Summary
The interferon-inducible transmembrane proteins (IFITMs) are a family of small transmembrane proteins belonging to the interferon (IFN)-stimulated gene (ISG) superfamily and strongly induced by IFNs. In this paper, we studied the expression profile of IFITMs in 32 organ tissues. The IFITM mRNA expression profile showed that IFITM1, IFITM2 and IFITM3 were expressed in each tissue, especially, in spermatophore, spermaduct, testicle and epididymis. The expression of IFITM1, IFITM2 and IFITM3 showed a trend from high to low. Except for IFITM3 and IFITM6, the others IFITMs were highly expressed in the bone marrow, and the expression level of them was higher in the tibia than that in other parts of the long bones. In liver, the relative expression of IFITM1 and IFITM3 was higher than that of other members. The expression level of IFITM5 was the highest in bone marrow, successively in pancreas, and it was low in skin, smooth muscle and fat. Interestingly, the expression profile of IFITM2 and IFITM7 in tissues was similar to IFITM5. The expression of IFITM2, IFITM5 and IFITM10 were higher in smooth muscle than that in skeletal muscle. IFITM2, IFITM5, IFITM7 and IFITM10 were both highly expressed in esophagus and trachea. In addition, the expression of IFITM6 in eyes was high, and also in pancreas, gallbladder and bone. In the present study, we systematically analyzed the mRNA expression profile of IFITMs in 32 organ tissues, providing the foundation for the study of the function of the IFITMs.
Keywords: Rat, IFITMs, gene expression
1. Introduction
Interferons (IFNs), a group of low molecular glycoproteins synthesized by host cells, was discovered by Isaacs, who studied the interference factor about influenza virus in chick embryo chorioallantoic membrane (1). The following interferons are classified as type I, including IFN-α and IFN-β, and type II including IFN-γ only (2). INFs are a family of proteins that are released by a variety of cells in response to infections caused by viruses. A high level of antiviral protection is achieved by IFN-α, IFN-β and IFN-γ. Antiviral activity of interferons is based on the induction and regulation of innate and acquired immune mechanisms. By binding to transmembrane receptors, INFs interact with target cells activating the JAK/STAT pathways, but also other signaling pathways (3). This leads to induction and activation of many antiviral agents, such as protein kinase RNA-activated (PKR), the expression of interferon inducible transmembrane proteins (ISGs), ribonuclease 2-5A pathway, and Mx proteins, as well as numerous apoptotic pathways. As a result of the protective effect of interferons, virus binding to cells is stopped. Interferon inducible transmembrane protein (IFITM) is a key class of proteins in the ISGs-regulated expression products (4). The IFITM family has different functions, including the control of cell proliferation, promotion of homotypic adhesion, prevention of viral infection, maturation of bone matrix mineralization, regulation of germ cell development, etc (5–7).
The human IFITM family consists of 5 members, namely IFITM1, IFITM2, IFITM3, IFITM5 and IFITM10, located on chromosome 11. Differently, the mice IFITM family includes IFITM1, IFITM2, IFITM3, IFITM5, IFITM6, IFITM7 and IFITM10. Except for IFITM7 located in chromosome 16, other genes in mice are located on chromosome 7. IFITM1, expressed in leukocytes and endothelial cells, has anti-proliferation functions and promotes homotypic adhesion (5,8,9). In many cell lines, IFITM2 could cause the cell cycle to stagnate and p53-dependent apoptosis (10). IFITM3 can inhibit the proliferation of cells (11–13). As a host immune barrier, IFITM1, IFITM2 and IFITM3 play an important role in the process of defense against respiratory virus invasion (14–17). Recently, they also have been reported as biomarkers of several tumors (11,18–20). IFITM5 is a IFITM-like protein, only expressed in human and mouse bone, especially in osteogenesis cells (21). The point mutation of IFITM5 causes osteogenesis imperfecta type V (22), and the overexpression of the mutant gene could promote Saos2 cell line apoptosis (23). IFITM6 was strongly expressed in spleen macrophages of tumor bearing mice. The IFITM10 sub-family is divided into two groups: aquatic and terrestrial types, suggesting that IFITM10 might be associated with adaptation to the aquatic environment. In addition, IFITM10 with CTSD can be used as a molecular marker for breast cancer (24). So far, no one has reported the function of IFITM7.
