Abstract
Vps34, a class III PtdIns3 lipid kinase involved in the control of both autophagic and endocytic systems, has been studied extensively in numerous fundamental cellular processes. Accumulating evidence indicates that Vps34 may also contribute to the development and progression of human cancers. However, the mechanism of Vps34 in tumorigenesis remains elusive. Here, we report an unanticipated role of Vps34 in the activation of p62 for cancer development. We identified that Vps34 is a transcriptional activator of p62 through competition of Nrf2 (nuclear factor erythroid 2-related factor 2) for Keap1 binding. Vps34 augments the association of PKC-δ with p62 for its phosphorylation at Serine 349, which leads to positive feedback on the Nrf2-dependent transcription of oncogenes. Additionally, we found that the expression of Vps34 is correlated with the tumorigenic activity of human breast cancer cells. Normally inactive in breast cancer, caspase 8 can cleave Vps34 at residue D285, which directly abolished its lipid kinase activity and dramatically altered cell invasion potential, colony formation, as well as tumorigenesis in orthotopic engraftments in mice. The cleavage at D285 blocks expression of LC3-II, Nrf2 and subsequently, p62, in addition to blocking tumor growth, indicating that the intact structure of Vps34 is essential for its activity. Moreover, either knockout of PKC-δ or knockdown of p62 by small interfering RNA in MCF-7 cells abrogates Vps34-dependent tumor growth. Data presented here suggested that Vps34 stimulates tumor development mainly through PKC-δ- activation of p62.
Introduction
Autophagy is a catabolic degradation process through which the cell recycles its own cytoplasmic components to maintain cellular homeostasis in response to various cellular stresses.1, 2 In mammalian cells, autophagy constitutes a multistep process coordinated by a wide range of molecular mechanisms in many cellular contexts and diseases, including cancer. However, the role of the autophagy pathway in tumorigenesis is controversial.3, 4, 5 Among autophagy related proteins, one protein that has attracted immense interest is the class III phosphoinositide 3-kinase (PI3K) vacuolar protein sorting 34 (VPS34), which forms numerous complexes with the core components p150 (Vps15), Beclin 1 and ATG14L to function in a variety of cellular and physiological events.6, 7, 8 It generates intracellular pools of phosphatidylinositol 3-phosphate (PtdIns(3)P) for protein trafficking. Another one is the well-known autophagic adaptor protein, sequestosome1 (SQSTM1, also known as p62 in humans). p62 is one of the selective substrates for autophagy, as well as a scaffold protein involved in multiple signaling pathways that govern important cellular events, including autophagosomes. Both Vps34 and p62 have important roles, separately, in vesicle nucleation and autophagy progression.7, 9, 10, 11 Furthermore, the expression of Vps34 is always at a high level in various cancer tissues relative to normal tissues,12 indicating that Vps34 could be a biomarker for breast cancer. Moreover, the expression and activity of p62 are frequently associated with human cancers. Recently, there has been increasing interest in exploring the link between Vps34 complexes and human diseases, including cancer.13, 14, 15, 16 However, the mechanisms responsible for Vps34-mediated cancer development remain largely unknown. Here, we report that defective autophagy stimulates cancer development via the Vps34 activation of p62. We further demonstrate that Vps34 is a transcriptional activator of p62 through competition of nuclear factor erythroid 2-related factor 2 (Nrf2) for Kelch-like ECH-associated protein 1 (Keap1) binding. In addition, we show that Vps34 augments the association of PKC-δ with p62 for its phosphorylation at Serine 349. Additionally, we demonstrate that, although normally inactive in breast cancer, caspase 8 directly abolishes the lipid kinase activity of Vps34 via the cleavage at residue D285, resulting in a dramatic alteration of Vps34 signaling in cell invasion potential, colony formation, as well as tumorigenesis. Moreover, knockout of PKC-δ by Crispr-cas9 and knockdown of p62 by small interfering RNA (siRNA) in MCF-7 cells abrogates Vps34-dependent tumor growth in vivo. These results constitute the first evidence that Vps34 contributes to the tumorigenesis of breast cancer through p62 phosphorylation at S349 by Vps34-recruitment of PKC-δ, and could be beneficial to elucidating the role of Vps34 and defective autophagy in cancer progress.
Results
Caspase 8 has important role in the fusion of autophagosome with lysosome
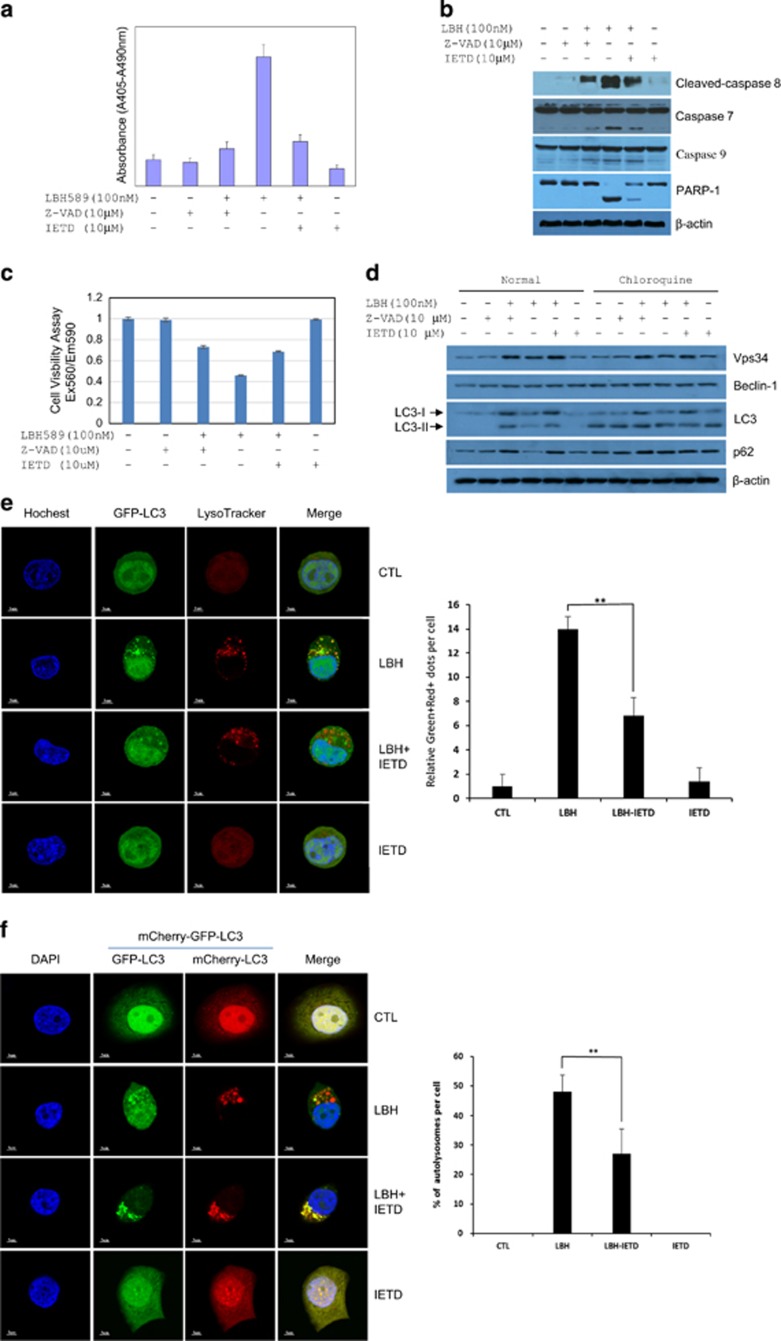
LBH589 is a hydroxamic acid inhibitor of histone deacetylases that is clinically used to treat multiple myeloma and is currently in clinical trials as a potential treatment for other cancers. Our previous studies suggest that LBH589 engenders both apoptosis and autophagy in breast cancer cells.17 To address whether LBH589 induces apoptosis mainly via the activation of caspase 8, the pan-caspase inhibitor z-VAD-fmk and caspase 8-specific inhibitor z-IETD-fmk were used, respectively, to treat MCF-7 cells with LBH589. Apoptotic cell death was completely prevented by z-VAD-fmk and significantly blocked by z-IETD-fmk (P<0.05) (Figure 1a). According to western analysis, z-VAD-fmk and z-IETD-fmk significantly inhibited the processing of caspase 7, caspase 8 and caspase 9, leading to reduced cleavage of PARP 1 induced by LBH589 (Figure 1b). Similarly, a large proportion of LBH589-treated cell viability loss was reduced by z-VAD-fmk and z-IETD-fmk (Figure 1c). Therefore, capspase-8 has a key role in LBH589-induced cell death.
Figure 1.
