Abstract
Background
A single subtype of canine influenza virus (CIV), A(H3N8), was circulating in the United States until a new subtype, A(H3N2), was detected in Illinois in spring 2015. Since then, this CIV has caused thousands of infections in dogs in multiple states.
Methods
In this study, genetic and antigenic properties of the new CIV were evaluated. In addition, structural and glycan array binding features of the recombinant hemagglutinin were determined. Replication kinetics in human airway cells and pathogenesis and transmissibility in animal models were also assessed.
Results
A(H3N2) CIVs maintained molecular and antigenic features related to low pathogenicity avian influenza A(H3N2) viruses and were distinct from A(H3N8) CIVs. The structural and glycan array binding profile confirmed these findings and revealed avian-like receptor-binding specificity. While replication kinetics in human airway epithelial cells was on par with that of seasonal influenza viruses, mild-to-moderate disease was observed in infected mice and ferrets, and the virus was inefficiently transmitted among cohoused ferrets.
Conclusions
Further adaptation is needed for A(H3N2) CIVs to present a likely threat to humans. However, the potential for coinfection of dogs and possible reassortment of human and other animal influenza A viruses presents an ongoing risk to public health.
Keywords: H3N2, canine influenza, CIV, ferrets, transmission
While influenza viruses exhibit species-specific determinants, sporadically, they cross the species barrier, rapidly adapt to a new host, and establish new lineages. Despite early reports showing the presence of antibodies against human influenza viruses in dogs [1, 2], there were no documented reports of canine influenza virus (CIV) infections until 2004, when the first CIV was identified in racing greyhound dogs in Florida [3]. Molecular and antigenic analyses revealed that all genes of the A(H3N8) virus isolated from infected dogs were closely related to equine influenza viruses [3]. The A(H3N8) virus spread to dogs in other states and continues to show limited circulation in the United States, especially among dogs housed in humane shelters [4].
The first evidence of A(H3N2) CIV in domestic dogs displaying respiratory symptoms occurred in South Korea in 2007. Unlike the A(H3N8) virus, all genes of the A(H3N2) CIV showed >95.5% nucleotide identity with Eurasian lineage avian influenza A viruses (IAVs) circulating in wild and domestic birds [5]. The virus spread widely among dogs in South Korea, parts of China, and Thailand. In April 2015, an A(H3N2) virus that was genetically similar to CIVs circulating in Asia was isolated from an infected golden retriever in Cook County, Illinois. According to the updates posted by Animal Health Diagnostic Center, Cornell University College of Veterinary Medicine, the virus has since spread to multiple states, causing respiratory disease in thousands of dogs across the United States [51].
Besides the 2 major subtypes of CIV, A(H3N8) and A(H3N2), other IAVs, including 2009 pandemic A(H1N1) [6, 7], A(H5N1) [8], A(H5N2) [9], and A(H9N2) [10], have been isolated from dogs. Further evidence that dogs can be infected with human influenza viruses comes from serological analyses [11, 12], as well as experimental infections [13, 14]. Influenza viruses have a segmented genome, which allows for reassortment in coinfected hosts and emergence of novel influenza virus strains. Since dogs are close human companions, the notion that new strains could arise in dogs is concerning from a public health standpoint. No human infections have been reported with CIVs to date; nonetheless, continuous surveillance and assessment of the antigenic and pathobiological features of novel viruses is important to minimize and mitigate the possibility of zoonotic infections. Pigs are susceptible to IAVs from a variety of different hosts and pose a risk of interspecies transmission of new reassortant viruses. A recent study showed that the A(H3N2) CIV, newly isolated in the United States, displayed limited replication in the lower respiratory tract of inoculated swine and was not transmitted to contact pigs [15]. Here, we analyzed the genetic, antigenic, and structural features of A(H3N2) CIVs isolated in the United States. Furthermore, in vitro replication in a human airway epithelial cell line (Calu-3), pathogenesis in mice and ferrets, and transmissibility in ferrets were assessed.
MATERIALS AND METHODS
Viruses
The A(H3N2) viruses A/canine/IL/12191/2015 and A/ Switzerland/9715293/2013 were propagated in eggs at 35°C for 48 hours. A/Brisbane/59/2007 (H1N1) was propagated in Madin-Darby canine kidney cells at 37°C for 48 hours. Details are included in the Supplementary Information.
Molecular and Phylogenetic Analysis
A/canine/IL/12191/2015 virus RNA was reverse transcribed with random hexamer primers and amplified by PCR using universal IAV primers [16]. Full-length open reading frames (ORFs) were generated by Illumina sequencing. Consensus sequences were assembled and BLAST analysis of each gene was performed in the GISAID database to identify the closest related viruses. Gene sequences of related viruses were aligned to A/canine/IL/12191/2015 virus sequences, using MUSCLE to create full-length ORF alignments. The full-length ORF sequences of A/canine/IL/11613/2015 virus were generated by Cornell University (kindly provided by Dr Amy Glaser) and included in phylogenetic analysis. Alignments were imported into MEGA 5.0, and phylogenetic trees were generated using the generalized time reversible model and maximum likelihood method with 1000 bootstrap replicates. Hemagglutinin (HA) protein sequence alignments included human A(H3N2) viruses for comparisons of the A(H3N2) CIV receptor-binding and putative antigenic sites.
Determination of HA and Neuraminidase (NA) Structures and Glycan Array Analyses
A/canine/IL/11613/2015 virus recombinant HA and NA crystallization, glycan microarray printing and recombinant HA (recHA) analyses have been described elsewhere [17, 18]. Details are included in Supplementary Information. Supplementary Table 2 lists glycans used in these experiments as well as a tabulated qualitative assessment of binding for each protein analyzed.