In the present study, we analyzed the expression pattern of IFITMs in different tissues of rats, providing a theoretical foundation for further research on their function.
2. Materials and Methods
2.1. Rat tissue collection
Rats (Wistar rats, male, 6-week-old) were purchased from the Animal Experimental Center of Shandong University. The study tissues compromised 32 organ tissues, including lung, heart, spleen, stomach, pancreas, liver, gallbladder, kidney, small intestine, large intestine, esophagus, trachea, cartilage, humerus, ulna, thighbone, tibia, eye, ear, skin, hair, spermatophore, spermaduct, epididymis, testis, skeletal muscle, smooth muscle, fat, brain, bone marrow and blood, etc. Each portion was frozen in liquid nitrogen and stored at −80°C until RNA was extracted. Total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's instructions.
2.2. Primer design
According to the IFITM family gene sequence on GenBank, primers were designed using https://lifescience.roche.com. Three pairs of primers were designed for each IFITM gene, according to the Ct value and the peak melting curve.
2.3. Reverse transcription (RT)-PCR analysis
Total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNAs from organ tissues were reverse-transcribed using PrimeScript™ RT Master Mix (Perfect Real Time) (Takara Biotechnology (Dalian) Co. Ltd.), and cDNAs of IFITMs were amplified by PCR using mice IFITMs specific primers. RT of RNA was performed in a final volume of 20 µL containing Random Primer (100 pmol/µL) of 1 µL, dNTP (10 mM) of 1 µL, 4 µg total RNA and diethyl pyrocarbonate (DEPC)-treated water. After incubation at 65°C for 5 min, 4 µL 5×PrimerScript Buffer, 0.5 µL RNase Inhibitor (40 U/µL) and 1 µL RNase (200 U/µL) were added, then the reaction was incubated at 30°C for 10 min, 42°C for 60 min, 75°C for 15 min and 4°C for 20 min. Quantitative PCR was performed using LightCycler 480 (Roche). Quantitative PCR was performed using LightCycler 480 (Roche). PCR was performed in a final volume of 40 µL containing 10 µL SYBR Green Master I (Roche), 1 µL upstream primers (10 mol/µL) and 1 µL downstream primers (10 mol/µL), 0.4 µL cDNA and 7.6 µL DEPC-treated water. PCR was performed for 40 cycles (denaturation at 94°C for 5 s, annealing at 55°C for 15s and extension at 72°C for 15 s). The IFITMs mRNA copy numbers in organ tissues were normalized to mRNA copy numbers of the house keeping gene, β-actin to give a value ΔCt. Other samples were then normalized to the lung sample giving ΔΔCt and finally the amount of IFITMs in organ samples relative to the lung sample was expressed as 2−ΔΔCt. This final value was used to determine the expression profile in 32 organ samples.
3. Results
3.1. RT-qPCR primer design and screening
The primers used for beta-actin (control) were 5′-CCAGAGCAAGAGAGGTAT-3′ (forward) and 5′-CTGTGGTGGTGAAGCTGTAG-3′ (reverse). The primer sequences of IFITMs are listed in supplementary Table S1. According to the Ct value and the peak melting curve, the best pairs we obtained are listed in Table 1 and used in our study.