Inhibition of caspase 8 promotes the fusion of autophagosome with lysosome. (a) Inhibition of LBH589-induced apoptosis by caspase inhibitors. Apoptotic cell death of MCF-7 cells induced by treatment with the indicated concentrations of LBH589 and/or caspase inhibitors for 48 h. For caspase inhibitor treatment, cells were pretreated with 10 μM z-VAD-FMK (Z-VAD) or z-IETD-FMK (IETD) for 4 h. (b) Z-VAD or IETD inhibited LBH589-induced cleavage of caspase 7, caspase 8, caspase 9 and PARP-1 in MCF-7 cells treated as indicated for 24 h. (c) Cell viability of MCF-7 cells treated as indicated for 24 h. (d) Accumulation of Vps34, LC3 and p62 in MCF-7 cells treated as indicated with or without 20 μM chloroquine for 24 h. (e) Confocal microscopic evaluation of autophagosome-lysosome fusion in MCF-7 cells. MCF-7 cells stably expressing EGFP-LC3 (Green) treated as indicated for 18 h were stained with lysosome tracker staining (Red). Immunofluorescence analyses were performed using confocal microscopic detection (63 × oil). Left, representative confocal images. Right, relative Green+red+ dots per cell calculated from 20 cells; bars, s.d. **P<0.01. (f) Disruption of autophagosome-lysosome fusion in MCF-7 cells. MCF-7 cells stably expressing tfLC3 were treated as indicated for 18 h. Immunofluorescence analyses were performed using confocal microscopic detection (60 × oil). Left, representative confocal images. Right, % of autolysosomes per cell was calculated from 20 cells by using the formula of (1-yellow dots/red dots) × 100; bars, s.d. **P<0.01. DAPI, 4', 6-diamidino-2-phenylindole.
The studies also revealed that autophagy has a protective role in LBH589-induced apoptosis.17 To confirm the induction of autophagy in this setting, autophagic flux was assayed by western blotting of autophagy markers. Exposure to LBH589 treatment triggered the maturation of autophagy, as indicated by increased levels of LC3 and decreased levels of p62, which is degraded in the autolysosome and rescued by lysosomal inhibitor chloroquine (Figure 1d). However, co-treatment with LBH589 and caspase inhibitors further increased the level of Vps34, LC3, and p62 (Figure 1d). The increased levels of p62 suggest that z-VAD-fmk and z-IETD-fmk might disrupt the autophagosome-lysosome fusion. As a specific inhibitor of caspase 8, z-IETD-fmk, was used to explore the involvement of caspase 8 in the fusion of autophagosome with lysosome. The colocalization of EGFP-LC3 (green dots) and lysosomotropic dye (LysoTracker RED, red dots) was induced to yield yellow dots and red-predominant yellow dots in response to LBH589 treatment in MCF-7 cells stably expressing EGFP-LC3. In addition, z-IETD-fmk prevented the colocalization of GFP-LC3 green dots with LysoTracker red dots, exhibiting increased green and red fluorescence signal separately in the cytoplasm, which was apparent in cells treated with the combination of LBH589 and z-IETD-fmk, in contrast to cells treated with LBH589 alone (Figure 1e). This was further confirmed in MCF-7 cells stably expressing mCherry-GFP-LC3 (tf-LC3) that LBH589 treatment induced the maturation of autophagolysosomes (mRFP+GFP−, red-predominant dots), but z-IETD-fmk blocked the fusion of autophagosome with lysosome (mRFP+GFP+, yellow dots) (Figure 1f). These results indicated that LBH589 likely exerts its cytotoxic effects predominantly through a caspase 8-dependent pathway, and activated caspase 8 enhances the maturation of autophagolysosome.
Active caspase-8 cleaves Vps34 at D285
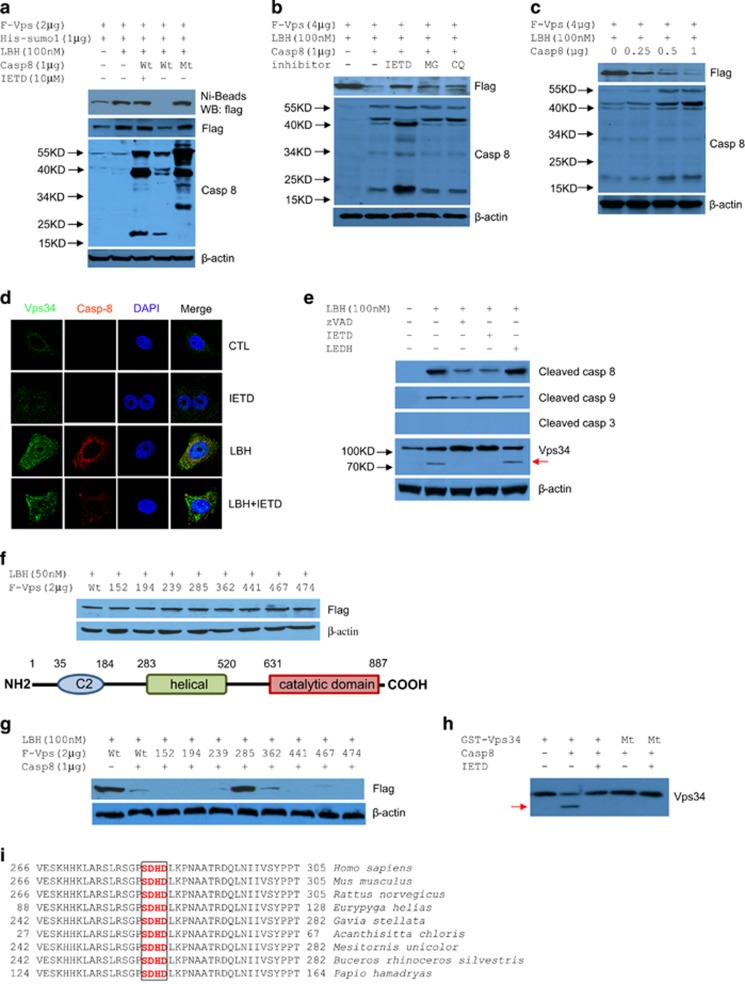
Interestingly, we also observed that the combination of caspase 8 inhibitor with LBH589 induced a stronger accumulation of Vps34 than LBH589 treatment alone (Figure 1d). This finding prompted us to investigate the correlation between caspase 8 and Vps34. Post-translational modification by Sumo 1 stabilizes and enhances the activity of Vps34.17 Hence, the effect of caspase 8 on the SUMOylation of Vps34 was initially assessed by Ni-bead precipitation of flag-tagged Vps34 in extracts of HEK293T cells co-expressing his-tagged Sumo 1, flag-Vps34, and caspase 8 (Wt) or its dominant negative mutant (Mt). Compared with control cells, in the presence of LBH589, active caspase 8 abolished a substantial amount of SUMOylated Vps34 and abrogated almost the total expressed flag-Vps34, which was rescued by z-IETD-fmk, but not in the case of catalytically inactive mutant of caspase 8 (Figure 2a). Moreover, unlike with the caspase 8 inhibitor z-IETD-fmk, neither the proteasome inhibitor MG132 nor the lysosome inhibitor chloroquine rescued the expression of Flag-Vps34 degraded by LBH589-activated caspase 8 (Figure 2b). Exogenous Vps34 was depleted in a dose-dependent manner by LBH589-activated caspase 8 (Figure 2c). These results indicated that active caspase 8 eliminates Vps34 through enzymatic cleavage.
Figure 2.
Vps34 is cleaved by active caspase 8. (a) Active caspase 8 eliminated SUMOylated Vps34 and total Vps34. Immunoblot of His-SUMO1-conjugated Vps34 and total Vps34 in HEK293T cells transfected as indicated for 24 h, and treated with LBH589 for 24 h. (b) Except z-IETD-FMK (20 μM), neither MG132 (10 μM) nor chloroquine (20 μM) blocked abrogation of F-Vps34 caused by active caspase 8. Immunoblot analyses were performed on transfected HEK293T cells, as indicated. (c) Active caspase 8 degraded Vps34 in a dose-dependent manner. Vps34 was extracted from HEK293T cells transfected as indicated. (d) Immunostaining of endogenous Vps34 (green) colocalized with active caspase 8 (red) in MCF7 cells treated with LBH589 and/or z-IETD-FMK, for 18 h. (e) Abrogation of endogenous Vps34 cleavage by caspase inhibitors in LBH589-treated MCF-7 cells. The red arrow denotes cleaved Vps34. (f) Upper, immunoblot analyses of variant mutant of Vps34 expression in HEK293T cells with the presence of LBH589 for 24 h. Lower, the location of predicted cleavage sites in Vps34 structure of scheme. (g) Caspase 8 cleavage of Vps34 at the site of Asp285. Immunoblot of the variant mutant effects on Vps34 expression in HEK293T cells cotransfected with caspase 8 treated with 50 nM LBH589 for 24 h. (h) In vitro cleavage of Vps34 by active caspase 8. The red arrow denotes cleaved Vps34. (i) Multiple alignment of Vps34 shows conservation of the caspase 8 cleavage site in the SDHD motif. DAPI, 4', 6-diamidino-2-phenylindole.