Antigenic Characterization
Virus-specific polyclonal antiserum against A/canine/ IL/12191/2015 virus was generated in serologically naive ferrets immunized intranasally with 106.0 50% egg infectious doses (EID50) of virus diluted in PBS. At 14 days post-inoculation, each ferret was boosted with 2048 HAU of virus mixed with Titermax Gold Adjuvant (Sigma-Aldrich, St. Louis, MO) and antiserum was collected 14 days post-boost. A/canine/ IL/12191/2015 virus was compared by hemagglutination inhibition (HI) assay to panels of antiserum against other A(H3N2) IAVs and corresponding reference viruses [19]. Human immune serum, pooled from adults (19–49 years old) who received an inactivated 2013–2014 seasonal influenza vaccine, was included in the analysis. The human sera were acquired through a contract and received as anonymous samples, and, thus, were exempt from review by the Centers for Disease Control and Prevention’s (CDC) Institutional Review Board.
Animal Experiments
Pathogenicity in mice and ferrets, and transmission efficiency between cohoused ferrets were evaluated as described elsewhere [20, 21]. Details are included in Supplementary Information.
Cell Culture and Viral Replication
Human airway epithelial Calu-3 cells, obtained from American Type Culture Collection (ATCC; Manassas, VA), were cultured on 12-mm transwell inserts as described previously [22]. Briefly, Calu-3 cells grown on transwells were inoculated apically in triplicate with A/canine/IL/12191/2015 (H3N2) virus, A/ Brisbane/59/2007 (H1N1) virus, or A/Switzerland/9715293/13 (H3N2) virus at a multiplicity of infection of 0.01 50% EID50/ cell for 1 hour, washed, and then incubated at 37°C in a 5% CO2 atmosphere. Viral titers in cell culture supernatant were determined by titration in eggs.
RESULTS
Phylogenetic and Genetic Characterization
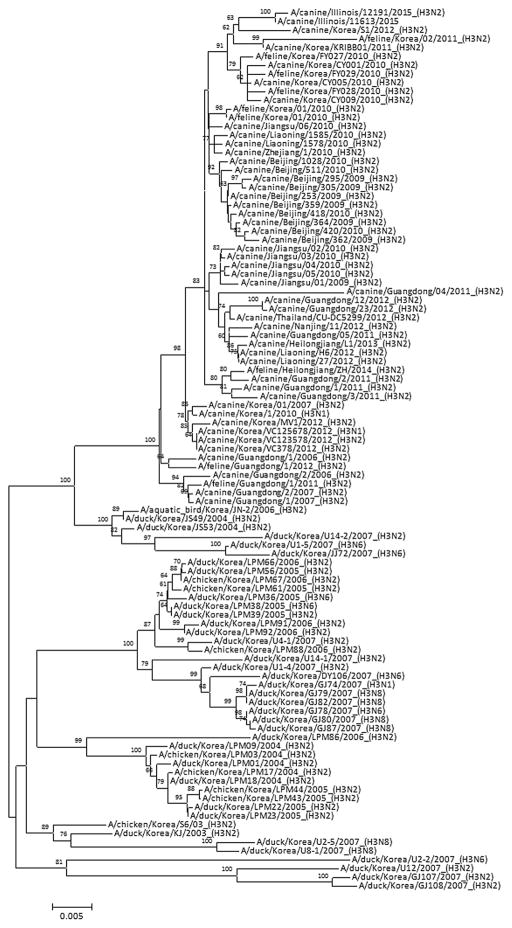
Phylogenetic analysis of A/canine/IL/12191/2015 and A/ canine/IL/11613/2015 virus genes showed that these viruses were closely related to A(H3N2) CIVs isolated in South Korea in 2011–2012 (Figure 1 and Supplementary Figures 1–7) and clustered in a larger group containing isolates from China and Thailand. While the US isolates of A(H3N2) CIVs showed 95% nucleotide identity to the duck A(H3N2) viruses, each gene segment of the US CIVs had >99% nucleotide identity to recently isolated South Korean CIVs, suggesting that the virus was recently transmitted between South Korean and US dogs.
Figure 1.
Phylogenetic tree of the canine influenza virus hemagglutinin genes. The phylogenetic tree was generated using a general time reversible model and maximum likelihood method with 1000 bootstrap replicates. Bootstrap values of ≥60 are shown at branch nodes. The scale bar represents nucleotide substitutions per site.
The NA gene segment of US CIVs had amino acid deletions at positions 76 and 77 that were also observed in many related CIVs from South Korea and China and did not possess any known markers of resistance to NA inhibitors. The matrix 2 protein contained 2 mutations (Asn30Asp and Thr215Ala) associated with increased virulence of avian IAVs in mice but no molecular markers associated with resistance to adamantane. The PB2, PB1-F2, and NS proteins each had mutations at positions previously reported to enhance polymerase activity, infectivity, and increased virulence of H5N1 avian IAVs in mice [23]. The PA and NP proteins did not have mutations of known significance.
HA Structure
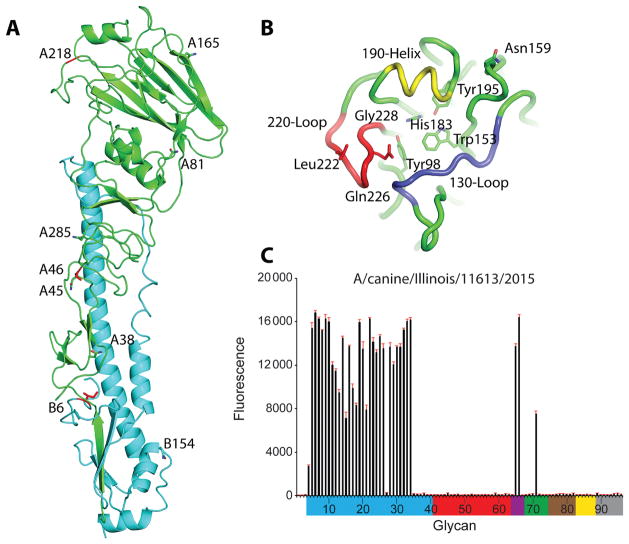
The 3-dimensional HA structure of the trimeric ectodomain from A/canine/IL/11613/2015 virus HA was determined by X-ray crystallography at a 3.0-Å resolution (Supplementary Table 1). Seven asparagine-linked glycosylation sites (NXS/T) were predicted in the CIV HA monomer; however, no interpretable carbohydrate electron density was observed at any location (Figure 2A). The HA of A/canine/IL/11613/2015 virus had 3 amino acid differences as compared to the HA of the A/ canine/IL/12191/2015 virus. The Ser46Pro difference, located near antigenic site C, could result in the loss of the glycosylation site at position 45 in A/canine/IL/12191/2015 virus HA (all positions stated indicate H3 structural numbering). Gly218Glu and Ile335Leu differences were identified near antigenic site D and in the HA2 portion of the protein, respectively (Figure 2A). The CIV HA was produced in the HA0 form, using a baculovirus expression system. For structural studies, the HA was digested with trypsin to remove the trimerization tag, which also cut the HA into the active HA1/HA2 form. Comparison of the CIV HA monomer to an avian H3 HA (PDB: 1MQL) [24] and a recent human H3 HA (PDB: 4WE8) [25] revealed a highly similar structure, with the Cα atoms superimposing to give a root mean square deviation of 0.94 Å and 1.03 Å, respectively.
