quantitative RT-PCR
4668 cells were grown on 100-mm dishes with or without doxycycline (1 μg/ml) for 24 hours followed by RNA isolation using the RNeasy kit (Qiagen, Valencia, CA). Samples for qRTPCR were prepared with 1× SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and primers against Usp9x (Real TimePrimers.com # VHPS-9861). The primers were optimized for amplification under the following reaction conditions: denaturing at 95°C for 10 minutes followed by 40 cycles of 95°C, 15 seconds at 60°C, 1 minute. Melting curves were analyzed for all samples after completion of the amplification protocol. GAPDH was used as the housekeeping gene expression control.
Invasion Assay
In vitro invasion was carried out in triplicates using Matrigel Invasion Chambers (BD Biosciences) according to the manufacturer's procedure. Invaded cells were counted under an inverted microscope after 24 hours.
Xenograft Studies
NSG [NOD/SCID/IL2r-g (null)] mice were injected mid-dorsally with 2 × 106 8041 or 5 × 106 MIAPACA2 cells in 0.1 ml of Matrigel/DMEM suspension. Tumors were allowed to establish to about 100 mm3, after which mice were tumor size matched and allocated to five per treatment group (vehicle or G9) for 8041 tumor-bearing mice and four per treatment group for MIAPACA2 tumor-bearing mice. G9 was administered in DMSO:PEG300 (1:1) by i.p injection every other day at 15 mg/kg. Tumor size was monitored by caliper measurements twice a week, and tumor volume was calculated using the following formula: volume = (width)2 × length × height/2. Animal weight was also recorded twice weekly. The tumors were isolated, and a small portion was sliced, minced, and snap frozen with liquid nitrogen followed by homogenization in DUB assay buffer. The DUB activity was determined following the DUB assay protocol as described above.
Statistical Analysis
All graphs were plotted using Graph Pad Prizm 6.0 software. The statistical significance was calculated by unpaired Student’s t test analysis.
Results
Usp9x Overexpression Increases 3D Growth and Invasion of Human Pancreatic Cancer Cells
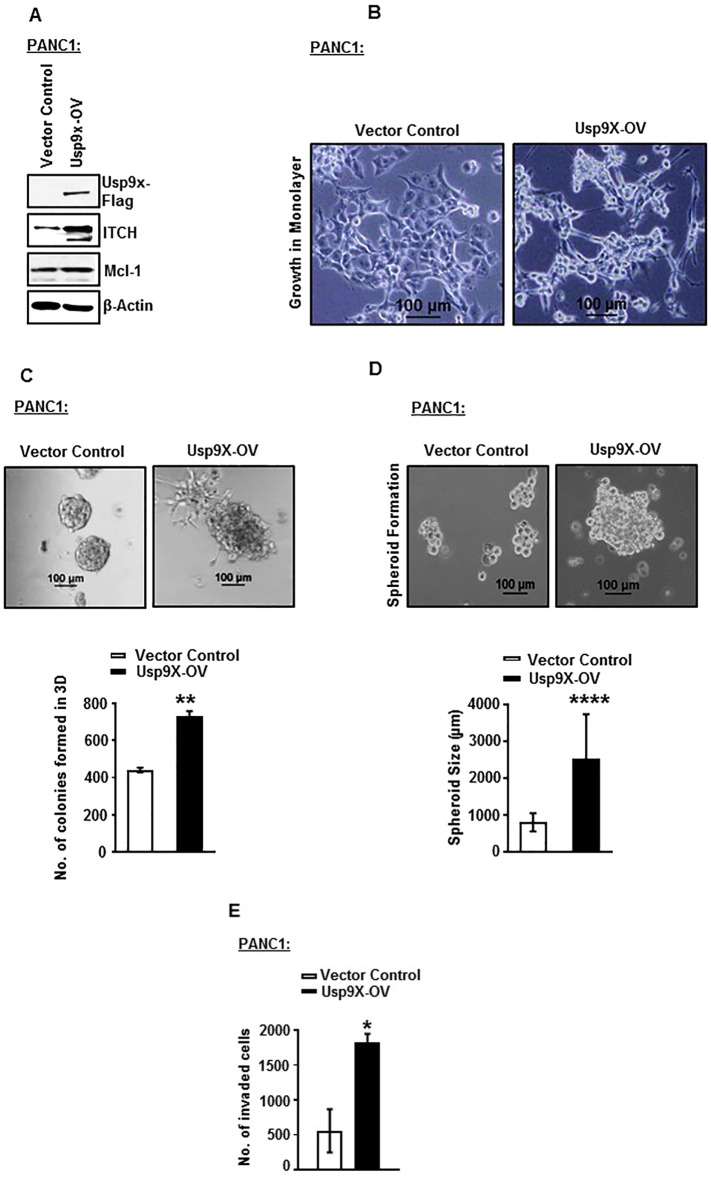
To examine the tumorigenic potential of Usp9x in human pancreatic cancer cells, we established a Flag-tagged-Usp9x overexpression (Usp9x-OV) system in the human PDAC cell line PANC1. We found that Flag-tagged-Usp9x overexpression induced the expression of two of the known cancer-associated proteins and substrates of Usp9x, ITCH, and Mcl-1 as compared to the vector control cells (Figure 1A, Supplementary Figure W1). Phenotypically PANC1-Usp9x-OV cells showed elongated spindle-like growth in monolayer, an increase in the size of spheroids grown in suspension culture on agarose; and a disorganized and malignant phenotype on 2% GFR Matrigel in 3D culture (Figure 1, B, C, and D). We also observed an increased number of colonies in 3D culture of PANC1-Usp9x-OV cells as compared to vector control cells (Figure 1C). Further, these PANC1-Usp9x-OV cells showed increased invasion with 3.5-fold increase in the invasion index in GFR Matrigel invasion assay as compared to the vector control cells (Figure 1E).
Figure 1.
Overexpression of Usp9x increases 3D colony formation and invasion of human PANC1 cells.
(A). Immunoblot analysis confirms expression of Flag-tagged Usp9x in human PDAC cell line PANC1. Usp9x-OV increases protein expression levels of Usp9x substrates ITCH and Mcl-1. β-Actin was used as the loading control.
(B) Phase contrast images show effect of Usp9x overexpression on the growth of PANC1 cells in monolayer. PANC1-Usp9x-OV cells show a fibroblast-like phenotype in monolayer cultures as compared to epithelial phenotype of PANC1-vector control cells.
(C) Phase contrast images show effect of Usp9x overexpression on the growth of PANC1 cells in 3D culture on 2% GFR Matrigel. PANC1-Usp9x-OV cells show a typical malignant phenotype with disruption of flat, round 3D colony growth as compared to PANC1-vector control cells. Representative column graph shows an increase in the number of colonies formed in 3D culture in PANC1-Usp9x-OV cells compared to PANC1-vector control cells.
