Abstract
Recent studies have shown the effect of microRNAs on HSC activation and transformation, which is essential for the pathogenesis of liver fibrosis. In our study, we explored the role of miR-185 in liver fibrosis. Plasma miR-185 was detected in hepatitis B virus-related liver fibrosis patients (S2/3, n = 10) by Illumina HiSeq sequencing, and healthy volunteers were selected (n = 8) as the control group. We found that the plasma miR-185 level in fibrosis patients was significantly downregulated. CCl4-induced fibrosis tissues in mouse livers and TGF-β1-activated HSCs also presented downregulated miR-185 concomitant with an increased expression of RHEB and RICTOR. To explore the correlations, LX-2 cells were transiently transfected with miR-185 mimics. The expression levels of α-SMA, collagen I, and collagen III were decreased as well as RHEB and RICTOR. Inhibition of endogenous miR-185 increased fibrogenic activity. Furthermore, dual-luciferase reporter assays indicated that miR-185 inhibited the expression of RHEB and RICTOR by directly targeting their 3′ UTRs. Moreover, silencing RHEB and RICTOR suppressed α-SMA and collagen expression levels. In conclusion, miR-185 prevents liver fibrogenesis by inhibiting HSC activation via inhibition of RHEB and RICTOR. These results provide new insights into the mechanisms behind the anti-fibrotic effect of miR-185.
Keywords: liver fibrosis, microRNA-185, RHEB, RICTOR, TGF-β1, HSC, mTOR pathway, extracellular matrix
Introduction
Liver fibrosis, a major characteristic of most chronic liver diseases, develops as a wound-healing response in the liver to cellular injury, reflecting the balance between liver repair and scar formation.1 With protracted damage, fibrosis can progress toward excessive scarring and organ failure, as in liver cirrhosis and primary liver cancer.2 During this process, the fibrous extracellular matrix (ECM) in the liver accumulates and increases. Various cell types are considered sources of ECM in liver fibrosis; however, among these, hepatic stellate cells (HSCs) are the major ECM-producing cells after liver injury.3, 4, 5 HSCs are present in the space of Disse in the liver and primarily store vitamin A in a quiescent or normal state. After activation, HSCs become proliferative and lead to increased expression of α-smooth muscle actin (α-SMA), loss of their vitamin A content, and acquisition of a myofibroblast-like phenotype (MF-HSC). They then lose their typical star shape and begin to produce excess ECM. This imbalance between ECM degradation and accumulation results in liver fibrosis.
Although liver fibrosis is a reversible disease, and effective treatment could prevent or reverse the fibrotic process, a drug has not yet been developed for treatment, and there is no sound diagnostic biomarker; therefore, treatment options are limited.4 A liver biopsy is the gold standard for identifying liver fibrosis, but the procedure is invasive and painful and carries the risk of complications. It is important that the process of fibrogenesis be determined to be able to develop effective antifibrotic therapies.
MicroRNAs (miRNAs) are short, regulating RNA molecules that interfere with gene expression at the post-transcriptional level by arresting gene translation, which, in turn, reduces or prevents protein synthesis.6, 7 Because of their gene-regulating ability, miRNAs are attractive therapeutic targets. Panels of misregulated miRNAs have been observed in a variety of human diseases,8 including liver fibrosis,9, 10, 11 suggesting the importance of maintaining homeostasis of miRNA expression.
miR-185, which was first described as a regulator of cancer progression, is located at chromosome 22q11.21. Accumulating evidence has since revealed the connection between miR-185 and liver cancer. Zhi et al.12 used an miRNA array profile to report that miR-185 is associated with venous metastasis of hepatocellular carcinoma (HCC) and then further investigated the clinical and pathological significance and prognostic value of miR-185 in early-stage HCC. The expression of miR-185 is decreased in human HCC tissues compared with non-neoplastic liver parenchyma. Qadir et al.13 showed that miR-185 is a tumor suppressor miRNA that inhibits HCC cell growth in vitro and in severe combined immunodeficiency mice by targeting the DNMT1/PTEN/Akt signaling pathway. We found that miR-185 induces potent autophagy in HCC by directly interacting with 3′ UTRs of the Ras homolog enriched in brain (RHEB) and the rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR) mRNAs14.
RICTOR and RHEB play important roles in regulating the mammalian target of rapamycin complex (mTORC1 and mTORC2) pathways. In the past decade, several studies have demonstrated that mTORC1 and mTORC2 participate in the process of transforming growth factor β1 (TGF-β1)-induced fibrogenesis in addition to canonical SMAD signaling15, 16, 17. Recent studies have shown that attenuated expression of miR-185 might be responsible for idiopathic pulmonary fibrosis,18, 19 hypertrophic scarring,20 and dilated cardiomyopathy (DCM);21 however, the relationship between miR-185 and liver fibrosis remains unclear, and there are still many unanswered questions regarding the roles of RHEB and RICTOR in liver fibrosis.
This study set out to assess whether aberrant expression of miR-185 exists in liver fibrosis. We found that miR-185 was significantly downregulated in the plasma of hepatitis B virus (HBV)-related liver fibrosis patients and in liver tissues of carbon tetrachloride (CCl4)-induced mouse fibrosis; therefore, the therapeutic potential for miR-185 in liver fibrogenesis was characterized in vitro by its overexpression or inhibition in HSCs. We revealed that miR-185 prevents hepatic fibrogenesis by attenuating HSC activation. In particular, we demonstrated that RHEB and RICTOR are direct targets of miR-185 in HSCs and that they are responsible for HSC activation and liver fibrosis.
Results
miR-185 Is Downregulated in the Plasma of Patients with HBV-Related Liver Fibrosis
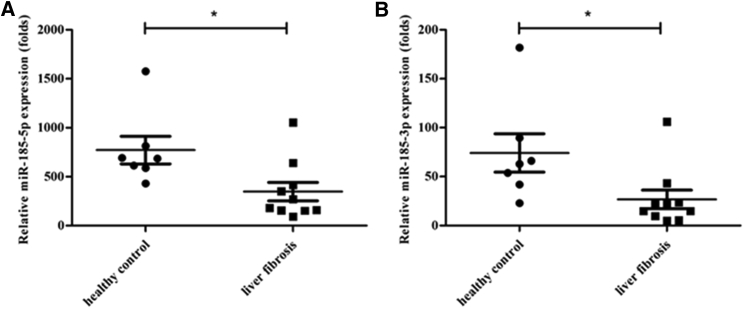
We first assessed the expression levels of miR-185 in different groups of human plasma, HBV-related liver fibrosis (n = 10), and healthy control (n = 8) by Illumina HiSeq sequencing. The clinical characteristics of the subjects are shown in Table 1. Cluster analysis of differentially expressed miRNAs was conducted. Compared with the healthy control group, 104 miRNAs were screened out from the liver fibrosis group, 72 miRNAs were upregulated, and 32 were downregulated. miR-185 was one of the 32 downregulated miRNAs. Plasma miR-185-5p (Figure 1A) and miR-185-3p (Figure 1B) expression levels were significantly decreased in fibrotic patients compared with those in the healthy controls (p = 0.02 and p = 0.0305, respectively). Because miR-185-5p is abundant and plays main functions compared with miR-185-3p, we selected miR-185-5p for further study.
Table 1.