Autophagy often occurs in cancer cells during chemotherapy, which induces apoptosis. Previous studies revealed a complex relationship between apoptosis-associated proteins and autophagy. Autophagy can be inhibited by antiapoptotic proteins BCL2, BCL-XL and MCL1;18, 19, 20 autophagy, however, can be triggered by proapoptotic proteins BAD, PMAIP1 and BBC3.21, 22 More importantly, autophagy-related proteins, such as p62,23 NBR123 and several ATG proteins, including ATG4D,24 ATG525 and Beclin-1,26, 27, 28 can be cleaved by calpain or caspases. Besides those proteins, Vps34 is also cleavable by caspase 3, caspase 7, caspase 8, and caspase 9 during apoptosis induced by growth factor or serum deprivation.27 In caspase 3-deficient MCF-7 cells, active caspase 8 (red) colocalized with endogenous Vps34 (green) induced by LBH589 treatment (Figure 2d). To test whether caspase 8 was responsible for the cleavage of endogenous Vps34, zVAD, IETD and caspase 9 inhibitor z-LEDH-fmk (LEDH) were used individually to treat cells together with LBH589. LBH589-induced cleavage of Vps34 was inhibited by both zVAD and IETD, but not LEDH in MCF-7 cells (Figure 2e). To verify the cleavage site of Vps34 by active caspase 8, eight Vps34 mutants were constructed according to the prediction of CASVM,29 in which certain aspartic acid residues (D) were substituted with glutamic acid (E) in different regions, except the catalytic domain as shown in the illustration (Figure 2f-lower), and all Vps34 mutants expressed well in HEK293T cells (Figure 2f-upper). Similar to the wild-type Vps34, seven of the eight mutants were degraded in the presence of LBH589 activated caspase 8, while only D to E mutation at site Asp-285 was resistant to the cleavage (Figure 2g), suggesting that D285 contributes to Vps34 cleavage. This was confirmed by in vitro caspase cleavage assay using GST-tagged Vps34 incubated with active caspases 8 (Figure 2h). These results support that caspase 8 is responsible for LBH589-induced cleavage of both endogenous and exogenous Vps34. Furthermore, multiple alignment of the proceeding SDHD motif of this Asp-285 was performed to investigate its relative importance to Vps34. The motif SDHD was highly conserved in a wide range of species, including other mammals (Figure 2i). Therefore, Vps34 is a direct substrate of caspase 8 and the conserved SDHD sequence (Vps34 amino acids 282–285) is the caspase-8 target site on this protein.
D285 structurally affects the activity and function of Vps34
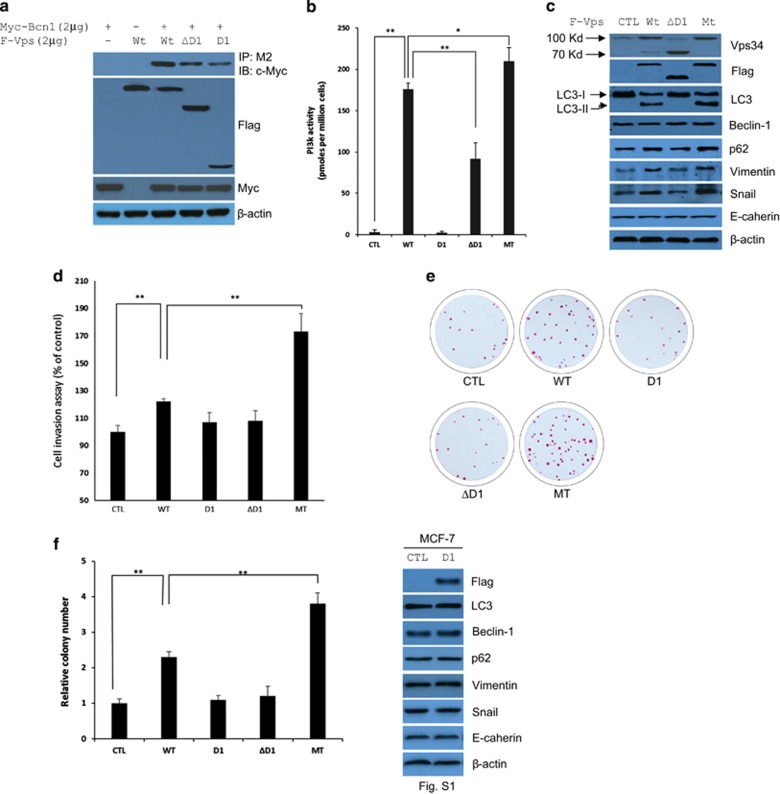
We next investigated whether blocking the cleavage of Vps34 affects its PI3K activity and relevant biological functions. Because the cleavage site (D285) by caspase 8 is located at the N-terminal of the helical region, we generated constructs to express flag-tagged Vps34-full length (Wt, 1-887 aa), Vps34-D1 (D1, 1-285 aa), Vps34-ΔD1 (ΔD1, 285-887 aa) and Vps34-Mt (Mt, D285E) for its functional analysis. As Beclin-1 is a critical component in class III PI3K complex (PI3KC3), which has a key role in the autophagy–lysosome pathway, Vps34-Wt, Vps34-D1 and Vps34-ΔD1 were transfected separately into HEK293T, together with Myc-tagged Beclin-1 to test the alteration of Beclin-1/Vps34 complex assembly. Immunoblotting results showed that cleavage of Vps34 at D285 decreased its affinity with Beclin-1 (Figure 3a). Consistent with this finding, biochemical analyses confirmed that N-terminal truncation reduced the output of PI3P levels more than threefold of wild-type Vps34, whereas D1 fragment did not exhibit any PI3K lipid kinase activity because of lacking PI3K kinase catalytic domain (Figure 3b). Therefore, N-terminal of Vps34 is essential for its PI3K lipid kinase activity.
Figure 3.
D285 structurally affects the activity and function of Vps34. (a) Cleavage of Vps34 at D285 weakened the interaction of Vps34 with Beclin1 (Bcn 1). Immunoblots of Myc-Beclin1 in M2 immunoprecipitates from HEK293T cells transfected as indicated. (b) Effect of blocking the cleavage on the in vitro lipid kinase activity of Vps34; bars, s.d. *P<0.05; **P<0.01. (c) and Supplementary Figure S1 Expression levels of EMT indicators and autophagic flux in parental cells or MCF-7 cells stably expressing Vps34, Vps34-D1, Vps34-ΔD1 and Vps34-Mt (D285E). (d) In vitro invasion of MCF-7 parental cells or MCF-7 cells stably expressing Vps34, Vps34-D1, Vps34-ΔD1 and Vps34-Mt (D285E). Column, average data of three independent experiments; bars, s.d. *P<0.05. (e) Cleavage of Vps34 at D285 diminishes its competence to promote colony formation. Clonogenic images of parental cells, MCF-7/Vps34, MCF-7/Vps34-ΔD1 and MCF-7/vsp34-Mt (D285E) photographed on day 14. (f) Colony numbers of cells as shown in Figure 3e were counted. Values plotted were means of three experiments plus or minus s.d. **P<0.01.
To determine the effect of Vps34 cleavage on its biological activities in breast cancer, we established stable transfectants of MCF-7 cells-expressing flag-tagged Vps34-Wt, Vps34-D1, Vps34-ΔD1 and Vps34-Mt to perform the Matrigel-based invasion assay and colony formation assay. In vitro evaluation showed that overexpression of Vps34-Wt markedly increased the invasiveness of MCF-7 cells, whereas both Vps34-D1 and Vps34-ΔD1 had no effect, and Vps34-Mt statistically and significantly intensified the increase (Figure 3d). Similar cases also extended to colongenic assessment (Figures 3e and f). Here, we also performed the assay for autophagic flux, and found that MCF-7 cells stably expressing Vps34-Wt dramatically increased the conversion of LC3 I to LC3 II forms, and the intracellular accumulation of p62 and the EMT markers Snail and Vimentin, but not Beclin1 and E-cadherin. As expected, Vps34-Mt stable cells further enhanced the accumulation of LC3, p62 as well as Snail and Vimentin relative to wild-type Vps34 cells, whereas neither D1 nor ΔD1-expressing cells did not (Figure 3c and Supplementary Figure S1). Taken together, these results suggested that intact structure is required for the function of Vps34 to support the invasiveness and colony formation.