**P < .005.
(D) Phase contrast images show increased spheroid size in suspension culture in PANC1-Usp9x-OV as compared with PANC1-vector control cells. Representative column graph shows an increase in the size of spheroids grown in suspension culture in PANC1-Usp9x-OV cells compared to PANC1-vector control cells. ****P < .00005.
(E) Representative column graph shows increased invasion in PANC1-Usp9x-OV cells through GFR Matrigel invasion chambers as compared to PANC1-vector control cells. *P < .05.
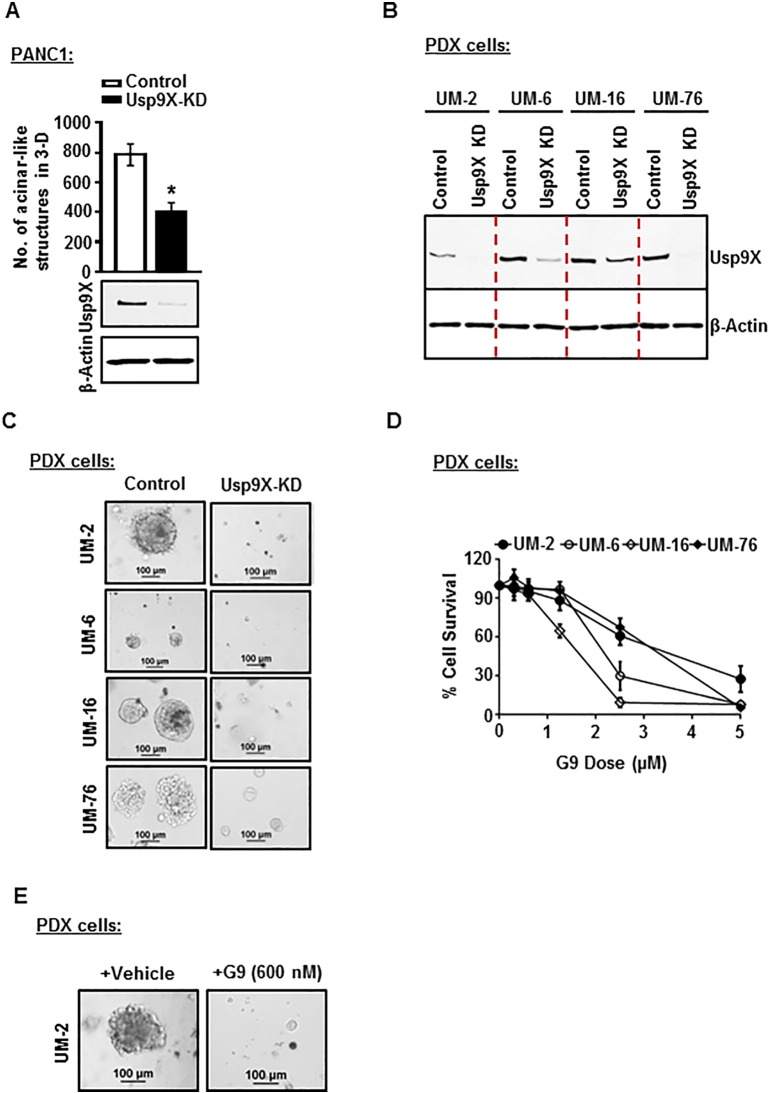
Usp9X Inhibition Inhibits 3D Colony Growth in Established and Patient Derived Human Pancreatic Cancer Cells
To assess the effect of Usp9x inhibition in human pancreatic cancers, Usp9x was first effectively knocked down in PANC1 cells by a previously described and characterized Usp9x targeted lentiviral-shRNA [16] (Figure 2A, Supplementary Figure W2). PANC1-Usp9x-KD cells showed decreased colony growth in 3D culture by 50% (Figure 2A). Adding clinical relevance to this observation, we also found a similar effect of Usp9x knockdown on four newly established human pancreatic cancer patient-derived (PDX) cell lines: UM2, UM6, UM16, and UM76. Usp9x KD in all four PDX cell lines completely inhibited 3D colony growth (Figure 2, B and C) as compared to their respective scrambled controls. Phenocopying this effect, we also found that treatment of UM-2, UM-6, UM-16, and UM-76 with Usp9x inhibitor G9 [30], [31] dose-dependently suppressed cell survival (Figure 2D), while 600 nM of G9 completely suppressed UM-2 3D colony growth when compared to untreated vehicle controls (Figure 2E).
Figure 2.
Inhibition of Usp9x inhibits 3D colony formation in established human PDAC cell line PANC1 and newly established human pancreatic patient-derived cell lines.
(A) Representative column graph shows a 50% decrease in number of colonies formed in 3D culture in PANC1-Usp9x-KD cells as compared to PANC1-vector control cells. *P < .05. Immunoblot analysis confirms Usp9x KD in PANC1 cells.
(B) Immunoblot analysis confirms Usp9x KD in 4 human pancreatic cancer cell lines derived from pancreatic tumor patient samples: UM-2, UM-6, UM-16, and UM-76.
(C) Phase contrast images of control and Usp9x-KD UM-2, UM-6, UM-16, and UM-76 cells grown on 2% GFR Matrigel in 3D culture for 8 days. UM-2-Usp9x KD, UM-6-Usp9x KD, UM-16-Usp9x KD, and UM-76-Usp9x KD show complete inhibition of colony formation in 3D culture as compared to respective scrambled control cells.
(D) MTT assay shows inhibition of cell growth and survival in UM-2, UM-6, UM-16, and UM-76 cells following treatment with small-molecule deubiquitinase inhibitor G9. Cells were grown in triplicates in 96-well culture clusters for 24 hours followed by treatment with serially diluted dose levels of G9. Cell growth/survival was determined after 48 hours of treatment. The average ± S.D. of triplicate assays is shown.
(E) Phase contrast images show inhibition of colony formation in G9-treated UM-2 cells. UM-2 cells were treated with vehicle or 600 nM G9 every 3 days while growing in 3D conditions for 8 days.