Clinical Characteristics of the Three Groups
| Parameter | Fibrosis Group (n = 10) | Cirrhosis Group (n = 8) | Healthy Controls Group (n = 8) |
|---|---|---|---|
| Age (years) | 37.38 ± 3.30 | 41.20 ± 3.43 | 45.42 ± 3.46 |
| Sex (male/female) | 7/3 | 5/3 | 5/3 |
| ALT (U/L) | 38.5 (24.68, 55.3) | 33.85 (20.20, 45.83) | <50.00 |
| AST (U/L) | 30.65 (24.75, 38.0) | 36.75 (27.55, 42.23) | <35.00 |
| HBV-DNA (IU/mL) | 2.47E+5 (3.94E+4,6.78E+5) | 1.79E+6 (5.48E+3,1.38E+8) | – |
| HBeAg+ (%) | 50 | 75 | – |
Figure 1.
miR-185 Is Downregulated in the Plasma of Patients with HBV-Related Liver Fibrosis
The expression of miRNAs in the plasma of patients (n = 10) and control (n = 8) groups was detected by Illumina HiSeq sequencing. (A) Levels of miR-185-5p were significantly lower in the patient group than in the healthy group. (B) The expression of miR-185-3p was downregulated compared with the control group. (*p < 0 0.05, **p < 0 0.01).
miR-185 Is Abundant in LX-2 Cells but Downregulated in TGF-β1-Activated LX-2 Cells
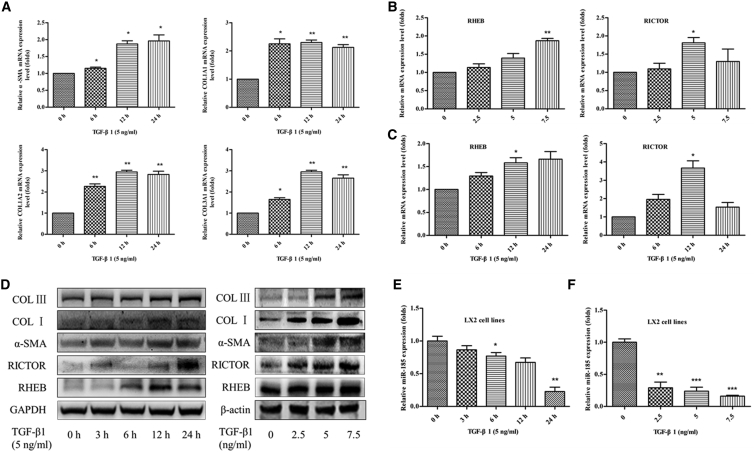
Activated HSCs secrete TGF-β1, the most potent fibrogenetic factor, to induce collagen production and ECM accumulation; therefore, we used recombinant human TGF-β1 protein to treat human HSC cell line LX-2 cells and then examined the expression levels of miR-185 and the key genes involved in the activation of HSCs, including α-SMA, collagen type I (COL I), and collagen type III (COL III) (Figures 2A and 2D). LX-2 cells were treated with 5.0 ng/mL TGF-β1 for 0, 3, 6, 12, or 24 hr, respectively. We found that miR-185 was abundant in LX-2 cells but significantly decreased in TGF-β1-activated LX-2 cells in a time-dependent manner (Figure 2E).We also observed a similar decrease in miR-185 expression in a dose-dependent manner when LX-2 cells were treated with different amounts of TGF-β1 for 24 hr (Figure 2F). The upregulated expression of α-SMA, COL I, and COL III levels confirmed that TGF-β1 could further stimulate LX-2 cell activation and that the cytokine TGF-β1 has a pivotal role in promoting liver fibrosis. During the process of HSC activation, the mRNA and protein expression of RICTOR and RHEB also remarkably increased when TGF-β1 further stimulates LX-2 cell activation (Figures 2B–2D). The expression of RHEB was upregulated in a time- and dose-dependent manner, and the highest expression of RICTOR was at a dose of 5 nM after added TGF-β1 for 12 hr. Thus, we hypothesized that miR-185, RHEB, and RICTOR might participate in regulating liver fibrosis.
Figure 2.
miR-185 Is Abundant in LX-2 Cells but Downregulated in Activated LX-2 Cells
The human hepatic stellate cell (HSC) cell line LX-2 was treated with 5.0 ng/mL transforming growth factor β-1 (TGF-β1) for 0, 3, 6, 12, or 24 hr or with 2.5, 5.0, or 7.5 ng/mL TGF-β1 for 24.0 hr. (A) mRNA expression of key genes involved in the activation of HSC α-SMA, COL1A1, COL1A2, and COL3A1 was analyzed by real-time PCR. All genes increased in a time-dependent manner. mRNA levels of Ras homolog enriched in brain (RHEB) and rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR) were assessed by real-time PCR. When HSCs were activated by TGF-β1 with a different dose (B) or time (C), RHEB and RICTOR were upregulated in mRNA levels. (D) Protein expressions of α-SMA, COL I, COL III, RHEB, and RICTOR was determined by western blotting; GAPDH or β-actin was used as a loading control. When HSCs were activated, all genes were upregulated in protein levels. miR-185-5p expression was reduced in activated HSCs in a dose-dependent manner (E) or time-dependent manner (F) examined by real-time PCR (n = 3, *p < 0 0.05, **p < 0.01). Data represent mean ± SEM.
miR-185 Suppressed HSC Activation and Basal/TGF-β1-Induced ECM Production of LX-2 Cells
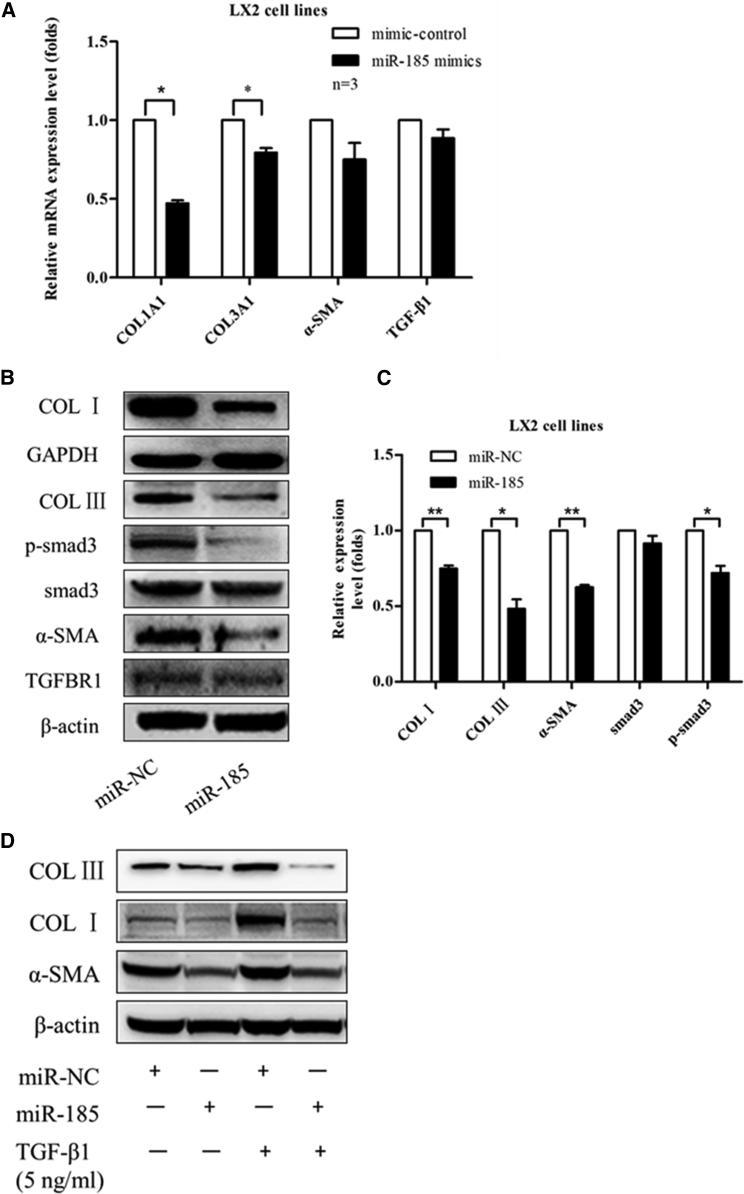
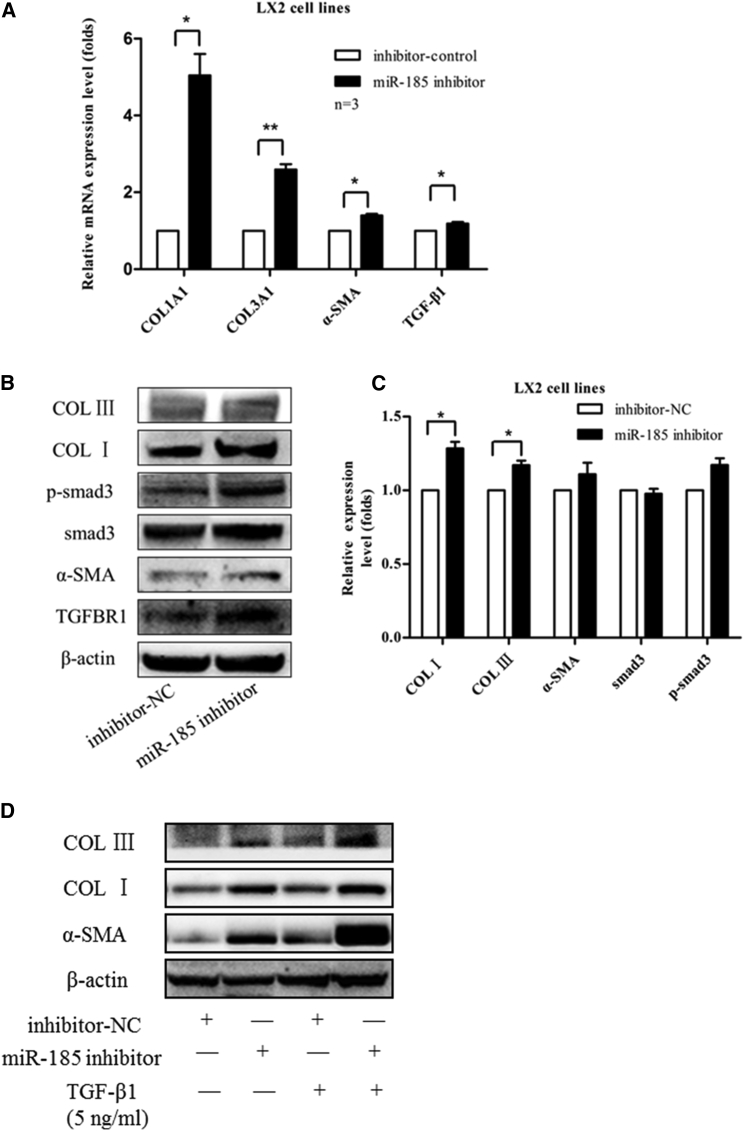