Vps34 enhances the expression of p62 transcriptionally via NRF2
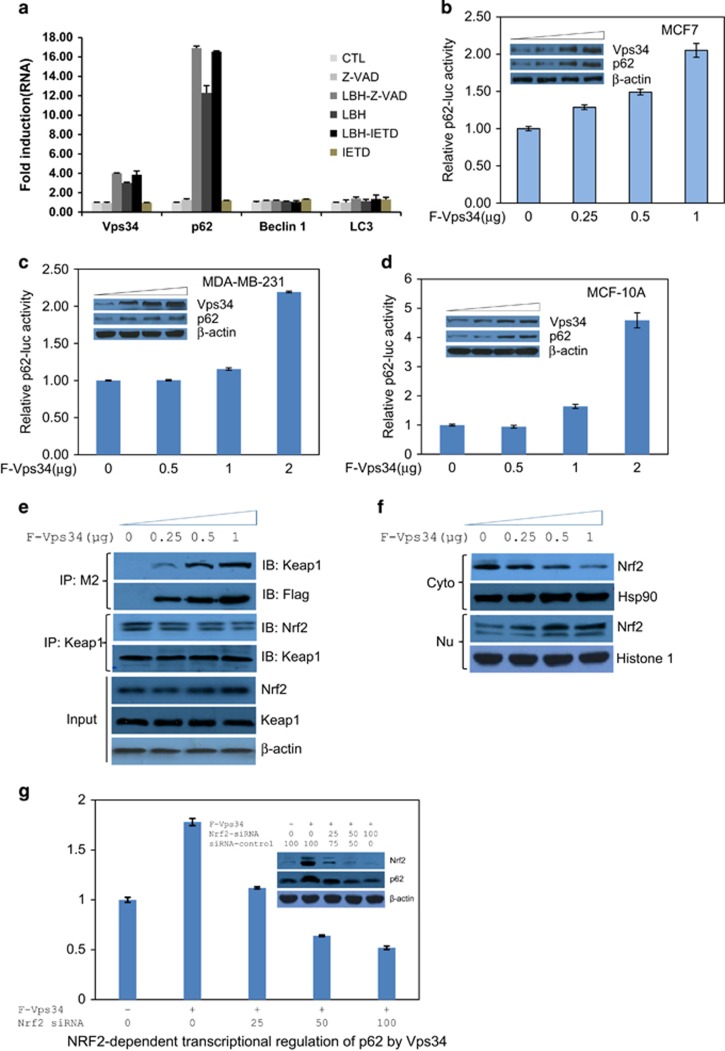
To further evaluate whether accumulation of Vps34, p62 and LC3, in response to co-treatment of LBH589 with caspase inhibitors, was regulated transcriptionally, quantitative expression analyses using Real-Time qRT–PCR were performed. Although little change in LC3 and Beclin 1 mRNA were detected, mRNA of p62 increased notably than Vps34 in MCF-7 cells treated with LBH589 and the caspase inhibitor (Figure 4a). Combined with the previous observation that Vps34 overexpression in MCF-7 cells augmented induction of p62, we speculated that Vps34 might stimulate the gene expression of p62. To test this hypothesis, p62-promoter luciferase assay was performed in three cell lines, that is, MCF-7, MDA-MB-231, and MCF10A. As shown in Figures 4b–d, transient ectopic expression of Vps34 dose-dependently stimulated the activity of p62 promoters, and the translational level of p62 was positively correlated to Vps34 alteration. These results supported that Vps34 contributes to the expression of p62. It was recently reported that the transcription factor Nrf2 affects p62 transcription and forms a positive feedback loop with Nrf2.30, 31, 32 Thus, we hypothesized that Nrf2 might function as a transcriptional regulator for Vps34-mediated upregulation of p62. When transfected into MCF-7 cells, we found that Vps34 docks directly onto Keap1 to release Nrf2 from the complex sequestered by Keap1, an adaptor of the Cul3–ubiquitin E3 ligase complex that degrades Nrf2. Upon overexpression of Vps34, the expression pattern of both Keap1 and Nrf2 exhibited no difference in MCF-7 cells (Figure 4e). Intriguingly, Vps34 dose-dependently extracted Keap1 from Keap1–Nrf2 complex (Figure 4e), and led to a reduction of Nrf2 in cytoplasm and enrichment in nucleus (Figure 4f). Nuclear accumulation is usually considered to be necessary for Nrf2 to function as a transcription factor for p62.31 Therefore, we next determined whether Nrf2 was involved in the Vps34-induction of p62. First, we tested whether Nrf2 was a key element for Vps34-upregulation of p62 by using siRNA to knockdown Nrf2, and then estimated the activity of p62 transcription by luciferase assay. Knockdown of Nrf2 effectively diminished Vps34-induced activation of the p62 promoter and, concomitantly, reduced the augmentation of p62 caused by Vps34 overexpression in MCF-7 cells (Figure 4g), which is consistent with the expression pattern of Vps34 and p62 (Figure 1d; Figures 4b–d). These results indicated that Vps34 can initiate Nrf2-activated expression of p62 and assembles a Vps34-Nrf2-p62 axis. Therefore, Vps34 has an intrinsic function to modulate p62 levels through a mechanism dissociating the Keap1–Nrf2 complex.
Figure 4.
Vps34 enhances the expression of p62 transcriptionally via NRF2. (a) Real-Time PCR analysis of Vps34, Beclin-1, LC3 and p62 in MCF-7 cells with treatment as indicated for 24 h. Vps34 dose-dependently increased the transcriptional activity of the p62-promoter in MCF-7 cells (b), MDA-MB-231 cells (c), and MCF-10 A cells (d). The transcriptional activity of p62-promoter was expressed as relative luciferase units (RLU) normalized to Renilla activity and the expression level of Vps34 and p62 were further confirmed by western blotting of cell lysates. (e), (f) Vps34 dose-dependently enhanced the expression and the binding with Keap1 to release Nrf2 for its nuclear translocation. MCF7 cells were transfected with increased FLAG-Vps34 constructs for 48 h, as indicated. Immunoblot analyses of Nrf2 in cytosolic extracts and nuclear extracts. (g) Knockdown of Nrf2 eliminated Vps34-induced p62-promoter activation. MCF-7 cells were transfected with FLAG-Vps34 and/or Nrf2 siRNA for 24 h prior, followed by luciferase assay. Cell lysates were analyzed by immunoblots with anti-Nrf2, anti-p62 and anti-β-actin antibodies.
Vps34 induces PKC-δ-dependent phosphorylation of p62
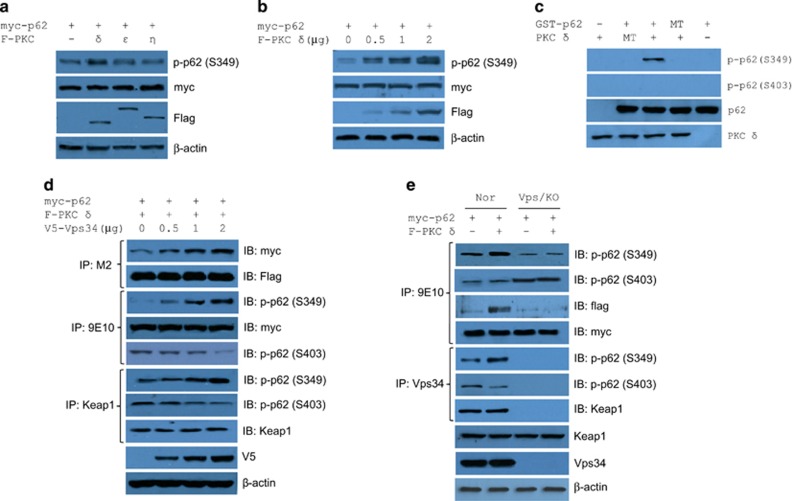
The selectivity of autophagy relies on adaptor proteins, such as p62, to attach the autophagic machinery with its specific substrates.33 The phosphorylation at S349 contributes to tumor growth through the disruption of the Keap1–Nrf2 complex due to the high affinity of S349-phosphorylated p62 to Keap1.34 Therefore, Vps34 may stimulate the S349-phosphorylation of p62, a protein demonstrated to promote tumorigenesis.35 Protein kinase C (PKC) is a serine/threonine protein kinase family, consisting of three subfamilies including classical (α, βI, βII and γ), novel (δ, ε, θ and η), and atypical (ξ and λ). p62 was initially identified as a binding partner of atypical PKC.36 Surprisingly, p62 was not found to be a substrate for classical and atypical PKCs (data not shown). To explore the possibility of p62 as a substrate for PKC, we next tested whether the S349-phosphorylation of p62 was by PKC δ, PKC ε or PKC η. We found that only PKC-δ enhanced the S349 phosphorylation in HEK293T cells (Figure 5a) and in a dose-dependent manner (Figure 5b). To determine whether the PKC-δ phosphorylates S349 of p62 in vitro, we performed in vitro phosphorylation assays using purified GST-fused p62 and His6-PKC-δ, showing that p62 was phosphorylated at S349 by PKC-δ, but not PKC-δ with K378M mutation, whereas S349 phosphorylation was not observed in the S349A mutant (Figure 5c). Therefore, PKC-δ is one of the protein kinases for the phosphorylation of p62 at S349. As expected, Vps34 dose-dependently enhanced the S349-phosphorylation of p62 by PKC δ in HEK293T cells, resulting in elevated Keap1–p62 (p-S349) complex and decreased Keap1–p62 (p-S403) complex (Figure 5d). More interestingly, Crispr-cas9 knockout of Vps34 abrogated the interaction between p62 and PKC δ, resulting in decreased PKC δ-dependent phosphorylation of p62 at S349 accompanied by increased PKC δ-independent phosphorylation of p62 at S403 in MCF-7 cells (Figure 5e). Vps34 enhanced the interaction between Keap1 and S349 phosphorylated p62 (Figure 5d) while endogenous Vps34 existed in a complex with Keap1 and preferentially formed complex with S349 phosphorylated, not S403 phosphorylated p62 (Figure 5e). Except for the transactivation of Nrf2 by disrupting Keap1–Nrf2 complex30, 34 in autophagy, p62 is also a multi-functional and multi-domain protein, participating in many other signal pathways and cellular functions, for example, the removal of damaged mitochondria in parkin-mediated mitophagy,37, 38, 39 and NF-κB activation by binding to and activating atypical PKC, RIP1 kinase or TRAF6 (TNFα receptor-associated factor 6).40, 41, 42
Figure 5.