Supplementary Table S1. the IFITM primer sequence of rat.
| Gene name | Upstream primer sequence (5′-3′) | Downstream primer sequence (5′-3′) |
|---|---|---|
| IFITM1-1 | TGAGATCTCCACGCCTGAC | CCACCATCTTCCTGTCCCTA |
| IFITM1-2 | GCTGAGATCTCCACGCCTG | ACCATCTTCCTGTCCCTAGA |
| IFITM1-3 | CGACAACCACCACAATCAAC | CCATCTTCCTGTCCCTAGACTTC |
| IFITM2-1 | CTGGTCCCTGTTCAATACGC | CATCTTCCTGTCCCTGGACTT |
| IFITM2-2 | GGTCTGGTCCCTGTTCAATA | CACCATCTTCCTGTCCCTGGA |
| IFITM2-3 | CGGTAGTCTTTCAGTCGCTTTC | TGTGGACAGATATACGAAGGT |
| IFITM3-1 | CTGGTCCCTGTTCAATACGC | ACCATCTTCCGATCCCTAGAC |
| IFITM3-2 | GTCTGGTCCCTGTTCAATAC | CCACCATCTTCCGATCCCTAG |
| IFITM3-3 | TCAACATGCCCAGAGAGGT | CCATCTTCCGATCCCTAGACT |
| IFITM5-1 | ATCTGTGCTGCCTTGGTTTC | GCTTTGGAGCCATACTGCTT |
| IFITM5-2 | CTGTGCTGCCTTGGTTTCCT | TTTGGAGCCATACTGCTTTG |
| IFITM5-3 | GCTGGTCCACTCTGTCAAGG | GTCCCAGGAGCAGCAATG |
| IFITM6-1 | CTGGTCCACATTCAGCACA | CATCTTCCTGTCCCTGGACTT |
| IFITM6-2 | TGTGGTTTACATCAACAGTGACA | CATCTTCCTGTCCCTGGACTT |
| IFITM6-3 | GGGTTTCATTGCCTATGTCTACTC | TGATAAGTATGATGAAGCAGACGAC |
| IFITM7-1 | GTGTTCAACGTGCCCAGAG | ATTGAACAGGGACCAGACCA |
| IFITM7-2 | ACGCTCTTCATGAACTTCTGC | TCTGCCGATCCCTACACTTC |
| IFITM7-3 | TGGTCTGGTCCCTGTTCAAT | TCTGCCGATCCCTACACTTC |
| IFITM10-1 | GACACCACAGAGGTGAACGA | GCTTCTTGTCCCGAACTTTG |
| IFITM10-2 | AGTAGGTGAGCGGTACTGGTG | GCCTGGGAATCCAGAAGG |
| IFITM10-3 | TGGGAACAAAGTGGAGTGCT | CAGCCAGAGCCTGACTCAC |
| β-Actin | GGCAATGAGCGGTTCCGAT | TGCCTGGGTACATGGTGGT |
Table 1. Ct value of IFITM genes by RT-QPCR.
| Name | Mean Ct | SD |
|---|---|---|
| IFITM1-1 | 33.90 | 0.211974841 |
| IFITM1-2 | 33.96 | 0.424617475 |
| IFITM1-3 | 34.76 | 0.21007935 |
| IFITM2-1 | 33.66 | 0.140118997 |
| IFITM2-2 | 33.00 | 0.146401275 |
| IFITM2-3 | 33.01 | 0.321299445 |
| IFITM3-1 | 32.92 | 0.130128142 |
| IFITM3-2 | 33.11 | 0.31214313 |
| IFITM3-3 | 36.37 | 0.482942371 |
| IFITM5-1 | 37.91 | 0.160934769 |
| IFITM5-2 | 37.74 | 0.972950838 |
| IFITM5-3 | 37.92 | 0.077674535 |
| IFITM6-1 | 32.71 | 0.176729549 |
| IFITM6-2 | 37.29 | 0.362261416 |
| IFITM6-3 | 33.53 | 0.167431578 |
| IFITM7-1 | 30.92 | 0.102632029 |
| IFITM7-2 | 32.11 | 0.317647603 |
| IFITM7-3 | 31.79 | 0.060277138 |
| IFITM10-1 | 40 | 0 |
| IFITM10-2 | 40.00 | 0 |
| IFITM10-3 | 31.53 | 0.17691806 |
| β-Actin | 26.61 | 0.060827625 |
Note: the marked primer was chosen to use in our study.