Usp9x Inhibition Promotes 3D Colony Growth Independent of Mutant Kras Activity in Mouse Pancreatic Tumor-Derived Cell Lines
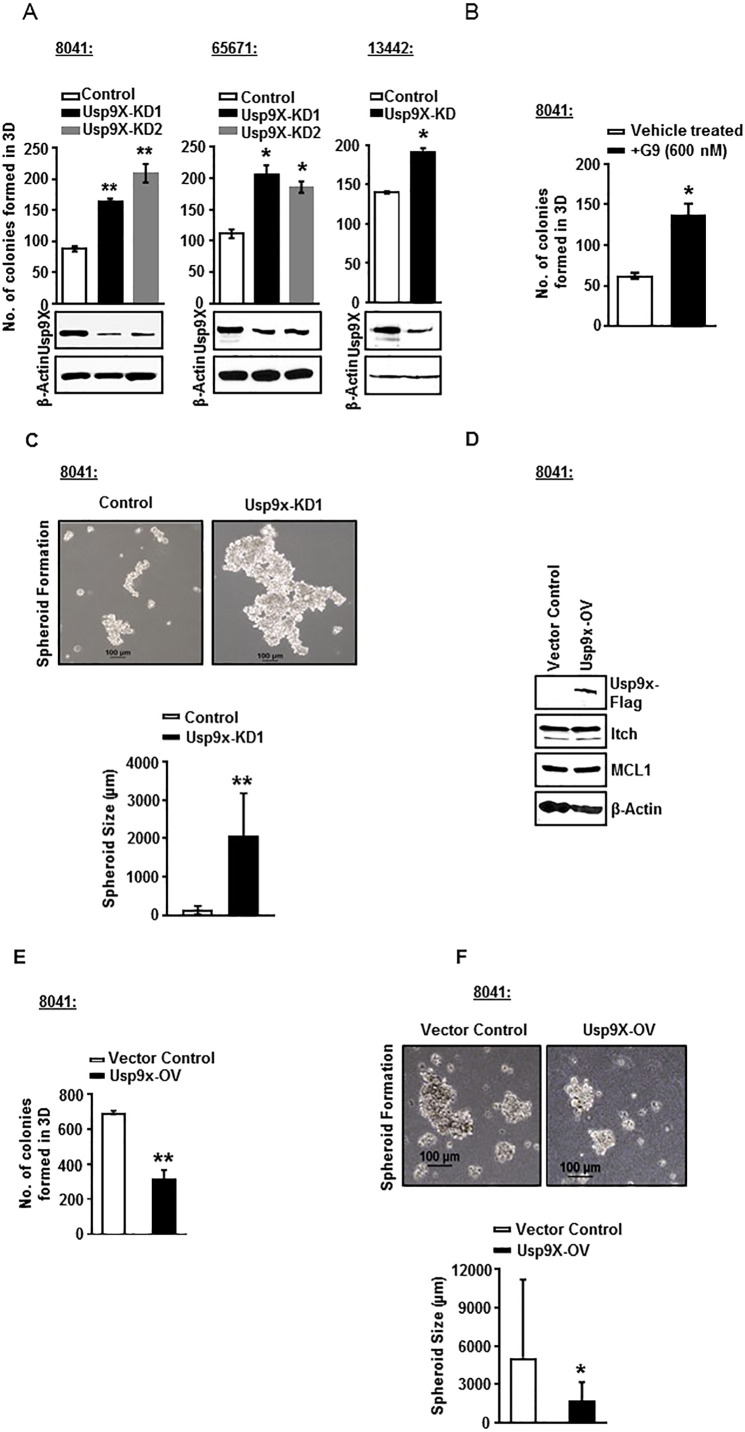
In order to clarify the role of Usp9x in mouse pancreatic cancer, we used a pancreatic cancer mouse model developed in the laboratory of Dr. Marina Pasca di Magliano. The 8041, 65671, and 13442 cell lines were established from mouse pancreatic tumors with constitutive expression of KrasG12D and Tp53R172H. The 4668 cell line was established from mice transgenic for inducible KrasG12D, which allows for DOX-inducible, pancreas-specific, and reversible expression of the oncogenic KrasG12D, with or without inactivation of one allele of the tumor suppressor gene p53 [11]. Two distinct Usp9x shRNA constructs induced ~70% to 80% Usp9x knockdown in all three murine pancreatic tumor cell lines (Figure 3A and Supplementary Figure W3). When equal numbers of 8041-Usp9x-KD1, 8041-Usp9x-KD2, 65671-Usp9x-KD1, 65671-Usp9x-KD2, and 13442-Usp9x-KD cells and their respective scrambled control cells were plated in 3D culture, all of the Usp9x-KD cells showed an increase in the number of colonies formed as compared to the scrambled control cells (Figure 3A). Similarly, treatment of 8041 cells with a low dose of Usp9x small molecule inhibitor G9 (600 nM) resulted in an increase in the number of colonies formed in 3D by two-fold (Figure 3B). In support, 8041-Usp9x-KD1 cells also showed increased spheroidal size as compared to 8041-control cells when grown in suspension on agarose plates (Figure 3C). Conversely, Flag-tagged overexpression of Usp9x in 8041 cells led to a decrease in the number of colonies formed in 3D and a reduction in the size of spheroidal structures formed in suspension culture on agarose plates (Figure 3, D, E, and F). Unlike the human pancreatic cancer cells, no change in the protein expression levels of Usp9x substrates Itch and Mcl-1 was observed in 8041-Usp9x-OV cells as compared to 8041-vector-control cells (Figure 3, D, E, and F and Supplementary Figure W4). These data show that in contrast to human pancreatic tumor cell line models, Usp9x suppresses cell growth, and its inhibition leads to increased colony formation in mouse pancreatic cancer cell lines.
Figure 3.
Usp9x KD promotes growth, while Usp9x overexpression reduces growth, in 3D culture of mutant KrasG12D pancreatic mouse xenograft cell lines with constitutive expression of KrasG12D and Tp53R172H.
(A) Immunoblot analysis confirms Usp9x KD in three mutant KrasG12D pancreatic mouse xenograft cell lines with constitutive expression of KrasG12D and Tp53R172H: 8041, 65671, and 13442. Representative column graphs show that 8041-Usp9x-KD1 and 8041-Usp9x-KD2, 65671-Usp9x-KD1 and 65671-Usp9x-KD2, and 13442-Usp9x-KD1 cells form increased numbers of colonies as compared to their respective scrambled control cells when grown on 2% Matrigel for 8 days in 3D culture. *P < .05, **P < .005.
(B) Representative column graph shows that treatment of 8041 cells with small-molecule inhibitor G9 at 600 nM lea to formation of an increased number of colonies as compared to vehicle-treated controls when grown on 2% Matrigel for 8 days in 3D culture. *P < .05.
(C) Phase contrast images show that 8041-Usp9x-KD1 cells form larger spheroidal structures as compared to scrambled control cells when grown in suspension culture on agarose plates. A total of 106 cells of 8041-control or 8041-Usp9x- KD1 cells were plated on agarose plates and allowed to grow in suspension for 5 days. Representative column graph shows an increase in the size of spheroids grown in suspension culture in 8041-Usp9x- KD1 cells as compared to 8041-control. **P < .005
(D) Immunoblot analysis confirms expression of Flag-tagged Usp9x in murine 8041 cells. The expression levels of ITCH and Mcl-1 remain unaltered in 8041-Usp9x-OV cells as compared to vector controls. β-Actin was used as the loading control.