Vps34 induces PKC-δ-dependent phosphorylation of p62. (a), (b) PKC δ is a specific protein kinase to p62 in cells. Cell lysates from MCF-7 cells transfected as indicated were analyzed by western blotting with antibodies recognizing phosphorylated p62 at S349, Myc-tag, falg-tag and β-actin. (c) In vitro phosphorylation of p62 by PKC-δ. (d) Vps34 dose-dependently enhanced the formation of p62-PKC δ machinery, and consequently the phosphorylation of p62. MCF7 cells were cotransfected with myc-p62, F-PKC δ and increasing number of V5-Vps34 constructs for 48 h followed by immunoprecipitation assays for the formation of p62-PKC δ machinery and the phosphorylation of p62. (e) Ablation of Vps34 decreased PKC-δ-dependent phosphorylation of p62. Myc-p62 with or without F-PKC-δ was cotransfected into MCF-7 or Vps34 knockout MCF-7 cells for 48 h followed by the detection, as indicated.
Intact structure is essential for Vps34-dependent tumor progress
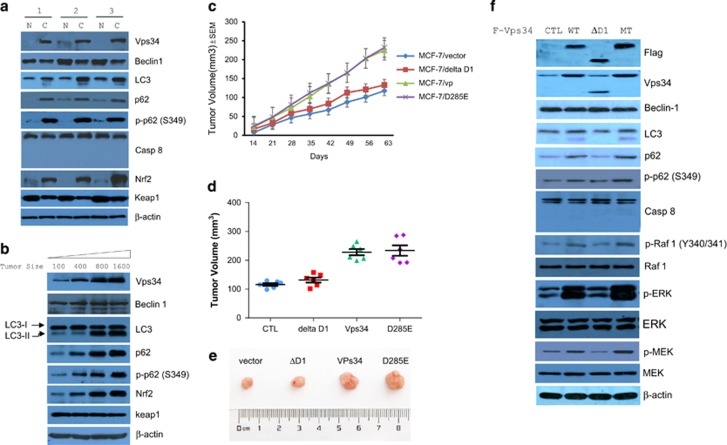
As p62 is also a transcriptional target gene for Nrf2, it creates a positive feedback loop during antioxidant response.32 Emerging evidence shows that abnormal expression of p62 has indeed been associated with several cancers, including prostate, kidney, liver, lung, breast and oral cancer cases.12, 43, 44, 45 Beyond p62 and S349-phosphorylated p62, we observed that the expression levels of various autophagy-related proteins, including Vps34, LC3, and Nrf2, were positively associated with cancer progression, whereas caspase 8 was inactivated in breast cancer tissues and MCF-7 carcinoma mouse xenografts (Figures 6a and b). Having observed the effects of Vps34 cleavage on tumor cell proliferation and invasion in vitro, we sought to investigate its role in an in vivo setting. MCF-7 cells stably expressing flag-tagged Vps34, Vps34-ΔD1 and Vps34-Mt (D285E) were inoculated individually into the mammary fat pads of immunodeficient mice with E2-supplementation to compare their overall growth rates. As shown in Figures 6c–e, MCF-7 cells stably expressing flag-tagged Vps34 or Vps34-Mt, but not Vps34-ΔD1, showed a significant increase (>2-fold) in tumor growth over parental MCF-7 cells (P<0.01). As with its expression in breast cancer tissues (Figure 6a), caspase 8 exists in inactive status in MCF-7 carcinoma mouse xenografts, and the expression of LC3, Vps34 and p62 were higher in Vps34-Mt-expressing cells than in Vps34-Wt-expressing cells. However, expression in neither Vps34-ΔD1-expressing cells nor parental MCF-7 cells was higher than that of wild-type Vps34-expressing cells developed tumors (Figure 6f). In particular, D285E mutation further increased Vps34 phosphorylation of Raf 1, ERK and MEK, but the removal of the N-terminal abolished the activity of Vps34 (Figure 6f), which is consistent with that PI3K inhibition leads to suppression of protein kinase MEK/ERK signaling.46, 47 Phosphorylation of Tyr 340/341 is involved in Raf 1 activation, leading to the phosphorylation and activation of the MEK/ERK cascade for tumor growth. These results strongly suggest that intact structure is important for Vps34-dependent phosphorylation of Raf 1, ERK and MEK, indicating a Vps34 activation of ERK pathway in tumor growth.
Figure 6.
Intact structure is essential for Vps34-dependent tumorigenesis. (a) Immunoblot analysis of lysates from matched pairs of breast cancer tissues, marked as C, and adjacent normal tissue, marked as N, was performed with antibodies as indicated. (b) Protein expression as indicated in a was detected in carcinoma tissue following increased tumor volumes of MCF-7 xenograft. (c) Assay of tumorigenicity. MCF-7/Vector, MCF-7/Vps34, MCF-7/Vps34-ΔD1, Vps34-Mt (D285E) cells (1.5 × 106) were injected into nude mice (six mice per group), separately. Growth rates of tumors were measured and volumes (mm3) were calculated on each of the indicated days after inoculation; each point represents the mean (±s.e.m.) calculated volume of six tumors. At 35-day post-inoculation, tumor volumes were measured (d), and tumors were photographed (e). (f) Immunoblot analysis of lysates from tumor tissues in d were performed with antibodies as indicated.
Vps34 stimulates tumor progress through PKC-δ activation of p62 and ERK pathway
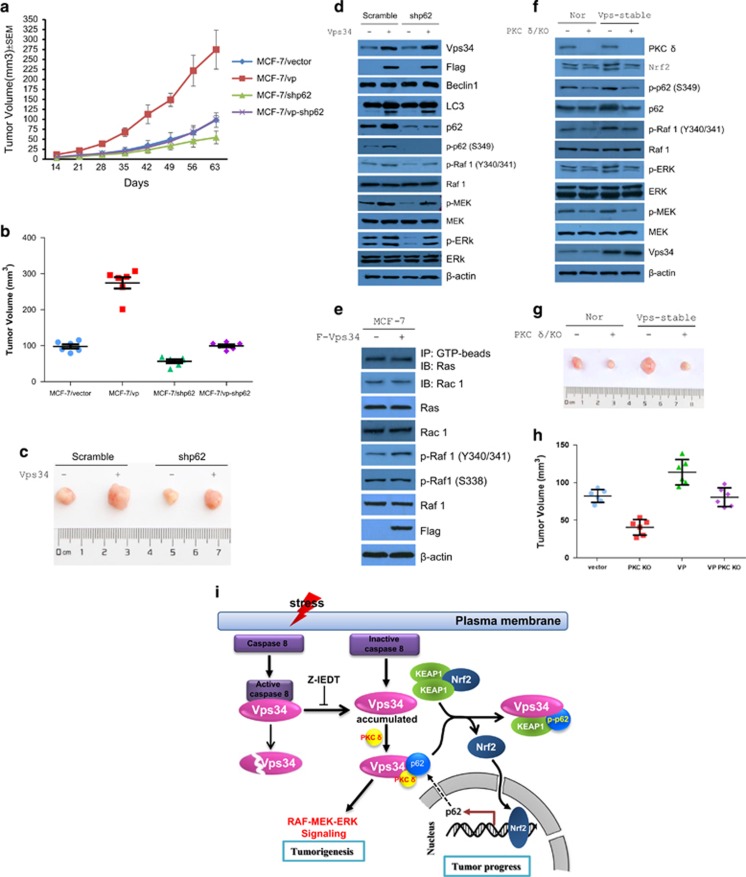
To test the hypothesis that Vps34 promotes tumor progress via p62 induction, we generated p62-knockdown MCF-7 and p62-knockdown MCF-7 cells stably expressing Vps34. These stable cells and their parental cells were next inoculated into immunodeficient mice with E2-supplementation. In contrast to p62-controls, the mice that received p62-short hairpin RNA (shRNA) or p62-shRNA-Vps34-expressing cells developed tumors that were slower in growth and volume than those receiving parental cells or Vps34-expressing cells (Figures 7a–c), indicating that Vps34 stimulation of tumor growth is majorly via p62. Immunoblotting data showed that knockdown of p62 decreased the phosphorylation levels of Raf 1, MEK and ERK induced by Vps34 overexpression in vivo (Figure 7d), indicating that p62 is partially involved in Vps34-dependent activation of Raf 1–MEK–ERK pathway. These data supported that p62 is necessary for Vps34 promotion of tumorigenesis, which is consistent with the role of p62 in tumorigenesis.48, 49
Figure 7.