3.2. mRNA expression profiles of IFITMs in different tissues
According to the RT-PCR, we obtained a result that the IFITM1, IFITM2 and IFITM3 genes of the rat were expressed in all 32 organ tissues examined (Figure 1). In bone marrow, the relative expression levels of IFITM1 and IFITM2 were higher, whereas IFITM3 was lower. In spleen, stomach, pancreas, liver, gallbladder and kidney, the relative expression levels of IFITM1 and IFITM3 were higher. Except for IFITM1, the expression levels of IFITM2 and IFITM3 were strong in skin and fat, while the relative expression levels of IFITM2 and IFITM3 were higher in skeletal muscle than in smooth muscle. In addition, the relative expression levels of IFITM1, IFITM2 and IFITM3 in bone were lower, however those in spermatophore were highest and in epididymis were lowest in tissues of the reproductive system.
Figure 1.
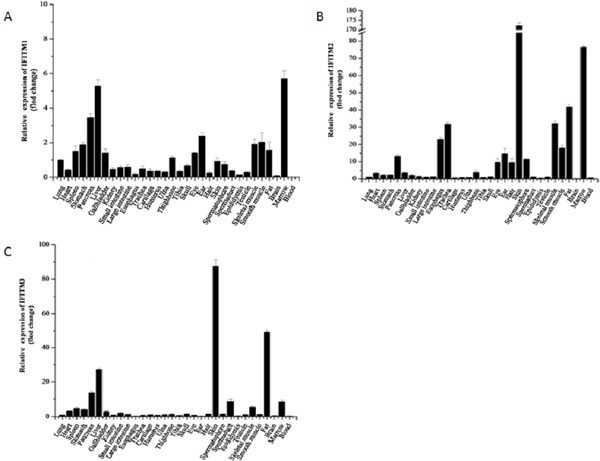
The relative expression of IFITM1(1A), IFITM2(1B) and IFITM3(1C) in different rat tissues.
The relative expression levels of IFITM5 and IFITM10 in the bone marrow were highest, and IFITM5 and IFITM10 were highly expressed only in esophagus, trachea, skull, bone, eye, ear, hair and skin, whereas they were expressed at a lower level in large intestine, small intestine, brain, blood and the tissues of the reproductive system. In smooth muscle, IFITM5 and IFITM10 expression was significantly higher than that in skeletal muscle. The relative expression level of IFITM5 was higher in pancreas and IFITM5 was also expressed in fat (Figure 2).
Figure 2.
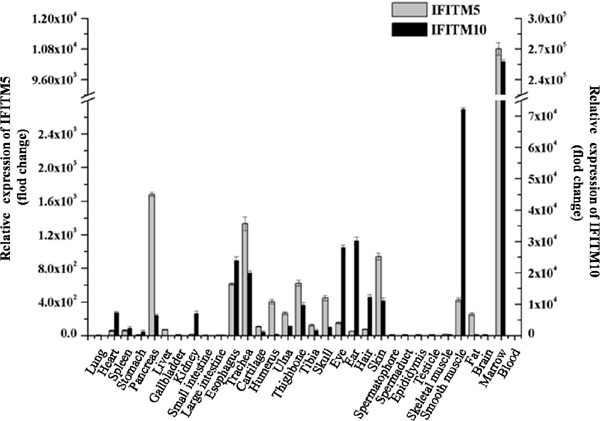
The relative expression of IFITM5 and IFITM10 in different rat tissues.