(E) Representative column graph shows that 8041-Usp9x-OV cells form a decreased number of colonies as compared to 8041-vector control cells when grown on 2% Matrigel for 8 days in 3D culture.
(F) Phase contrast images show the effect of Usp9x overexpression on the growth of 8041 cells as spheroids in suspension culture. Usp9x expression in 8041 cells shows decreased growth as compared to vector controls. Representative column graph shows a decrease in the size of spheroids grown in suspension culture in 8041-Usp9x-OV cells as compared to 8041-Vector Control cells. **P < .005.
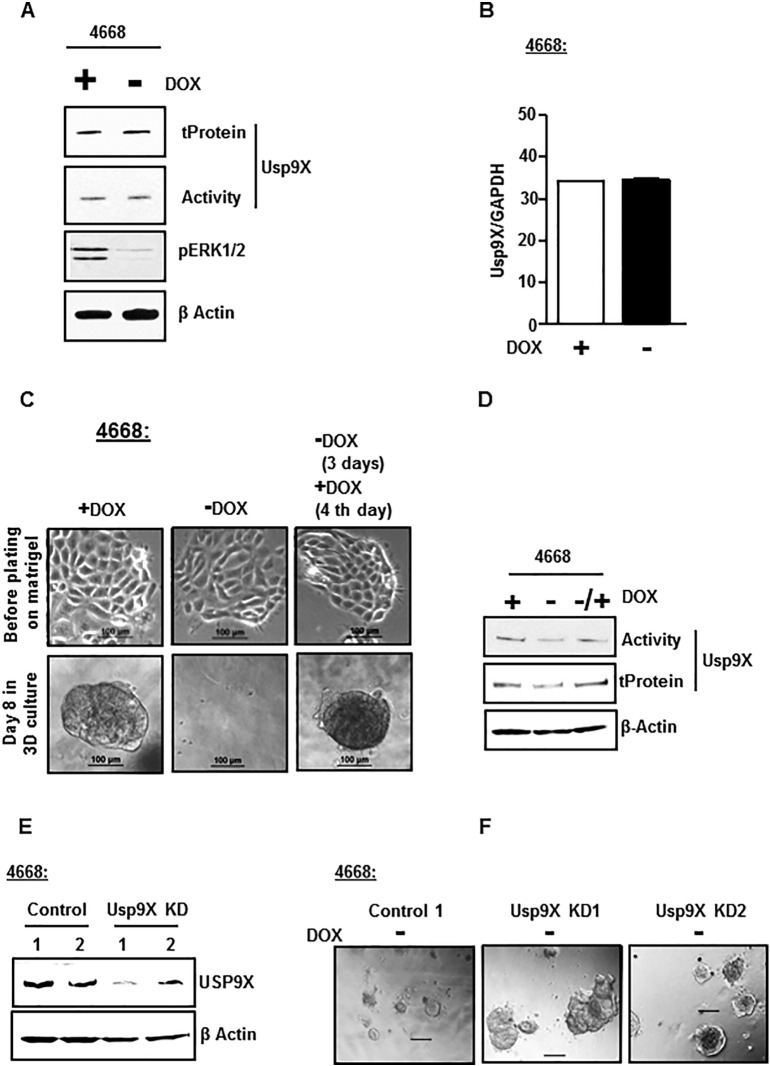
To further confirm the tumor suppressor role of Usp9x in mouse model of pancreatic cancer and to determine if the tumor suppressor function of Usp9x is dependent on mutant Kras, we first sought to determine whether the transcript or protein expression of Usp9x was regulated by KrasG12D activity. DOX depletion in KrasG12D inducible 4668 cells for 48 hours was sufficient to inhibit KrasG12D activity as measured by phospho-ERK1/2 expression levels (Figure 4A). However, the inhibition of KrasG12D did not alter Usp9x activity or expression as assessed by DUB activity assay and total protein levels assessed by immunoblot analysis (Figure 4A and Supplementary Figure W5), and the transcript levels assessed by RT-PCR analysis (Figure 4B). Thus, the Usp9x transcript and protein levels in these mouse pancreatic tumor cell lines were found to be independent of KrasG12D activity.
Figure 4.
Usp9x promotes growth in 3D culture independent of mutant Kras in the mutant KrasG12D pancreatic mouse xenograft cell line with inducible expression of KrasG12D and Tp53R172H.
(A) Immunoblot analysis shows no change in the activity and total protein levels of Usp9x in DOX depleted 4668 cells with withdrawn (−) KrasG12D support as compared to controls with sustained (+) DOX support. 4668 cells with DOX-inducible KrasG12D expression were DOX depleted for 48 hours followed by DUB activity assessment by DUB activity assay using HA-UbVS. The samples were subjected to immunoblot analysis with anti-HA antibody to detect HA-UbVS–labeled Usp9x (Activity). Inhibited phospho-ERK1/2 levels confirm withdrawal of KrasG12D support in DOX-depleted 4668 cells. β-Actin was used as the loading control.
(B) RT-PCR analysis of 4668 cells show no change in the transcript levels of Usp9x in the absence (−) or presence (+) of DOX (KrasG12D expression) for 48 hours. Average ± S.D. of triplicate assays is shown.
(C) Phase contrast images show that the inhibition of mutant KrasG12D activity mediated by DOX depletion in inducible 4668 mouse pancreatic tumor cells inhibits colony formation on 2% GFR Matrigel in 3D culture. The 3D colony formation was rescued by retreatment with DOX. No effect of DOX depletion was observed on cell growth in 2D monolayers. 4668 cells were grown with or without DOX as 2D monolayer (top) for 48 hours before plating on 2% Matrigel with continued or depleted DOX condition in 3D (bottom). At day 4, DOX depletion was rescued by adding DOX back for another 4 days. Phase contrast images were acquired at day 8 of 3D culture.
(D) Immunoblot analysis shows that Usp9x activity and protein levels are only nominally affected in 4668 cells in the sustained presence or absence of KrasG12D support by DOX addition or depletion.
(E) Immunoblot analysis confirms Usp9x KD in 4668-Usp9x-KD1 and 4668-Usp9x-KD2 cells as compared to scrambled 4668-control cells.
(F) Phase Contrast images show that DOX-depleted and Kras activity-inhibited 4668-Usp9x-KD1 and 4668-Usp9x-KD2 cells form aggressive colonies in 3D culture as compared to DOX-depleted 4668-control cells that show disintegration of colony formation.
