Abstract
Background
Mathematical models offer the possibility to investigate the infectious disease dynamics over time and may help in informing design of studies. A systematic review was performed in order to determine to what extent mathematical models have been incorporated into the process of planning studies and hence inform study design for infectious diseases transmitted between humans and/or animals.
Methods
We searched Ovid Medline and two trial registry platforms (Cochrane, WHO) using search terms related to infection, mathematical model, and study design from the earliest dates to October 2016. Eligible publications and registered trials included mathematical models (compartmental, individual-based, or Markov) which were described and used to inform the design of infectious disease studies. We extracted information about the investigated infection, population, model characteristics, and study design.
Results
We identified 28 unique publications but no registered trials. Focusing on compartmental and individual-based models we found 12 observational/surveillance studies and 11 clinical trials. Infections studied were equally animal and human infectious diseases for the observational/surveillance studies, while all but one between humans for clinical trials. The mathematical models were used to inform, amongst other things, the required sample size (n = 16), the statistical power (n = 9), the frequency at which samples should be taken (n = 6), and from whom (n = 6).
Conclusions
Despite the fact that mathematical models have been advocated to be used at the planning stage of studies or surveillance systems, they are used scarcely. With only one exception, the publications described theoretical studies, hence, not being utilised in real studies.
Electronic supplementary material
The online version of this article (10.1186/s12879-017-2874-y) contains supplementary material, which is available to authorized users.
Keywords: Study design, Research design, Mathematical models, Infectious diseases, Systematic review
Background
Infectious diseases contribute substantially to the global burden of disease and are major public health issues worldwide [1]. The value of mathematical models for infectious diseases is widely recognised in various fields such as ecology or epidemiology [2–4]. These models have been used with different objectives such as understanding infectious disease dynamics, informing public health policies or guidelines through, for example, the modelling of potential additional interventions [5], or computing key indicators [6]. In the context of public health surveillance in particular, mathematical models have been also used to detect potential epidemics making use of (temporal) surveillance system data [7], evaluate the performances of a surveillance system [8], or monitor programmes [9].
Mathematical models may also help in informing design of studies, including cross-sectional studies, clinical trials, or surveillance systems, and have been advocated to be used at the planning stage of studies to inform their design [5, 10–13]. Well-designed clinical trials and observational studies are needed to investigate the impact of interventions before they can be used on large scale. Furthermore, public health authorities also need efficient tools for monitoring infectious diseases. Each improvement in design and monitoring of clinical trials and observational studies will allow a more efficient usage of resources crucial to current and later implementation, monitoring, and evaluation of promising interventions.
The current use of mathematical models in planning studies has to our knowledge never been systematically summarised. The objective of our study was to systematically review mathematical models used to inform the design of a study related to an infectious disease transmitted between humans, between animals, or between animals and humans (zoonosis). We documented to which extent and how mathematical models have been incorporated into the process of planning studies or surveillance systems. Finally, by performing this review, we hope to trigger more attention to the use of mathematical models in planning studies and to more explicitly document design considerations in mathematical modelling studies in abstract and keywords.
Methods
We used a protocol to describe the methods in detail for our systematic review (Additional file 1).
Eligibility criteria
We searched for publications and registered trials in which mathematical models were described and used to inform the design of infectious disease studies, i.e. inform study design in the context of sample sizes and/or selecting the (number of) sampling times and/or power calculation and/or from whom to collect samples and/or what should be monitored. We were interested in implemented studies as well as in methodological papers.
We included studies that used individual-based models (IBMs, including agent-based models, microsimulation, etc.), compartmental models, or Markov models [14, 15]. IBMs are models in which the infection process for every individual in the population is tracked; compartmental models are models in which individuals in the population are subdivided into ‘compartments’ and the models track the infection process for these individuals collectively; and Markov models predict how an individual moves from one health state to another over time, assuming that the individual is always in one of a finite number of states and that the transition to the next state depends only on the values of the current state.
We excluded publications and study protocols of registered trials if the mathematical model was not used to design a study, for example, the model was only used for data analysis or only used for investigating the potential impact of new interventions on infectious disease spread.
Information source
We searched Ovid Medline, Cochrane Central Register of Controlled Trials, and WHO International Clinical Trials Registry Platform from the earliest date of the database to October 2016 without language restrictions. Search strategies used subject headings specific to each database and free text search that combined terms for: infection, mathematical models, and study designs (see Appendix 1 in Additional file 1 for the detailed search strategies). Reference lists of included publications and study protocols of registered trials were screened to identify additional relevant publications.
Selection
Two reviewers (SH, SB) screened titles and abstracts of retrieved publications and description of registered trials in the database. Discrepancies were solved by discussion or by consulting a third reviewer (NH). Any publication or registered trial selected as being potentially eligible was retained for review of the full text; for registered trials we made three attempts to retrieve the study protocol by contacting electronically the principal investigator (listed in the trial registry).