The expression level of IFITM6 was the highest in eye, higher in pancreas, gallbladder and bone, and very low in spleen, intestine, brain and the tissues of the reproductive system (Figure 3). The expression level of IFITM7 was similar to IFITM2, with the highest in skin, higher in the esophagus, eye, ear, hair, skeletal muscle, smooth muscle and bone marrow, and lower in other tissues (Figure 4).
Figure 3.
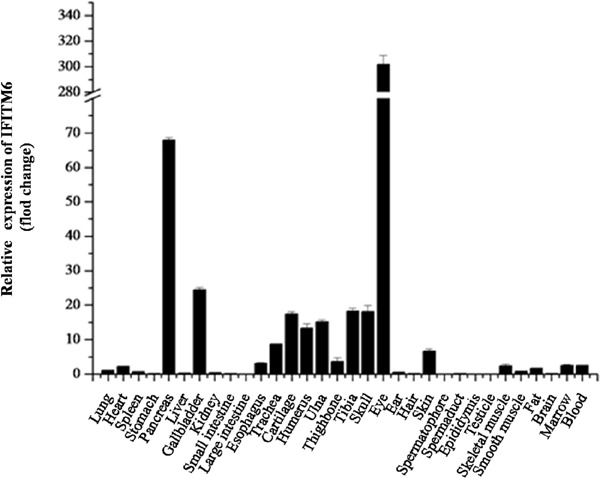
The relative expression of IFITM6 in different rat tissues.
Figure 4.
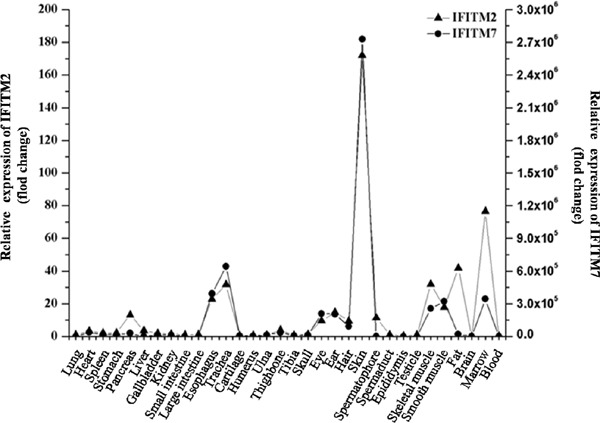
The relative expression trends of IFITM2 and IFITM7 in different rat tissues.
4. Discussion
Interferon inducible transmembrane (IFITM) proteins, playing an important role in cell proliferation, adhesion and immune system regulation, are a recently discovered family of cellular anti-viral proteins that restrict the replication of a number of enveloped and non-enveloped viruses. IFITM proteins are located in the plasma membrane and endosomal membranes, the main portals of entry for many viruses. The interferon-inducible transmembrane proteins (IFITMs) are a family of small transmembrane proteins belonging to the ISG superfamily and strongly induced by IFNs (25). The IFITM proteins are the distinct restriction factors known to block viral entry, including restriction of virus fusion with the late endosomal or lysosomal compartments and penetration into the cytoplasm (4,26). It has been shown that gene cluster IFTIM1, 2, and 3, the immune-related genes, are critically involved in immune defense against a broad variety of viruses, including influenza virus, dengue virus, filoviruses, coronavirus, hepatitis C virus, lyssaviruses, and West Nile virus (14,27–29). Bailey showed that IFITM3 was expressed in the epithelial cells of the airway and visceral pleura, while IFITM1 and IFITM2 had low expression in other parts of the body (30). In our study, they were expressed in lung and trachea, but also expressed in other tissues. These findings are consistent with previous research that IFITM1, IFITM2 and IFITM3 were widely expressed in the human body (4), and had a differential expression profile in the development of primordial germ cells (13). The relative expression levels of IFITM2 and IFITM3 in skin were significantly higher than those in other tissues, suggesting that IFITM2 and IFITM3 may be related to antiviral defense. In testis and epididymis, spermatophore and spermaduct, IFITM1, IFITM2 and IFITM3 expression showed a trend from high to low, while IFITM5, IFITM6, IFITM7 and IFITM10 expression is very low. IFITM7 expression is similar to that of IFITM2, indicating that they have a similar function. Moffatt reported that IFITM5 is related to calcium deposition in bone (21). We found that IFITM5 was not only expressed in bone, but also expressed in bone marrow, skin, pancreas and smooth muscle, and this is inconsistent with previous studies that IFITM5 was only expressed in bone. The reason for this difference may be that the specific expression of IFITM5 in bone is only in certain mammals (21). In the zebrafish model, IFITM5 was expressed in brain, muscle and liver tissues with no expression in bone; in the chicken model, its expression was high in muscle and liver, low in brain and ovary, but none in bone (31). Therefore, the expression level of IFITM5 in bone is different in various species, indicating that the IFITM5 gene may have other functions other than bone mineralization.