To investigate whether miR-185 regulates liver fibrosis, first miR-185 and miR-negative control (NC) were transiently transfected into LX-2 cells. After 48 hr, expression levels of liver fibrogenesis-related genes were detected by qRT-PCR (Figure 3A) and western blotting (Figure 3B). We found that miR-185 overexpression downregulated the mRNA and protein levels of COL I α1 (COL1A1), COL III α1 (COL3A1), and α-SMA. In addition, to document the physiological relevance of miR-185 on liver fibrosis, we inhibited the expression of endogenous miR-185 and repeated the above validation assays in LX-2 cells. Compared with anti-NC, anti-miR-185 upregulates the mRNA and protein expression levels of COL I, COL III, and α-SMA (Figures 4A and 4B). Hence, these results demonstrate the physiological relevance of endogenous miR-185 in the regulation of liver fibrosis.
Figure 3.
miR-185-Suppressed HSC Activation and Basal/TGF-β1-Induced Fibrous ECM Production of LX-2 Cells
LX-2 cells were seeded into six-well plates at a density of 1.0 × 106 cells per well, and cells were transiently transfected overnight with miR-185 mimics (miR-185) or negative mimic controls (miR-NC). (A and B) After 48 hr, (A) real-time qPCR was performed to analyze the mRNA levels, and (B) western blotting was performed to analyze the protein levels of liver fibrogenesis-related genes. miR-185 mimics downregulated mRNA and protein expression levels of COL I, COL III, and α-SMA. (C) Western blotting data were quantified using the ImageJ software. (D) To detect the effects of miR-185 on TGF-β1-induced profibrogenic gene expression, LX-2 cells were transfected with miR-185 mimics for 24 hr and then replaced with new culture medium treated with or without 5.0 ng/mL TGF-β1 for another 24 hr. α-SMA, COL I, and COL III protein levels were detected by western blotting. miR-185 can repress phenotypes associated with TGF-β1-induced transition of resident fibroblasts. The results are representative of three independent experiments and are presented as mean ± SEM (n = 3, *p < 0 0.05, **p < 0 0.01).
Figure 4.
Knockdown of Endogenous miR-185 Promoted Basal/TGF-β1-Induced Liver Fibrosis
LX-2 cells were transfected with inhibitor-NC or miR-185 inhibitor for 48 hr; inhibition of miR-185 in LX-2 cells induced liver fibrogenesis-related gene transcription (A) and translocation (B). (C) Western blot data were quantified. (D) TGF-β1-induced liver fibrosis, and miR-185 inhibitors (anti-miR-185) upregulated protein expression levels of COL I, COL III, and α-SMA after TGF-β1 further induced HSC activation. Three independent experiments gave similar results (mean ± SEM, n = 3, *p < 0 0.05, **p < 0 0.01).
To determine whether phenotypes associated with TGF-β-induced fibrosis are also influenced by miR-185, when LX2 cells were treated with TGF-β1 for 24 hr, we transiently transfected with miR-185/miR-negative control (NC) or anti-miR-185/anti-miR-NC and then focused on the changes in expression levels of active myofibroblast structural protein markers COL I, COL III, and α-SMA. The western blotting results suggest that miR-185 can repress phenotypes associated with TGF-β-induced transition of resident fibroblasts. An miR-185 inhibitor can induce COL I, COL III, and α-SMA expression at the transcriptional and translocational levels (Figures 3D and 4D).
miR-185 Directly Suppresses RHEB and RICTOR Expression and Indirectly Attenuates the Expression of Fibrogenesis-Related Genes
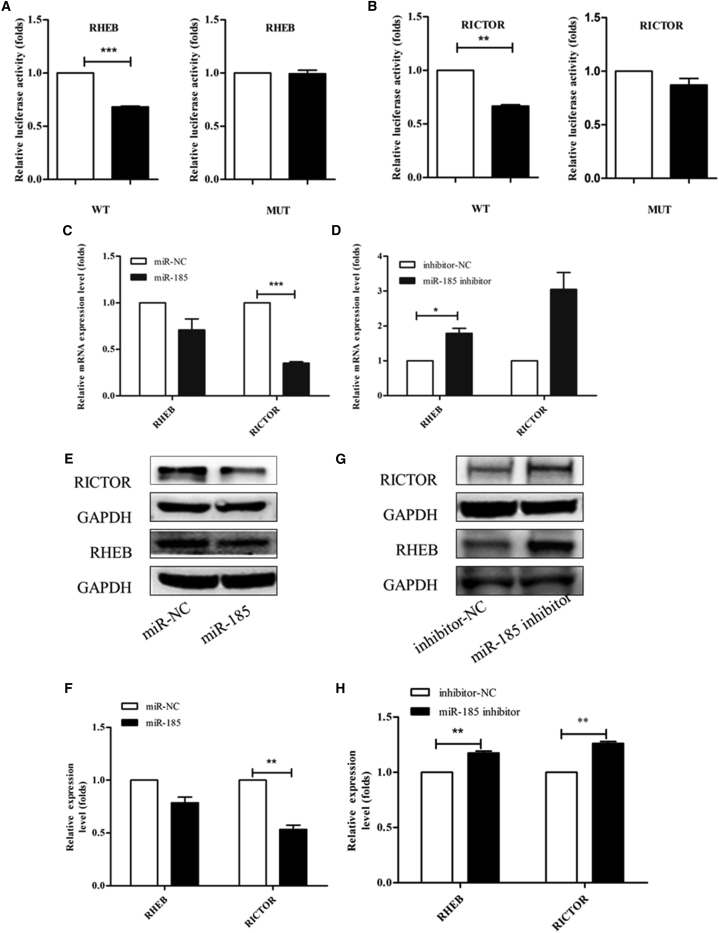
We previously reported that miR-185 induces potent autophagy in HCC by directly interacting with the 3′ UTRs of the RICTOR and RHEB mRNAs.14 We evaluated whether the downregulation of fibrogenesis-related genes in liver fibrosis was mediated by RHEB and RICTOR and their potential interaction with miR-185. Plasmid construction, cloning, and mutagenesis of 3′ UTR seed regions were described in detail in a published paper.14 To determine whether RHEB and RICTOR were also miR-185 target genes in LX-2 cells, a luciferase reporter assay was performed as described previously.14 Treatment with miR-185 mimics significantly reduced the activity of firefly luciferase with the wild-type but not mutant 3′ UTRs of RHEB and RICTOR RNAs (Figures 5A and 5B). The protein and mRNA levels of RHEB and RICTOR were downregulated in LX-2 cells transfected with miR-185 mimics (Figures 5C, 5E, and 5F). When LX-2 cells were transfected with miR-185 inhibitors, the mRNA and protein levels of RHEB and RICTOR were upregulated (Figures 5D, 5G, and 5H). These results strongly indicate that RHEB and RICTOR are also direct target genes of miR-185 in LX-2 cells.