Vps34 stimulates tumor progress through PKC-δ activation of p62 and ERK signaling. Tumorigenicity of MCF-7 or MCF-7/Vps34 stably infected with lentiviral control or p62 shRNA. In all, 1.5 × 106 cells of each transfectant were injected into nude mice (six mice per group), separately. (a) Growth rate of tumors was measured, and volumes (mm3) were calculated on each of the indicate days after inoculation; each point represents the mean (±s.e.m. calculated volume of six tumors. At 35-day post-inoculation, tumor volumes were measured (b), and tumors from mice were photographed (c). (d) Immunoblot analysis of lysates from tumor tissues in c was performed with antibodies as indicated. (e) Ras and Rac1 were not involved in Vps34 activation of Raf 1. GST-beads pulldown and immunoblot analysis of lysates from MCF-7 or MCF-7/Vps34 was performed with antibodies as indicated. (f) PKC-δ is a key effector in Vps34 activation of p62 and Raf 1 signaling. Analysis of cell lysates from MCF-7 or MCF-7/Vps34 cells and PKC-δ knockout MCF-7 or MCF-7/Vps34 cells performed with antibodies as indicated. (g, h) Ablation of PKC-δ blocked Vps34-dependent tumorigenicity. At 35-day post-inoculation, tumors from mice inoculated MCF-7 or MCF-7/Vps34 and PKC-δ knockout MCF-7 or MCF-7/Vps34 cells were photographed and measured. (i) Working model of Vps34-dependent tumorigenicity.
To determine the activation mechanism of ERK pathway by Vps34, GTP-Ras, GTP-Rac 1 and active Raf 1 were detected in parental cells or Vps34-expressing cells. Immunoblotting data showed that the mechanism of Vps34 activation of Raf 1 was not dependent of GTP-Ras or GTP-Rac (Figure 7e), which is different from class I PI3K.46, 47 Because Vps34 induces PKC-δ-dependent phosphorylation of p62, we sought to test whether Vps34 promotes the phosphorylation of Raf 1 via PKC-δ. We generated PKC-δ-knockout MCF-7 and PKC-δ-knockout MCF-7 cells stably expressing Vps34. Immunoblotting data showed that ablation of PKC-δ decreased the phosphorylation levels of Raf 1, MEK and ERK in both MCF-7 and MCF-7 cells stably expressing Vps34, consistent with the phosphorylation of p62 at S349 (Figure 7f). Importantly, ablation of PKC-δ altered Vps34-dependent-expression of Nrf2 and p62. These results suggest that PKC-δ is necessary for activation of Nrf2 and p62. To test the importance of Vps34 and PKC-δ in tumor progress, Vps34-stable MCF-7 cells and their parental cells as well as PKC-δ-knockout MCF-7 cells and PKC-δ-knockout MCF-7 cells stably expressing Vps34 were next inoculated into immunodeficient mice with E2-supplementation. The mice that received PKC-δ-knockout MCF-7 cells or PKC-δ-knockout Vps34-expressing MCF-7 cells developed tumors that were slower in volume than those receiving parental cells or Vps34-expressing MCF-7 cells (Figures 7g and h), indicating that PKC-δ has key role in Vps34 stimulation of tumor growth. These observations are of substantial interest given the potential importance of simultaneously inhibiting Vps34 and PKC-δ to control the p62 and MEK/ERK pathways in tumor cells.
PI3K isoforms are divided into three classes based on the structure and lipid substrate preference.50 Among them, Vps34 forms different complexes for autophagy machinery and endosomal trafficking, resulting in specific localizations and functions.51, 52 Usually, autophagy exists in many tumors, helping tumor cell growth and survival under nutritional and hypoxic stress during tumor development. However, Vps34 was observed to be induced in response to the stress of tumor development, and initiated autophagosome creation with a lower level of p62 turnover (Figures 6a and b). This observation suggested that the potential function of Vps34 is not restricted to autophagy pathways during tumor progress.
S349-phosphorylation in response to stress enables p62 to bind Keap1 with higher affinity than the non-phosphorylated state.34 Evidence that we provided supports that Vps34 has an intrinsic function to induce Nrf2-dependent expression and recruit PKC-δ for S349-phosphorylation of p62. p62 has been shown to have an important role in tumorigenesis35 through the activation of Nrf2-targeted oncogenes by S349-phosphorylation. Therefore, our results suggest that Vps34 has pivotal roles in the progress of tumor, except its role in autophagy machinery and vesicle trafficking. In conclusion, elevation of Vps34 results in tumor progression via the PKC-δ-phosphorylation of p62 at S349 and PKC-δ involved phosphorylation of Raf 1 at Y340/341. The major way is Vps34 activation of Nrf2-targeted genes through the higher affinity of Keap1 for S349-phosphorylated p62 and the minor way is PKC-δ-dependent Vps34 activation of Raf 1 for Raf 1–MEK–ERK signaling (Figure 7i).
Discussion
Autophagy is an evolutionarily conserved lysosomal degradation pathway that occurs in response to stress, which induces cell apoptosis. Induction of autophagy prevents cells from apoptosis. On the contrary, inhibition of autophagy would promote apoptotic cell death and inhibit tumorigenesis. The PI3K pathway is critical in both normal physiological regulations and pathological conditions, including tumorigenesis. Data presented here constitute important evidence that p62 is necessary for Vps34-stimulated tumor development.
p62 functions as an adaptor protein for autophagic clearance of ubiquitinated and misfolded proteins, and then p62 itself is degraded by autophagy. Increased levels of the p62 protein indicate that autophagic flux is impaired. Hence, it seems difficult to determine the reason that p62 expression is elevated accompanied by defective autophagy. In cells, p62 can be phosphorylated at various sites, and is in dynamic equilibrium between different phosphorylated states. Recently, p62 was reported to be phosphorylated at S403 in the UBA domain for increasing the affinity of p62 with misfolded proteins for autophagic degradation.53, 54 This specific function confers to p62 sensitivity to the accumulation of ubiquitinated proteins and the responsibility to their autophagic degradation.
We found that p62 protein levels are induced transcriptionally in MCF-7 cells by elevated Vps34. Upon Vps34-recruitment of PKC-δ, the induced p62 molecules remain mainly in the S349-phosphorylated state, which functions as an oncoprotein driver of tumorigenesis (Figures 5d and e; Figures 6a and b). This assists in understanding the high p62 expression accompanied by defective autophagy. In addition, activation of p62 by accumulated Vps34 is impaired when NRF2 is ablated in breast cancer cells (Figure 4g), which is consistent with that the accumulation of p62 due to autophagy deficiency promotes tumorigenesis through activation of the NRF2 pathway.5, 30, 48, 55 Although Vps34 has been well-characterized to have a pivotal role in autophagy, our studies suggest that Vps34 also functions as one of the main effector pathways of MEK-ERK, regulating cancer cell growth. Further investigation reveals that p62 also has a role in regulation of MEK-ERK activation, implying that p62 is a key mediator of Vps34-dependent cancer development. On the basis of our findings, we hypothesize that Vps34 enhances the activation of p62 protein for tumor development via a novel mechanism. Therefore, our data indicate that Vps34 serves not only as a key autophagic regulator, but also as a potential therapeutic target in cancer. In shaping future approaches toward personalized medicine, we propose that Vps34-p62-PKC δ protein machinery may constitute an especially promising target for breast cancer therapy.
Materials and methods
Reagents
Antibodies were commercially obtained: Nrf2 (ab62352), PKC-δ (ab182126) and Vimentin (ab92547) from Abcam (Cambridge, UK); E-cadherin (610404) from BD Biosciences (San Jose, CA, USA), Caspase3 (#9662), Cleaved Caspase3 (#9661), caspase 7 (#9492), caspase 8 (#9746), cleaved caspase 8 (#9496), caspase 9 (#9502), Cleaved Caspase9 (#9501), ERK (#9107), p-ERK (#4370), MEK (#4694), p-MEK (#9154), PARP (#9542), Snail (#9891 s), Vps34 (#3811) and p-p62 (S349) (#95697 s) from Cell Signaling Technology (Danvers, MA, USA); p-p62 (S403) (GTX128171) and Vps34 (GTX129528) for western blot from Genetex (Irvine, CA, USA); Hsp90α (ADI-SPA-840- F) from Enzo (Farmingdale, NY, USA); Ras (05-516), Rac1 (05-389) from Millipore (Billerica, MA, USA) and Histone H1 (sc-8030), keap1 (sc-33569), Raf-1 (sc-52827), p-Raf-1 (Ser338) (sc-12358), p-Raf-1 (Tyr340/341) (sc-16806) from Santa Cruz Biotechnology (Dallas, TX, USA). Other antibodies for Flag-tag, Myc-tag, Vps34 (for IP), Beclin-1, LC3, p62, and β-actin were as previously described.17 Panobinostat (LBH589, cat: S1030) was purchased from Selleck Chemicals (Houston, TX, USA). z-VAD-FMK, Z-IETD-FMK and Z-LEDH-FMK were purchased from R&D R&D Systems (Minneapolis, MN, USA). An in situ Cell Death Detecting Kit (cat: 11684795910) was purchased from Roche diagnostic (Mannheim, Germany). Nrf2 siRNA (sc-37030) and P62/SQSTM1 shRNA (sc-29679-SH) were obtained from Santa Cruz Biotechnology. All other chemicals were obtained from Sigma-Aldrich (St Louis, MO, USA).