Outcomes
The primary outcomes were the description of the characteristics of the mathematical models incorporated, and the description of the design part considered in the process of planning studies or surveillance systems.
Data collection and analysis
The two reviewers (SH, SB) independently extracted data and discrepancies were solved by discussion or by consulting the third reviewer (NH). An extraction sheet was developed using Microsoft Excel and piloted to extract information about the investigated infection, population, model characteristics, and study design.
The characteristics of mathematical models used in planning studies were summarised stratified by study type, i.e. observational and surveillance studies, and clinical trials. We described the infections and populations studied, the main characteristics of the mathematical model used, the main outcome of the study, and the design outcomes (see Table 1) investigated by the authors. If there were multiple publications using the same model for the same setting to investigate the same study design, the earliest publication was considered to be the original (see further). We also documented model reporting items (e.g. diagram of the model or parameters values) and how the infection transmission was modelled.
Table 1.
Description of design outcomes
| Design outcome | Description |
|---|---|
| Follow-up | The model was used to determine/inform the follow-up time of the study. |
| Timing of sampling | The model was used to determine/inform at which time point(s) sampling should be performed. |
| Frequency | The model was used to determine/inform the frequency at which sampling has to be collected (over time) during the study. |
| Number | The model was used to determine/inform the number of sampling to collect over time during the study. |
| Monitoring | The model was used to identify parameters or indicators that should be monitored during the study. |
| Sample size | The model was used to determine/inform the sample size. |
| Whom | The model was used to determine/inform which subgroups of the population studied should be sampled. |
| Power | The model was used to perform statistical power calculations. |
Results
Our literature searches identified 571 unique publications/registered trials; 68 full-text publications were screened and 6 principal investigators of registered trials were contacted; 30 eligible publications were included [16–45]. Reasons for exclusion are summarised in Fig. 1. Of these 30 publications, 28 were considered to be unique, i.e. different models were used for different settings and study designs. There were two publications referring to the same randomised controlled trial (RCT) and reporting the same model (Cori 2014 [35] and Hayes 2014 [40]), and one publication re-stated the results from a previous publication (Wu 2005 [39] and Wu 2002 [30]).With only one exception (Cori 2014 [35]), the publications described theoretical studies, hence, not being utilised in real trials.
Fig. 1.
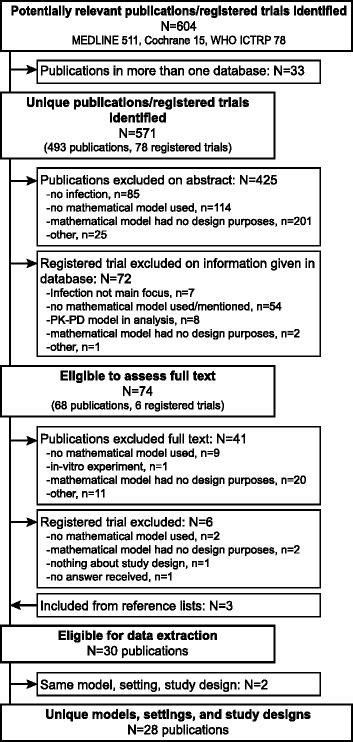
Flow diagram of included and excluded publications and registered trials
We focused on the characteristics of the 23 studies which used IBMs or compartmental models to inform the study design because, in the context of infectious diseases, these types of models are most used and allow infectious disease dynamics (e.g. herd immunity) to be directly incorporated. There were 12 observational or surveillance studies [16–27] (Table 2, Additional file 2: Table S1) and 11 clinical trials [28–38] (Table 3, Additional file 2: Table S2). The characteristics of the 5 studies which used Markov models [41–45] (all clinical trials for humans) are given in Additional file 2 (Table S3 and Table S4).
Table 2.