Next, we established 3D cultures of the 4668 cell line. Growth on a reconstituted basement membrane like 2% GFR Matrigel is a physiologically more relevant in vitro system that captures key features of the tumorigenic phenotype not detectable with a simple monolayer culture. This 3D system can be used effectively to interrogate the malignant phenotype and to decipher key signaling pathways involved in cellular transformation [32]. The validity of the inducible system and the dependence of tumor formation on mutant KrasG12D expression were confirmed by growth in 3D culture. DOX-treated 4668 cells formed colonies in 3D culture which disintegrate following withdrawal of Kras support by DOX depletion. The colony-forming ability is recovered when Kras support is reestablished with DOX replenishment (Figure 4C). The activity and total protein levels of Usp9x were just nominally affected by sustained DOX depletion and replenishment (Figure 4D and Supplementary Figure W6). Next, the 4668 cells were subjected to lentiviral shRNA-mediated knockdown of Usp9x. The two Usp9x shRNA constructs induced >80% Usp9x knockdown (Figure 4E). Unlike the scrambled control colonies which disintegrate in the absence of DOX (as shown also in Figure 4C), DOX-depleted 4668-Usp9x KD cells with inhibited Kras activity form very aggressive colonies in 3D culture (Figure 4F), indicating that Usp9x inhibition can promote tumorigenicity even in the absence of persistent KrasG12D activity.
Usp9x Inhibition Reduces 3D Colony Growth in Mutant Kras Cell Line MIAPACA2 But Not in w/t BXPC3 Cells
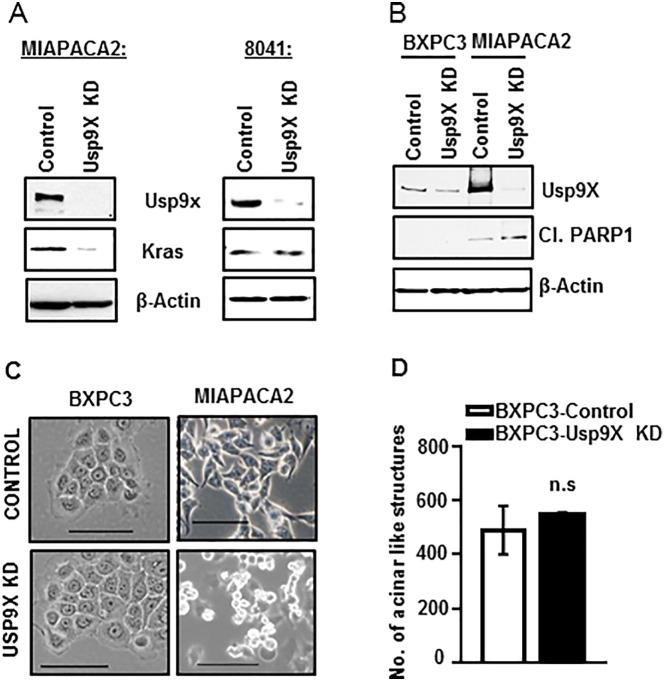
In our effort to assess whether Usp9x plays a growth promoter role in human pancreatic tumors independent of mutant Kras as we observed for mouse pancreatic cancer cell lines, we knock down Usp9x in Kras mutant human pancreatic cancer cell line MIAPACA2 using a previously described and characterized Usp9x-targeted lentiviral shRNA [16]. First, we examined Kras protein expression levels in MIAPACA2-Usp9x-KD versus 8041-Usp9x-KD cells. As shown previously (in Figure 3A), the murine pancreatic cancer 8041-Usp9x-KD cells have increased colony growth in suspension and 3D culture. Kras expression levels remained unaltered in 8041-Usp9x-KD cells as compared to 8041-control cell (Figure 5A and Supplementary Figure W7). Contrary to this observation, MIAPACA2-Usp9x-KD cells showed decreased Kras protein expression levels (Figure 5A and Supplementary Figure W7). We also observed that Usp9x KD decreased Mcl-1 levels in MIAPACA2 cells but increased Mcl-1 levels in 8041 cells (Supplementary Figure W8). Concomitantly, MIAPACA2-Usp9x-KD showed a rapid induction of apoptosis with PARP cleavage (Figure 5B). Phenotypic changes like nuclear fragmentation further confirmed apoptosis induction in MIAPACA2-Usp9xKD cells as compared to scrambled control cells (Figure 5C). However, Usp9x KD in the w/t Kras cell line BxPC3 did not show any effect on cellular phenotype and growth in 2D or 3D condition (Figure 5, B, C, and D and Supplementary Figure W9).
Figure 5.
Usp9x inhibition induces apoptosis in Kras mutant human cell line MIAPACA2, but Kras w/t BxPC3 cells remain unaffected.
(A) Immunoblot analysis shows that Usp9x KD in human mutant Kras PDAC cell line MIAPACA2 reduces mutant Kras expression levels, whereas Usp9x KD in the mouse mutant Kras cell line 8041 has no effect on Kras.
(B) Immunoblot analysis confirms Usp9x KD in human PDAC cell lines MIAPACA2 and BxPC3 with mutant Kras and w/t Kras, respectively. Usp9x KD in MIAPACA2 cells led to rapid apoptosis induction as indicated by PARP cleavage, but no such effect was observed in the w/t Kras cell line BxPC3.
(C) Phase contrast images further confirm apoptosis induction in MIAPACA2-Usp9x KD cells as compared to MIAPACA2-Control cells. No effect of Usp9x KD on the phenotype of BxPC3 cells grown in monolayer was observed.
(D) Representative column graph shows that BxPC3-Usp9x-KD cells show no effect on colony formation on 2% Matrigel in 3D culture when compared to BxPC3-Control cells. n.s., not significant.