Quantitative real-time PCR (qPCR) analysis and constructs
MCF-7 cells were harvested following treatment. RNA was isolated with RNeasy Plus Mini kits (QIAGEN, Hilden, Germany) according to the supplier's protocol. Total RNAs (500 ng) was reverse-transcribed in 25 μl using TaqMan Reverse Transcription Reagents (Applied Biosystems Inc., Foster City, CA, USA. Each qPCR reaction was performed with 2.5 μl cDNA in 20 μl total volume using the Power SYBR Green PCR Master Mix and subjected to real-time PCR on a CFX Connect Detection System (Bio-Rad).
Primers used were followed as Vps34 (5′-GCT GTC CTG GAA GAC CCA AT-3′ and 5′-TTC TCA CTG GCA AGG CCA AA-3′), p62/SQSTM1 (5′-GCACCC CAA TGT GAT CTG C-3′, 5′-CGC TAC ACA AGT CGT AGT CTG G-3′), Beclin-1 (5′-GGT TGA GAA AGG CGA GAC AC-3′, 5′-ATT GTG AGG ACACCC AAG CA-3′) and LC3 (5′-CAG CAT GGT GAG TGT GTC CA-3′, 5′-AGAAGC CGA AGG TTT CCT GG-3′). The results were normalized to GAPDH (5′-GGA GCG AGA TCC CTC CAA AAT-3′, 5′-GGC TGT TGT CAT ACT TCT CATGG-3′).
The vector encoding mRFP-GFP-LC3 (tfLC3) was excised from ptfLC3 (Addgene, 21074) and subcloned into the lentiviral vector pCDH1-MCS1-EF1-puro (System Biosciences, CD510A-1). EGFP-LC3 (Addgene,11546) p62-promoter luciferase reporter was developed according to a previous report.12 The other constructs were obtained as follows: pcDNA3-Casp8 (Addgene, 11817), pcDNA3-Casp8-Mt (Addgene, 11818). N-terminal flag-tagged Vps34, V5-tagged Vps34, myc-tagged Beclin-1 and His-Sumo1 constructs were developed as previously described.17 The vectors encoding Vps34-D1 (aa 1-285), Vps34-ΔD1 (aa 285-887) were derived from flag-Vps34 and subcloned into the pcDNA3.1 N-flag vector. Vps34 mutants containing truncation or point mutation were generated by PCR-based site-directed mutagenesis.
p62 and PKC coding sequences were amplified from the RNAs above using primers as follows:
p62-F 5′-GGCGGCGGTACCATGGCGTCGCTCACCGTGAAG-3′
p62-R 5′-GGCGGCCTCGAGAACGGCGGGGGATGCTTTGAATAC-3′
PKC η-F 5-′GGCGGCGAATTCTGCAGATATCCGATGTCGTCTGGC-3′
PKC η-R 5′-GGCGCTCTAGACTATGGTTGCAATTCTGGAGACAC-3′
PKC-δ-F 5′-GGCGGCGAATTCTGCAGATATCCGATGGCGCCGTTC-3′
PKC-δ-R 5′-GGCGGCTCTAGATCAATCTTCCAGGAGGTGCTCG-3′
PKC ε-F 5′-GGCGGCGAATTCTGCAGATATCCGATGGTAGTGTTC-3′
PKC ε-R 5′-GGCGGCTCTAGATCAGGGCATCAGGTCTTCACC-3′
p62 and PKC expression vectors were generated by subcloning PCR products into the KpnI/XhoI site of pcDNA3.1-myc-his (B) or the EcoRI/XhoI site of Flag-pcdna3.1 (invitrogen). All constructs and mutants used in this study were verified by nucleotide sequencing and restriction digestion.
Cell culture
HEK-293 T (Cat. #HCL4517) was from GE lifesciences (Buckinghamshire, UK), MCF-10 A (CRL-10317), MCF-7 (HTB-22) and MDA-MB-231 cells (HTB-26) were from the American Type Culture Collection (ATCC). HEK-293 T, MCF-10 A, MCF-7 cells and stable transfectants were maintained in Dulbecco's Modified Eagle's Medium with 10% fetal bovine serum (vol/vol). MDA-MB-231 cells were maintained in RPMI 1640 with 10% fetal bovine serum (vol/vol). Cells were passaged two to three times-per-week, and exponentially growing cells were used for all described experiments.
Immunoprecipitation and Ni-beads pull-down assays
Immunoprecipitation of flag-Vps34 or myc-Beclin-1 from cell lysates was performed as described.17 To detect the effects of caspase 8 on flag-Vps34 conjugation with His–SUMO1, constructs were transfected into HEK-293 T cells for 48 h, followed by the purification of His-tagged proteins. The eluted proteins were immunoblotted with anti-Flag antibody.
Immunofluorescence confocal microscopy
MCF7 cells were grown on coverslips and treated with drugs for 18 h. Immunofluorescence confocal microscopy was performed as described. Briefly, permeabilized cells were incubated with mouse anti-cleaved caspase 8 and rabbit anti-Vps34 antibodies for 2 h at room temperature. Cells were washed with 1 × PBS, and incubated with Alexa Fluor 594-conjugated anti-rabbit and Alexa Fluor 488-conjugated anti-mouse secondary antibodies (Invitrogen) in the dark for 1 h at room temperature. The slides were washed with 1 × PBS and mounted with Vectashield containing 4', 6-diamidino-2-phenylindole. Images were acquired with an LSM510 confocal microscope with a 63 × objective (Carl Zeiss). To monitor autophagic flux, recombinant lentivirus encoding tfLC3 was produced and transduced into MCF-7 control or flag-tagged Vps34-expressing MCF-7 cells as described.17 For lysosome tracker staining, cells stably expressing EGFP-LC3 were incubated with 75 nM LysoTracker Deep Red (Thermo) for 30 min at 37 °C. Images were obtained using a FV3000 Olympus confocal microscope with a 60 × objective (Olympus, Tokyo, Japan).
PI (3) KC3 lipid kinase activity assays
Extracts from Flag-tagged Vps34, Vps34-ΔD1 and Vps34-D285E-expressing cells were immunoprecipitated with M2 beads (Sigma), followed by extensive wash, and pre-incubated at room temperature for 10 min in 60 μl of reaction system containing 2 μg of sonicated phosphatidylinositol. The reaction was performed as described.56 The lipid kinase activity was assayed using the class III ELISA kit (Echelon) according to the manufacturer’s instructions.
DNA fragmentation and cell death assays
To determine the effects of z-VAD-FMK or z-IETD-FMK on LBH589-induced apoptosis, MCF7 cells were treated with drugs for 48 h, and then analyzed by Cell Death Detection ELISAPLUS (Roche) according to the manufacturer’s instructions.
CRISPR/Cas9-mediated Vps34 knockout in MCF-7 cells
CRISPR/Cas9 system was used to establish a stable Vps34 or PKC-δ knockout MCF-7 cells. The target sequences for human Vps34 and human PKC-δ were 5′-GACCCAATGTTGAAGTTCTC-3′ and 5′-GACATCAACACCTTGGCCTG-3′ (703-684 exon 3), respectively. Two complementary oligonucleotides with BpiI restriction sites for guide RNAs were cloned into guide RNA-Cas9 vector (SyngenTech, #BG13402) for the generation of Vps34-Cas9 construct. MCF-7 cells were transfected with Vps34-Cas9 construct. The genome sequences of the edited locus in selected colonies were confirmed by sequencing analyses and the protein level of Vps34 in the colonies were tested by western blot analyses.
Viability assays
Cell viability was assessed using the CellTiter-Blue Cell Viability Assay (Promega, Fitchburg, WI, USA) following manufacturer's protocol. Briefly, MCF-7 cells were seeded in 96-well plates and treated with drugs for 48 h. Then, 20 μl of CellTiter-Blue reagent was added to each well, shaken for 10 s, and incubated at 37 °C for 4 h. Fluorescence was measured at an excitation wavelength of 579 nm and emission of 584 nm.
Colony-formation assays
A clonogenic assay was performed as described. Briefly, parental cells or stable transfectants were plated in triplicate at densities of 200 cells per well in a 6-well plate. After 14 days of incubation at 37 °C, the plates were fixed and stained with 0.25% crystal violet, and colonies were counted manually.
In vitro invasion assays
In vitro invasion assay were performed, as previously described57 by using the Cultrex Cell Invasion Assay kit (RandD Systems). In brief, serum-starved cells in 50 μl serum-free medium were placed in the top chamber and allowed to invade for 24 h. The lower chambers (assay chamber) were filled with 10% fetal bovine serum medium. After incubation, migrated cells on the upper chamber of the membrane were dissociated with cell dissociation solution containing Calcein AM at 37 °C for 1 h, and read the plate at 485 nm excitation and 520 nm emissions.