Characteristics of publications – Observational and surveillance studies (N = 12)
| First author, year | Infection | Population | Model | Main outcome | Design outcome(s) | Remarks | ||
|---|---|---|---|---|---|---|---|---|
| Epidemiological category | Name | Typea | Structured/Networkb | |||||
| Graat, 2001 [16] | Animal | Bovine herpesvirus 1 | Cattle farming | Compartmental - deterministic | Yes/No | Reproduction ratio between herds | - Frequency - Sample size |
− |
| Michael, 2006 [17] | Human, vector-borne | Lymphatic filariasis | Not described | Compartmental – deterministicc | No/No | Prevalence of microfilaraemia | - Frequency - Sample size - Monitoring - Power |
− |
| Savill, 2008 [18] | Animal | Avian influenza | Commercial poultry flocks (The Netherlands) | IBM | Yes/No | False alarm rate | - Monitoring | − |
| Arnold, 2013 [19] | Animal | Avian influenza | Poultry farming | IBM | Yes/Yes | Size and duration of an outbreak | - Sample size - Whom |
Spatial model |
| Smieszek, 2013 [20] | Human, respiratory |
Influenza | An US high school (teachers, students, staff) | IBM | Yes/Yes | Performance of collocation ranking | - Sample size - Whom |
− |
| Ciccolini, 2014 [21] | Human, nosocomial | Nosocomial pathogens | Acute hospitals (England, The Netherlands) | Compartmental - stochastic | Yes/Yes | Time to detection and number of infected hospitals | - Sample size - Whom |
− |
| Gonzales, 2014 [22] | Animal | Avian influenza | Layer chickens (The Netherlands) | Compartmental - deterministic | Yes/No | Required sample size and frequency for early detection | - Frequency - Number - Sample size |
− |
| Leslie, 2014 [23] | Animal | Classical swine fever | Wild pig, Kimberley region (Australia) | IBMc | Yes/Yes | Epidemic length, number of days to complete the surveillance, number of cells sampled, number of groups to be sampled | - Sample size - Whom |
A within-herd model combined with a spatial between-herd model |
| Mizumoto, 2014 [24] | Human, vector borne | Dengue virus | Not described | Compartmental - deterministic | Yes/No | Relative risk of severe dengue and ‘dengue hemorrhagic fever’/ ‘dengue shock syndrome’ during secondary infection | - Timing of sampling | − |
| Pinsent, 2014 [25] | Animal | Avian influenza | Commercial poultry barns | Compartmental - deterministic | No/No | Estimates of basic reproduction number and time of virus introduction | - Frequency - Sample size |
− |
| van Bunnik, 2015 [26] | Human, nosocomial | Meticillin-resistant Staphylococcus aureus | Hospitals (Scotland) | Compartmental - stochastic | Yes/Yes | Time until first detection of new health-care associated infection | - Sample size - Whom |
Similar model as Ciccolini, 2012 |
| Vinh, 2015 [27] | Human, respiratory | Influenza | General population | Compartmental - deterministic | No/No | Statistical identifiability of antibody generation, antibody waning, and reinfection | - Frequency - Sample size - Power |
− |
amodel type: IBM – individual based model; b structured: population structure is reflected in model, network: network of contacts between individuals is explicitly modelled; c model type obtained from the original article
Table 3.
Characteristics of publications – Clinical trials (N = 11)
| First author, year | Infection | Population | Model | Main outcome | Design outcome(s) | Remarks | ||
|---|---|---|---|---|---|---|---|---|
| Epidemiological category | Name | Type a | Structured/Network b | |||||
| Atlas, 1993 [28] | Human, water-borne | Cryptosporidium | Volunteer subjects | Compartmental - deterministicc | No/No | Probability of infection | - Sample size | Exprimental study |
| Lipsitch, 2001 [29] | Human, respiratory | Streptococcus pneumoniae | Not described | Compartmental - deterministic | No/No | Simple and conditional odds-ratios | - Timing of sampling | − |
| Wu, 2002 [30]; Wu, 2005 [39] | Human, STI | HIV | Within-host (cells) | Compartmental - deterministic | No/No | HIV viral load change | - Timing of sampling - Frequency - Number - Sample size - Power |
Statistical model used for fitting data |
| Clermont, 2004 [31] | Human, bacterial | Generic Gram-negative pathogen | Within-host (virtual infected patients) | Compartmental - deterministic | Yes/No | Identify people who will well respond to the anti-tumor necrosis factor | - Whom | − |
| Hallett, 2008 [32] | Human, STI | HIV | Heterosexual population | Compartmental - deterministic | Yes/Yes | HIV incidence rate ratio | - Follow-up - Sample size - Power |
− |
| Dimitrov, 2013 [33] | Human, STI | HIV | Heterosexual population representative of sub-saharan Africa | Compartmental - deterministic | Yes/No | HIV incidence | - Sample size - Power |
− |
| Nishiura, 2013 [34] | Animal | influenza A viruses | Ferret in cages | Compartmental - stochastic | No/No | Number of pairs to include in 1-to-1 transmission studies | - Sample size - Power |
− |
| Cori, 2014 [35]; Hayes, 2014 [40] | Human, STI | HIV | Adults, 18-44y, South Africa and Zambia | Compartmental - deterministic | Yes/Yes | HIV incidence | - Monitoring - Power |
Effectively used to plan a three-arm cluster RCT |
| Cuadros, 2014 [36] | Human, STI | HIV | Serodiscordant couples; male population | IBM | No/Yes | HIV incidence | - Power | − |
| Scott, 2014 [37] | Human, respiratory | Streptococcus pneumoniae | Infants | Compartmental - deterministic | Yes/No | Vaccine efficacy against acquisition and/or duration | - Follow-up - Timing of sampling - Monitoring |
− |
| Herzog, 2015 [38] | Human, STI | Chlamydia trachomatis | Women | Compartmental - deterministic | No/No | Pelvic inflammatory disease incidence | - Follow-up - Sample size - Power |
− |
amodel type: IBM – individual based model; b structured: population structure is reflected in model, network: network of contacts between individuals is explicitly modelled; c model seen as compartmental model
Results of the considered 23 studies show that compartmental models were the most commonly used models (8 observational/surveillance studies, 10 clinical trials). Infections studied were equally animal and human infectious diseases for the observational or surveillance studies, while all but one between humans for clinical trials. Epidemiological categories of infections transmitted between humans were sexually transmitted infections (STIs, mainly HIV; 6 clinical trials), respiratory infections (2 observational/surveillance studies, 2 clinical trials), nosocomial infections (2 observational/surveillance studies), vector-borne infections (2 observational/surveillance studies), water-borne infections (1 observational/surveillance study), hypothetical bacterial infection (1 observational/surveillance study). Influenza (avian, n = 4, or in ferrets, n = 1) was the infection the most studied among infections transmitted between animals. A population structure was reflected in 14 models (9 observational/surveillance studies, 5 clinical trials) and a network of contacts between individuals was explicitly modelled in 8 models (5 observational/surveillance studies, 3 clinical trials).
We observed diverse patterns of model reporting across the publications (Additional file 2: Table S1 and Table S2). Almost all publications described - to a certain extent - the model structure, some reported equations, figures, and how the course of infection was implemented, while others reported only one or even none of those. For three publications [17, 23, 28] we had to obtain the model type from the referenced original publication [46–48]. About a third of the publications reported the software in which the mathematical model was implemented but code is available, either as a supplementary material or through request to the authors, in only two publications [36, 38]. The sources for the model parameters used were mostly a mixture of calibration, assumptions by authors, and estimations from other data.
In observational or surveillance studies, the design outcome most studied was sample size (n = 10), followed by frequency of sampling and population from whom to sample (n = 5), monitoring and power (n = 2), and number of samples and timing of sampling (n = 2). In clinical trials, the most studied design outcome was power (n = 7), followed by sample size (n = 6), timing of sampling and follow-up (n = 3), monitoring (n = 2), frequency of sampling, number of samples and population from whom to sample (n = 1). Seven research question categories were identified among the studies included: detect infection early, estimate epidemiological parameters, compare different trial arms, include potentially good responders in an RCT, follow trial progression, detect changes in infection values over time, and determine appropriate time point(s) to estimate a parameter. Table 4 shows for each design outcome which research questions were investigated. More details about the study designs can be found in Additional file 2: Table S1 and Table S2.
Table 4.
Design outcomes and corresponding research question
| Design outcomes | References by main research questions |
|---|---|
| Follow-up | Determine appropriate time point to estimate a parameter: Mizumoto [24]; Lipsitch [29]; Wu [30]; Scott [37]; Herzog [38]; Hallett [32] |
| Timing of sampling | Determine appropriate time point to estimate a parameter: Mizumoto [24]; Lipsitch [29]; Wu [30]; Scott [37]; Herzog [38]; Hallett [32] |
| Frequency | Detect infection early: Graat [16]; Michael [17]; Gonzales [22] Estimate epidemiological parameters: Pinsent [25]; Vinh [27] Compare different trial arms: Wu [30] |
| Number | Detect infection early: Gonzales [22] Compare different trial arms: Wu [30] |
| Monitoring | Detect infection early: Michael [17]; Savill [18] Follow trial progression: Cori [35]; Scott [37] |
| Sample size | Detect infection early: Graat [16]; Michael [17]; Arnold [19]; Smieszek [20]; Ciccolini [21]; Gonzales [22]; Leslie [23]; van Bunnik [26] Estimate epidemiological parameters: Atlas [28]; Pinsent [25]; Vinh [27] Compare different trial arms: Wu [30]; Hallett [32]; Dimitrov [33], Nishiura [34]; Herzog [38] |
| Whom | Detect infection early: Arnold [19]; Smieszek [20]; Ciccolini [21]; Leslie [23]; van Bunnik [26] Include potentially good responders in a RCT: Clermont [31] |
| Power | Estimate epidemiological parameters: Vinh [27] Compare different trial arms: Wu [30]; Hallett [32]; Dimitrov [33]; Nishiura [34]; Cori [35]; Cuadros [36]; Herzog [38] Detect changes in infection values over time: Michael [17] |
Discussion
We found in this systematic review 28 unique publications but no registered trials in which mathematical models were described and used to inform the design of infectious disease studies. Only one mathematical model was effectively used to plan a study (a three-arm cluster RCT) whereas all others described theoretical studies. Focusing on the 23 compartmental and individual-based models, we found almost equal amount of observational or surveillance studies and clinical trials whereby compartmental models were most commonly used. Various infection categories have been investigated with equal numbers of animal and human infections studied among the observational or surveillance studies and mainly human infections among the clinical trials. Enough details are provided for the compartmental models, except for two [17, 22], to replicate the model. For IBMs more details are needed in order to replicate the model if the source code is not available. For example, the ‘ODD’ (Overview, Design concepts, and Details) protocol has been proposed to standardize reporting of individual-based and agent-based models [49, 50]. None of the five IBM publications followed or mentioned this protocol, however, one provides the model source code [36]. The mathematical models were utilised to inform, amongst other things, the following design outcomes: required sample size, statistical power, frequency at which samples should be taken, and from whom.
One explanation of the scarcity of mathematical modelling to design real studies, despite the anticipated gain in study efficiency, is arguably the lack of fundamental research in the sense that there are no existing databases or software to access mathematical models which are already implemented in a study design framework in contrast to classical sample size calculation with freely-available or chargeable software. In our systematic review, only a third of the publications reported the software used for the mathematical model and only two mentioned availability of code. Additionally, it is even difficult to ascertain which mathematical models already exist for a specific infection in order to extend them to the study design framework. Account should be taken of the fact that building a (well-validated) model is time-consuming and modelling expertise specific to the infectious disease of interest is needed. On the other hand, finding the optimal sample size, frequency of sampling, etc. using a mathematical model can lead to computer intense processing steps.
Sample size is not the only design outcome which can be investigated by the mathematical models as shown in our results. Interestingly, mathematical models have been used to determine the appropriate time point(s) at which a parameter such as the effect size of the intervention should be estimated. For example, a publication in the field of vaccination investigated two different estimates of vaccine efficacy (prevalence odds ratio and prevalence ratios) and their change over time. The authors found that the timing of sample collection can affect the interpretation of results about vaccine efficacy against bacterial carriage in an RCT [37].
We observed that the same research question was investigated looking at different design outcomes. For example, the question about early detection of an infection was studied by exploring the frequency at which samples should be taken, the number of samples to be collected, and the sample size but also from whom samples should be taken, and what parameters or indicators should be monitored during a study [16–23, 26].
There is no guarantee that a study would not fail to show the expected effect size in case the study team used a mathematical model at the planning stage of a study. A mathematical model always implies underlying assumptions by its structure and parameter values. However, those assumptions for mathematical models can be taken into account in uncertainty and sensitivity analyses for the effect size of interest. The estimated effect size and its uncertainty, in turn, can be considered in, for example, sample size calculations. Most importantly, mathematical models can be used to deal with the complexity of infectious diseases like the dependency between individuals and give insights how this can influence the study design.
The strength of our study is that the search of publications and registered trials had no restriction about the type of infection or the transmission route. Additionally, this is, to our knowledge, the first systematic review of the current use of mathematical models in planning infectious disease studies. A weakness of this review is that the literature search might have missed some relevant publications and registered trials because there is neither a single MeSH term for mathematical models nor a general usage of keywords to describe mathematical models, a problem observed also in other systematic reviews [51, 52]. In addition, for some infections, like HIV, only the abbreviation is commonly stated. Hence, no variation of the word ‘infection’ is used in the title, abstract, or especially in the description of the registered trial on the trial registry platform. We tested the inclusion of specific infections (HIV, malaria, and tuberculosis) which resulted in a limited number of additional hits, of which only two would meet our inclusion criteria [53, 54]. We tried to overcome the limitations of keywords and MeSH terms used by building a search strategy which uses different terms to identify models and infections and by searching for additional publications in the reference lists of included publications. However, we do not present mathematical models that are only described in grey literature like reports or that are not publicly available. Similarly, publications that only used indirect modelling results without describing the mathematical model were not included.
Conclusion
Despite the fact that mathematical models have been advocated to be used at the planning stage of studies or surveillance systems [5, 10–13], they are used scarcely as shown by this systematic review. With only one exception, the publications described theoretical studies, hence, not being utilised in real studies. Generic statistical approaches for sample size calculations, most often assuming independence between individuals, do not capture the complex nature of infectious disease epidemiology. This is an oversimplification as e.g. treating infected persons may affect the people around them by reducing the spreading of the infection; mathematical modelling could thus be used to account for such characteristics. The results of this systematic review offer an overview of the current use of mathematical models in the context of study design and indicate that future research is needed.
Additional files
Study protocol and search strategy. (PDF 74 kb)
Additional results tables. (PDF 54 kb)
Acknowledgements
Authors SB and NH acknowledge support of the Antwerp Study Centre for Infectious Diseases (ASCID) at the University of Antwerp.
Funding
Funding SH: Part of this work has been funded by the Special Research Fund of Hasselt University and by the Mobility Grant of Medical University of Graz. Funding SB, NH: This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement 682,540 — TransMID). NH further acknowledges support of the Chair in Evidence-based Vaccinology at the University of Antwerp financed by a gift from Pfizer (2009–2017) and GSK (2016).
Availability of data and materials
Not applicable.
Abbreviations
- IBM
Individual-based model
- RCT
Randomised controlled trial
- STI
Sexually transmitted infection
Authors’ contributions
SAH and NH conceived the systematic review. SAH and SB selected studies, extracted and analysed data. NH was the third reviewer to solve discrepancies. SAH and SB wrote the first draft of the manuscript. All authors contributed to the interpretation of the results, commented on the manuscript and approved the final version.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12879-017-2874-y) contains supplementary material, which is available to authorized users.
Contributor Information
Sereina A. Herzog, Email: herzog.sereina@gmail.com
Stéphanie Blaizot, Email: Stephanie.Blaizot@uantwerpen.be.
Niel Hens, Email: niel.hens@uhasselt.be.
References
- 1.World Health Organization . Research for universal health coverage. Geneva, Switzerland: World Health Organization; 2013. [Google Scholar]
- 2.Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford, New York: Oxford University Press; 1991. [Google Scholar]
- 3.Keeling MJ, Rohani P. Modeling infectious diseases in humans and animals. Princeton, NJ: Princeton University Press; 2008. [Google Scholar]
- 4.Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347:aaa4339. doi: 10.1126/science.aaa4339. [DOI] [PMC free article] [PubMed]
- 5.Garnett GP, Cousens S, Hallett TB, Steketee R, Walker N. Mathematical models in the evaluation of health programmes. Lancet. 2011;378:515–525. doi: 10.1016/S0140-6736(10)61505-X. [DOI] [PubMed] [Google Scholar]
- 6.Hens N, Shkedy Z, Aerts M, Faes C, Van Damme P, Beutels P. Modeling infectious disease parameters based on serological and social contact data: a modern statistical perspective. New York: Springer; 2012. [Google Scholar]
- 7.Mohtashemi M, Szolovits P, Dunyak J, Mandl KD. A susceptible-infected model of early detection of respiratory infection outbreaks on a background of influenza. J Theor Biol. 2006;241:954–963. doi: 10.1016/j.jtbi.2006.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckeridge DL, Burkom H, Moore A, Pavlin J, Cutchis P, Hogan W. Evaluation of syndromic surveillance systems--design of an epidemic simulation model. MMWR supplements. 2004;53:137–143. [PubMed] [Google Scholar]
- 9.Cauchemez S, Boelle P, Thomas G, Valleron A. Estimating in real time the efficacy of measures to control emerging communicable diseases. Am J Epidemiol. 2006;164:591–597. doi: 10.1093/aje/kwj274. [DOI] [PubMed] [Google Scholar]
- 10.Boily M, Masse B, Alsallaq R, Padian NS, Eaton JW, Vesga JF, Hallett TB. HIV treatment as prevention: considerations in the design, conduct, and analysis of cluster randomized controlled trials of combination HIV prevention. PLoS Med. 2012;9:e1001250. doi: 10.1371/journal.pmed.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koopman JS, Jacquez G, Chick SE. New data and tools for integrating discrete and continuous population modeling strategies. Ann N Y Acad Sci. 2001;954:268–294. doi: 10.1111/j.1749-6632.2001.tb02756.x. [DOI] [PubMed] [Google Scholar]
- 12.Michael E, Malecela-Lazaro MN, Kazura JW. Epidemiological modelling for monitoring and evaluation of lymphatic filariasis control. Adv Parasit. 2007;65:191–237. doi: 10.1016/S0065-308X(07)65003-9. [DOI] [PubMed] [Google Scholar]
- 13.Restif O, Hayman DT, Pulliam JR, Plowright RK, George DB, Luis AD, et al. Model-guided fieldwork: practical guidelines for multidisciplinary research on wildlife ecological and epidemiological dynamics. Ecol Lett. 2012;15:1083–1094. doi: 10.1111/j.1461-0248.2012.01836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vynnycky E, White R. An introduction to disease modelling. Oxford: Oxford University Press; 2010. [Google Scholar]
- 15.Sonnenberg FA, Gregory P, Yomtovian R, Russell LB, Tierney W, Kosmin M, Carson JL. The cost-effectiveness of autologous transfusion revisited: implications of an increased risk of bacterial infection with allogeneic transfusion. Transfusion. 1999;39:808–817. doi: 10.1046/j.1537-2995.1999.39080808.x. [DOI] [PubMed] [Google Scholar]
- 16.Graat EA, de Jong MC, Frankena K, Franken P. Modelling the effect of surveillance programmes on spread of bovine herpesvirus 1 between certified cattle herds. Vet Microbiol. 2001;79:193–208. doi: 10.1016/S0378-1135(00)00356-4. [DOI] [PubMed] [Google Scholar]
- 17.Michael E, Malecela-Lazaro MN, Maegga BT, Fischer P, Kazura JW. Mathematical models and lymphatic filariasis control: monitoring and evaluating interventions. Trends Parasitol. 2006;22:529–535. doi: 10.1016/j.pt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Savill NJ, St Rose SG, Woolhouse ME. Detection of mortality clusters associated with highly pathogenic avian influenza in poultry: a theoretical analysis. J R Soc Interface. 2008;5:1409–1419. doi: 10.1098/rsif.2008.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold ME, Irvine RM, Tearne O, Rae D, Cook AJ, Breed AC. Investigation into sampling strategies in response to potential outbreaks of low pathogenicity notifiable avian influenza initiated in commercial duck holdings in great Britain. Epidemiol Infect. 2013;141:751–762. doi: 10.1017/S0950268812001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smieszek T, Salathe M. A low-cost method to assess the epidemiological importance of individuals in controlling infectious disease outbreaks. BMC Med. 2013;11:35. doi: 10.1186/1741-7015-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciccolini M, Donker T, Grundmann H, Bonten MJ, Woolhouse ME. Efficient surveillance for healthcare-associated infections spreading between hospitals. P Natl Acad Sci USA. 2014;111:2271–2276. doi: 10.1073/pnas.1308062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzales JL, Boender GJ, Elbers AR, Stegeman JA, de Koeijer AA. Risk based surveillance for early detection of low pathogenic avian influenza outbreaks in layer chickens. Prev Vet Med. 2014;117:251–259. doi: 10.1016/j.prevetmed.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Leslie E, Cowled B, Graeme Garner M, Toribio JA, Ward MP. Effective surveillance strategies following a potential classical swine fever incursion in a remote wild pig population in North-Western Australia. Transbound Emerg Dis. 2014;61:432–442. doi: 10.1111/tbed.12044. [DOI] [PubMed] [Google Scholar]
- 24.Mizumoto K, Ejima K, Yamamoto T, Nishiura H. On the risk of severe dengue during secondary infection: a systematic review coupled with mathematical modeling. J Vector Dis. 2014;51:153–164. [PubMed] [Google Scholar]
- 25.Pinsent A, Blake IM, White MT, Riley S. Surveillance of low pathogenic novel H7N9 avian influenza in commercial poultry barns: detection of outbreaks and estimation of virus introduction time. BMC Infect Dis. 2014;14:427. doi: 10.1186/1471-2334-14-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Bunnik BAD, Ciccolini M, Gibbons CL, Edwards G, Fitzgerald R, McAdam PR, et al. Efficient national surveillance for health-care-associated infections. BMC Public Health. 2015;15:832. doi: 10.1186/s12889-015-2172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinh DN, Boni MF. Statistical identifiability and sample size calculations for serial seroepidemiology. Epidemics. 2015;12:30–39. doi: 10.1016/j.epidem.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atlas RS, DuPont HL, Overall JE. Computer simulation of alternative sampling strategies to estimate risk of infection from cryptosporidium. Comput Biol Med. 1993;23:283–94. [DOI] [PubMed]
- 29.Lipsitch M. Measuring and interpreting associations between antibiotic use and penicillin resistance in Streptococcus Pneumoniae. Clin Infect Dis. 2001;32:1044–1054. doi: 10.1086/319604. [DOI] [PubMed] [Google Scholar]
- 30.Wu HL, Ding AA. Design of viral dynamic studies for efficiently assessing potency of anti-HIV therapies in AIDS clinical trials. Biom J. 2002;44:175–196. doi: 10.1002/1521-4036(200203)44:2<175::AID-BIMJ175>3.0.CO;2-3. [DOI] [Google Scholar]
- 31.Clermont G, Bartels J, Kumar R, Constantine G, Vodovotz Y, Chow C. Silico design of clinical trials: a method coming of age. Crit Care Med. 2004;32:2061–2070. doi: 10.1097/01.CCM.0000142394.28791.C3. [DOI] [PubMed] [Google Scholar]
- 32.Hallett TB, Garnett GP, Mupamberiyi Z, Gregson S. Measuring effectiveness in community randomized trials of HIV prevention. Int J Epidemiol. 2008;37:77–87. doi: 10.1093/ije/dym232. [DOI] [PubMed] [Google Scholar]
- 33.Dimitrov DT, Masse BR, Boily M. Beating the placebo in HIV prevention efficacy trials: the role of the minimal efficacy bound. J Acq Immun Def Synd. 2013;62:95–101. doi: 10.1097/QAI.0b013e3182785638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiura H, Yen H, Cowling BJ. Sample size considerations for one-to-one animal transmission studies of the influenza a viruses. PLoS One. 2013;8:e55358. doi: 10.1371/journal.pone.0055358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cori A, Ayles H, Beyers N, Schaap A, Floyd S, Sabapathy K, et al. HPTN 071 (PopART): a cluster-randomized trial of the population impact of an HIV combination prevention intervention including universal testing and treatment: mathematical model. PLoS One. 2014;9:e84511. doi: 10.1371/journal.pone.0084511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuadros DF, Abu-Raddad LJ, Awad SF, Garcia-Ramos G. Use of agent-based simulations to design and interpret HIV clinical trials. Comput Biol Med. 2014;50:1–8. doi: 10.1016/j.compbiomed.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Scott P, Herzog SA, Auranen K, Dagan R, Low N, Egger M, Heijne JCM. Timing of bacterial carriage sampling in vaccine trials: a modelling study. Epidemics. 2014;9:8–17. doi: 10.1016/j.epidem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Herzog SA, Low N, Berghold A. Sample size considerations using mathematical models: an example with Chlamydia trachomatis infection and its sequelae pelvic inflammatory disease. BMC Infect Dis. 2015;15:233. doi: 10.1186/s12879-015-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu HL. Statistical methods for HIV dynamic studies in AIDS clinical trials. Stat Methods Med Res. 2005;14:171–92. [DOI] [PubMed]
- 40.Hayes R, Ayles H, Beyers N, Sabapathy K, Floyd S, Shanaube K, et al. HPTN 071 (PopART): rationale and design of a cluster-randomised trial of the population impact of an HIV combination prevention intervention including universal testing and treatment - a study protocol for a cluster randomised trial. Trials. 2014;15:57. doi: 10.1186/1745-6215-15-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen HH, Prevost TC, Duffy SW. Evaluation of screening for nasopharyngeal carcinoma: trial design using Markov chain models. Brit J Cancer. 1999;79:1894–1900. doi: 10.1038/sj.bjc.6690301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longini IM, Hudgens MG, Halloran ME, Sagatelian KA. Markov model for measuring vaccine efficacy for both susceptibility to infection and reduction in infectiousness for prophylactic HIV vaccines. Stat Med. 1999;18:53–68. doi: 10.1002/(SICI)1097-0258(19990115)18:1<53::AID-SIM996>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 43.Hoad KA. van't Hoog AH, Rosen D, Marston B, Nyabiage L, Williams BG, et al. Modelling local and global effects on the risk of contracting tuberculosis using stochastic Markov-chain models. Math Biosci. 2009;218:98–104. doi: 10.1016/j.mbs.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Tuite AR, Fisman DN. Spectrum bias and loss of statistical power in discordant couple studies of sexually transmitted infections. STD. 2011;38:50–56. doi: 10.1097/OLQ.0b013e3181ec19f1. [DOI] [PubMed] [Google Scholar]
- 45.Auranen K, Rinta-Kokko H, Goldblatt D, Nohynek H, O'Brien KL, Satzke C, et al. Design questions for Streptococcus Pneumoniae vaccine trials with a colonisation endpoint. Vaccine. 2014;32:159–164. doi: 10.1016/j.vaccine.2013.06.105. [DOI] [PubMed] [Google Scholar]
- 46.Chan MS, Srividya A, Norman RA, Pani SP, Ramaiah KD, Vanamail P, et al. Epifil: a dynamic model of infection and disease in lymphatic filariasis. Am J Trop Med Hyg. 1998;59:606–614. doi: 10.4269/ajtmh.1998.59.606. [DOI] [PubMed] [Google Scholar]
- 47.Cowled BD, Garner MG, Negus K, Ward MP. Controlling disease outbreaks in wildlife using limited culling: modelling classical swine fever incursions in wild pigs in Australia. Vet Res. 2012; doi: 10.1186/1297-9716-43-3. [DOI] [PMC free article] [PubMed]
- 48.Auranen K, Rinta-Kokko H, Goldblatt D, Nohynek H, O'Brien KL, Satzke C, et al. Colonisation endpoints in Streptococcus Pneumoniae vaccine trials. Vaccine. 2013;32:153–158. doi: 10.1016/j.vaccine.2013.08.061. [DOI] [PubMed] [Google Scholar]
- 49.Grimm V, Berger U, DeAngelis DL, Polhill JG, Giske J, Railsback SF. The ODD protocol: a review and first update. Ecol Model. 2010;221:2760–2768. doi: 10.1016/j.ecolmodel.2010.08.019. [DOI] [Google Scholar]
- 50.Grimm V, Berger U, Bastiansen F, Eliassen S, Ginot V, Giske J, et al. A standard protocol for describing individual-based and agent-based models. Ecol Model. 2006;198:115–126. doi: 10.1016/j.ecolmodel.2006.04.023. [DOI] [Google Scholar]
- 51.Willem L, Verelst F, Bilcke J, Hens N, Beutels P. Lessons from a decade of individual-based models for infectious disease transmission: a systematic review (2006-2015) BMC Infect Dis. 2017;17:612. doi: 10.1186/s12879-017-2699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herzog SA, Heijne JCM, Althaus CL, Low N. Describing the progression from Chlamydia trachomatis and Neisseria gonorrhoeae to pelvic inflammatory disease: systematic review of mathematical modeling studies. Sex Transm Dis. 2012;39:628–637. doi: 10.1097/OLQ.0b013e31825159ff. [DOI] [PubMed] [Google Scholar]
- 53.Boily MC, Mâsse BR, Desai K, Alary M, Anderson RM. Some important issues in the planning of phase III HIV vaccine efficacy trials. Vaccine. 1999;17:989–1004. doi: 10.1016/S0264-410X(98)00316-8. [DOI] [PubMed] [Google Scholar]
- 54.Cohen T, Hedt BL, Pagano M. Estimating the magnitude and direction of bias in tuberculosis drug resistance surveys conducted only in the public sector: a simulation study. BMC Public Health. 2010;10:355. doi: 10.1186/1471-2458-10-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study protocol and search strategy. (PDF 74 kb)
Additional results tables. (PDF 54 kb)
Data Availability Statement
Not applicable.