In addition, IFITM10 was highly expressed in the bone marrow; in smooth muscle, its expression was significantly higher than that in skeletal muscle; in the tissues of the intestinal and reproductive systems, it was unexpressed. Thus, the expression profile of IFITM10 is similar to that of IFITM5. Their distribution may be related to their gene homology (31), suggesting that IFITM10 and IFITM5 have similar functions. It is reported that IFITM10 was expressed in the brain and spleen of mice, whereas not expressed in bone (31). The expression level of IFITM10 in chicken was decreased with the continuous development of embryonic germ cells (32). IFITM10 had been reported in spleen tissues, and we found that IFITM10 was also expressed in esophagus, trachea, bone and muscle. IFITM6 was highly expression in macrophages (7), however, our results showed that IFITM6 was highly expressed in eye but not in spleen. There are still a few studies on IFITM6, IFITM7 and IFITM10. Currently, researchers pay more attention to the mechanism of their resistance to virus invasion and their relationship with tumors (30,33). In the present study, mRNA expression profile of IFITMs in different tissues was systematically compared and analyzed in rats, providing a favorable reference for revealing the functions of IFITMs. Next, we will analyze the protein expression profile of the IFITM family members.
Acknowledgements
This work was supported by a grant from the State Major Infectious Disease Research Program (China Central Government, 2017ZX10103004-007), Shandong Provincial Natural Science Foundation of China (2015ZRC03171) and Major Major Scientific and Technological Program of Shandong Province (2016ZDJS07A10)
References
- 1. Isaacs A, Lindenmann J. Pillars Article: Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957; 147:258-267. [PubMed] [Google Scholar]
- 2. Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004; 202:8-32. [DOI] [PubMed] [Google Scholar]
- 3. Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: A current view. Int Immunol. 2005; 17:1367-1378. [DOI] [PubMed] [Google Scholar]
- 4. Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013; 13:46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Evans SS, Collea RP, Leasure JA, Lee DB. IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J Immunol. 1993; 150:736-747. [PubMed] [Google Scholar]
- 6. Lange UC, Saitou M, Western PS, Barton SC, Surani MA. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003; 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith RA, Young J, Weis JJ, Weis JH. Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun. 2006; 7:113-121. [DOI] [PubMed] [Google Scholar]
- 8. Deblandre GA, Marinx OP, Evans SS, Majjaj S, Leo O, Caput D, Huez GA, Wathelet MG. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J Biol Chem. 1995; 270:23860-23866. [DOI] [PubMed] [Google Scholar]
- 9. Evans SS, Lee DB, Han T, Tomasi TB, Evans RL. Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood. 1990; 76:2583-2593. [PubMed] [Google Scholar]
- 10. Brem R, Oraszlan-Szovik K, Foser S, Bohrmann B, Certa U. Inhibition of proliferation by 1-8U in interferon-alpha-responsive and non-responsive cell lines. Cell Mol Life Sci. 2003; 60:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tirosh B, Daniel-Carmi V, Carmon L, Paz A, Lugassy G, Vadai E, Machlenkin A, Bar-Haim E, Do MS, Ahn IS, Fridkin M, Tzehoval E, Eisenbach L. ‘1-8 interferon inducible gene family’: putative colon carcinoma-associated antigens. Br J Cancer. 2007; 97:1655-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joung I, Angeletti PC, Engler JA. Functional implications in apoptosis by interferon inducible gene product 1-8D, the binding protein to adenovirus preterminal protein. J Microbiol. 2003; 41:295-299. [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka SS, Matsui Y. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev. 2002; 119:S261-S267. [DOI] [PubMed] [Google Scholar]