GST-fusion proteins
Plasmids for expressing GST fusion proteins of Vps34 and p62 by inserting complementary DNA fragments of Vps34 and Mt (D285E), p62 and MT (S349A) into pGEX-2TK vector (GE Healthcare). GST-fused proteins were expressed in BL21 (DE3) E. coli cells by induction with 0.1 mM isopropyl thiogalactoside for 12 h at 30 °C. GST fusion proteins were affinity purified with glutathione-Sepharose resin (GE Healthcare) according to the manufacturer’s protocol.
In vitro caspase cleavage assays
Vps34 proteins (Wt and D285E) were incubated with six units of caspase 8 (Sigma) in caspase buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 10% sucrose, 1 mM EDTA, 0.1% CHAPS and 10 mM dithiothreitol) at 37 °C for 45 min. The reaction was stopped by addition of 2 × SDS sample loading buffer. Samples were detected by western blot analysis using antibody against Vps34.
In vitro kinase assay
His6-tagged PKC-δ or dominant negative PKC-δ (K378M) was purified from baculovirus infected Sf9 cells and suspended in 20 μl kinase buffer (20 mM HEPES pH 7.2, 10 mM MgCl2, 5 mM dithiothreitol, 20 nM ATP, 0.05%NP-40) containing purified GST or GST-p62 recombinant proteins (GST-p62 or GST-p62 S349A). After incubation at 30 °C for 30 min, the reactions were terminated by addition of 2 × SDS sample loading buffer. The phosphorylated products were detected by western blot analysis using antibody against p-S349 p62 or p-S403 p62.
Luciferase reporter assays
MCF-7, MDA-MB-231 or MCF10A cells were plated in 12-well plates in dulbecco’s modified eagle’s medium containing 10% fetal bovine serum at 1.5 × 105 cells/well. MDA-MB-231 cells were plated in RPMI 1640 at 1.5 × 105 cells/well. At 1 day after plating, cells were transfected by Lipofectamine 2000. After 24 h, cell lysates were prepared, and luciferase activity was determined using dual luciferase assay system from Promega Corporation. Luciferase activity was normalized to Renilla luciferase activity or to β-gal activity.
Tumorigenicity assay in vivo
To assess in vivo tumor growth for parental cells and stable transfected MCF-7 cells, 4–6-week-old female Balb/c nude mice (Sino-British SIPPR/BK Lab. Animal Ltd, China) were used with subcutaneous inoculation with 1.5 × 106 tumor cells as previously described.58 The use and care of experimental animals was approved by the Institutional Animal Care and Use Committee (IACUC), Roche R & D Center (Shanghai, China). Briefly, Mice (six mice in each group) were administered 17-β-estradiol and observed for increases in tumor volume over 35 days to assess tumor progress. Tumor diameters were measured at regular intervals, and tumor volumes were calculated as volume=(width)2 × length/2.
Statistical analyses
Data were presented as the mean±s.e.m. and analyzed using the Graphpad software (La Jolla, CA, USA). Differences between the means from the groups were determined using Student t-test or one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Significance
Vps34 and p62 are well known for autophagy pathways. Both of them may also contribute to the development and progression of human cancers. We show here that Vps34 functions in activation of p62 for cancer development beyond autophagy. The expression of Vps34, documented for the phosphorylation of MEK and ERK, is strongly correlated with tumorigenic activity of human breast cancer cells. Vps34 augments PKC-δ to p62 for its phosphorylation at Serine 349, leading to transcriptional expression of p62 through its affinity with Keap1. The intact structure of Vps34 is essential for its lipid kinase activity, expression of LC3-II and Nrf2 as well as p62, in addition to tumor growth. Moreover, ablation of PKC-δ or p62 expression in MCF-7 cells abrogates Vps34-dependent tumor growth. Targeting Vps34-p62-PKC-δ machinery may be useful as a therapy to block breast cancer progression.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81572721 and 81272391 to YY).
Author contributions
XJ co-designed the research concept, planned the overall experimental design, and performed experiments. YB, ZQ and XL contributed to the study design, provided scientific knowledge, contributed to the interpretation and analysis of the data. HL, XK, ZZ and FS performed experiments on mice, and contributed to the interpretation and analysis of the data. XL contributed to the interpretation and analysis of the data, and provided breast cancer patient samples. LJ co-designed the research concept and contributed to the discussion of the data. YY designed the research concept, planned the overall experimental design, supervised the study and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic control of autophagy. Cell 2014; 159: 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Lotze MT, Zeh HJ 3rd. AGER/RAGE-mediated autophagy promotes pancreatic tumorigenesis and bioenergetics through the IL6-pSTAT3 pathway. Autophagy 2012; 8: 989–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Wei Y, Zou Z, Tucker K, Rakheja D, Levine B et al. Genetic inhibition of autophagy promotes p53 loss-of-heterozygosity and tumorigenesis. Oncotarget 2016; 7: 67919–67933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137: 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2008; 105: 19211–19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11: 385–396. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Liu X, Klionsky DJ. SnapShot: selective autophagy. Cell 2013; 152: 368–368.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 2015; 282: 4672–4678. [DOI] [PubMed] [Google Scholar]
- Thompson HG, Harris JW, Wold BJ, Lin F, Brody JP. p62 overexpression in breast tumors and regulation by prostate-derived Ets factor in breast cancer cells. Oncogene 2003; 22: 2322–2333. [DOI] [PubMed] [Google Scholar]
- Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci USA 2012; 109: 2003–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell 2010; 38: 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch DS, Shen Y, Dokmanovic M, Wu WJ. pp60c-Src phosphorylates and activates vacuolar protein sorting 34 to mediate cellular transformation. Cancer Res 2010; 70: 5974–5983. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013; 19: 983–997. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK et al. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci USA 2013; 110: 6841–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122: 927–939. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 2007; 26: 2527–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J 2011; 30: 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 2008; 27(Suppl 1): S137–S148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Fung G, Piesik P, Zhang J, Luo H. Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ 2014; 21: 1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 2009; 122: 2554–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006; 8: 1124–1132. [DOI] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010; 29: 1717–1719. [DOI] [PubMed] [Google Scholar]
- Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang P, Sun Q, Ding WX, Yin XM, Sobol RW et al. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res 2011; 71: 3625–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LJ, Tan TW, Ranganathan S. CASVM: web server for SVM-based prediction of caspase substrates cleavage sites. Bioinformatics 2007; 23: 3241–3243. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 2010; 12: 213–223. [DOI] [PubMed] [Google Scholar]
- Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci USA 2011; 108: 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 2010; 285: 22576–22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 2013; 51: 618–631. [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. P62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009; 137: 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol 1998; 18: 3069–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 2010; 6: 1090–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12: 119–131. [DOI] [PubMed] [Google Scholar]
- Tang F, Wang B, Li N, Wu Y, Jia J, Suo T et al. RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS One 2011; 6: e24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J 2000; 19: 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J 1999; 18: 3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP et al. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell 2004; 6: 303–309. [DOI] [PubMed] [Google Scholar]
- Sakakura K, Takahashi H, Kaira K, Toyoda M, Oyama T, Chikamatsu K. Immunological significance of the accumulation of autophagy components in oral squamous cell carcinoma. Cancer Sci 2015; 106: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdelski C, Reiswich V, Hube-Magg C, Kluth M, Minner S, Koop C et al. Cytoplasmic accumulation of sequestosome 1 (p62) is a predictor of biochemical recurrence, rapid tumor cell proliferation, and genomic instability in prostate cancer. Clin Cancer Res 2015; 21: 3471–3479. [DOI] [PubMed] [Google Scholar]
- Li L, Shen C, Nakamura E, Ando K, Signoretti S, Beroukhim R et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell 2013; 24: 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro G, Rodon J, Bedell C, Kwak E, Baselga J, Braña I et al. Correction: Phase i safety, pharmacokinetic, and pharmacodynamic study of SAR245408 (XL147), an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. Clin Cancer Res 2015; 21: 2192–0432CCR-15-0185. [DOI] [PubMed] [Google Scholar]
- Ebi H, Costa C, Faber AC, Nishtala M, Kotani H, Juric D et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc Natl Acad Sci USA 2013; 110: 21124–21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell 2008; 13: 343–354. [DOI] [PubMed] [Google Scholar]
- Cai-McRae X, Zhong H, Karantza V. Sequestosome 1/p62 facilitates HER2-induced mammary tumorigenesis through multiple signaling pathways. Oncogene 2015; 34: 2968–2977. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010; 11: 329–341. [DOI] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci 2006; 119: 605–614. [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J Cell Sci 2011; 124: 2692–2701. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 2011; 44: 279–289. [DOI] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 2011; 193: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 2015; 520: 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Rao R, Shen J, Tang Y, Fiskus W, Nechtman J et al. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res 2008; 68: 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus P, Stanley ER, Schafer R, Abraham D, Aharinejad S. Colony-stimulating factor-1 antibody reverses chemoresistance in human MCF-7 breast cancer xenografts. Cancer Res 2006; 66: 4349–4356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.