Figure 2.
Structure and glycan array binding of canine influenza virus hemagglutinin (HA). A, Cartoon representation of the HA monomer. HA1 is green, and HA2 is cyan. The predicted glycosylation sites on HA1 (A38, A45, A81, A165, and A285) and HA2 (B6 and B154) are labeled and shown as sticks. The amino acid differences between A/canine/ IL/12191/2015 and A/canine/IL/11613/2015 viruses are shown in red sticks. B, Binding site elements. The 130-loop is purple, the 190-helix is yellow, and the 220-loop is red. Conserved residues are shown in green sticks. The amino acids discussed in the text are labeled and shown in sticks. C, Glycans on the microarray are grouped according to sialic acid linkage, as follows: α2–3 SA, blue; α2–6 SA, red; α2–6/ α2–3 mixed SA, purple; N-glycolyl SA, green; α2–8 SA, brown; β2–6 and 9-O-acetyl SA, yellow; and asialoglycans, gray (Supplementary Table 2). Error bars reflect the standard error in the signal for 6 independent replicates on the array.
Receptor-Binding Site Analysis
Similar to other IAVs, the consensus receptor-binding site (RBS) was composed of three structural elements: a 190-helix (residues 188–94), 220-loop (residues 220–8), and 130-loop (residues 131–9). Highly conserved residues Tyr98, Trp153, His183, and Tyr195 were identified at the base of the pocket (Figure 2B). The HA sequences from A/canine/IL/12191/2015 and A/canine/IL/11613/2015 viruses primarily retained amino acids conserved among Eurasian lineage low-pathogenicity avian H3N2 IAVs. Amino acids in and around the RBS were typical of avian-origin IAVs, with signature Gln226 and Gly228 suggesting preferential binding to avian-like α2,3-linked sialic acid (SA) receptors. The HA proteins had a single mutation of Ser159Asn, which is an adaptation potentially resulting in increased binding to α2,6-linked SA receptors. This mutation was found in the majority of A(H3N2) CIVs in the databases. Similar to viruses detected in eastern Asia, the US CIVs had a Leu at position 222, differentiating them from avian H3 IAVs, which typically have a highly conserved Trp at this position. Interestingly, Leu at position 222 was also shared among A(H3N8) CIVs circulating in the United States, suggesting its potential role in adaptation of these viruses to dogs [26–28]. Glycan-binding analyses of recombinant CIV HA revealed a strong binding preference for the α2–3-linked SAs and mixed α2–3/α2–6 branched SAs (numbers 65 and 66). It also showed a relatively strong binding to N-glycolylneuraminic acid–containing glycans (number 71; Figure 2C and Supplementary Table 2).
NA Structure
The crystal structure was determined to a 1.8-Å resolution (Supplementary Table 1), with good electron density for most of the residues, except the 150-loop area. The virus strain used for recombinant NA studies, A/canine/IL/11613/2015, had an NA identical to that of A/canine/IL/12191/2015 virus. The CIV NA structure was a typical box-shaped tetrameric association of identical monomers, containing six 4-stranded, antiparallel β-sheets that formed a propeller-like arrangement (Figure 3), as previously described [29]. One calcium ion-binding site, conserved in all known influenza A and B virus NA loops [30, 31], was observed in CIV NA. Calcium ions were previously shown to be critical for the thermostability and activity of influenza virus NAs, and this conserved metal site was proposed to be important in stabilizing a reactive conformation of the active site by otherwise flexible loops [30, 31]. Although CIV NA had 6 potential N-linked glycosylation sites at Asn86, Asn146, Asn200, Asn234, Asn313, and Asn402 in the final model, interpretable glycan density was only observed at Asn146, Asn200, and Asn234. Residue Asn146, which was situated on the membrane-distal surface close to the active site, was the only glycosylation site conserved among all other influenza A and B virus NAs [32, 33].
Figure 3.
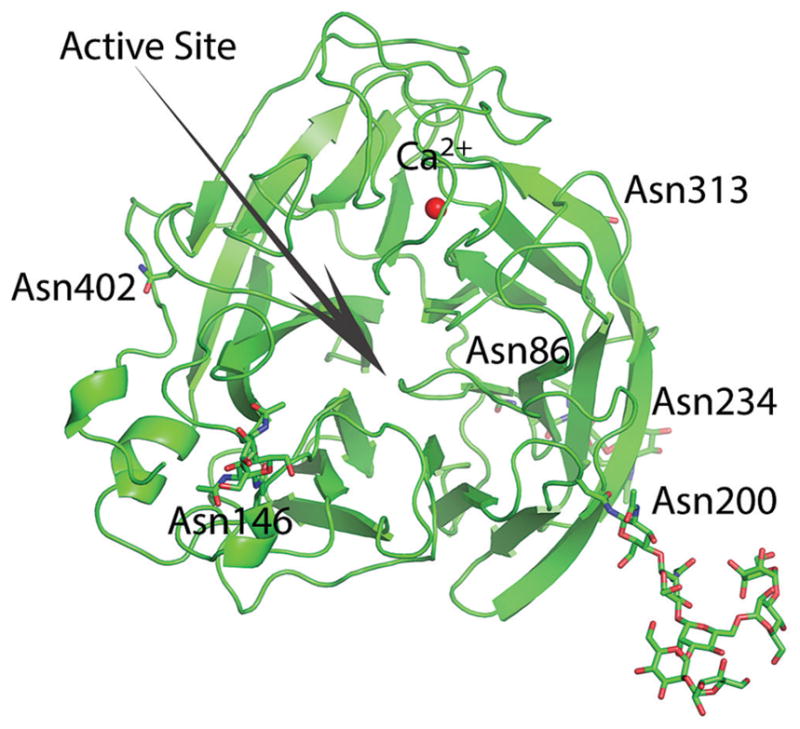
Overall structure of canine influenza virus neuraminidase. The active site is shown with an arrow. Predicted glycosylation sites are shown in sticks.
Antigenic Characterization
HI assay demonstrated that the HA of A/canine/IL/12191/2015 virus was antigenically distinct from the HA of A(H3N8) CIVs previously isolated from dogs in the United States (Table 1). Although weak reactivity was detected between A/canine/ IL/12191/2015 virus–generated antisera and A(H3N8) CIV, heterologous HI titers were 32-fold lower as compared to the homologous virus titer. Likewise, a 16-fold reduction in the heterologous titer of antisera produced against A/canine/FL/43/2004 (H3N8) virus was observed when tested with A/canine/IL/12191/2015 (H3N2) virus, as compared to the homologous titer, indicating 2-way specificity of antiserum produced against these canine viruses. The HA of A/canine/IL/12191/2015 virus shared about 78% amino acid similarity with A(H3N8) CIVs currently circulating in the United States and had on average >70 amino acid differences in the mature HA protein as compared to recent A(H3N8) viruses, including at least 15 residues identified in putative antigenic sites. Compared with closely related South Korean viruses, A/canine/IL/12191/2015 virus had <6 amino acid changes in the HA protein, suggesting the likelihood of antigenic relatedness of these viruses. Interestingly, antiserum generated against a recent avian-like A(H3N8) isolate, A/harbor seal/NH/179629/2011, was more broadly cross-reactive with both CIVs, suggesting antigenic relatedness between this avian lineage and genetically distant canine strains. Ferret antiserum produced against the seasonal vaccine IAV strain, A/Switzerland/9715293/2013, as well as human serum pooled from adults who received an inactivated 2013–2014 seasonal influenza vaccine, did not react with any of the animal-origin viruses.
Table 1.
Hemagglutination Inhibition Reactions of Animal-Origin and Seasonal H3 Influenza A Viruses
| Reference Antigens | Reference Ferret Antisera | |||||
|---|---|---|---|---|---|---|
| Subtype | Canine/FL | Switz/9715293 | Harbor Seal/NH | Canine/IL | Human Poola | |
| A/canine/FL/43/2004 | H3N8 | 1280 | 5 | 640 | 80 | 5 |
| A/Switzerland/9715293/2013 | H3N2 | 5 | 1280 | 5 | 5 | 80 |
| A/harbor seal/NH/179629/2011 | H3N8 | 40 | 5 | 160 | 40 | 5 |
| A/canine/IL/12191/2015 | H3N2 | 80 | 5 | 160 | 2560 | 5 |
| Test Antigen | ||||||
| A/canine/NY/118491/2011 | H3N8 | 320 | 5 | 320 | 80 | 5 |
Post-vaccination immune serum pooled from adults (19–49 years old) who received an inactivated 2013–2014 seasonal influenza vaccine.
Pathogenicity in Mice
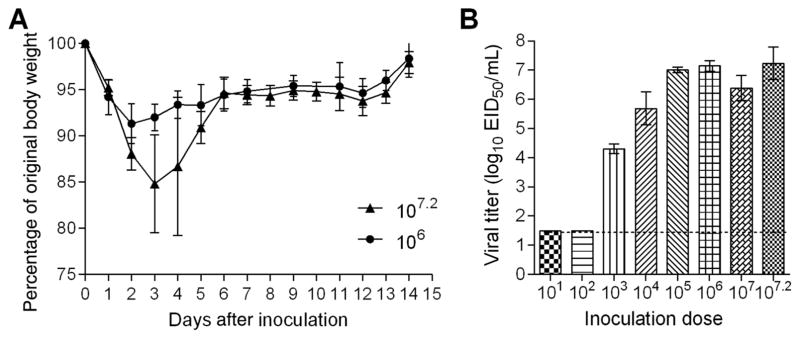
Unlike the majority of seasonal A(H1N1) and A(H3N2) viruses, most avian influenza viruses that can bind to α2,3-linked SA receptors, do not require prior adaptation to cause disease in mice [34]. Mice inoculated with 107.2 EID50 of A/canine/ IL/12191/2015 virus displayed signs of infection, including severe weight loss, ruffled hair, and hunched posture. Except for 1 mouse, which was euthanized on day 4 after inoculation because of excessive weight loss, all of the animals recovered from the infection. Mice inoculated with 106.0 EID50 of virus displayed transient weight loss (up to 9%) and no other signs of morbidity (Figure 4A). Groups of 3 mice were inoculated with serial dilutions of virus, and virus titers in lungs collected on day 3 after inoculation were used to calculate the 50% mouse infectious dose (MID50). A/canine/IL/12191/2015 virus efficiently replicated in mouse lungs without prior adaptation. Detectable virus was observed in mice inoculated with doses >102 EID50, with the highest mean lung titer reaching 107.1 EID50/mL (Figure 4B). By 6 days after inoculation, mice inoculated with 107.2 EID50 still had detectable virus in lungs (mean titer, 104.7 EID50/mL; data not shown). The MID50 for the A(H3N2) CIV was 102.5 EID50.
Figure 4.
Pathogenicity of A(H3N2) canine influenza virus in mice. A, Groups of 5 mice were intranasally inoculated with 107.2 or 106.0 50% egg infective doses (EID50) of A/canine/IL/12191/2015 (H3N2) virus and observed for signs of morbidity and mortality for 14 days. The percentage weight loss (±SD) is shown. B, Additional groups of 3 mice were inoculated with 107.2 EID50 or serial 10-fold dilutions ranging from 107.0 to 101.0 EID50 of virus and were euthanized 3 days after inoculation, when lung tissues were collected for viral titer determination. Viral titers are presented as log10 EID50/mL (±SD). The limit of detection is 1.5 log10 EID50/mL.
Pathogenicity and Transmissibility in Ferrets
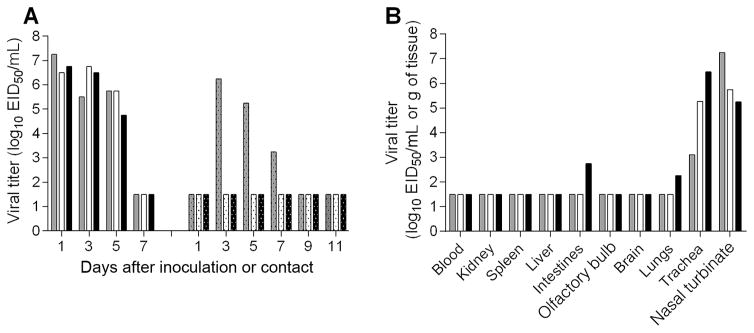
Ferrets inoculated with A/canine/IL/12191/2015 (H3N2) virus exhibited minimal weight loss (mean maximum, 3.1%) and an increase in body temperature (mean maximum, 1°C above baseline), and 2 of 3 infected ferrets exhibited mild lethargy (data not shown). None of the infected ferret had respiratory symptoms, such as sneezing or nasal discharge; however, ocular drainage was observed in 1 ferret on days 2–9 after inoculation. A mean peak titer of 106.8 EID50/mL was found in nasal washes on day 1 after inoculation, and virus was cleared in all animals after 7 days (Figure 5A). All inoculated ferrets seroconverted (HI titers on day 27 after inoculation ranged from 320 to 640). At day 3 after inoculation, the virus was detected in the nasal turbinates and trachea of all inoculated ferrets, with average titers of 106.1 EID50/mL and 105.0 EID50/g of tissue, respectively (Figure 5B). Only 1 ferret had low levels of virus in the lungs (102.3 EID50/g). No virus was detected in rectal swab samples collected for up to 5 days after inoculation (data not shown), but an intestine sample from one of the ferrets had low levels of virus (102.7 EID50/g) on day 3 after inoculation. No virus was recovered from other extrapulmonary tissues, indicating that the canine A(H3N2) virus did not replicate systemically.
Figure 5.
Pathogenicity and transmission of A(H3N2) canine influenza virus in ferrets. Six ferrets were intranasally inoculated with 107.1 50% egg infective doses (EID50) of A/canine/IL/12191/2015 virus. A, The following day, a serologically naive ferret was placed in the same cage with an inoculated ferret for the assessment of virus transmission between 3 ferret pairs in direct contact. Nasal wash titers from individual ferrets are presented. Viral titers are presented as log10 EID50/mL. B, Three inoculated ferrets were euthanized 3 days after inoculation, and tissues were collected for assessment of viral titers. Blood and nasal turbinate viral titers are presented as log10 EID50/mL, and kidney, spleen, liver, intestines (pooled duodenum, jejunoileal loop, and descending colon), olfactory bulb, brain (pooled anterior and posterior brain), lung, and trachea are presented as log10 EID50/g of tissue. The limit of detection is 1.5 log10 EID50 per g or mL.
The fact that the CIV replicated very efficiently in the upper respiratory tract prompted us to evaluate the transmissibility of the virus in a direct-contact setting. The virus was transmitted in 1 of 3 pairs of cohoused animals within 3 days, as evidenced by a high virus load in nasal wash samples (106.3 EID50/mL; Figure 5A) and seroconversion (HI titer on day 26 after contact, 640). The infected ferret displayed minimal weight loss (4.9%) and cleared the virus by day 9 after contact.
Replication in Calu-3 Cells
The human bronchial epithelial cell line (Calu-3), when grown on transwell inserts, resembles the human airway epithelium, as the cells form tight junctions to achieve transepithelial resistance [35]. Calu-3 cells were inoculated with 0.01 EID50/cell of A/ canine/IL/12191/2015 virus, A/Brisbane/59/2007 (H1N1) virus, or A/Switzerland/9715293/2013 (H3N2) virus. All viruses productively infected Calu-3 cells at 37°C (Figure 6). The A(H3N2) CIV replicated to similar titers as the seasonal A(H1N1) virus, reaching titers of ≥108.5 EID50/mL at 72 hours after inoculation, with no statistically significant differences between these viruses at any of the time points. In comparison, the seasonal A(H3N2) virus replicated less efficiently, reaching a mean average titer of 107.8 EID50/mL by 72 hours after inoculation; however, statistical significance between the seasonal A(H3N2) virus and CIV was only observed at the 24-hour time point.
Figure 6.
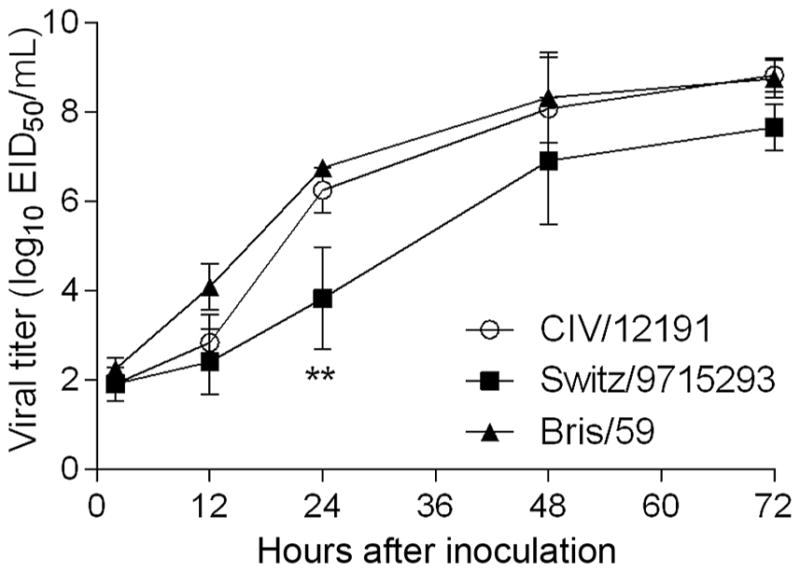
Replication kinetics of canine A(H3N2) virus in human airway epithelial (Calu-3) cells. Calu-3 cells were grown on transwell inserts and were inoculated apically in triplicate with a multiplicity of infection of 0.01 of A/canine/ IL/12191/2015 virus (CIV/12191), A/Brisbane/59/2007 (H1N1) virus (Bris/59), or A/ Switzerland/9715293/2013 (H3N2) virus (Switz/9715293). The cells were incubated at 37°C, and culture supernatants were collected 2, 12, 24, 48, and 72 hours after inoculation for viral titer determination in eggs. Statistical significance of the difference between the titers of the canine and each seasonal virus at each time point was determined by 2-way analysis of variance with the Bonferroni post hoc test. No statistically significant difference was observed between CIV/12191 and Bris/59 viruses. **P < .01 for comparison between CIV/12191 and Switz/9715293 viruses.
DISCUSSION
The emergence of a new IAV in domestic animals represents a major public health risk because it provides the opportunity for zoonotic infections to occur in pet owners or persons with high levels of exposure to animals, potentially allowing novel IAVs to adapt to humans. High nucleotide similarity between the A(H3N2) CIVs isolated in the United States and those recently detected in South Korea and China is suggestive of a direct transmission event or introduction of this virus into the United States in early 2015. Generally, avian IAVs bind preferentially to cells expressing α2,3-linked SAs, while human IAVs preferentially bind to α2,6-linked SAs found on cells in the upper respiratory tract of humans [36] and ferrets [37]. Upper and lower respiratory tracts of dogs largely express α2,3-linked SA receptors [5, 38], which likely facilitated the transmission of avian A(H3N2) influenza virus to dogs. The HA of the A/ canine/IL/12191/15 and A/canine/IL/11613/2015 viruses possessed the key residues (Gln226 and Gly228) necessary for α2,3-linked SA binding. Despite a few HA changes associated with mammalian adaptation (ie, Ser159Asn and Trp222Leu), these CIV HAs exhibited an avian receptor-binding preference. In addition, few markers of enhanced virulence were identified in the NA or internal proteins of this virus, indicating a lack of key mutations associated with increased pathogenicity for avian influenza viruses or adaptation to humans.
Dogs infected with A(H3N2) CIVs typically develop signs of infection, including fever, lethargy, anorexia, nasal/ocular discharge, sneezing, and cough, and transmission of virus between dogs is efficient [39]. Interspecies transmission of A(H3N2) CIV has been demonstrated from dogs to cats, while transmission from dogs to ferrets was not observed in an experimental setting [40, 41]. Ferrets are naturally susceptible to human and avian influenza viruses and develop clinical signs similar to those seen in infected humans [34]. In this study, inoculated ferrets displayed minimal morbidity and no respiratory signs. A/canine/IL/12191/15 (H3N2) virus was not transmitted between all cohoused pairs of ferrets. It is possible that the lack of respiratory symptoms may have limited the quantity of virus expelled from the infected animals and contributed to the lack of efficient transmission [42, 43]. Despite the lack of overt respiratory symptoms, A/canine/ IL/12191/15 (H3N2) virus replicated most efficiently in the nasal turbinates and trachea, but low levels of virus were detected in the lungs. Previous studies of earlier strains of A(H3N2) CIVs (A/canine/Korea/01/2007 and A/canine/Korea/LBM412/2008) in ferrets demonstrated some differences in phenotypes as compared to the virus evaluated here. The 2007 A(H3N2) CIV replicated less efficiently in ferret nasal samples but was transmitted more frequently between paired ferrets in direct contact (2 of 3 pairs [40] and 3 of 3 pairs [44]). The 2008 A(H3N2) CIV replicated more efficiently, was transmitted between animals in 3 of 6 ferret pairs, and caused substantially greater morbidity (15% weight loss) in inoculated ferrets [45] as compared to the 3.1% weight loss found using the A/canine/IL/12191/2015 virus reported here.
Antigenic differences between A(H3N8) and A(H3N2) CIVs reported in this study and the results of a recent study in mice [46] suggest that dogs previously vaccinated with A(H3N8) CIV vaccine may not be protected from infection or disease caused by the A(H3N2) CIV. Unless dogs are vaccinated with one of the currently available A(H3N2) CIV vaccines, the lack of immunity to the new A(H3N2) CIV may allow for additional opportunities for coinfection of this subtype with other influenza viruses. Serological analysis of dog serum samples showed that, in some cases, dogs tested positive for both canine A(H3N2) and 2009 pandemic A(H1N1) viruses, suggesting the possibility of coinfection with both viruses [47]. In fact, reassortants of A(H3N2) CIV and 2009 pandemic A(H1N1) viruses have been reported. An A(H3N1) virus with an HA gene from an A(H3N2) CIV and 7 genes homologous to 2009 pandemic A(H1N1) virus was isolated from dogs in South Korea [48]. This new reassortant virus was less pathogenic than classical CIV in experimentally infected dogs. An A(H3N2) CIV isolate containing a matrix gene from 2009 pandemic A(H1N1) virus (CIV/H3N2mv) was also isolated from dogs [49]. Ferrets and dogs experimentally infected with CIV/H3N2mv displayed signs of respiratory infection, and the virus had the capacity of efficient transmission between cohoused dogs and cohoused ferrets.
Overall, A/canine/IL/12191/2015 virus was capable of efficient replication in vitro in human airway epithelial cells and in the upper airways of ferrets and mice but was unable to be transmitted efficiently, likely owing to its avian receptor-binding properties. Receptor specificity is considered a major hurdle for influenza virus to adapt to humans and acquire the ability to sustain transmission in the population [50]. Evidence indicating that dogs can be infected with both canine and human influenza viruses raises the concern that a reassortant virus could emerge that is capable of infecting humans. The emergence of this new A(H3N2) CIV and the continued antigenic drift and reassortment [28] that it demonstrates in mammals warrants global surveillance and a better understanding of the pathogenesis and the potential for transmission of these viruses. Risk assessments such as these will improve our pandemic preparedness and will help mitigate the risk of zoonotic influenza virus infections and the threat to public health.
Supplementary Material
Acknowledgments
We thank Kerry Franzen, Leo Koster, and Katie Mozingo with the National Veterinary Services Laboratories, US Department of Agriculture Animal and Plant Health Inspection Service, for support in the growth and testing of the viruses characterized here; and Dr Amy Glaser and Cornell University, for proving the A/canine/ IL/11613/2015 virus genomic sequences used for this study.
Financial support. This work was supported by the Oak Ridge Institute for Science and Education (to J. A. P.-P. and H. M. C.).
Footnotes
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Supplement sponsorship. This work is part of a supplement sponsored by the Centers for Disease Control and Prevention.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nikitin A, Cohen D, Todd JD, Lief FS. Epidemiological studies of A-Hong Kong-68 virus infection in dogs. Bull World Health Organ. 1972;47:471–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Kilbourne ED, Kehoe JM. Demonstration of antibodies to both hemagglutinin and neuraminidase antigens of H3N2 influenza A virus in domestic dogs. Intervirology. 1975;6:315–8. doi: 10.1159/000149485. [DOI] [PubMed] [Google Scholar]
- 3.Crawford PC, Dubovi EJ, Castleman WL, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–5. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 4.Pecoraro HL, Bennett S, Huyvaert KP, Spindel ME, Landolt GA. Epidemiology and ecology of H3N8 canine influenza viruses in US shelter dogs. J Vet Intern Med. 2014;28:311–8. doi: 10.1111/jvim.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song D, Kang B, Lee C, et al. Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis. 2008;14:741–6. doi: 10.3201/eid1405.071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin D, Sun S, Du L, et al. Natural and experimental infection of dogs with pandemic H1N1/2009 influenza virus. J Gen Virol. 2012;93:119–23. doi: 10.1099/vir.0.037358-0. [DOI] [PubMed] [Google Scholar]
- 7.Promed-mail. Archive no. 20091222.4305. 22 December 2009.
- 8.Songserm T, Amonsin A, Jam-on R, et al. Fatal avian influenza A H5N1 in a dog. Emerg Infect Dis. 2006;12:1744–7. doi: 10.3201/eid1211.060542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan GJ, Ling ZS, Zhu YL, Jiang SJ, Xie ZJ. Genetic characterization of a novel influenza A virus H5N2 isolated from a dog in China. Vet Microbiol. 2012;155(2–4):409–16. doi: 10.1016/j.vetmic.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Xu X, Liu Q, et al. Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect Genet Evol. 2013;20:471–5. doi: 10.1016/j.meegid.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Su S, Chen J, Jia K, et al. Evidence for subclinical influenza A(H1N1)pdm09 virus infection among dogs in Guangdong Province, China. J Clin Microbiol. 2014;52:1762–5. doi: 10.1128/JCM.03522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houser RE, Heuschele WP. Evidence of prior infection with influenza A/Texas/77 (H3N2) virus in dogs with clinical parainfluenza. Can J Comp Med. 1980;44:396–402. [PMC free article] [PubMed] [Google Scholar]
- 13.Todd JD, Cohen D. Studies of influenza in dogs. I. Susceptibility of dogs to natural and experimental infection with human A2 and B strains of influenza virus. Am J Epidemiol. 1968;87:426–39. doi: 10.1093/oxfordjournals.aje.a120833. [DOI] [PubMed] [Google Scholar]
- 14.Song D, Kim H, Na W, et al. Canine susceptibility to human influenza viruses (A/ pdm 09H1N1, A/H3N2 and B) J Gen Virol. 2015;96(Pt 2):254–8. doi: 10.1099/vir.0.070821-0. [DOI] [PubMed] [Google Scholar]
- 15.Abente EJ, Anderson TK, Rajao DS, Swenson S, Gauger PC, Vincent AL. The avian-origin H3N2 canine influenza virus that recently emerged in the United States has limited replication in swine. Influenza Other Respir Viruses. 2016;10:429–32. doi: 10.1111/irv.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B, Donnelly ME, Scholes DT, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, Carney PJ, Donis RO, Stevens J. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol. 2012;86:8645–52. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens J, Blixt O, Glaser L, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–55. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Klimov A, Balish A, Veguilla V, et al. Influenza virus titration, antigenic characterization, and serological methods for antibody detection. Methods Mol Biol. 2012;865:25–51. doi: 10.1007/978-1-61779-621-0_3. [DOI] [PubMed] [Google Scholar]
- 20.Maines TR, Lu XH, Erb SM, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maines TR, Chen LM, Matsuoka Y, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–6. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng H, Pappas C, Katz JM, Tumpey TM. The 2009 pandemic H1N1 and triple-reassortant swine H1N1 influenza viruses replicate efficiently but elicit an attenuated inflammatory response in polarized human bronchial epithelial cells. J Virol. 2011;85:686–96. doi: 10.1128/JVI.01568-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prevention CfDCa. H5N1 Genetic Changes Inventory: a tool for influenza surveillance and preparedness. Atlanta, GA: Centers for Disease Control and Prevention; [Accessed 28 December 2015]. http://www.cdc.gov/flu/pdf/avianflu/h5n1-inventory.pdf. [Google Scholar]
- 24.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–18. doi: 10.1016/s0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Carney PJ, Chang JC, Guo Z, Villanueva JM, Stevens J. Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins. Virology. 2015;477:18–31. doi: 10.1016/j.virol.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang G, Li S, Blackmon S, et al. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol. 2013;94(Pt 12):2599–608. doi: 10.1099/vir.0.054692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins PJ, Vachieri SG, Haire LF, et al. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A. 2014;111:11175–80. doi: 10.1073/pnas.1406606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Hughes J, Murcia PR. Origins and evolutionary dynamics of H3N2 canine influenza virus. J Virol. 2015;89:5406–18. doi: 10.1128/JVI.03395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Qi J, Liu Y, et al. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol. 2011;85:8431–5. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong AK, Pegg MS, Taylor NR, von Itzstein M. Evidence for a sialosyl cation transition-state complex in the reaction of sialidase from influenza virus. Eur J Biochem. 1992;207:335–43. doi: 10.1111/j.1432-1033.1992.tb17055.x. [DOI] [PubMed] [Google Scholar]
- 31.Burmeister WP, Cusack S, Ruigrok RW. Calcium is needed for the thermostability of influenza B virus neuraminidase. J Gen Virol. 1994;75(Pt 2):381–8. doi: 10.1099/0022-1317-75-2-381. [DOI] [PubMed] [Google Scholar]
- 32.Russell RJ, Haire LF, Stevens DJ, et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–9. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 33.Janakiraman MN, White CL, Laver WG, Air GM, Luo M. Structure of influenza virus neuraminidase B/Lee/40 complexed with sialic acid and a dehydro analog at 1. 8-A resolution: implications for the catalytic mechanism. Biochemistry. 1994;33:8172–9. doi: 10.1021/bi00193a002. [DOI] [PubMed] [Google Scholar]
- 34.Thangavel RR, Bouvier NM. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng H, Goldsmith C, Thawatsupha P, et al. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J Virol. 2007;81:12439–49. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–6. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 37.Leigh MW, Connor RJ, Kelm S, Baum LG, Paulson JC. Receptor specificity of influenza virus influences severity of illness in ferrets. Vaccine. 1995;13:1468–73. doi: 10.1016/0264-410x(95)00004-k. [DOI] [PubMed] [Google Scholar]
- 38.Daly JM, Blunden AS, Macrae S, et al. Transmission of equine influenza virus to English foxhounds. Emerg Infect Dis. 2008;14:461–4. doi: 10.3201/eid1403.070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song D, Lee C, Kang B, et al. Experimental infection of dogs with avian-origin canine influenza A virus (H3N2) Emerg Infect Dis. 2009;15:56–8. doi: 10.3201/eid1501.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H, Song D, Moon H, et al. Inter- and intraspecies transmission of canine influenza virus (H3N2) in dogs, cats, and ferrets. Influenza Other Respir Viruses. 2013;7:265–70. doi: 10.1111/j.1750-2659.2012.00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song DS, An DJ, Moon HJ, et al. Interspecies transmission of the canine influenza H3N2 virus to domestic cats in South Korea, 2010. J Gen Virol. 2011;92:2350–5. doi: 10.1099/vir.0.033522-0. [DOI] [PubMed] [Google Scholar]
- 42.Maines TR, Belser JA, Gustin KM, et al. Local innate immune responses and influenza virus transmission and virulence in ferrets. J Infect Dis. 2012;205:474–85. doi: 10.1093/infdis/jir768. [DOI] [PubMed] [Google Scholar]
- 43.Gustin KM, Katz JM, Tumpey TM, Maines TR. Comparison of the levels of infectious virus in respirable aerosols exhaled by ferrets infected with influenza viruses exhibiting diverse transmissibility phenotypes. J Virol. 2013;87:7864–73. doi: 10.1128/JVI.00719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyoo KS, Kim JK, Kang B, et al. Comparative analysis of virulence of a novel, avian-origin H3N2 canine influenza virus in various host species. Virus Res. 2015;195:135–40. doi: 10.1016/j.virusres.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Lee YN, Lee DH, Park JK, et al. Experimental infection and natural contact exposure of ferrets with canine influenza virus (H3N2) J Gen Virol. 2013;94(Pt 2):293–7. doi: 10.1099/vir.0.042473-0. [DOI] [PubMed] [Google Scholar]
- 46.Willis E, Parkhouse K, Krammer F, Hensley SE. Canine H3N8 influenza vaccines partially protect mice against the canine H3N2 strain currently circulating in the United States. Vaccine. 2016;34:5483–7. doi: 10.1016/j.vaccine.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Shen Y, Zhang X, et al. A serological survey of canine H3N2, pandemic H1N1/09 and human seasonal H3N2 influenza viruses in dogs in China. Vet Microbiol. 2014;168:193–6. doi: 10.1016/j.vetmic.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Song D, Moon HJ, An DJ, et al. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J Gen Virol. 2012;93(Pt 3):551–4. doi: 10.1099/vir.0.037739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moon H, Hong M, Kim JK, et al. H3N2 canine influenza virus with the matrix gene from the pandemic A/H1N1 virus: infection dynamics in dogs and ferrets. Epidemiol Infect. 2015;143:772–80. doi: 10.1017/S0950268814001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Wu Y, Zhang W, Qi J, Gao GF. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol. 2014;12:822–31. doi: 10.1038/nrmicro3362. [DOI] [PubMed] [Google Scholar]
- 51.Animal Health Diagnostic Center, Cornell University. Canine influenza H3N2 updates. [Accessed 20 October 2016];Test results by State and over time. 2016 Sep 30; https://ahdc.vet.cornell.edu/news/civchicago.cfm.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.