Figure 5.
miR-185 Directly Binds to the 3′ UTRs of the RHEB and RICTOR mRNAs
pmirGLO vectors containing either the wild-type (WT) or mutant (mut) binding sites of miR-185-5p in RHEB (A) and RICTOR (B) mRNAs were constructed to conduct luciferase reporter assays as described previously. Dual-luciferase reporter assays were performed to verify binding between miR-185-5p and RHEB and RICTOR mRNA. The HSC line LX-2 was co-transfected with a pmirGLO vector containing either the WT or mut target sites plus either the miR-185-5p mimics or the mimic control oligonucleotide. Results of relative luciferase activity are shown as the mean ± SEM obtained from triplicate experiments. (C–E and G) RHEB and RICTOR mRNA and protein levels were analyzed by real-time qPCR (C and D) and western blotting (E and G), respectively. The mRNA and protein levels of RHEB and RICTOR were downregulated in LX-2 cells transfected with miR-185 mimics but upregulated when transfected with miR-185 inhibitors. (F and H) Western blotting data were quantified using the ImageJ software. Data shown represent the means ± SEM of independent experiments (unpaired two-sample Student’s t test, n = 3, **p < 0 0.01, *p < 0 0.05).
High Expression of RHEB and RICTOR Mediates Fibroblast Activation
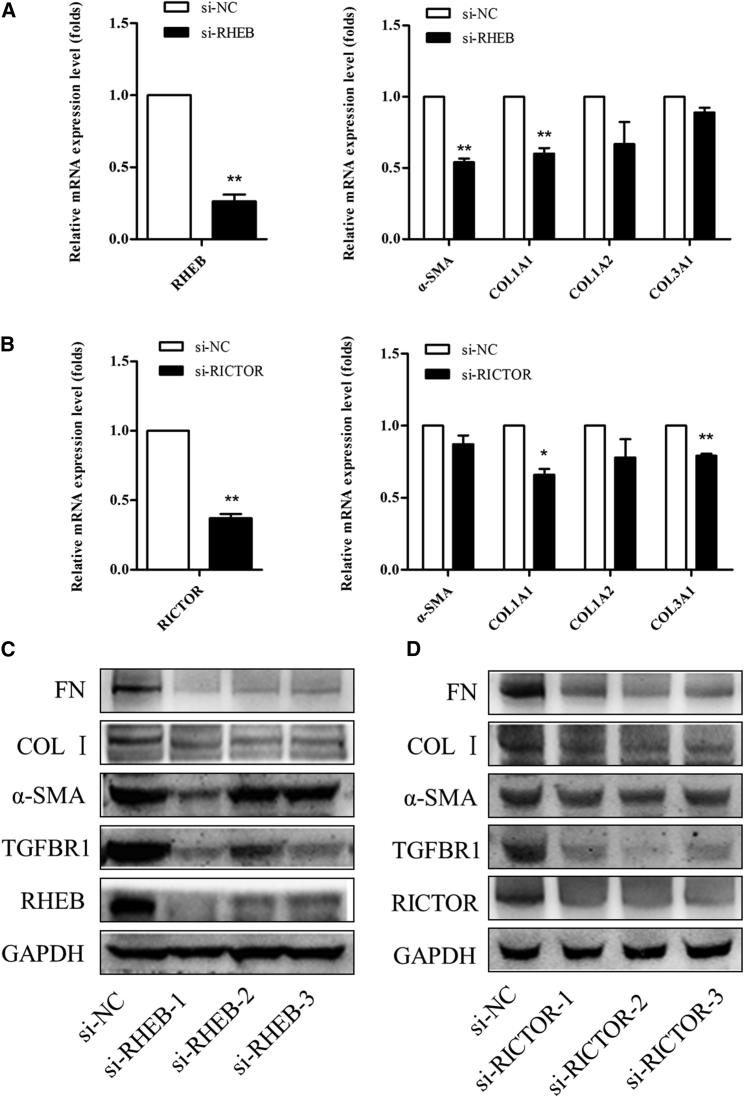
TGF-β1-mediated epithelial-mesenchymal transition (EMT) induces RHEB and RICTOR expression. To further evaluate the roles of RHEB and RICTOR in liver fibroblast activation, a small interfering RNA (siRNA) that reduces RHEB and RICTOR mRNA and protein expression was used to reduce their expression in LX-2 cells. Three siRNA oligos interfering with RHEB/RICTOR were designed by GenePharma. Taking the off-target effect into consideration, we analyzed the interference effect by RT-PCR and western blot during the experiment. siRNA-NC was used as the negatively control to ensure the specificity of siRNA-RHEB/RICTOR. The reduction of RHEB and RICTOR expression also blocked HSC activation (Figure 6). Taken together, these results suggest that high expression of RHEB and RICTOR is necessary for HSC activation.
Figure 6.
RHEB and RICTOR Mediated the Effects of miR-185 on LX-2 Cell Activation
Quantitative real-time PCR determined α-SMA, COL1A1, COL1A2, and COL3A1 mRNA expression in LX-2 cells transfected with si-RHEB (A) and si-RICTOR (B). The reduction of RHEB and RICTOR expression also blocked HSC activation at the mRNA level. Three siRNA oligos interfering with the same specific target genes (RHEB/RICTOR) to produce a similar effect were designed by GenePharma. Western blotting was performed to confirm the knockdown of RHEB (C) and RICTOR (D) expression and fibrogenesis-related gene protein expression levels in LX-2 cells after transfection with si-RHEB and si-RICTOR. The first si-RHEB seems to be the most effective si-RNA oligo, and three si-RICTOR oligos are all effective. The reduction of RHEB and RICTOR expression also blocked HSC activation at the protein level.