Usp9x Inhibition Reduces Tumor Burden in Human MIAPACA2 Xenografts vs No effect in Mouse 8041 Xenografts
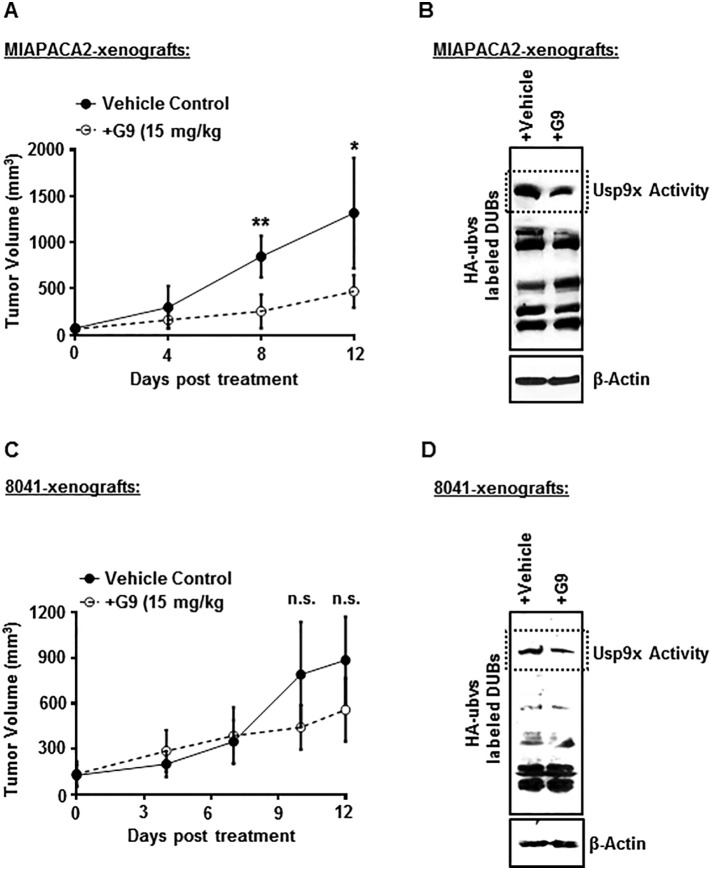
Our data established a distinct role for Usp9x in human PDAC as compared to genetically engineered mouse pancreatic cancer. This implied that Usp9x may promote tumorigenicity in human pancreatic cancer and its inhibition could potentially inhibit tumor growth. To examine this potential, we investigated the effect of G9 treatment on human MIAPACA2 tumor xenografts. We injected human MIAPACA2 cells subcutaneously into NSG mice. Primary tumor development was monitored by caliper measurements, and once measurable, mice were separated into two groups and were treated with either vehicle control (PEG300/DMSO) or G9 at 15 mg/kg. Tumor growth, animal weight, behavior, and mobility were monitored during treatment. In parallel, we also established murine 8041 tumors and subjected them to similar G9 treatment and tumor monitoring regimen as the human MIAPACA2 xenografts. Consistent with the in vitro findings, Usp9x inhibition resulted in the suppression of tumor growth in human tumor xenografts, but we did not observe any significant effect on the growth of 8041 tumors xenografts (Figure 6, A and C), although the Usp9x activity was inhibited effectively by G9 treatment in both human MIAPACA2 and mouse 8041 xenograft tumors (Figure 6, B and D). These results support the in vitro data shown here and strongly suggest that Usp9x inhibition can effectively reduce tumor burden in human pancreatic cancer. Weight loss was not significantly different between control and G9-treated mice in either MIAPACA2 or 8041 xenografts (Supplementary Figure W10).
Figure 6.
Small-molecule inhibitor G9 decreases tumor size in human MIAPACA2 xenografts but has no effect on murine 8041 xenografts.
(A) G9 treatment significantly reduces tumor size in human MIAPACA2 xenografts. N = 4 for each group. *P < .05, **P < .005. A total of 5 × 106 MIAPACA2 cells were injected subcutaneously, mid-dorsally in NSG mice. When measurable, tumors were size matched and treated i.p. with either vehicle or 15 mg/kg of G9 every other day for 12 days. Tumor size was monitored, and tumor measurements were recorded.
(B) Immunoblot analysis shows inhibited activity of Usp9x in G9-treated human MIAPACA2-xenograft tumors. Tumor tissue was extracted from vehicle- or G9-treated mice and lysed in DUB assay buffer before incubation with HA-UbVS and immunoblot analysis with anti-HA antibody to detect HA-UbVS–labeled DUBs as a measure of DUB activity (Usp9x activity is denoted). β-Actin was used as the loading control.
(C) G9 treatment showed no significant effect on growth of murine 8041 xenograft tumors. Tumors were established with 2 × 106 8041 cells as described in A. N = 5 for each group.
(D) Immunoblot analysis shows inhibition of Usp9x activity in G9-treated murine 8041-xenograft tumors. β-Actin was used as the loading control.
Discussion
Usp9x targeting agents may have therapeutic potential in the treatment of a broad range of tumors, and it has been predicted that a small-molecule Usp9x inhibitor will have a major impact on cancer therapy. We have been interested in developing specific and pan-deubiquitinase small molecule inhibitors with activity against Usp9x. Continuing our efforts, we recently reported a new compound, EOAI3402143 or G9, with greater solubility and inhibitory activity against Usp9x than our original WP1130 compound [30], [31]. Considering the broad applicability of Usp9x inhibitors as anticancer agents, it is imperative to examine the utility of G9 as a therapeutic agent in the treatment of pancreatic cancer, a deadly disease with very poor prognosis and few treatment options.
Since the role of Usp9x in human pancreatic cancers has not yet been fully defined, we examined whether Usp9x has tumor-promoting activity in human pancreatic cancers, as has been reported for various other human cancers [14], [15], [16], [17], [18], [19]. Our data clearly show a tumor promoter function for Usp9x in cell-based assays. Usp9x expression increases cell survival and 3D growth, and induces invasion in human PDAC cell line PANC1. ShRNA-mediated Usp9x knockdown suppresses 3D growth in PANC1 cells and induces apoptosis in MIAPACA2 cells. Adding clinical relevance to our findings, Usp9x KD completely diminished 3D growth in patient-derived PDX cells. G9-mediated inhibition of Usp9x in pancreatic cancer cells not only reduced the in vitro 3D growth and invasion but also inhibited tumor growth of human pancreatic cancer MIAPACA2 cells in vivo.
Although these studies implicate Usp9x as a therapeutic target in the treatment of human pancreatic cancer, they also need to be cautiously interpreted in the light of the reported tumor suppressive role of Usp9x in genetically engineered mouse models of PDAC [13]. An enquiry into the role of Usp9x in murine cell lines using cell lines established from mouse pancreatic tumors with constitutive or inducible expression of KrasG12D and Tp53R172H supports a tumor suppressor role for Usp9x in that its absence maintains or amplifies mouse pancreatic tumors even when KrasG12D contribution is attenuated. Dox withdrawal de-induced the activity of KrasG12D in 4668 cells leading to regression of the colonies formed in 3D culture. Following colony regression, it is possible that a few surviving cells stayed as monolayer, and when KrasG12D activity was re-induced with Dox replacement or Usp9x was inhibited by shRNA mediated knockdown, colonies reestablish. In a previous study using a model system to mimic oncogene dependence in 3D culture, Jechlinger et al. showed that Dox-dependent induction of mutant Kras can cause single cells to develop into solid, polarized acini [33]. Upon de-induction of KrasG12D, the solid structures regress, leaving a repolarized monolayer of viable cells. These cells were found to display a phenotype consistent with progenitors of mammary epithelium; they excluded Hoechst dye 33342, and reformed acini in 3D cultures and repopulated mammary fat pads more efficiently than cells harvested from uninduced acini [33]. It is quite possible that 4668 cells act in a similar manner in response to de-induction and re-induction of KrasG12D. As a result of de-induction, the murine PDAC cells acquire progenitor characteristics where Usp9x acts to suppress tumor growth but Usp9x inhibition promotes tumorigenesis even in the absence of KrasG12D. Though it is beyond the scope of current publication, it will be important to further investigate if induction and then de-induction of Kras can cause similar phenotypic changes in murine pancreatic cells as has been reported in mouse mammary epithelial cells [33], and to further assess whether these acquired characteristics are Usp9x activity dependent. Considering this, it is not surprising that, unlike human MIAPACA2 tumors which showed reduced tumor burden in response to G9 treatment, mouse 8041 tumors showed no significant effect on tumor burden. Thus, G9 treatment inhibits tumor growth only in human pancreatic tumors and not those attempting mimicry in mouse pancreatic tumor models.