- 14. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009; 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, Brass AL. The IFITMs inhibit Zika virus replication. Cell Rep. 2016; 15:2323-2330. [DOI] [PubMed] [Google Scholar]
- 16. Muñoz-Moreno R, Cuesta-Geijo MÁ, Martinez-Romero C, Barrado-Gil L, Galindo I, Garcia-Sastre A, Alonso C. Antiviral role of IFITM proteins in african swine fever virus infection. PloS One. 2016; 11:e0154366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui K, Wang H, Zai S, Feng Y. Expression of IFITM3 in colorectal carcinoma and its clinical significance. Zhonghua Zhong Liu Za Zhi. 2015; 37:352-355. (in Chinese) [PubMed] [Google Scholar]
- 18. Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007; 26:594-603. [DOI] [PubMed] [Google Scholar]
- 19. Fumoto S, Shimokuni T, Tanimoto K, Hiyama K, Otani K, Ohtaki M, Hihara J, Yoshida K, Hiyama E, Noguchi T, Nishiyama M. Selection of a novel drug-response predictor in esophageal cancer: A novel screening method using microarray and identification of IFITM1 as a potent marker gene of CDDP response. Int J Oncol. 2008; 32:413-423. [PubMed] [Google Scholar]
- 20. Li D, Peng Z, Tang H, Wei P, Kong X, Yan D, Huang F, Li Q, Le X, Li Q, Xie K. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 2011; 17:3558-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, Godin E, de Oliveira PT, Atkins GJ, Nanci A, Thomas G. Bril: A novel bone-specific modulator of mineralization. J Bone Miner Res. 2008; 23:1497-1508. [DOI] [PubMed] [Google Scholar]
- 22. Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, Iden S, Wirth B, Eysel P, Koerber F, Schoenau E, Bohlander SK, Wollnik B, Netzer C. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012; 91:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu BY, Lu YQ, Han F, Wang Y, Mo XK, Han JX. Effects of the overexpression of IFITM5 and IFITM5 c.-14C>T mutation on human osteosarcoma cells. Oncol Lett. 2017; 13:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Z, Liu J, Li M, Yang H, Zhang C. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. Plos One. 2012; 7:e49265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013; 425:4937-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu SL. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013; 9:e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang IC, Bailey CC, Weyer JL, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011; 7:e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. The IFITM proteins inhibit HIV-1 infection. J Virol. 2011; 85:2126-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton N, Kellam P, Fife M. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. J Virol. 2013; 87:12957-12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu F, Ng SS, Chow BK, Sze J, Lu G, Poon WS, Kung HF, Lin MC. Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J Neurooncol. 2011; 103:187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hickford D, Frankenberg S, Shaw G, Renfree MB. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics. 2012; 13:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okuzaki Y, Kidani S, Kaneoka H, Iijima S, Nishijima KI. Characterization of chicken interferon-inducible transmembrane protein-10. Biosci Biotechnol Biochem. 2017; 81:914-921. [DOI] [PubMed] [Google Scholar]
- 33. He J, Li J, Feng W, Chen L, Yang K. Prognostic significance of INF-induced transmembrane protein 1 in colorectal cancer. Int J Clin Exp Pathol. 2015; 8:16007-16013. [PMC free article] [PubMed] [Google Scholar]