CCl4-Induced Liver Fibrosis in Mice Downregulates miR-185 and Upregulates RHEB and RICTOR Expression
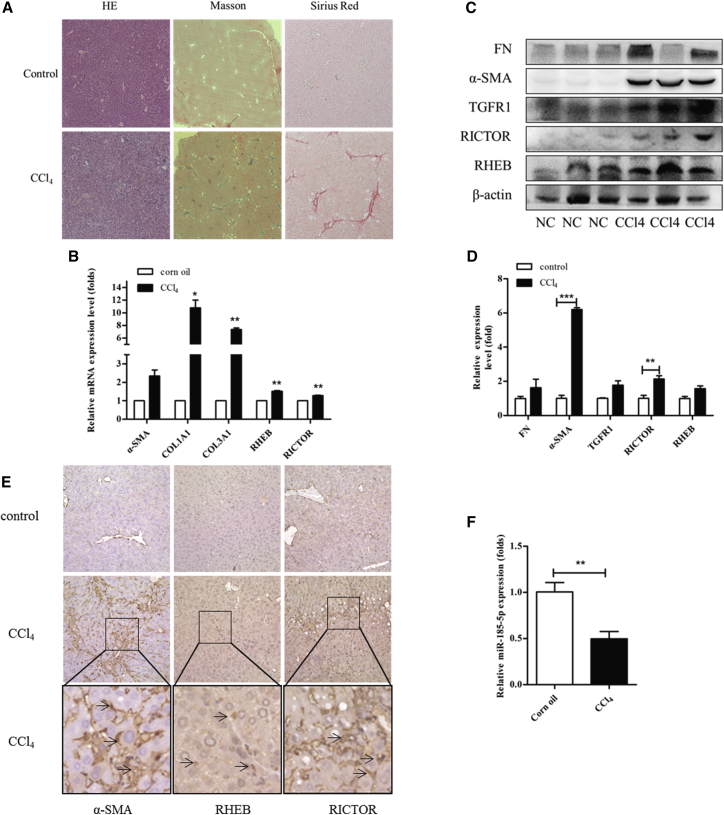
To further assess the in vivo effect of miR-185 on liver fibrogenesis, a liver fibrosis model was first established by injecting mice with CCl4 three times a week for 4 weeks. The histopathological changes in the liver were visualized by H&E staining, and collagen deposition was assessed by Masson staining and Sirius red staining (Figure 7A). As reported, continuous CCl4 treatment resulted in hepatic necrosis and led to liver fibrosis (Figures 7B–7D). Furthermore, a significant downregulation of miR-185 was observed in fibrotic livers collected from CCl4-treated mice (Figure 7F) compared with non-fibrotic livers isolated from the vehicle-treated group. Consistently, miR-185was decreased in human fibrosis and cirrhotic livers compared with normal livers, as described previously. Conversely, RHEB and RICTOR expression in the liver significantly increased after CCl4 treatment, as indicated by qRT-PCR, western blotting, and immunohistochemical staining (Figures 7B–7E), suggesting that miR-185 might contribute to the regulation of RHEB and RICTOR expression during liver fibrogenesis.
Figure 7.
CCl4-Induced Liver Fibrosis in Mice Downregulates miR-185 and Upregulates RHEB and RICTOR
(A) The histopathological changes in livers 1 month after injection of CCl4 are shown by H&E staining, Masson’s trichrome staining, and Sirius red staining of sections from two representative livers. (B) Real-time qPCR analysis for α-SMA, COL1A1, COL1A2, COL3A1, RHEB, and RICTOR in two groups. mRNA levels increased in all genes. All results of relative expression values are shown as the mean ± SEM. (C) Western blotting analysis for α-SMA, fibronectin (FN), TGFR1, RHEB, RICTOR, and GAPDH in livers of representative mice from each group. The protein levels were upregulated in the CCl4 group. α-SMA and collagen are obviously increased, and the changes in RHEB and RICTOR levels are modest. (D) Data represent one of three experiments with similar results. Western blotting data were quantified using the ImageJ software. (E) Immunohistochemistry for α-SMA, RHEB, and RICTOR in liver sections from representative NC and CCl4 livers. In the model group, protein levels of α-SMA, RHEB, and RICTOR increased. (F) Real-time qPCR measured miR-185-5p expression in livers from NC and CCl4-treated mice. miR-185-5p expression was reduced in the CCl4 group.
Discussion
Liver fibrosis is a wound-healing response by the liver to cellular injury and is characterized by deposits of collagen fibers. These fibers continue to contribute to the deterioration of normal liver function. Upon injury, activated HSCs undergo a transition into proliferative, profibrogenic, contractile myofibroblasts that strongly produce TGF-β1. In turn, TGF-β1 stimulates HSCs to generate fibrosis not only by increasing cell numbers but also by increasing matrix production per cell.22
In this study, miR-185 expression was downregulated in TGF-β1-induced activated HSCs compared with their relative quiescent phenotype (Figures 2E and 2F). LX-2 cells were treated with different doses of TGF-β1 for 24 hr or with 5.0 ng/mL at different times, and α-SMA, COL I, and COL III were upregulated, which represented the activation of HSCs. miR-185 decreased in a dose-dependent or time-dependent manner, which was consistent with the results in fibrotic liver tissues from human and rodent models. Compared with the healthy control group, 104 miRNAs were screened out from the liver fibrosis group, 72 miRNAs were upregulated, and 32 were downregulated. miR-185 (miR-185-5p and miR-185-3p) was one of the 32 downregulated miRNAs (Figures 1A and 1B) by Illumina HiSeq sequencing, which is a high-throughput sequencing technology producing highly accurate, reproducible, and quantitative readouts of small RNAs, including those expressed at low levels. The same results were identified in fibrotic liver tissues in mice compared with normal liver tissues in mice (Figure 7F). All of these data suggest that miR-185 might play an inhibiting role in liver fibrogenesis and that its downregulation might be associated with HSC activation.
Several reports have demonstrated that miRNAs play a critical role in the control of various HSC function diseases.23, 24 Csak et al.25 demonstrated that miR-155 deficiency can reduce steatosis and fibrosis without decreasing inflammation in steatohepatitis. Hyun et al.26 found that miR-378a-3p suppresses the activation of HSCs by targeting Gli3. The expression of Gli3 is regulated by smoothened-dependent nuclear factor κB (NF-kB) signaling. miR-29b prevents liver fibrogenesis by inhibiting HSC activation and induces HSC apoptosis by inhibiting the phosphatidylinositol 3-kinase (PI3K)/AKT pathway.27 miR-101 promotes the reversal of activated HSCs to a quiescent state, as indicated by suppression of proliferation and migration.28, 29 Several reports have shown that downregulation of miR-185 has been implicated in various fibrotic diseases, and Xiao et al.20 found that miR-185 was markedly suppressed in hypertrophic scar (HS) tissue. In vitro, miR-185 can regulate TGF-β1 and COL I through the predicted binding sites in its 3′ UTR. The findings of Lei et al.18 suggested that attenuated expression of miR-185 and miR-186 may be responsible for COL V overexpression during idiopathic pulmonary fibrosis. In the study by Yu et al.,21 patients with DCM could be divided into miR-185high and miR-185low groups. During the 1-year observation period, a total of four (16.7%) miR-185low patients died, whereas none of the miR-185high patients died, These findings suggest that high miR-185 levels might be associated with a favorable prognosis by repressing B cell function in DCM. All of these results are consistent with our results showing that miR-185 plays a protective role in tissue fibrosis. However, a paper by Li et al.30 appears to demonstrate the exact opposite of our result. They show upregulation of miR-185 in blood from patients with HBV-related liver fibrosis, with the increase being fibrotic stage dependent, and increased miR-185 in rats with liver fibrosis induced by dimethylnitrosamine (DMN) and bile duct ligation (DBL). Why do we have the exact opposite results? Maybe because of the following three reasons. First, maybe we have different inclusion and exclusion criteria. In vitro, we used different cell lines. Second, we used a different detection method. Third, the sample size is small, and the statistical efficiency effect is perhaps low. The findings of this study need to be confirmed with a larger sample size and longer duration of observation. In the current experiment, we speculated that miR-185 might play an essential role in liver fibrosis. To test this hypothesis, an in vitro study of cultured HSCs was warranted to determine the direct effect by which miR-185 protects against fibrosis; therefore, the mechanism of miR-185 downregulation in liver fibrosis was evaluated. miR-185 and NC miRNA were transiently transfected into human HSCs. In keeping with the hypothesis, overexpression of miR-185 inhibited the expression of α-SMA, COL I, COL III, and p-Smad3 in HSC lines (Figure 3). In addition, to document the physiological relevance of miR-185 in liver fibrosis, we inhibited the expression of endogenous miR-185 and repeated the above validation assays using LX-2 cells. Compared with NC miRNA inhibitors, anti-miR-185 upregulated the mRNA and protein expression of COL I, COL III, and α-SMA (Figure 4). In addition, miR-185 can repress the phenotypes associated with TGF-β1-induced transition of resident fibroblasts (Figure 3D), suggesting that miR-185 negatively regulates fibrosis by targeting collagen matrix synthesis by inhibiting the activation of HSCs.