Usp9x may suppress tumor progression in a very specific context during early stages of tumorigenesis, before the tumor is established. Defining that context may be essential to understanding pancreatic tumor development in a genetically engineered mouse model. Since development of pancreatic tumors requires “reprogramming” or dedifferentiation, Usp9x-mediated deubiquitination within that program may underlie the distinctions noted in Usp9x facilitating or disengaging tumorigenicity. In addition to consideration given to the potential role of Usp9x in the reprogramming of pancreatic premalignancy, additional consideration may be applied to potential species specificity target distinctions as well as partial functional dependence on Kras mutational status. Usp9x KD did not affect the Kras levels in mouse 8041 cells but reduced Kras levels in human MIAPACA2 cells. Similar distinction was also observed for Mcl-1, one observed substrate of Usp9x, where Usp9x KD increased Mcl-1 levels in 8041 cells but decreased Mcl-1 in MIAPACA2 cells. In addition to these apparent species-dependent differences, context- as well as Kras status–dependent distinctions were also seen within human cell lines. Usp9xKD in mouse 8041 cells showed no change in the growth of cells as monolayer in the standard 2D system (Supplementary Figure W11), but 3D growth was increased. In human PDAC Usp9x KD induced apoptosis in mutant kras homozygous condition (in MIAPACA2) while heterozygous Kras mutant (PANC1) cells showed more rapid (38%) growth in 2D and reduced colony formation in 3D culture (>50%). Usp9x inhibition in the wild-type Kras BxPC3 cells showed no effect on growth in 2D or in 3D culture. Thus, the presence of a wild-type Kras allele may influence Usp9x function.
Usp9x has a clear role in tumor promotion in human pancreatic cancers which is clearly distinct from activity in murine models. Considering the ever-increasing number of substrates that have been reported for Usp9x and the associated cellular and physiological functions regulated by Usp9x-mediated deubiquitination, these findings substantiate how dynamically Usp9x orchestrates its influence over various biological processes.
In conclusion, our results highlight that, in advanced human pancreatic cancers, Usp9x promotes tumorigenicity. Thus, inhibition of Usp9x in established pancreatic tumors, particularly in those with elevated Usp9x expression, may provide a therapeutic opportunity. However, caution must also be exercised in treating other forms of cancer as loss of Usp9x suppressor function may promote cryptic pancreatic tumors with preexisting genetic lesions. Additional monitoring and application of additional chemotherapy (i.e., gemcitabine) may alleviate major concern in targeting Usp9x in a therapeutic setting.
Financial Support
We would like to acknowledge financial support by the Leukemia Lymphoma Society Therapeutic Acceleration Program (N.D.), the John S. & Suzanne C. Munn Cancer Research Fund of the University of Michigan Comprehensive Cancer Center (N.D.), Allen H. Blondy Research Fund for Melanoma (M.T., H.P.), The Harry J. Lloyd Charitable Trust (N.D.), and the Michigan Translational Research and Commercialization (MTRAC) program (N.D.).
Acknowledgement
We would like to thank the patients who provided specimens for establishment of the PDX cell lines and agreed to be part of an IRB-approved study. We also thank Dr. Dzwokai Ma (University of California, Santa Barbara) for kindly providing vectors used in this study. We would also like to thank Yi Liu for assistance with animal studies.
Footnotes
Disclaimer: This article was prepared while Nicholas J. Donato was employed at the University of Michigan. The opinions expressed in this article are the author's own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2017.11.007.
Appendix A. Supplementary data
Supplementary Figures
References
- 1.Muniraj T, Jamidar PA, Aslanian HR. Pancreatic cancer: a comprehensive review and update. Dis Mon. 2013;59(11):368–402. doi: 10.1016/j.disamonth.2013.08.001. https://doi.org/10.1016/j.disamonth.2013.08.001 Epub 2013/11/05. [S0011-5029(13)00153-3 [pii], PubMed PMID: 24183261] [DOI] [PubMed] [Google Scholar]
- 2.Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7(4):266–273. [Epub 2001/09/20] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [PubMed PMID: 23335087] [DOI] [PubMed] [Google Scholar]
- 4.Ducreux M, Boige V, Goere D, Deutsch E, Ezra P, Elias D, Malka D. The multidisciplinary management of gastrointestinal cancer. Pancreatic cancer: from pathogenesis to cure. Best Pract Res Clin Gastroenterol. 2007;21(6):997–1014. doi: 10.1016/j.bpg.2007.10.025. https://doi.org/10.1016/j.bpg.2007.10.025 Epub 2007/12/12. doi: S1521-6918(07)00132-1 [pii] [PubMed PMID: 18070700] [DOI] [PubMed] [Google Scholar]
- 5.Westphalen CB, Olive KP. Genetically engineered mouse models of pancreatic cancer. Cancer J. 2012;18(6):502–510. doi: 10.1097/PPO.0b013e31827ab4c4. https://doi.org/10.1097/PPO.0b013e31827ab4c4 Epub 2012/11/29. [00130404-201212000-00005 [pii], PubMed PMID: 23187836; PMCID: 3594661] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. https://doi.org/10.1101/gad.1415606 Epub 2006/05/17. doi: 20/10/1218 [pii] [PubMed PMID: 16702400] [DOI] [PubMed] [Google Scholar]
- 7.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. https://doi.org/10.1126/science.1164368 Epub 2008/09/06. [1164368 [pii], PubMed PMID: 18772397; PMCID: 2848990] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143(2):545–554. [Epub 1993/08/01, PubMed PMID: 8342602; PMCID: 1887038] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. https://doi.org/10.1101/gad.1158703 Epub 2003/12/19. [1158703 [pii], PubMed PMID: 14681207; PMCID: 305262] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [Epub 2004/01/07, doi: S153561080300309X [pii], PubMed PMID: 14706336] [DOI] [PubMed] [Google Scholar]
