Abstract
Exosomes are extracellular vesicles released by cells which carry proteins, lipids and nucleic acids and function in intercellular communication. Previously we determined that exosomes released from Mycobacterium tuberculosis infected macrophages carry mycobacterial proteins and lipids. However, the RNA composition within these exosomes has not been defined. In this study we characterized the exosomes released from M .tuberculosis-infected macrophages and identified a cohort of mouse mRNA and miRNA. Quantitative RT-PCR analysis showed less abundance of miRNAs in exosomes released from infected compared to uninfected macrophages. Moreover, over 100 transcripts were found to be enriched or unique to exosomes from infected cells including transcripts involved in regulating an immune response. The exosomal RNA could be transferred and expressed in naïve macrophages and was biological active, stimulating production of inflammatory mediators and inducing apoptosis in recipient cells. Interestingly, we also identified mycobacterial transcripts in exosomes released from infected macrophages as well as from exosomes isolated from TB patient serum. To our knowledge this is the first study to identify bacterial-derived RNA in exosomes. Our results suggest that exosomal RNA released from M. tuberculosis-infected macrophages may have functional and diagnostic potential in the context of a mycobacterial infection.
Keywords: Mycobacterium tuberculosis, macrophage, exosomes, microRNA, mRNA
Introduction
The number of studies concerning bioactive vesicles has significantly increased in recent years with much of this increase stemming from work on exosomes. This class of bioactive vesicles is formed through the fusion of multivesicular bodies (MVBs) with the plasma membrane and release of intra-luminal vesicles as exosomes (1). Exosomes are lipid bilayer vesicles of 30 to 100 nm in size, as defined by electron microscopy, with a density of 1.13 g/ml to 1.19 g/ml (2). They can be distinguished from other extracellular vesicles by size, composition and cellular origin (3). Exosomes are released from various cell types including DCs (4), mast cells (5), platelets (6), macrophages (7) as well as cells of non-hematopoietic origin such as astrocytes (8) neurons (9) and epithelial cells (10) and are found in various body fluids (11, 12, 13, 14, 15, 16). Biophysically, exosomes are equivalent to cytoplasm enclosed in a lipid bilayer with the external domains of transmembrane proteins exposed to the extracellular environment. Originally observed as a mechanism to remove transferrin receptor during reticulocyte maturation (17, 18), more recent studies have focused on the role of exosomes in antigen presentation (19, 20, 21), immune surveillance (22) and induction of tolerogenic responses (23).
The lipid and protein composition of exosomes has been extensively analyzed by various techniques including western blotting, fluorescence-activated cell sorting (FACS), immuno-EM and mass spectrometry. Exosome composition can vary depending on the cell type of origin but some common protein components have been defined (24). Pioneering studies by Valadi and coworkers determined that exosomes are also enriched in mRNA and miRNA. The exosomes derived from human (HMC-1) and mouse (MC/9) mast cell lines were found to transport RNA to neighboring mast cells and the mRNA was subsequently translated, indicating that it was biologically active (25). Recent studies also suggest that exosomes have a selective subset of miRNA that can be functionally transferred as a consequence of fusion with recipient cells (26, 27). The sources of these exosomes/extracellular vesicles were from cultured cells (28) and from body fluids (29, 15). Together, the data suggest that exosome’s function as carriers of genetic information and that this genetic material plays a role in cell-cell communication.
Exosomes released from macrophages infected with M. tuberculosis (M.tb) or treated with mycobacterial culture filtrate proteins as well as exosomes isolated from M.tb-infected mice have been characterized for their protein cargo and have been shown to promote both innate and acquired immune responses in vitro and in vivo (30, 31, 32). However, it is not known whether the RNA present in these exosomes contributes to this immune response. To address this question requires characterization of the RNA content within the exosomes. In the present study, we profiled the exosomes released from M.tb-infected macrophages and showed the exosomes to contain host miRNAs and messenger RNAs. While we observed a general diminished level of host miRNAs in exosomes from infected cells, we also observed a subset of miRNAs and mRNA transcripts unique to these exosomes. Our results suggest that there is selective packaging of RNA content in exosomes following an M.tb infection. The exosomal RNA could be transferred to and translated in recipient cells and could elicit a biologically response in these cells. Surprisingly, we also detected mycobacterial transcripts in exosomes released from M.tb-infected macrophages and from extracellular vesicles derived from TB patient serum. To our knowledge this is the first study to show the presence of pathogen-derived RNA in exosomes released during a bacterial infection.
Results
Defining the miRNA content within exosomes derived from M. tuberculosis-infected and uninfected RAW264.7 cells
Exosomes were isolated from the culture supernatants of uninfected and M.tb-infected RAW264.7 macrophages by filtration and differential centrifugation (33, 34). We obtained approximately 25μg of exosomes from 1×107 cells. Exosomes were analyzed as previously described (34) and were of the expected size and composition (data not shown). Total RNA was isolated from 400 µg of exosomes and analyzed for quality and size distribution on an Agilent bioanalyzer. As shown in figure 1A, we observed predominantly small RNA in the exosomes which was expected since we size-selected on a urea polyacrylamide gel for small RNA. To eliminate any potential contaminating RNA attached to exosomes, the exosomes were treated with RNAse A prior to RNA isolation. This was done for all exosomes prior to RNA isolation. To further characterize the small RNA content in exosomes, cDNA libraries were constructed using the small RNA population derived from exosomes or from donor macrophages (Figure 1b and c). Since we wished to maximize coverage of miRNA sequences within exosomes, we biased our cDNA loading onto the 454 Sequencer to include 25% volume for each exosome library, 20% for each donor cell library and 10% for negative control. Therefore, we did not interpret quantitative differences between miRNAs from the sequencing data. After trimming of adaptor and primer sequences from our reads, a megablast was performed against the mature mouse miRNAs in mirbase. Using 1 as the cutoff E value we identified 52 and 57 miRNAs in exosomes released from uninfected and infected cells respectively. We also identified 57 and 80 miRNAs present in uninfected and infected RAW264.7 cells respectively (Table I). Approximately 60% of the miRNAs were present in exosomes from both infected and uninfected macrophages including the miRNAs Mmu 99b-5p, Mmu 30c, Mmu 30a, Mmu 191, Mmu 378, Mmu 210, Mmu 423-5p & Mmu 486-5p which have been previously reported to be expressed following mycobacterial infection both in vitro and in vivo (37, 38, 39, 40, 41, 42). From the cohort of miRNAs identified in exosomes, a subset was further selected for validation by PCR or by SYBR Green based quantitative PCR. For these experiments, RNA was isolated from independent preparations of exosomes and results were drawn from three experimental replicates. Total RNA was polyadenylated and converted to cDNA and PCR amplified using miRNA specific primer and PerfeCTa universal primer (Figure 2a). In order to compare miRNA levels between the different exosome preperations we needed an endogenous control and therefore we tested the miRNAs U6, Sno202, Sno135 and Sno 234 which have been used previously in this regard (43, 44, 45). Of these different controls, only Sno234 showed similar band intensities on an agarose gel. To further evaluate whether Sno234 could be used as an endogenous control we compared Ct values across all exosome cDNAs. The Exosomal cDNA Ct values for Sno234 in triplicate for replicate experiments was: H37Rv infected macrophage (25.89+/−0.051 and 26.8+/−0.099) and uninfected macrophages (26.04+/−0.249 and 26.12+/−0.186). Therefore Sno234 was used as an endogenous control for all subsequent experiments. Quantitative PCR on selected miRNAs showed an overall suppression of these miRNAs ranging from 2 to over 1000 fold in exosomes from infected compared to uninfected cells, although the level of suppression varied between experiments (Figure 2b). However, we failed to observe any down-regulation of this subset of miRNAs in RAW264.7 cells following a mycobacterial infection (Figure 2c). To determine if the limited incorporation of miRNAs into exosomes was related to pathogenicity, we quantified expression of select miRNAs in exosomes released from RAW264.7 cells infected with non-pathogenic M. smegmatis. We observed a similar decrease in abundence of miRNAs relative to uninfected exosomes (Unpublished observation). Our results suggest that mycobacterial infection of macrophages results in the general inhibition of miRNA incorporation into exosomes. To understand the significance behind this suppression we evaluated the quantified miRNAs for mRNA targets using the miRDB and functional KEGG pathway analysis. The analysis showed that potential gene targets for these miRNAs included those associated with immune surveillance and inflammation (Table II).
Figure 1. Exosomes derived from cell culture supernatants of M.tb-infected macrophages contain small RNA.
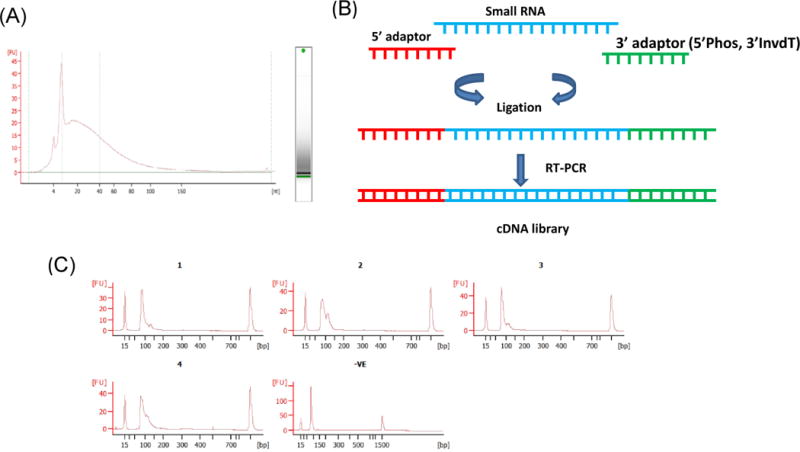
Exosomes were isolated from cell culture supernatants of uninfected or M.tb-infected RAW264.7 macrophages. RNA was isolated from exosomes and tested on a Bioanalyzer small RNA pico chip (A). A Cartoon showing the steps involved in producing a cDNA library containing ligated Illumina adaptors used for RT-PCR (B). The cDNA libraries were analyzed on a Bioanalyzer for size distribution (C). 1, exosomes from uninfected cells; 2, exosomes from infected cells; 3, uninfected RAW 264.7 cells; 4, infected RAW 264.7 cells; 5, ligation control.
Table I.
Mouse miRNAs identified in exosomes and RAW 264.7 macrophages by sequencing of the small RNA libraries (BLASTn against mature mir Base). Unique to each group is marked in red (E = 1)
| Un Exo | Rv Exo | Un RAW | Rv RAW |
|---|---|---|---|
| 1224-3p, , , 182-5p, , 100-5p, , , 101a-3p, 101c, 101b-3p, 101a-5p, 183-5p, , 140-3p, , 140-5p, 191-5p, 152-5p, 210-3p, , 27b-3p, 27a-3p, , , 2182, 3473b, 3473, 3473d, , , , , 341-3p, 30c-2-3p, 30c-1-3p, , , 378-3p, 378b, 378-5p, 345-5p, 423-5p, 5100, 5128, , 709, 99b-5p, 99a-5p, , , | 152-5p, 101b-3p, 101a-3p, 101c, 101a-5p, , 100-5p, , , 1224-3p, 191-5p, 140-3p, 140-5p, , 182-5p, 183-5p, 210-3p, 27b-3p, 27a-3p, 24-3p, , 2182, , 378-3p, 378b, 378-5p, 345-5p, 3473b, 3473, 3473d, , , 341-3p, , 30c-2-3p, 30c-1-3p, 423-5p, , , , , , , 5100, , 5128, , , , , 709, , 99a-5p, 99b-5p, , , | 151-3p, 191-5p, 140-3p, 146b-5p, 146a-5p, 146a-3p, 1196-5p, 152-5p, , , 182-5p, 193a-3p, 193b-3p, 183-5p, 1195, 1935, 140-5p, , , 210-3p, 27b-3p, 27a-3p, 29a-3p, 24-3p, 29c-3p, , 29b-3p, , 2182, 378b, 378-3p, 378-5p, 345-5p, 3074-5p, 30a-3p, 30e-3p,30d-3p, , 3473b, 3473, , 471-5p, , 5100,5097, 532-5p, , 5109, 5621-5p, 5128, 709, , , 93-5p, , let7a-5p, let7c-5p | 146b-5p, 146a-5p, 146a-3p, 182-5p, , , , , , 183-5p, 151-3p, , 152-5p, 140-3p, 191-5p, , 140-5p, 193a-3p, 193b-3p, , , , , , , , 1935, 1196-5p, 1195, , 210-3p, 2182, , 27a-3p, 27b-3p, , 24-3p, , , , , , 29a-3p, 29c-3p, 29b-3p, 30e-3p, 30a-3p, 3473b, 3473, 378-3p, 378b, 345-5p, , , 30d-3p, 3074-5p, , 378-5p, , , 471-5p, , 5100, , 5128, 532-5p, 5097, , 5109, , 5621-5p, , 709, , , , 93-5p, let7c-5p, , let7a-5p |
Figure 2. Exosomes released from M.tb-infected macrophages show limited incorporation of host miRNAs.
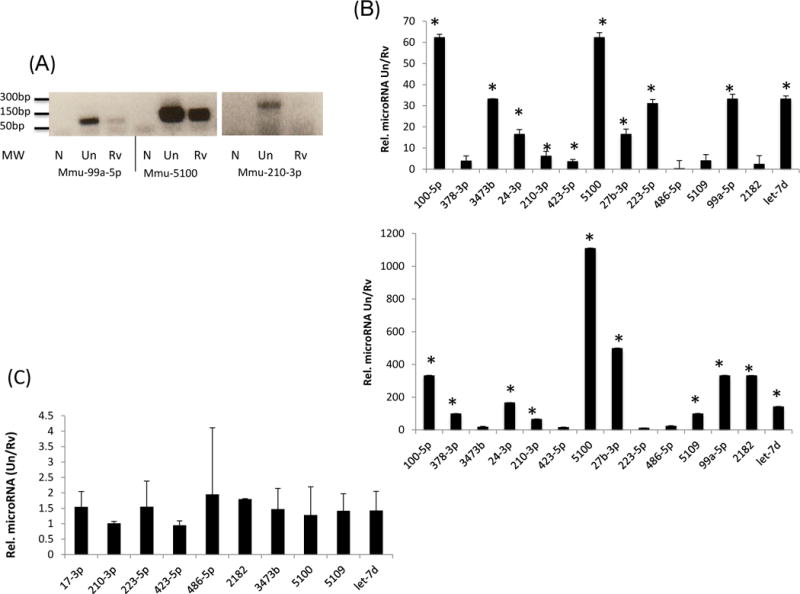
Total RNA was isolated from exosomes and reverse transcribed using miR-Script cDNA synthesis kit. PCR was performed for selected miRNAs using miRNA specific forward primers and a universal reverse primer (A). Quantitative RT-PCR in triplicate was performed on selected miRNAs using sno234 as the endogenous miRNA control and results are shown from two independent experiments with standard deviations and p value<0.05 denoted by an asterick (*) (B). Selected miRNAs that showed limited incorporation into exosomes released from infected RAW264.7 cells were analyzed for expression levels in both infected and uninfected cells (C). Results were drawn from three independent experiments with standard deviations. N; no template control, Un; exosomes from uninfected cells, Rv; exosomes from infected cells.
Table II.
Functional pathways identified in mRNA targets for the selected miRNAs
| Pathway | Number of genes | Representative genes | Gamma corrected p value |
|---|---|---|---|
| Calcium signaling pathway | 8 | itpr2, Cacna1c,Ntsr1, Pdgfra,Camk2b | 0.00157555 |
| Neuroactive ligand-receptor interaction | 9 | Gria3,Adora1, Sstr1,Oxtr, Adcyap1r1,Thrb | 0.00213164 |
| MAPK signaling pathway | 7 | Atf2,Atf4,Acvr1b, CD14,Map2k1,M apk14 | 0.02555087 |
| ErbB signaling pathway | 9 | Mapk9,Pik3cg,C amk2b,Pak4 | 5.05E-05 |
| Natural killer cell mediated cytotoxicity | 8 | Pik3cg,Vav1,Ptpn11 | 0.0011577 |
| Jak-STAT signaling pathway | 8 | Il15ra, Pim1, Pik3cg, Csf3r, Ptpn11 | 0.01062047 |
| Ubiquitin mediated proteolysis | 7 | Ube2k, Prpf19, Anapc7, Pml | 0.02275327 |
Profiling the mRNA transcripts within exosomes released from infected and uninfected RAW 264.7 cells
A microarray analysis was undertaken to identify the host mRNA signatures present in exosomes. In order to generate the 1 μg of total RNA needed for the gene expression studies we used approximately 400 µg of exosomes. To determine enrichment of specific transcripts in exosomes following mycobacterial infection, exosomal RNA from uninfected cells and cellular RNA from uninfected or M.tb-infected donor macrophages were used for comparison. We identified 2428 and 2638 transcripts in exosomes from infected and uninfected cells respectively (Figure 3a; Datasets S1 and S2). However, it is important to note that the results are drawn from a mixed population of exosomes and that individual exosomes will differ in the transcripts present and will contain only a subset of the total transcripts. This is likely also true for the miRNAs identified in exosomes. A large number of transcripts detected in exosomes from infected cells encoded for ribosomal proteins and we could also detect transcripts encoding for RNA binding proteins such as rbed1, csde1, rbmxrt, tarbp2 etc. However, we also observed genes involved in MAPK signaling (tnfrsf1a, mapkapk2, phospholipaseA2, ly96) T cell activation (cd14, cd40, cd80 and cd86) apoptosis (bcl2l1, irak2, nfkbia) and proteasome, (psmd13, psmb8, psmb9). From the total transcripts identified, 65 showed 2 fold or higher expression with a p value ≤ 0.05 in exosomes released from infected compared to uninfected cells (Dataset S3). However, only 16 of these 65 transcripts showed greater than 2 fold higher expression in infected compared to uninfected RAW264.7 cells. A subset of genes from the group of 65 more abundant transcripts were validated by SYBR Green based quantitative PCR including those previously characterized as functioning in immune responses such as prss15, slc15a3 and ncf4 (Figure 3b). Transcription levels were normalized to GAPDH which had Ct values of 24.52+/−0.442 and 24.08+/−0.956 for exosomes from uninfected and infected macrophages respectively. We also identified 69 transcripts that were less abundant in exosomes from infected cells relative to uninfected cells including hmgb2, traf3, psmd3 and ctsf. Interestingly, we identified a subset of transcripts which appeared unique to each exosome population (Tables S1 and S2). A subset of the 46 transcripts present in exosomes from M.tb-infected cells included those involved in immune surveillance such as ccl21b, lcn2, traf1 and procr were further tested by PCR. As predicted from our array data, we did not detect these transcripts in exosomes from uninfected cells, confirming that they are absent or present at very low levels in this exosome population (Figure 3c). To determine whether selective incorporation of these transcripts into exosomes relates to their enrichment in host cells following a mycobacterial infection, we analyzed the expression patterns of these transcripts in M.tb-infected cells compared to uninfected cells. We found that most transcripts were enriched in macrophages infected with M.tb compared to uninfected cells suggesting that the presence of these unique transcripts in exosomes stem, at least in part, from their higher expression in infected cells (data not shown). A KEGG Pathway analysis was undertaken on the total transcripts identified in exosomes. We observed a few functional pathways which were specific to exosomes from infected or uninfected cells but the majority of pathways were defined in both exosome populations including regulation of actin cytoskeleton, TLR signaling and MAPK signaling among others (Tables S3 and S4). For the 46 unique transcripts identified in exosomes from infected cells, only the cytokine-cytokine receptor interaction emerged as significant and included genes such as ccl4, ccl3,ccl5 & cxcl10. For the 65 transcripts that were more abundant in exosomes from infected compared to uninfected cells, the pathways identified included proteasome (psmb9, psmd3 & psme2), Ag processing and presentation (cd74, lgmn & psme3) and Systemic lupus erythematosus which consisted of genes such as hist1h2ab, hist1h2bc etc.
Figure 3. Unique set of host transcripts are detected in exosomes isolated from M.tb-infected macrophages.
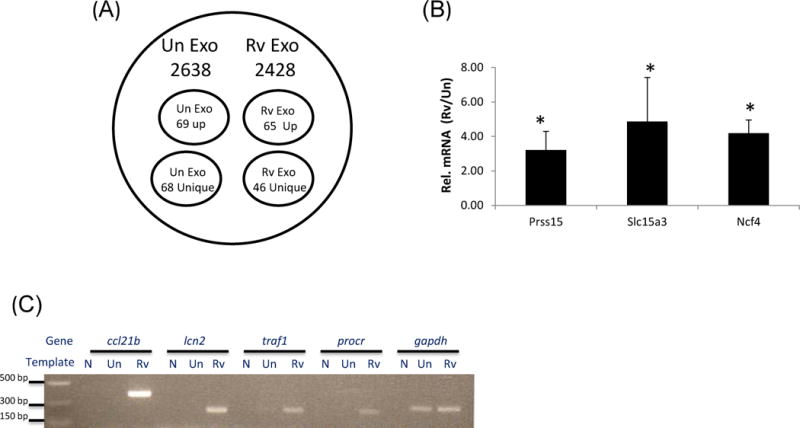
Total RNA was isolated from exosomes and cells, reverse transcribed, labeled with Cy3 and hybridized onto whole mouse genome arrays. The signal intensities were converted to expression values and transcripts were identified that had expression values above empty well values for all three experimental replicates (A). Three representative transcripts from the cohort of genes defined by array analysis as highly expressed in exosomes from infected RAW264.7 cells were validated for quantitative differences compared to uninfected exosomes by SYBR Green based qRT-PCR (B). Five transcripts that were unique to exosomes from infected cells were further validated by RT-PCR. Primers were designed for specific transcripts and exosomal RNA was derived from independent exosome preparations (C). Exo; exosomes, N; no template control, Un; exosomes from uninfected cells, Rv; exosomes from infected cells. Results are representative of three independent experiments for both the microarray analysis and qRT-PCR validation which includes standard deviations.
Exosomes released from M. tuberculosis infected macrophages contain mycobacterial transcripts
A BLAST analysis against the whole M.tb transcriptome was performed using the sequence data from the small RNA library. We identified mycobacterial transcripts Rv3809c, Rv3533, Rv0243, Rv1101c and Rv2024c multiple times in the RNA library when using exosomes isolated from infected macrophages. Although our sequence data indicated that exosomes contain fragments of these mycobacterial genes, the length of the transcripts were unclear as we only selected for small RNA fragments when generating the library. Therefore we designed multiple primers for Rv3809, Rv3533 and Rv0243 to provide maximum coverage for each gene. RNA was isolated from independent exosome preparations and converted to cDNA which was used as template for the PCR. We detected PCR products for each of these three mycobacterial transcripts. No products were identified when using the cDNA generated from exosomes released from uninfected cells (Figure 4a). Although we were able to detect products for 2 or 3 primer sets for each transcript, we could not detect full length transcripts using primers that extend the whole gene suggesting that only fragments of mycobacterial transcripts get incorporated into exosomes released from M.tb-infected macrophages. To detect other potential mycobacterial transcripts in exosomes released from infected cells, total RNA was converted to ds cDNA, labeled with Cy3 and hybridized onto a custom whole MTB genome array. RNA from uninfected exosomes was used as negative control and M.tb RNA as a positive control. These gene expression studies identified an additional 13 mycobacterial transcripts in exosomes from infected macrophages. Independent preparations of exosomal RNA were used to validate the gene expression results by RT-PCR. These results confirmed the presences of 9 of the 13 mycobacterial transcripts which included Rv0740, Rv0288, Rv1344, Rv0968, Rv1942c, Rv0664, Rv0190, Rv1757c and Rv1369c. A few representative transcripts are shown in figure 4b. As expected, the mycobacterial transcripts were not detected in exosomes released from uninfected macrophages. A randomly selected set of mycobacterial transcripts detected in exosomes were tested for their expression in RAW264.7 macrophages following infection with M. tuberculosis. The presence of these transcripts in host cells suggested their expression in host following infection and subsequent incorporation in vesicles.
Figure 4. Exosomes released from M.tb-infected macrophages contain mycobacterial RNA.
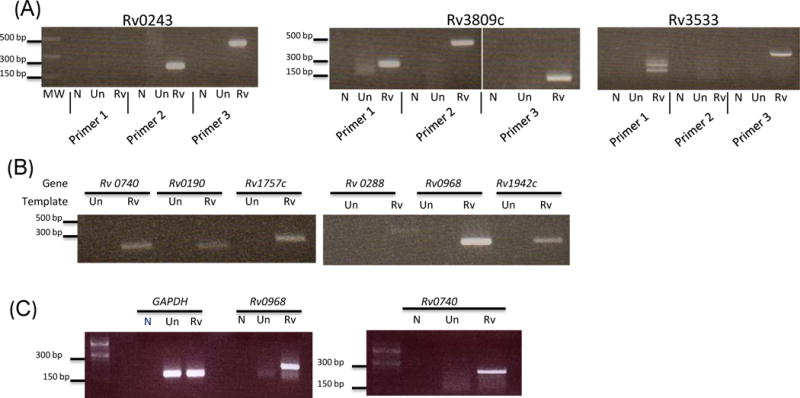
Multiple sets of primers were designed for selected mycobacterial transcripts that were identified in sequencing of the exosome small RNA library. PCR amplifications were performed for each primer pair in an attempt to amplify different regions of each transcript (A). Exosomal RNA was reverse transcribed to double stranded cDNA, labeled with Cy3 and hybridized to whole M.tb genome arrays. Selected mycobacterial transcripts identified in the gene expression analysis were validated by PCR (B). RAW264.7 macrophages were infected with M. tuberculosis or left uninfected. Cellular RNA was isolated and converted to single stranded cDNA and gene specific primers were used for PCR amplification (C).
To rule out that the RNA transcripts were derived from vesicles released from mycobacteria during the course of the infection, we purified exosomes by sucrose gradient to remove any potential contaminating vesicles. The sucrose-purified exosomes contained the exosomal markers LAMP-1 and Tsg101. However, only the exosomal RNA from infected macrophages were positive for mycobacterial transcripts. In contrast, RNA from infected and uninfected cells were positive for GAPDH (Fig. S1). To further confirm that the RNA was not from vesicles released mycobacteria, RAW264.7 cells were either infected with M. tuberculosis as described above or equal number of bacteria were added to media alone (i.e. no macrophages) and flasks were incubated for 72 hours. Supernatants were harvested, centrifuged at 3000g for 10 minutes followed by filtration through 0.22 micron twice (remove intact bacteria). The supernatants were further centrifuged at 100,000g to pellet down exosomes/extracellular vesicles. Protein quantification using BCA showed a protein concentration of 20ug/flask from vesicles isolated from infected macrophages whereas vesicles isolated from the media with bacterial alone (i.e. no macrophages) showed a protein concentration of 4.5ug/flask. The isolated vesicles was treated with RNaseA and RNA was extracted as described in Material and Methods. We failed to detect any RNA in the material derived from similar volume of culture media to which equal amounts of bacteria had been added (data not shown). In contrast, as indicated above, RNA was present in exosomes isolated from infected macrophage culture media and these exosomes contained mycobacterial RNA. This indicates that if any vesicles were released from the mycobacteria when present in culture media they did not contain detectable RNA.
Exosomal RNA can be transferred to recipient cells
To evaluate whether RNA from exosomes can be transferred to uninfected macrophages, purified exosomes were labeled with SYTO RNA select stain and added to RAW 264.7 cells that were stained with wheat germ agglutinin (WGA) to visualize cell membranes. As observed by fluorescent microscopy, the RNA in exosomes was transferred to recipient macrophages and appeared primarily cytoplasmic (Figure 5). To confirm that the dye incorporation into recipient cells was exosome dependent, the SYTO RNA Select dye was centrifuged in buffer alone (i.e. no incubation with exosomes) and any material that was pelleted at 100,000×g was added to the RAW 264.7 cells. No visible staining of macrophages with the SYTO RNA dye was observed after incubation (Fig. S2).
Figure 5. Exosomal RNA can be transferred to recipient macrophages.
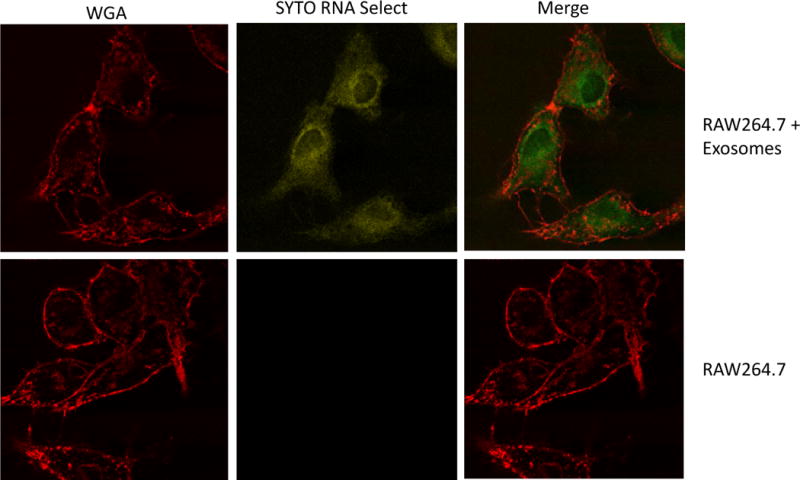
RAW264.7 cells were treated with Alexa Fluor 594-labeled Wheat Germ Agglutinin (WGA) to visualize cell membranes. Purified exosomes were labeled with the SYTO Select RNA stain and these labeled exosomes were added to RAW264.7 cells or cells were left untreated. The monolayers were washed and the uptake of exosomal RNA was visualized using a BioRad MRC 1024 Scanning Confocal coupled to a Nikon Diaphot 200 microscope using LaserSharp 2000 acquisition software. The images were acquired at 40× with 2× digital zoom with x,y dimensions of 0.24 μm per pixel and the images were processed using Image J software. Shown is a representative picture from two independent experiments.
Exosomal RNA can be translated to protein upon delivery to recipient cells
In view of the transfer of exosomal RNA to recipient cells, we evaluated whether exosomal RNA delivered to recipient cells was translated. We targeted the mRNA encoding mouse lipocalin-2 since it was detected exclusively in exosomes derived from M.tb infected macrophages but not from uninfected macrophages. Further, lipocalin-2 is known to inhibit mycobacterial growth in vitro through sequestration of iron uptake implicating its role in the mycobacterial innate immune response (46). Human THP-1 cells were incubated with mouse exosomes derived from M.tb infected cells or uninfected cells for 4 hours and the monolayers were washed to remove any remaining extracellular exosomes. Untreated THP-1 cells were used as a control. The cells were incubated for 24 hours, lysed and probed for mouse lipocalin-2 protein expression by western blot. As shown in figure 6, we observed the expression of murine lipocalin-2 upon treatment of human THP-1 cells with mouse exosomes from infected cells. No lipocalin-2 was observed in THP-1 cells treated with exosomes from uninfected cells or in control untreated THP-1 cells. We used a mouse monoclonal antibody made to a recombinant peptide consisting of amino acids 20 to 200 of mouse lipocalin-2 which was indicated to be specific to mouse lipocalin-2. To confirm the specificity, the mouse monoclonal antibody was tested against a human lipocalin-2 peptide consisting of amino acids 20 to 198. The antibody did not recognize the recombinant human lipocalin-2 (Figure S3). Our results indicate that exosomal mRNA can be transferred into recipient cells and translated into protein.
Figure 6. Exosomal RNA leads to synthesis of protein upon delivery to recipient cells.
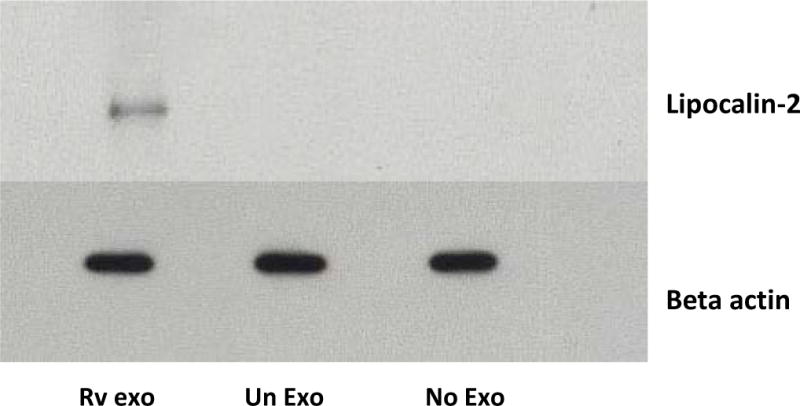
PMA differentiated THP-1 cells were treated with exosomes derived from M.tb infected or uninfected RAW264.7 macrophages or left untreated. After 24 hours, cells were harvested, lysed in RIPA buffer and probed with mouse lipocalin-2/NGAL antibody (A) or rabbit monoclonal antibody for beta actin (B).
Exosomal RNA induces a pro-inflammatory response in recipient cells
As our data supports the transfer of exosomal RNA to recipient macrophages, we next evaluated the macrophage’s biological response to this RNA. Exosomal RNA was transfected into RAW264.7 cells using the Hiperfect transfection reagent which is a mixture of cationic and neutral lipids and facilitates the cellular uptake of RNA. The RNA dosage corresponded to approximately 50 exosomes per cell in our experiments. 24 hours post transfection, the cell culture supernatants were profiled for 40 different cytokine and chemokines using a mouse cytokine array. As a control, some cells were treated with Hiperfect alone. Exosomal RNA from both uninfected and M.tb-infected macrophages induced significantly higher levels of sICAM-1, RANTES and I-TAC compared to untreated cells or cells treated with the Hiperfect transfection reagent. However, only RNA isolated from exosomes released by infected cells induced a significant increase in the secretion of CCl2, MIP-2, TNF-α and IL-1ra by RAW264.7 macrophages compared to resting cells or cells treated with Hiperfect alone (Figure 7a and b).
Figure 7. Transfection of recipient cells with exosomal RNA stimulates cytokine and chemokine secretion and induces significant apoptosis.
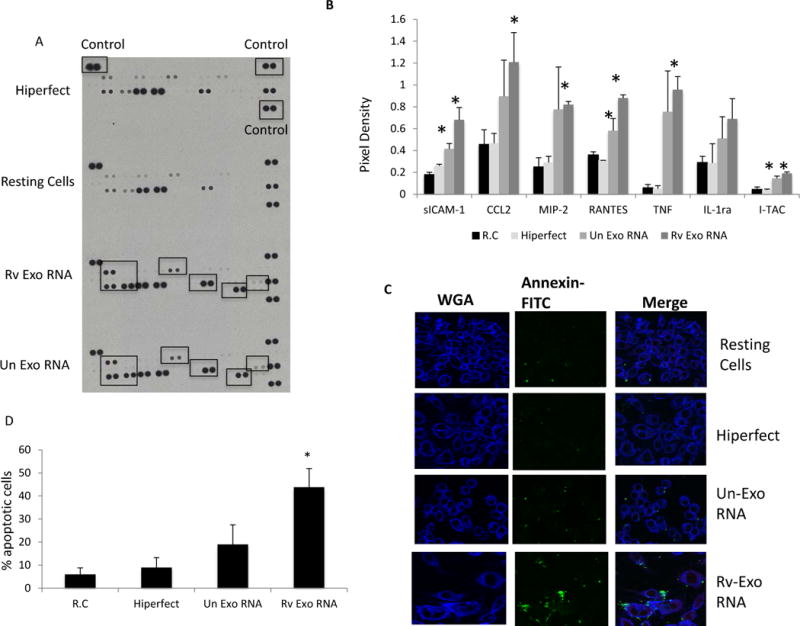
RNA was isolated from exosomes released from uninfected or M.tb infected RAW264.7 macrophages. 250ng of RNA was mixed with 3μl of Hiperfect transfection reagent and added to RAW264.7 macrophages. Untreated macrophages or macrophages treated with Hiperfect reagent alone were used as controls. Following incubation in growth media for 24 hours, cell culture supernatants were harvested and probed for the presence of cytokines or chemokines using a Mouse Cytokine Antibody Array Panel (A). Pixel densities for spots corresponding to differentially expressed proteins were defined using Imaje J software and plotted (B). In separate experiments, cells were stained with FITC-Annexin V and fixed with 2% paraformaldehyde. FITC-labeled cells were visualized on a confocal microscope (C) and the percentage of Annexin-V positive cells defined by visual counting (D). Graphs results are a combination of two independent experiments +SD. *indicates a P < 0.5 compared to untreated cells.
Since mycobacterial transcripts were detected in the exosomal RNA and previous reports indicate that mycobacterial RNA can induce apoptosis in human macrophages (47) we addressed the possibility that the RNA from exosomes derived from infected cells could induce apoptosis in recipient cells. As shown in figure 7c transfection of naïve cells with this exosomal RNA resulted in elevated levels of phosphatidyl serine on the outer leaflet of the plasma membrane as defined by annexin-V staining. Increased phosphatidyl serine exposure is associated with early events in apoptosis. Quantitation of the number of annexin-V positive cells +/−transfection with exosomal RNA is shown in figure 7d.
Mycobacterial transcripts are present in extracellular vesicles (EVs) derived from serum of human TB patients
With the detection of mycobacterial transcripts in exosomes released from M.tb-infected macrophages, we explored the possibility of detecting M.tb transcripts in exosomes isolated from human TB patient serum. RNA was isolated from extracellular vesicles purified from the serum of culture positive TB patients or from a healthy control. Since we were limited in the amount of human serum and therefore vesicles, we pre-amplified the exosomal RNA using a WTA amplification kit to obtain sufficient quantity of cDNA for hybridization onto the whole M.tb genome arrays. From the hybridization experiments we putatively identified five mycobacterial transcripts in exosomes from TB patients. To further evaluate whether the genes defined in the array experiments were present in TB patient exosomes, primers were designed for each of these mycobacterial transcripts and the cDNA, produced from pre-amplified RNA, was used as a template for PCR. We confirmed that Rv2796 and Rv1369c were present in serum exosomes from TB infected individuals as shown by PCR and SYBR Green based qRT-PCR (Figure 8a and b).
Figure 8. RNA isolated from EVs derived from serum of TB-infected individuals contains mycobacterial transcripts.
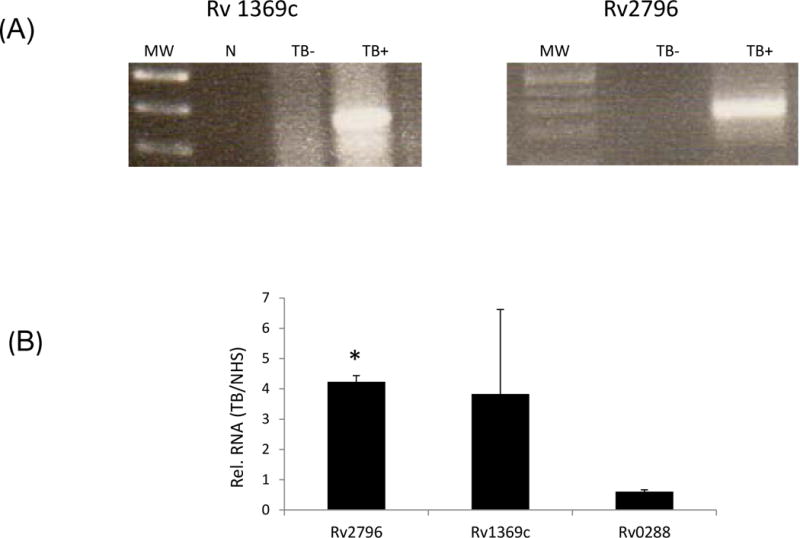
RNA was isolated from exosomes derived from the serum of culture-positive TB patients and from health controls. The RNA was initially amplified and then reverse transcribed to double stranded cDNA, labeled with Cy3 and hybridized to whole M.tb genome arrays. The M.tb transcripts that were identified in the array analysis were selected for validation by RT-PCR (A) and SYBR Green based qRT-PCR (B). N; no template control, Un; exosomes from uninfected cells, Rv; exosomes from infected cells, TB−; exosomes isolated from non-TB, healthy control serum, TB+; exosomes isolated from TB patient serum. Results are from the average of two separate experiments + SD.
Discussion
Recent studies have shown that exosomes, as vehicles of intercellular communication, not only transport proteins and lipids, but also biologically active RNA. The RNA encapsulated in exosomes is termed “exosomal shuttle RNA” (esRNA) and consists of functional miRNA and mRNA that can be transferred to recipient cells and modulate their transcriptome. Exosomal RNA has been studied in diverse fields such as cell biology, immunology, cancer biology, neurobiology and has been characterized in several cell types including mast cells (25), T and B cell lines (26), bone marrow derived dendritic cells (27), macrophages (48) and carcinoma cell lines (45). These studies indicate that exosomes provide a natural delivery system for RNA both in vitro and in vivo and also highlight the potential use of exosomal RNA as molecular biomarkers against several diseases. However, the role of exosomes in transporting genetic material, specifically RNA in a background of a bacterial infection has not been undertaken. We addressed this question in the context of a mycobacterial infection. In present study we have defined the RNA content of exosomes released from murine RAW264.7 macrophages following infection with M.tb and showed these exosomes to contain miRNA, mRNA as well as mycobacterial transcripts. Further, the detection of mycobacterial transcripts in exosomes was not limited to an in vitro infection as extracellular vesicles derived from TB patient serum also contained TB transcripts.
The role of miRNAs in responding to bacterial infections is poorly understood. The miRNAs could be involved in regulating gene expression for pathways that are important in the immune response to pathogens. Differences in circulating miRNAs in serum of TB patients compared to controls have been observed and these differences may provide signatures to distinguish active from latent TB (42). Although the authors did not specifically look at exosomes, it is known that the majority of miRNAs in human serum or saliva are encapsulated in exosomes allowing for increased stability of the RNA (49). Recently, exosome-enclosed miRNAs in exhaled breath have been suggested as potential biomarkers for patients with pulmonary diseases such as tuberculosis (50). In our initial experiments we focused on the miRNAs present in exosomes released from M.tb-infected macrophages and how this compares to exosomes from uninfected cells. To explore this question, we adopted a sequencing approach to identify the miRNAs in exosomes. Although we identified a subset of miRNAs that were specific to exosomes from infected cells, the majority of miRNAs identified were present in exosomes from both infected and uninfected macrophages, suggesting a general conservation in the trafficking and incorporation of miRNAs into exosomes. In total, we identified 57 miRNAs in exosomes released from infected macrophages including Mmu 223 and 486-5p which belong to the cohort of differentially expressed miRNAs in the serum of TB patients (40, 42). Previous studies have shown that miRNA 99b is highly up-regulated in M. tuberculosis infected dendritic cells (DCs) and it targets TNF-α and TNFRSF-4 receptor gene transcripts and that less abundance of this miRNA leads to a significant loss in bacterial survival in DCs (38). Mycobacterial secreted protein ESAT6 is also a known effector of miR-155 whose up-regulation following an M.tb infection modulates the expression of a subset of proteins that benefit the establishment of an infection (51). These results suggest that cellular miRNAs which are more abundant following infection with M.tb may provide a mechanism of immune evasion by the pathogen. Surprisingly we found in our quantitative RT-PCR studies that the level of these as well as other miRNAs was significantly diminished in exosomes released from M.tb-infected compared to uninfected cells. The identified miRNAs were involved in various pathways including Calcium signaling, MAPK signaling, Natural killer cell mediated cytotoxicity, and Jak-STAT signaling all of which are involved in the immune response to infection. This difference in miRNAs between the exosome from infected and uninfected cells was not reflected in the concentration observed in whole cells. However, we only defined their cellular concentration 72 hours post-infection and therefore it is unclear how they compared at earlier time points. The mechanism for miRNA incorporation into exosomes is still being defined. However, a recent study indicates that the RNA binding protein Heterogeneous Nuclear Ribonucleoprotein A2B1 (hnRNPA2B1) can bind a specific subset of miRNAs through their EXOmotifs and control their loading into exosomes (52). This suggests that the repertoire of RNA binding proteins present within an exosome may affect which miRNAs are trafficked and incorporated into exosomes. Interestingly, we found an increased expression of Annexin II in exosomes released from infected cells compared to uninfected cells (data not shown). Recently Annexin II has been identified as a novel RNA binding protein that binds directly to both ribonucleotide homopolymers and human c-myc RNA and it has been hypothesized to be involved in the recruitment of appropriate cellular and/or viral components to generate HCV-RNA-containing exosomes (53).
Our study also defined the mRNA signatures within exosomes and found distinct subsets of transcripts enriched in each group suggesting selective incorporation of host mRNA into exosomes. Analysis of the data identified a unique group of transcripts present only in exosomes from infected cells which included genes such as traf, lcn2 that are known to play an important role in innate immunity. A number of transcripts were also identified that were present at significantly higher or lower concentration in exosomes from infected compared to uninfected cells. A GO analysis indicated that the affected pathways included cytokine-cytokine receptor interaction, proteasome, T cell activation and systemic lupus erythmatosus. The observation that within exosomes released from infected cells we observe diminished levels of miRNAs which block translation of genes involved in the host immune response while genes involved in promoting inflammation show relatively higher concentration suggest that the RNA content of these exosomes is primed to stimulate the host immune response to a mycobacterial infection. Since exosomes exert their affect beyond the infected cell, it is possible that the export of these RNA molecules work in concert with cytokines and other factors to stimulate the immune response against invading pathogens. Future studies are needed to understand the mechanisms that determine incorporation of select mRNAs into exosomes.
Unexpectedly we detected mycobacterial transcripts in exosomes derived from M.tb-infected macrophages. We identified the mycobacterial transcripts through sequencing of the small RNA library as well as in our expression analysis using M.tb whole genome arrays. Previous studies have shown the presence of viral trans-activator response element (TAR) RNA in exosomes isolated from cell culture supernatants of HIV-1 infected cells and from patient sera. This TAR miRNA was not associated in the Ago2 complexes outside the exosomes but was enclosed within exosomes (54). Epstein Barr virus encoded miRNAs have also been shown to be secreted by EBV infected B cells via exosomes (55). Beyond viral RNA, our study is the first to show the presence of pathogen associated RNA in exosomes released from infected cells. How mycobacterial RNA is incorporated into exosomes is presently unclear. However, previous reports have shown that mycobacterial DNA gains access to cyosolic receptors likely through perforation of the phagosome membrane mediated by the ESX-1 secretion system (56). Also, previous studies have shown SecA2-mediated secretion of bacterial nucleic acids by Listeria monocytogenes that enables infected macrophages to detect viable bacteria in cytosol via immune sensory receptors RIG-I, MDA5 and STING (57). There are likely additional mechanisms that contribute to the presence of prokaryote RNA in cytosol of infected cells. These include leakage of bacterial debris that contains nucleic acids from phagosomal compartments (58) autolysis within the cytosolic compartment (59) or nucleic acid release from viable bacteria in the cytosol (60).
Since exosomes released from M.tb infected macrophages contain a distinct repertoire of miRNAs, mRNAs as well as pathogen derived mycobacterial RNA, we hypothesized that the exosomal RNA would elicit a unique host response upon its delivery to recipient cells. Our study indicated that exosomal RNA can be transferred to recipient RAW264.7 cells and that the mRNA for mouse lipocalin-2 present in exosomes from infected cells could be translated to protein upon delivery to human THP-1 cells. These results show the functionality of the RNA encapsulated in exosomes Since it was necessary to distinguish the activity of the exosomal RNA in modulating the host immune response from that played by the pathogen associated molecular patterns (PAMPs), also present in exosomes released from infected macrophages, it was necessary to purify the exosomal RNA away from the other exosome components. The purified RNA was transfected into naïve RAW264.7 cells and the treated cells evaluated for expression of various cytokines and chemokines. We observed certain commonalities in the cellular response when using exosomal RNA from infected and uninfected cells. However, exosomal RNA from infected cells was more potent in not only inducing higher secretion of TNF-α, MIP-2 and CCL2 but also in driving apoptosis. At present it is unclear if the increased apoptosis is due to the presences of mycobacterial RNA in these exosomes. Nevertheless, our results indicate that specific host-or mycobacterial-derived RNA molecules present in exosomes released from infected cells could contribute to a pro-inflammatory response and apoptotic signals in cells recruited to the site of an M.tb infection.
In conclusion, our study shows that M.tb-infected macrophages secrete exosomes that contain a unique subset of host miRNAs and mRNA as well as mycobacterial RNA. This unique composition leads to a differential response by the recipient cells. We hypothesize that the host immune response would benefit from the exosomes released from infected macrophages due to the specific composition of host miRNAs and mRNAs; however, a test of this hypothesis awaits further study. Finally, we also report the detection of mycobacterial RNA in exosomes isolated from TB patient serum. These results point to the potential use of exosomal RNA in TB diagnostics. This is the first study to characterize the RNA content of exosomes in the context of a bacterial infection and adds an additional layer of complexity to the function of exosomes during an M.tb infection.
Materials & Methods
Ethic Statement for use of human material
Only publicly available, de-identified or unidentified? serum samples were used for this project and the Notre Dame Institutional review board under protocol # 13-09-1221 gave exempt status to the isolation of exosomes from human serum as performed in this study.
Cell culture, infection and exosome isolation
RAW264.7 cells were cultured in complete DMEM media with 10% fetal bovine serum. . Exosomes were isolated from cell culture supernatants of confluent uninfected or M.tb infected RAW264.7 macrophages. 10 flasks each with 1×107 cells in 25 ml of complete DMEM were infected with M.tb so as to achieve approximately 80% infectivity as determined previously by an uptake assay (33, 34). The monolayers were washed three times with 1X PBS after 4 hours following infection with M.tb and exosome free media was added to the cells and the cells were incubated for 72 hours. The conditioned media was harvested and centrifuged at 3000×g for 10 minutes to remove cells debris followed by filteration twice through 0.22μm and the supernatant was further centrifuged at 100,000g for 1 hour. For some experiments, exosomes were resuspended in 100μl of 1X PBS and layered on top of a linear sucrose gradient. (0.25–2M sucrose, 20mM Hepes/NaOH pH 7.2). The exosomes formed a distinct ring based on their density (1.13 and 1.18 g/ml) and were carefully recovered from the gradient.
Serum of TB patients was kindly provided by the foundation of New and Innovative Diagnostics (FIND) and by Luke Davis, UCSF and included serum from HIV positive and HIV negative TB patients. Extracellular vesicles were isolated from a total of 4.8 ml of TB patient serum and 8 ml of serum obtained from a healthy volunteer (Normal Human Serum; NHS) by successive centrifugation steps. Briefly, the serum was passed through 0.4 µm filter and centrifuged at 1,500×g for 15 minutes followed by 17,000×g for 30 minutes. The supernatant was finally centrifuged at 100,000×g for 2 hours to pellet the extracellular vesicles.
RNA isolation & cDNA synthesis
RNA was isolated from exosomes derived from cell culture supernatants or human serum using MirVANA kit (Ambion) following manufacturer’s instructions. Prior to RNA isolation exosomes were treated with RNAse A at 10µg/ml at 37°C for 30 minutes to confine the analysis to RNA encapsulated within the exosomes. Exosomes were also treated with DNAse1 (Invitrogen) following manufacturer’s instructions. The RNA isolated from serum exosomes was pre-amplified using Whole Transcriptome Amplification kit WTA-2 (Sigma). From 400 µg of exosomes we obtained approximately 1.0µg of total RNA.
Construction of small RNA libraries
The RNA was size fractionated on a 15% tris-borate-EDTA (TBE) urea polyacrylamide gel (Biorad) and small RNA libraries were constructed as described previously (35). The concentration of cDNA libraries for small RNA were determined with a fluorescence based quantitation method (PicoGreen) and the samples were run on Roche 454 Genome Sequencer FLX instrument following manufacturer’s instructions.
The raw reads were filtered to eliminate adaptor and primer sequences and the sequences in fasta format were run against the mature mouse miRNA sequences available in miRBase using BLAST software under linux operating system. MicroRNAs were identified using E value 0.01 or 1.
qPCR validation of miRNAs
Total RNA from exosomes was polyadenylated and reverse transcribed to cDNA using qScript miRNA cDNA synthesis kit (Quanta Biosciences). QPCR was performed using PerfeCTa SYBR Green SuperMix and samples were run on AB7500 Fast Cycler following manufacturer’s instructions. The relative miRNA expression was normalized to the endogenous reference gene and was quantitated using the comparative Ct method with the formula 2−ΔΔCT.
Gene Expression Studies
Total RNA from exosomes or from RAW264.7 cells was converted to double stranded cDNA using Super Script ds cDNA synthesis kit (Invitrogen), labeled and hybridized onto Nimblegen arrays (mouse or M.tuberculosis genomes) as previously described (33). Pathway analysis was performed with the Pathway-Express program (36) of the Onto-tools Suite.
SYTO RNA select staining of exosomes
Purified exosomes were labeled with SYTO RNA Select green fluorescent stain (Molecular Probes) following the manufacturer’s instructions and excessive stain was washed from exosomes by centrifugation in 1X PBS. The cell monolayers (1×105/well) were treated with labeled exosomes (25μg/well) for 2 hours. The monolayers were washed with I×PBS and the cells were stained with Alexa Flour-594-labeled Wheat Germ Agglutinin stain per manufacturer’s instructions. Images were acquired on a BioRad MRC 1024 Scanning Confocal coupled to a Nikon Diaphot 200 microscope using LaserSharp 2000 acquisition software. The settings included laser power (30%), Iris diameter in Airy units (2.5), gain (1386) and an offset (24) with PMT off. The images were acquired at 40× with 2× digital zoom with x,y dimensions of 0.24 μm per pixel and the images were processed using Image J software.
In vivo translation
Human THP-1 cells (1×106) were differentiated with 20ng/ml PMA for 48 hours. The monolayers were washed with PBS and incubated in complete RPMI media for an additional 24 hours. Exosomes (125μg) derived from M.tb infected or uninfected mouse RAW264.7 macrophages were added to the cells or the cells were left untreated for 4 hours. The cells were subsequently washed to remove remaining exosomes and fresh culture media was added. 24 hours later cells were lysed in RIPA buffer and probed for mouse lipocalin-2 expression using monoclonal Mouse Lipocalin-2/NGAL antibody (R&D Systems) which does not cross react with human Lipocalin 2. Samples were also probed with Rabbit monoclonal antibody for beta actin (Cell Signaling) as a loading control.
Transfections
RAW264.7 macrophages (2×105 cells per well) were seeded in a 24 well plate in DMEM complete growth medium. 250 ng of exosomal RNA was mixed with 3μl of Hiperfect transfection reagent (Qiagen) and added drop wise to the cells following manufacturer’s instructions. Resting macrophages or macrophages treated with the transfection reagent Hiperfect alone were used as controls. Following incubation in normal growth conditions for 24 hours, the cell culture supernatants were harvested and tested for presence of cytokines and chemokines using Mouse Cytokine Array Panel A (R&D Systems) following manufacturer’s instructions. In separate experiments, RAW264.7 macrophages were seeded on coverslips and transfected with exosomal RNA as described above. The cells were stained with FITC conjugated Annexin V (BioLegend) following manufacturer’s instructions and fixed with 2% paraformaldehyde. The cells were visualized on a confocal microscope and the percentage of apoptotic cells was determined by counting cells in 10 independent fields for each treatment.
Statistical analyses
Data was analyzed by a one-tailed or paired Student's t test. Statistical significance was assumed at p ≤0.05. Each experiment was conducted 2 or 3 times and error bars represent standard deviation values.
Supplementary Material
Figure 1. Exosomes purified on a sucrose gradient were positive for mycobacterial transcripts. Exosomes were isolated from cell culture supernatants from uninfected or M.tb-infected RAW264.7 cells and purified on a linear sucrose gradient. Exosomes were tested for exosomal markers by western blot using anti-mouse LAMP-1 and anti-Tsg101 (A). Exosomes were treated with RNAseA and RNA was extracted from exosomes and treated with DNAse1. RNA was converted to single stranded cDNA and PCR amplified for selected mycobacterial transcripts using GAPDH as a PCR control (B).
Figure 2. Ultracentrifugation of SYTO RNA-Select Dye in buffer alone does not result in RNA fluorescence. SYTO RNA Select dye was resuspended in 1× PBS and centrifuged at 100,000g for 1 hour. Pelleted aggregates were resuspended in buffer and added to cell monolayers. Cells were washed with 1X PBS and the cells were stained with Alexa Flour-594-labeled Wheat Germ Agglutinin stain per manufacturer’s instructions. Images were acquired on a BioRad MRC 1024 Scanning Confocal coupled to a Nikon Diaphot 200 microscope using LaserSharp 2000 acquisition software.
Figure 3. The mouse lipocalin-2/NGAL antibody does not recognize human lipocalin-2. Recombinant human lipocalin-2 peptide (5ug) was used to test the specificity of anti mouse lipocalin-2 antibody used in the transfer experiments. The antibody did not react with recombinant human lipocalin-2, although it recognized murine lipocalin-2 in cell lysate derived from RAW264.7 macrophages infected with M. tuberculosis.
Synopsis.
Our study shows a signature of host derived miRNAs and mRNA transcripts as well as mycobacterial RNA in exosomes derived from M. tuberculosis infected macrophages. The exosomal mRNA was translated upon delivery to recipient cells and was biological active suggesting a role in modulation of host response to M. tuberculosis. Mycobacterial transcripts were also detected in extracellular vesicles (EVs) derived from the serum of TB infected individuals pointing to the significance of exosomal RNA in TB diagnostics.
Acknowledgments
The studies were funded, in part, through a grant to J.S. from the National Institute of Health (RO1AI052439). We kindly thank the Notre Dame Genomics Facility especially John Tan and Melissa Pullins for their technical expertise.
Footnotes
There was no conflict of interest for any of the authors on this manuscript.
References
- 1.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. Journal of Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buschow SI, Nolte-’t Hoen EN, van Niel G, Pols MS, ten Broeke T, Lauwen M, Ossendorp F, Melief CJ, Raposo G, Wubbolts R, et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10:1528–1542. doi: 10.1111/j.1600-0854.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- 5.Al-Nedawi K, Szemraj J, Cierniewski CS. Mast cell-derived exosomes activate endothelial cells to secrete plasminogen activator inhibitor type 1. Arterioscler Thromb Vasc Biol. 2005;25:1744–1749. doi: 10.1161/01.ATV.0000172007.86541.76. [DOI] [PubMed] [Google Scholar]
- 6.Azevedo LC, Janiszewski M, Pontieri V, Pedro Mde A, Bassi E, Tucci PJ, Laurindo FR. Platelet-derived exosomes from septic shock patients induce myocardial dysfunction. Crit care. 2007;11:R120. doi: 10.1186/cc6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. The Journal of Biol Chem. 2007;282:25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and Glioblastoma cells release exosomes carrying mt DNA. J Neur Trans. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 9.Faure J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V, et al. Exosomes are released by cultured cortical neurones. Mol and Cell Neurosciences. 2006;31:642–648. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Buning J, von Smolinski D, Tafazzoli K, Zimmer KP, Strobel S, Apostolaki M, Kollias G, Heath JK, Ludwig D, Gebert A. Multivesicular bodies in intestinal epithelial cells: responsible for MHC class II-restricted antigen processing and origin of exosomes. Immunology. 2008;125:510–521. doi: 10.1111/j.1365-2567.2008.02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 12.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc of the Natl Acad of Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. Journal of Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 14.Admyre C, Grunewald J, Thyberg J, Gripenback S, Tornling G, Eklund A, Scheynius A, Gabrielsson S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. The Eur Res J. 2003;22:578–583. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 15.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasser C, O’Neil SE, Ekerljung L, Ekstrom K, Sjostrand M, Lotvall J. RNA-containing exosomes in human nasal secretions. Am J of Rhinology & Allergy. 2011;25:89–93. doi: 10.2500/ajra.2011.25.3573. [DOI] [PubMed] [Google Scholar]
- 17.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) The Journal of Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 18.Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur J of Cell Biol. 1984;35:256–263. [PubMed] [Google Scholar]
- 19.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. The Journal of Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaput N, Thery C. Exosomes: immune properties and potential clinical implementations. Sem in immunopathol. 2011;33:419–440. doi: 10.1007/s00281-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 21.Qazi KR, Gehrmann U, Domange Jordo E, Karlsson MC, Gabrielsson S. Antigen-loaded exosomes alone induce Th1-type memory through a B-cell-dependent mechanism. Blood. 2009;113:2673–2683. doi: 10.1182/blood-2008-04-153536. [DOI] [PubMed] [Google Scholar]
- 22.Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, Paris L, Abalsamo L, Colone M, Molinari A, et al. Immune surveillance properties of human NK cell-derived exosomes. Journal of Immunol. 2012;189:2833–2842. doi: 10.4049/jimmunol.1101988. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson M, Lundin S, Dahlgren U, Kahu H, Pettersson I, Telemo E. “Tolerosomes” are produced by intestinal epithelial cells. Eur J of Immunol. 2001;31:2892–2900. doi: 10.1002/1521-4141(2001010)31:10<2892::aid-immu2892>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 24.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 25.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 26.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Comm. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, et al. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giri PK, Kruh NA, Dobos KM, Schorey JS. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics. 2010;10:3190–3202. doi: 10.1002/pmic.200900840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giri PK, Schorey JS. Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PloS ONE. 2008;3:e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Schorey JS. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur J of Immunol. 2013;43:3279–3290. doi: 10.1002/eji.201343727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh PP, LeMaire C, Tan JC, Zeng E, Schorey JS. Exosomes released from M. tuberculosis infected cells can suppress IFN-gamma mediated activation of naive macrophages. PloS ONE. 2011;6:e18564. doi: 10.1371/journal.pone.0018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh PP, Smith VL, Karakousis PC, Schorey JS. Exosomes isolated from mycobacteria-infected mice or cultured macrophages can recruit and activate immune cells in vitro and in vivo. Journal of Immunol. 2012;189:777–785. doi: 10.4049/jimmunol.1103638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin RD, O’Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh PK, Singh AV, Chauhan DS. Current understanding on micro RNAs and its regulation in response to Mycobacterial infections. Journal of Biomed Sci. 2013a;20:14. doi: 10.1186/1423-0127-20-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, Suar M, Van Kaer L, Bishai WR, Das G. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. The Journal of Biol Chem. 2013b;288:5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J of Clin Microbiol. 2011;49:4246–4251. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miotto P, Mwangoka G, Valente IC, Norbis L, Sotgiu G, Bosu R, Ambrosi A, Codecasa LR, Goletti D, Matteelli A, et al. miRNA Signatures in Sera of Patients with Active Pulmonary Tuberculosis. PloS ONE. 2013;8:e80149. doi: 10.1371/journal.pone.0080149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X, Yang D, Yu H, Cui L, Shan Y, et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis. 2012;12:384–392. doi: 10.1186/1471-2334-12-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Yang S, Sun G, Tang X, Lu S, Neyrolles O, Gao Q. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PloS ONE. 2011;6:e25832. doi: 10.1371/journal.pone.0025832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer SU, Pfaffl MW, Ulbrich SE. Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotech Letts. 2010;32:1777–1788. doi: 10.1007/s10529-010-0380-z. [DOI] [PubMed] [Google Scholar]
- 44.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucl Acids Res. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saiga H, Nishimura J, Kuwata H, Okuyama M, Matsumoto S, Sato S, Matsumoto M, Akira S, Yoshikai Y, Honda K, Yamamuto M, Takeda K. Lipocalin2 dependent inhibition of mycobacterial growth in alveolar epithelium. J Immunol. 2008;181:8251–8257. doi: 10.4049/jimmunol.181.12.8521. [DOI] [PubMed] [Google Scholar]
- 47.Obrego´ n-Henao A, Duque-Correa MA, Rojas M, Garcı´a LF, Brennan PJ, et al. Stable Extracellular RNA Fragments of Mycobacterium tuberculosis Induce Early Apoptosis in Human Monocytes via a Caspase-8 Dependent Mechanism. PLoS ONE. 2012;7:e29970. doi: 10.1371/journal.pone.0029970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald MK, Tian Y, Qureshi RA, Gormley M, Ertel A, Gao R, Lopez EA, Alexander GM, Sacan A, Fortina P, Ajit SK. Functional significance of macrophage-derived exosomes in inflammation and pain. PAIN. 2014:1–13. doi: 10.1016/j.pain.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS ONE. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha A, Yadav AK, Chakraborty S, Kabra SK, Lodha R, Kumar M, Kulshreshtha A, Sethi T, Pandey R, Malik G, et al. Exosome-enclosed microRNAs in exhaled breath hold potential for biomarker discovery in patients with pulmonary diseases. The Journal of Allergy and Clin Immunol. 2013;132:219–222. doi: 10.1016/j.jaci.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 51.Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, Ghosh Z, Sharma P, Kundu M, Basu J. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol. 2012;14:1620–1631. doi: 10.1111/j.1462-5822.2012.01827.x. [DOI] [PubMed] [Google Scholar]
- 52.Villaroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrum M, Sanchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitler-Bauer C, Wieland S, Chisari FV. Short range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host & Microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narayanan A, Iordanskiy S, Das R, Van Duyne R, Santos S, Jaworski E, Guendel I, Sampey G, Dalby E, Iglesias-Ussel M, et al. Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. The Journal of Biol Chem. 2013;288:20014–20033. doi: 10.1074/jbc.M112.438895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhovena MAJ, Hopmansa ES, et al. Functional delivery of viral miRNAs via exosomes. Proc of the Natl Acad of Sci USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host & Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. The EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host & Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1. Exosomes purified on a sucrose gradient were positive for mycobacterial transcripts. Exosomes were isolated from cell culture supernatants from uninfected or M.tb-infected RAW264.7 cells and purified on a linear sucrose gradient. Exosomes were tested for exosomal markers by western blot using anti-mouse LAMP-1 and anti-Tsg101 (A). Exosomes were treated with RNAseA and RNA was extracted from exosomes and treated with DNAse1. RNA was converted to single stranded cDNA and PCR amplified for selected mycobacterial transcripts using GAPDH as a PCR control (B).
Figure 2. Ultracentrifugation of SYTO RNA-Select Dye in buffer alone does not result in RNA fluorescence. SYTO RNA Select dye was resuspended in 1× PBS and centrifuged at 100,000g for 1 hour. Pelleted aggregates were resuspended in buffer and added to cell monolayers. Cells were washed with 1X PBS and the cells were stained with Alexa Flour-594-labeled Wheat Germ Agglutinin stain per manufacturer’s instructions. Images were acquired on a BioRad MRC 1024 Scanning Confocal coupled to a Nikon Diaphot 200 microscope using LaserSharp 2000 acquisition software.
Figure 3. The mouse lipocalin-2/NGAL antibody does not recognize human lipocalin-2. Recombinant human lipocalin-2 peptide (5ug) was used to test the specificity of anti mouse lipocalin-2 antibody used in the transfer experiments. The antibody did not react with recombinant human lipocalin-2, although it recognized murine lipocalin-2 in cell lysate derived from RAW264.7 macrophages infected with M. tuberculosis.