We also found that TGF-β1 downregulates miR-185 in HSC cells, which is associated with a marked upregulation of not only α-SMA and collagens, which are the markers for fibrosis, but also RHEB and RICTOR, which could upregulate the mammalian targets of mTORC1 and mTORC2 signaling. Both mTORC1 and mTORC2 are structurally and functionally distinct multiprotein complexes.31 The activity of MTORC1 is controlled by RHEB, a GTP-binding protein;32, 33 RICTOR is a scaffold protein that regulates the assembly and substrate binding of MTORC2. Resent findings support a role for both mTORC1 and mTORC2 in the pathogenesis of fibrosis.15, 17 We previously reported that miR-185 directly interacts with the 3′ UTRs of RICTOR and RHEB and induces potent autophagy in hepatocellular carcinoma.14 To determine whether miR-185 acting as an antifibrotic member has a relationship with RHEB and RICTOR, a luciferase reporter assay was performed that demonstrated that RHEB and RICTOR were also miR-185 target genes in LX-2 cells. As confirmed by luciferase reporter assay, increased expression of RHEB and RICTOR was the result of downregulation of miR-185 (Figure 5); therefore, it is possible that miR-185 could regulate the expression of RHEB and RICTOR to play a different role in different types of cells.
miR-185 induced the suppression of activated HSCs, including ECM production and α-SMA expression, which was confirmed by RHEB and RICTOR siRNAs (Figure 6). In vitro application reduced the expression of RHEB and RICTOR, leading to a reversal of fibrosis. These results indicated that loss of RHEB and RICTOR expression had potential protective effects on liver fibrosis, which was in accordance with the findings of Jiang et al.34 That study revealed that TGF-β1 could induce the activation of RHEB/mTORC1 and RICTOR/mTORC2 signaling and that the two pathways are associated with the perpetuation of fibroblast activation and fibrosis, which promote the activation of kidney fibroblasts and contribute to the development of interstitial fibrosis.
It is well known that CCl4-induced liver fibrosis is a robust and widely used model in liver fibrosis. Sustained/chronic inflammation leads to fibrosis; therefore, we hypothesized that miR-185 would also attenuate in CCl4-induced inflammatory response and fibrosis livers. Collectively, our results indicate that inflammatory and fibrogenic changes were observed by H&E staining, and collagen deposition was assessed by Masson staining and Sirius red staining (Figure 7A). Consistently, miR-185 was decreased in human fibrosis and cirrhotic livers compared with normal livers, as described previously. Conversely, RHEB and RICTOR expression in diseased livers significantly increased after CCl4 treatment (Figures 7B–7F). It would be important to establish that miR-185 overexpression ameliorates the severity of liver fibrosis to determine whether miR-185 plays a key role in liver fibrogenesis. To test this, miR-185 induced in a mouse model of liver fibrosis will be studied further.
Many miRNAs are expressed in a tissue- or organ-specific manner, suggesting that miRNA biomarkers are more likely to have high specificity. Matsuura et al.10 reported that circulating let-7 might serve as a surrogate marker for hepatic fibrosis progression in chronic hepatitis C.
Conclusions
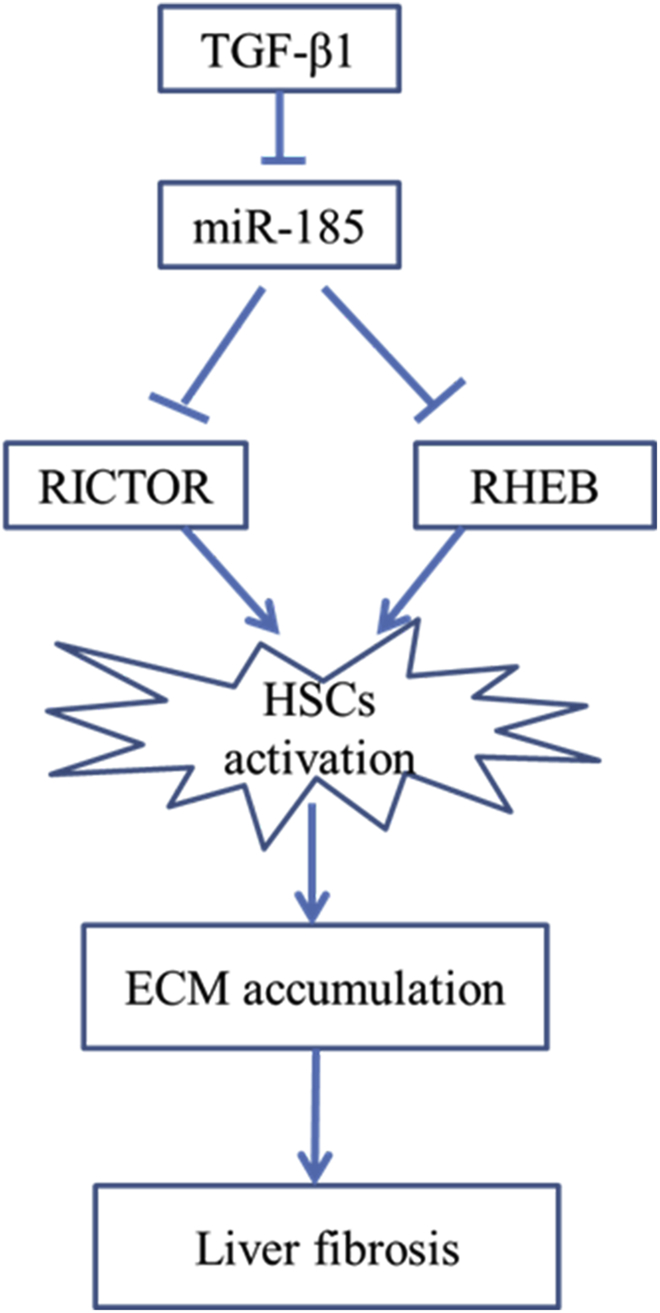
Our study demonstrates that miR-185 suppresses the activation of HSCs by targeting RHEB and RICTOR (Figure 8), and the observed negative correlation between fibrosis and the expression of miR-185 indicates that this molecule could be useful in assessing the severity of fibrosis regardless of etiology. To further investigate whether miR-185 could be identified as a candidate predictor of disease progression, a larger cohort with comparable clinicopathological features and complete follow-up data would be necessary.
Figure 8.
Schematic
TGF-β1-induced activated HSCs also downregulate miR-185 concomitant with increased expression of RHEB and RICTOR. miR-185 prevents liver fibrogenesis by inhibiting HSC activation through inhibiting RHEB and RICTOR.
Study Limitations
Our analysis was limited by the small sample sizes of the various etiologic groups; therefore, further studies are warranted to reveal whether miR-185 correlations are also characteristic of the various etiology groups or whether these relationships are only summed characteristics of the fibrosis samples by reason of the various etiologies.
Materials and Methods
Cell Cultures
LX-2 cells (a human HSC cell line) were purchased from Xiangya Central Laboratory (Xiangya School of Medicine, Hunan, China). The cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Grand Island, NY, USA), 100 U/mL penicillin G, and 100 μg/mL streptomycin (SV30010; Thermo Scientific, Rockford, IL, USA), and incubated in 5.0% CO2 at 37°C. Recombinant human TGF-β1 from R&D Systems (240-B-010/CF; Minneapolis, MN, USA) was added to the supernatant at 2.5, 5.0, or 7.5 ng/mL for 24 hr, respectively.
Cell Transfection
Cells were cultured to 50%–60% confluence and transiently transfected with 50.0 nM NC miRNA (miR-NC), miRNA mimics (miR-185), NC miRNA inhibitors (anti-NC), or miRNA inhibitors (anti-miR-185) using Polyplus transfection reagent (n 06Y0312E9; Polyplus-transfection, NY, USA) according to the manufacturer’s protocols (https://wenku.baidu.com/view/2540f5a0910ef12d2bf9e724.html). For experiments involving the transfection of RNAi, siRNAs were also transfected using Polyplus-transfection reagent with a final concentration of 50 nM according to the manufacturer’s protocols (http://www.docin.com/p-203375956.html). miRNAs mimics, inhibitors, and NCs were purchased from RiboBio (Guangzhou, China), and siRNAs were purchased from GenePharma (Shanghai, China). For RHEB/RICTOR, the company designed three siRNA oligos interfering with the same specific target genes (RHEB/RICTOR) to produce a similar effect.
Target Prediction and Luciferase Reporter Assay
The wild-type 3′ UTR of RHEB and RICTOR mRNA and its target-site mutant 3′ UTR were amplified as described previously.14 Luciferase activities were measured in a microplate luminometer according to the instructions for the Dual-Luciferase Reporter Assay System (E1910; Promega, Madison, WI, USA).
RNA Isolation and Real-Time qPCR
RNA was extracted according to the manufacturer’s instructions using the R6834 Total RNA Kit (Omega Bio-Tech, Norcross, GA, USA) and analyzed by real-time qPCR using 1206352 SYBR Green qPCR Master Mix (Applied Biosystems, Warrington, UK) on an ABI 7500 system (Applied Biosystems, Waltham, MA, USA). The optimized primers used for real-time PCR are listed in Table 2.
Table 2.
Primers Used for Real-Time PCR
| Genes | Species | Sense (5′–3′) | Antisense (5′–3′) |
|---|---|---|---|
| RHEB | Hum | GGAATCTTCTGCTAAAGAAA | GCATGAAGACTTGCCTTGTG |
| RICTOR | Hum | AGTGAATCTGTGCCATCGAG | AAGGAAAAATGTGCTAGTAGAGCT |
| α-SMA | Hum | GGGAATGGGACAAAAAGACA | CTTCAGGGGCAACACGAA |
| COLL1A1 | Hum | GGGATTCCCTGGACCTAAAG | GGAACACCTCGCTCTCCA |
| COLL1A2 | Hum | CTGGAGAGGCTGGTACTGCT | AGCACCAAGAAGACCCTGAG |
| COLL3A1 | Hum | CTGGACCCCAGGGTCTTC | GACCATCTGATCCAGGGTTTC |
| β-actin | Mus | CTAAGGCCAACCGTGAAAAG | ACCAGAGGCATACAGGGACA |
| RHEB | Mus | TTTTTGGAATCTTCTGCTAAAGAAA | TGAAGCTGCTCCATCAATCTT |
| RICTOR | Mus | ACATTCAGCAGAGCAACGAG | GGAAACAAGGAAGCATTCACA |
| COLL1A1 | Mus | TTCTCCTGGCAAAGACGGAC | CGGCCACCATCTTGAGACTT |
| COLL1A2 | Mus | TAGCCAACCGTGCTTCTCAG | TCTCCTCATCCAGGTACGCA |
| COLL3A1 | Mus | AAGGCTGCAAGATGGATGCT | GTGCTTACGTGGGACAGTCA |
| α-SMA | Mus | GAGACTCTCTTCCAGCCATCTT | TGATCTCCTTCTGCATCCTGTC |
Hum, human; Musc, mouse.
miRNA Isolation and Quantitation
Total RNAs were prepared using the Total RNA Kit (R6834; Omega, Norcross, GA, USA), and total miRNAs were isolated using the AM1560 mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Shanghai, China) according to the manufacturer’s instructions. The high-capacity 4368814 cDNA Reverse Transcription Kit (Applied Biosystems, Warrington, UK) and MQP-0105 Bulge-Loop miRNA qRT-PCR Primer Set (RiboBio, Guangzhou, China) were used to convert selected miRNAs to cDNAs. The expression of selected miRNAs was analyzed by qRT-PCR using 4367659 Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). RNU6B was used to normalize the mature miRNA expression levels (MQP-0105; RiboBio, Guangzhou, China).
Western Blotting
An equal amount of protein from cell lysate was loaded into each well of a 12% or 10% SDS-PAGE gel after denaturation with SDS loading buffer. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane, incubated with blocking buffer (5% fat-free milk) for 1.0 hr at room temperature and blotted with the following antibodies overnight: anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5174, Cell Signaling Technology, USA), anti-β-actin (sc-47778, Santa Cruz Biotechnology, USA), anti-mTOR (ab32028, Abcam, USA), anti-p-mTOR (ab109268, Abcam, USA), or anti-RHEB (ab92313, Abcam, USA) and anti-RICTOR (53A2, CST, USA), anti-α-SMA (BM0002, Boster, Wuhan, China), anti-collagen I (ab84956, Abcam, USA), anti-collagen III (ab7778, Abcam, USA), anti-Smad3 (9523, CST, USA), or anti-p-Smad3 (9520, CST, USA). Immunoreactive bands were detected using an enhanced chemiluminescence system (32209, Thermo Scientific, USA). Chemiluminescent signals were visualized and analyzed using the Fusion Solo system (Vilber, France). Western blotting data were quantified using ImageJ software.
Animal Studies
To induce liver fibrosis, 4-week-old C57BL/6J male mice purchased from Vital River Laboratory Animal Technology (Beijing, China) were intraperitoneally injected with CCl4 (0.5 mL/kg body weight, mixed with corn oil at 1:50) three times a week for 4 weeks. Age-matched mice were treated with corn oil only as the vehicle control. All experiments using mice were conducted in accordance with the Institutional Animal Care and Use Committee of the Institute of Zoology (Chinese Academy of Sciences).
Patients and Tissues
The patient inclusion criteria were a diagnosis of chronic HBV infection or a sustained positive result in hepatitis B antigen (HBsAg) of more than 6 months and/or HBV DNA ≥ 500 IU/mL without nucleoside (NAs) of antiviral therapy in 6 months. Diagnosis was made according to the Guidelines of Prevention and Treatment of Chronic HBV Infection.35 The exclusion criteria included HIV or other hepatitis infection, alcohol consumption of more than 30 g/day, hepatic injury because of metabolism or other causes, history of treatment for hepatic diseases, or a liver biopsy not suitable for analysis. All patients with chronic HBV infection underwent a liver biopsy followed by pathological analysis, and the degree of hepatic fibrosis was determined according to Scheuer’s criteria36 (S0, no liver fibrosis; S1, expansion of portal fibrotic area; S2, fibrosis observed in and around portal area, may have formed fibrous septum; S3, clear fibrosis and hepatic lobule structure disorder but no cirrhosis; S4, early cirrhosis). Ten patients with chronic HBV and representing various etiologic backgrounds of fibrosis were enrolled in the study; eight healthy subjects from Beijing Ditan Hospital, Capital Medical University, were also enrolled. Written consent was obtained before sample collection, and the study was approved by the Human Ethics Committee of the Institute Research Ethics Committee of Beijing Ditan Hospital.
Illumina HiSeq Sequencing
A small RNA library is from human plasma. Briefly, the 18- to 30-nt fraction from 1.5 μg of plasma total RNA from a single donor was isolated by PAGE. Purified miRNAs were then 3′− and 5′−ligated to single-stranded oligonucleotides that contained universal primer sequences for reverse transcription and PCR. Reverse transcription and PCR generated a library of small RNA cDNA molecules. Sequences were compared with a reference database of known miRNA sequences (miRDeep2).
Cluster analysis and target gene prediction of differentially expressed miRNAs was carried out, and gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed miRNAs target genes were performed. The genes defined as differentially expressed were those with a false discovery rate (FDR) ≤ 0.01 and fold change (FC), |log2(FC)| > = 1.
Statistical Analyses
Each experiment was performed at least three times. Data were analyzed by paired Student’s t test with the SPSS 13.0 software (IBM, USA). p < 0.05 was considered statistically significant.
Author Contributions
J.C. designed the research study. M.H. and S.L. performed language editing. Y.M., S.F., J.Z., H.L., and X.Y. provided technical assistance. L.Z. performed the research, analyzed the data, and wrote the paper.
Conflicts of Interest
We declare no competing financial interests.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81470863/81670547) and Beijing Municipal Administration of Hospitals (ZYLX201402 and DFL2015170).
References
- 1.Friedman S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popov Y., Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology. 2009;50:1294–1306. doi: 10.1002/hep.23123. [DOI] [PubMed] [Google Scholar]
- 3.Schuppan D., Kim Y.O. Evolving therapies for liver fibrosis. J. Clin. Invest. 2013;123:1887–1901. doi: 10.1172/JCI66028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman S.L., Bansal M.B. Reversal of hepatic fibrosis -- fact or fantasy? Hepatology. 2006;43(2, Suppl 1):S82–S88. doi: 10.1002/hep.20974. [DOI] [PubMed] [Google Scholar]
- 5.Li H., You H., Fan X., Jia J. Hepatic macrophages in liver fibrosis: pathogenesis and potential therapeutic targets. BMJ Open Gastroenterol. 2016;3:e000079. doi: 10.1136/bmjgast-2016-000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Desmet V.J., Gerber M., Hoofnagle J.H., Manns M., Scheuer P.J. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513–1520. [PubMed] [Google Scholar]
- 9.Ji J., Zhang J., Huang G., Qian J., Wang X., Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 10.Matsuura K., De Giorgi V., Schechterly C., Wang R.Y., Farci P., Tanaka Y., Alter H.J. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology. 2016;64:732–745. doi: 10.1002/hep.28660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng C., Wang Y.L., Xie C., Sang Y., Li T.J., Zhang M., Wang R., Zhang Q., Zheng L., Zhuang S.M. Identification of a novel TGF-β-miR-122-fibronectin 1/serum response factor signaling cascade and its implication in hepatic fibrogenesis. Oncotarget. 2015;6:12224–12233. doi: 10.18632/oncotarget.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhi Q., Zhu J., Guo X., He S., Xue X., Zhou J., Hu B., Li H., Chen S., Zhao H., Kuang Y. Metastasis-related miR-185 is a potential prognostic biomarker for hepatocellular carcinoma in early stage. Biomed. Pharmacother. 2013;67:393–398. doi: 10.1016/j.biopha.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Qadir X.V., Han C., Lu D., Zhang J., Wu T. miR-185 inhibits hepatocellular carcinoma growth by targeting the DNMT1/PTEN/Akt pathway. Am. J. Pathol. 2014;184:2355–2364. doi: 10.1016/j.ajpath.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L., Liu S., Han M., Feng S., Liang J., Li Z., Li Y., Lu H., Liu T., Ma Y., Cheng J. MicroRNA-185 induces potent autophagy via AKT signaling in hepatocellular carcinoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317694313. 1010428317694313. [DOI] [PubMed] [Google Scholar]
- 15.Benaya Rozen-Zvi T.H., Susan C., Hubchak C.H. TGF-β/Smad3 Activates Mammalian Target of Rapamycin Complex-1 to Promote Collagen production by Increasing HIF-1α Expression. J. Physiol. Renal Physiol. 2013;10:1–37. doi: 10.1152/ajprenal.00215.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano I., McDonald P.C., Lock F.E., Dedhar S. Role of the integrin-linked kinase (ILK)/Rictor complex in TGFβ-1-induced epithelial-mesenchymal transition (EMT) Oncogene. 2013;32:50–60. doi: 10.1038/onc.2012.30. [DOI] [PubMed] [Google Scholar]
- 17.Chang W., Wei K., Ho L., Berry G.J., Jacobs S.S., Chang C.H., Rosen G.D. A critical role for the mTORC2 pathway in lung fibrosis. PLoS ONE. 2014;9:e106155. doi: 10.1371/journal.pone.0106155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei G.S., Kline H.L., Lee C.H., Wilkes D.S., Zhang C. Regulation of Collagen V Expression and Epithelial-Mesenchymal Transition by miR-185 and miR-186 during Idiopathic Pulmonary Fibrosis. Am. J. Pathol. 2016;186:2310–2316. doi: 10.1016/j.ajpath.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding X., Yu C., Liu Y., Yan S., Li W., Wang D., Sun L., Han Y., Li M., Zhang S. Chronic obstructive sleep apnea accelerates pulmonary remodeling via TGF-β/miR-185/CoLA1 signaling in a canine model. Oncotarget. 2016;7:57545–57555. doi: 10.18632/oncotarget.11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao K., Luo X., Wang X., Gao Z. MicroRNA-185 regulates transforming growth factor-β1 and collagen-1 in hypertrophic scar fibroblasts. Mol. Med. Rep. 2017;15:1489–1496. doi: 10.3892/mmr.2017.6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M., Liang W., Xie Y., Long Q., Cheng X., Liao Y.H., Yuan J. Circulating miR-185 might be a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. Sci. Rep. 2016;6:33580. doi: 10.1038/srep33580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyugos M., Lendvai G., Kenessey I., Schlachter K., Halász J., Nagy P., Garami M., Jakab Z., Schaff Z., Kiss A. MicroRNA expression might predict prognosis of epithelial hepatoblastoma. Virchows Arch. 2014;464:419–427. doi: 10.1007/s00428-014-1549-y. [DOI] [PubMed] [Google Scholar]
- 24.Appourchaux K., Dokmak S., Resche-Rigon M., Treton X., Lapalus M., Gattolliat C.H., Porchet E., Martinot-Peignoux M., Boyer N., Vidaud M. MicroRNA-based diagnostic tools for advanced fibrosis and cirrhosis in patients with chronic hepatitis B and C. Sci. Rep. 2016;6:34935. doi: 10.1038/srep34935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csak T., Bala S., Lippai D., Kodys K., Catalano D., Iracheta-Vellve A., Szabo G. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS ONE. 2015;10:e0129251. doi: 10.1371/journal.pone.0129251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyun J., Wang S., Kim J., Rao K.M., Park S.Y., Chung I., Ha C.S., Kim S.W., Yun Y.H., Jung Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat. Commun. 2016;7:10993. doi: 10.1038/ncomms10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Chu E.S., Chen H.Y., Man K., Go M.Y., Huang X.R., Lan H.Y., Sung J.J., Yu J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell. Oncotarget. 2014;6:7325–7338. doi: 10.18632/oncotarget.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu X., Zhang H., Zhang J., Zhao S., Zheng X., Zhang Z., Zhu J., Chen J., Dong L., Zang Y., Zhang J. MicroRNA-101 suppresses liver fibrosis by targeting the TGFβ signalling pathway. J. Pathol. 2014;234:46–59. doi: 10.1002/path.4373. [DOI] [PubMed] [Google Scholar]
- 29.Zhao S., Zhang Y., Zheng X., Tu X., Li H., Chen J., Zang Y., Zhang J. Loss of MicroRNA-101 Promotes Epithelial to Mesenchymal Transition in Hepatocytes. J. Cell. Physiol. 2015;230:2706–2717. doi: 10.1002/jcp.24995. [DOI] [PubMed] [Google Scholar]
- 30.Li B.B., Li D.L., Chen C., Liu B.H., Xia C.Y., Wu H.J., Wu C.Q., Ji G.Q., Liu S., Ni W. Potentials of the elevated circulating miR-185 level as a biomarker for early diagnosis of HBV-related liver fibrosis. Sci. Rep. 2016;6:34157. doi: 10.1038/srep34157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster K.G., Fingar D.C. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J. Biol. Chem. 2010;285:14071–14077. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 33.Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang L., Xu L., Mao J., Li J., Fang L., Zhou Y., Liu W., He W., Zhao A.Z., Yang J., Dai C. Rheb/mTORC1 signaling promotes kidney fibroblast activation and fibrosis. J. Am. Soc. Nephrol. 2013;24:1114–1126. doi: 10.1681/ASN.2012050476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu. European Association for the Study of the Liver EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Romanelli R.G., Stasi C. Recent Advancements in Diagnosis and Therapy of Liver Cirrhosis. Curr. Drug Targets. 2016;17:1804–1817. doi: 10.2174/1389450117666160613101413. [DOI] [PubMed] [Google Scholar]