- 11.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653. doi: 10.1172/JCI59227. https://doi.org/10.1172/JCI59227 Epub 2012/01/11. [59227 [pii], PubMed PMID: 22232209; PMCID: 3266788] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Mol Oncol. 2013;7(2):232–247. doi: 10.1016/j.molonc.2013.02.002. https://doi.org/10.1016/j.molonc.2013.02.002 Epub 2013/03/20. [S1574-7891(13)00026-4 [pii], PubMed PMID: 23506980] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grutzmann R, Aust D, Rummele P. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486(7402):266–270. doi: 10.1038/nature11114. https://doi.org/10.1038/nature11114 Epub 2012/06/16. [nature11114 [pii], PubMed PMID: 22699621; PMCID: 3376394] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, Australian Pancreatic Cancer Genome I Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109(16):5934–5941. doi: 10.1073/pnas.1202490109. [PubMed PMID: 22421440; PMCID: PMC3341075] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–107. doi: 10.1038/nature08646. [PubMed PMID: 20023629] [DOI] [PubMed] [Google Scholar]
- 16.Sun H, Kapuria V, Peterson LF, Fang D, Bornmann WG, Bartholomeusz G, Talpaz M, Donato NJ. Bcr-Abl ubiquitination and Usp9x inhibition block kinase signaling and promote CML cell apoptosis. Blood. 2011;117(11):3151–3162. doi: 10.1182/blood-2010-03-276477. [PubMed PMID: 21248063] [DOI] [PubMed] [Google Scholar]
- 17.Cox JL, Wilder PJ, Gilmore JM, Wuebben EL, Washburn MP, Rizzino A. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. PLoS One. 2013;8(5):e62857. doi: 10.1371/journal.pone.0062857. [PubMed PMID: 23667531; PMCID: 3647065] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng J, Hu Q, Liu W, He X, Cui L, Chen X, Yang M, Liu H, Wei W, Liu S. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn Pathol. 2013;8(1):177. doi: 10.1186/1746-1596-8-177. https://doi.org/10.1186/1746-1596-8-177 [PubMed PMID] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Kollipara RK, Srivastava N, Li R, Ravindranathan P, Hernandez E, Freeman E, Humphries CG, Kapur P, Lotan Y. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc Natl Acad Sci U S A. 2014;111(11):4251–4256. doi: 10.1073/pnas.1322198111. https://doi.org/10.1073/pnas.1322198111 Epub 2014/03/05. [1322198111 [pii], PubMed PMID: 24591637; PMCID: 3964108] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104(1):21–30. doi: 10.1007/s10549-006-9393-7. [PubMed PMID: 17004105] [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Cai T-Y, Zhu H, Yang L-Q, Jiang H, Dong X-W, Hu Y-Z, Lin N-M, He Q-J, Yang B. Synergistic antitumor activity of gemcitabine and ABT-737 in vitro and in vivo through disrupting the interaction of USP9X and Mcl-1. Mol Cancer Ther. 2011;10(7):1264–1275. doi: 10.1158/1535-7163.MCT-10-1091. [DOI] [PubMed] [Google Scholar]
- 22.Trivigno D, Essmann F, Huber SM, Rudner J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia. 2012;14(10):893–904. doi: 10.1593/neo.12598. [PubMed PMID: 23097624; PMCID: 3479835] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potu H, Peterson LF, Kandarpa M, Pal A, Sun H, Durham A, Harms PW, Hollenhorst PC, Eskiocak U, Talpaz M. Vol. 8. 2017. Usp9x regulates Ets-1 ubiquitination and stability to control NRAS expression and tumorigenicity in melanoma; p. 14449. [https://www.nature.com/articles/ncomms14449#supplementary-information] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peddaboina C, Jupiter D, Fletcher S, Yap JL, Rai A, Tobin RP, Jiang W, Rascoe P, Rogers MK, Smythe WR. The downregulation of Mcl-1 via USP9X inhibition sensitizes solid tumors to Bcl-xl inhibition. BMC Cancer. 2012;12:541. doi: 10.1186/1471-2407-12-541. [PubMed PMID: 23171055; PMCID: 3543233] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris DR, Mims A, Bunz F. Genetic disruption of USP9X sensitizes colorectal cancer cells to 5-fluorouracil. Cancer Biol Ther. 2012;13(13):1319–1324. doi: 10.4161/cbt.21792. [PubMed PMID: 22895071; PMCID: 3493440] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins MA, Brisset JC, Zhang Y, Bednar F, Pierre J, Heist KA, Galban CJ, Galban S, di Magliano MP. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One. 2012;7(12):e49707. doi: 10.1371/journal.pone.0049707. https://doi.org/10.1371/journal.pone.0049707 Epub 2012/12/12. [PONE-D-12-09212 [pii], PubMed PMID: 23226501; PMCID: 3513322] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson LF, Mitrikeska E, Giannola D, Lui Y, Sun H, Bixby D, Malek SN, Donato NJ, Wang S, Talpaz M. p53 stabilization induces apoptosis in chronic myeloid leukemia blast crisis cells. Leukemia. 2011;25(5):761–769. doi: 10.1038/leu.2011.7. [DOI] [PubMed] [Google Scholar]
- 28.Lo MC, Peterson LF, Yan M, Cong X, Hickman JH, Dekelver RC, Niewerth D, Zhang DE. JAK inhibitors suppress t(8;21) fusion protein-induced leukemia. Leukemia. 2013;27(12):2272–2279. doi: 10.1038/leu.2013.197. https://doi.org/10.1038/leu.2013.197 Epub 2013/07/03. [leu2013197 [pii], PubMed PMID: 23812420; PMCID: 3987672] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [PubMed PMID: 12798140] [DOI] [PubMed] [Google Scholar]
- 30.Peterson LF, Sun H, Liu Y, Potu H, Kandarpa M, Ermann M, Courtney SM, Young M, Showalter HD, Sun D. Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood. 2015;125(23):3588. doi: 10.1182/blood-2014-10-605584. [DOI] [PubMed] [Google Scholar]
- 31.Potu H, Peterson LF, Kandarpa M, Pal A, Sun H, Durham A, Harms PW, Hollenhorst PC, Eskiocak U, Talpaz M. Usp9x regulates Ets-1 ubiquitination and stability to control NRAS expression and tumorigenicity in melanoma. Nat Commun. 2017;8:14449. doi: 10.1038/ncomms14449. [PubMed PMID: 28198367; PMCID: PMC5316860] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal A, Kleer CG. Three dimensional cultures: a tool to study normal acinar architecture vs. malignant transformation of breast cells. J Vis Exp. 2014;86 doi: 10.3791/51311. https://doi.org/10.3791/51311 Epub 2014/05/07. [PubMed PMID: 24797513] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jechlinger M, Podsypanina K, Varmus H. Regulation of transgenes in three-dimensional cultures of primary mouse mammary cells demonstrates oncogene dependence and identifies cells that survive deinduction. Gene Dev. 2009;23(14):1677–1688. doi: 10.1101/gad.1801809. [PubMed PMID: WOS:000268011800010] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures