Abstract
Background:
Pseudomonas aeruginosa is a severe challenge for antimicrobial therapy, due to the chromosomal mutations or exhibition of intrinsic resistance to various antimicrobial agents such as most β-lactams. We undertook this study to evaluate the existence of SME, IMP, AIM, and VIM metallo-β-lactamases (MBL) encoding genes among P. aeruginosa strains isolated from Intensive Care Unit (ICU) patients in Al-Zahra Hospital in Isfahan, Iran.
Materials and Methods:
In a retrospective cross-sectional study that was conducted between March 2012 and April 2013, a total of 48 strains of P. aeruginosa were collected from clinical specimens of bedridden patients in ICU wards. Susceptibility test was performed by disc diffusion method. All of the meropenem-resistant strains were subjected to modified Hodge test for detection of carbapenemases. Multiplex polymerase chain reaction was performed for detection of blaVIM, blaIMP, blaAIM, and blaSME genes.
Results:
In disk diffusion method, imipenem and meropenem showed the most and colistin the least resistant antimicrobial agents against P. aeruginosa strains. Of the 48 isolates, 36 (75%) were multidrug resistant (MDR). Amplification of β-lactamase genes showed the presence of blaVIM genes in 7 (%14.6) strains and blaIMP genes in 15 (31.3%) strains. All of the isolates were negative for blaSME and blaAIM genes. We could not find any statistically significant difference among the presence of this gene and MDR positive, age, or source of the specimen.
Conclusion:
As patients with infections caused by MBL-producing bacteria are at an intensified risk of treatment failure, fast determination of these organisms is necessary. Our findings may provide useful insights in replace of the appropriate antibiotics and may also prevent MBLs mediated resistance problem.
Keywords: Antimicrobial resistance, metallo-β-lactamases, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa is generally known as an opportunistic invader rather than a cause of initial infection in healthy issues. It has now emerged as an important nosocomial pathogen in immunosuppressed patients and also leads to outbreaks of 9%–10% hospital-acquired infections with a high mortality rate.[1] The complete sequencing of a wild-type P. aeruginosa strain has prepared useful information about its pathogenicity and potential for resistance to many antibiotic agents and the development of increased multidrug resistance (MDR) in health-care settings.[2]
The carbapenems have been the selective drugs for the treatment of infections caused by penicillin or cephalosporin-resistant Gram-negative and nonfermenting bacilli such as P. aeruginosa and Acinetobacter spp.[1] Besides, their resistance to all β-lactams, the strains producing metallo-β-lactamases (MBL) are often resistant to aminoglycosides and fluoroquinolones. However, these generally stay susceptible to polymyxins.[3]
P. aeruginosa isolates that produce MBLs have caused many nosocomial outbreaks and crucial infections such as septicemia and pneumonia related to failure of cure with carbapenems in tertiary centers throughout the world.[4]
Nevertheless, carbapenem resistance can reduce outer membrane permeability, increase energy-dependent efflux, porin downregulation, and the productivity of carbapenem-hydrolyzing enzymes in P. aeruginosa.[1] In addition, many carbapenemase genes are carried out by plasmids and are easily transferable among various bacterial species and genera.[5]
Carbapenems (meropenem and imipenem) are effective against Gram-negative MDR strains.[6] Current data propose that carbapenem resistance is expanding among isolates of P. aeruginosa.[7] MBL-producing strains hydrolyze all carbapenems and deficiency of these enzymes can result in imipenem resistance and reduced meropenem sensitivity.[8]
In the last decade, various Classes A, B, and D β-lactamases have been discovered in P. aeruginosa.[9] The carbapenemases found are mostly different types of MBLs such as AIM, SME, GIM, IMP, NDM, SPM, and VIM, which their frequency has been increased in P. aeruginosa throughout the world.[1] The emergence of MBL-producing strains makes treatment hard and occasionally inefficient that cause high mortality.[2]
Nosocomial outbreaks caused by MBL-producing P. aeruginosa have been reported in different hospital units of our country such as many countries worldwide.[10,11] Carbapenemases that hydrolyze carbapenems and make them inactive have been progressively reported in Asia and Europe, Canada, and the United States.[12]
Detection of carbapenemases is difficult. Among phenotypic tests, the modified Hodge test (MHT) is a comparatively easy and simple test, but it is not recommended in nonfermenters.[3,12]
Identification of genes coding for carbapenem resistance by polymerase chain reaction PCR is usually reliable and accurate despite its limited application due to the cost.[1] Single or multiplex PCR assays require expert knowledge of carbapenemases epidemiology and is normally focused only on prevalent carbapenemase genes.[13]
Assessment of phenotypic and genotypic characterization of these isolates would be necessary for understanding the resistance mechanisms and its potential spread. There is not much data available on MBL-producing P. aeruginosa isolates from Iran. Therefore, we designed this study to determine the prevalence of MBL-producing P. aeruginosa (the existence of AIM, SME, IMP, and VIM) among P. aeruginosa strains isolated from Intensive Care Unit (ICU) patients in Al-Zahra Hospital in Isfahan, Iran.
Materials and Methods
Sampling and bacterial isolation
In a cross-sectional study, 48 P. aeruginosa isolates were collected from clinical specimens of bedridden patients in ICU wards of Al-Zahra Hospital of Isfahan city (March 2012 to April 2013). These isolates were obtained from cultures of specimens from tracheal aspirate, blood, urine, wound, catheter, and peritoneal fluid of the patients.
Bacteria were determined as P. aeruginosa by biochemical tests or if they had following characteristics: Gram-negative bacilli, citrate positive, nonfermentative, triple sugar iron Alk/Alk, motile, hydrogen sulfide (H2S) negative, urease negative, oxidase positive, and catalase positive. The confirmed isolates were kept preserved at −70°C. Standard strain P. aeruginosa ATCC 27853 was used as a control.[11]
Antimicrobial susceptibility tests
Antibiotic susceptibility testing of P. aeruginosa isolates was performed by the disc diffusion method on Mueller-Hinton agar (Merck, Germany). Susceptibility was defined according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.[13] Antibiogram discs containing meropenem (MEM 10 μg), imipenem (IPM 10 μg), ceftazidime (CAZ 30 μg), cefepime (FEP 30 μg), amikacin (AK 30 μg), ciprofloxacin (CIP 5 μg), colistin sulphate (10 μg), ampicillin/sulbactam (20 μg), and cefotaxime (CTX 30 μg) discs (MAST, Bootle, Merseyside, UK). P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as reference strains in susceptibility test.[11,14]
Modified Hodge test
All of the meropenem resistant strains (zones, i.e., 16–21 mm) were subjected to MHT for detection of carbapenemases. Besides, combined disk tests with boronic acid for the recognition of Class A carbapenemases or with ethylenediaminetetraacetic acid (EDTA) for the detection of MBLs. The existence of a zone of inhibition due to carbapenemase production by the test strain was considered as positive.[15,16]
Extraction of total DNA
For molecular detection, the total DNA content of each isolate was extracted with boiling method. In brief, a single colony from a 16 h culture on the nutrient agar (Hi-Media, India) was suspended in 50 μl of TES (50 mM Tris hydrochloride [pH 8.0], 5 mM EDTA, and 50 mMNaCl), and the suspension was heated at 95°C for 10 min and centrifuged at 15,000 ×g for 2 min. DNA containing supernatant was transferred to new sterile DNase free-RNase free microtubes.[17]
Rapid detection of VIM, SME, AIM, and IMP genes by multiplex polymerase chain reaction
Multiplex PCR was performed for amplification of P. aeruginosa VIM, IMP, SME, and AIM genes, using the primers listed in Table 1 according to the previous protocols.[18]
Table 1.
Oligonucleotides used in this study

Two microliters of total DNA was included to multiplex PCR in a 50 μL reaction mixture. The mix for the detection of blaIMP, blaVIM, blaAIM, and blaSME genes contains 1× PCR buffer (10 mmol/L Tris–HCl [pH 8.3]), 1.5 mmol/L of MgCl2, 0.125 mmol/L of each deoxynucleotide triphosphate, 10 μmol/L of each primer, and 2 U of AmpliTaq Polymerase (Fermentas R, Korea).
Amplification was carried out with the following thermal cycling conditions: initial DNA denaturation at 94°C for 10 min, then 36 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 40 s, and extension at 72°C for 50 s, followed by final extension at 72°C for 5 min. Agarose gel electrophoresis of the amplified DNA with 100 bp size marker (Fermentas R, Korea) was done for 1 h at 80 V in a 2% agarose gel in 1 × TAE (40 mmol/L Tris–HCl [pH 8.3], 2 mmol/L acetate, and 1 mmol/L EDTA) containing 0.05 mg/L ethidium bromide to detect the specific band.[18]
Statistical analysis
For the statistical analyses, the statistical software SPSS version 18.0 for Windows (SPSS Inc., Chicago, IL, USA) was utilized. Continuous variables were described as a mean ± standard deviation and compared using the Student's t-test or described as the median as well as range. Categorical variables were compared with Chi-square test or Fisher's exact test if the expected values were < 10. All tests were two-tailed, and P < 0.05 was considered statistically significant.
Results
Demographic characterization of carriages of Pseudomonas aeruginosa
The medical records of the 48 index patients were reviewed. The mean age was 49.5 ± 19.2 years, with a male to female ratio of 1.82:1. Seventeen (35.4%) of them were female and 31 (64.6%) were male.
The specimens included urine (n = 11, 30.5%), blood (n = 12, 25%), tracheal aspirate (n = 11, 22.9%), wound (n = 4, 8.3%), catheter (n = 10, 29.08%), and peritoneal fluid (n = 1, 9%). The most common underlying diseases were urinary and blood infections.
Antimicrobial susceptibilities of clinical strains
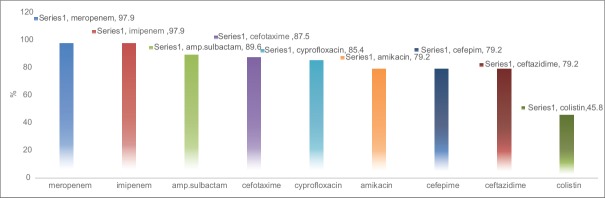
The antibiotic resistance of these strains is shown in Figure 1. In disk diffusion method, imipenem and meropenem showed the most and colistin the least resistant antimicrobial agents against P. aeruginosa strains. Susceptibility rates of the isolated P. aeruginosa strains in different samples are represented in Figure 1.
Figure 1.
Antimicrobial resistance rates among Pseudomonas aeruginosa isolates
Of the 48 isolates, 36 (75%) were MDR. Our results did not show any significant relation between the MDR-positive and MDR negative isolates and gender using Chi-square analysis (P = 0.99). Furthermore, difference between MDR and non-MDR isolates with a source of the specimen and mean age was not statistically significant (P > 0.05). Frequency of MDR and non-MDR isolates with demographic characteristics of patients is listed in Table 2.
Table 2.
Frequency of multidrug-resistant and nonmultidrug-resistant isolates according to demographic characteristics of patients
Bacterial isolates tested for the presence of VIM, AIM, IMP, and SME genes
All of the 48 P. aeruginosa isolates were evaluated with amplification of the gyrB gene. PCR screening was performed for all of these strains.
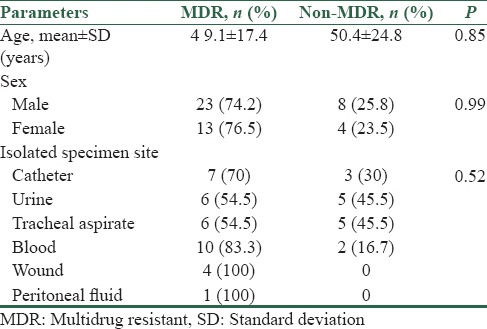
blaIMP and blaVIM genes were detected in 15 (31.3%) and 7 (14.6%) of the isolates, respectively. In one isolate of P. aeruginosa with the source of urine, both blaIMP and blaVIM genes were detected. Positive controls of blaIMP and blaVIM used individually or combined and produced anticipated bands and approved the specificity of the PCR primers used in simplex and multiplex PCR. Expected fragments were seen in multiplex PCR method [Figures 2–4]. MHT or combined-disk tests were positive for isolates with blaIMP or blaVIM genes, and all of them were MDR. Fisher's exact test showed a statistically significant difference between the presence of blaIMP gene and MDR positive (P = 0.009). Frequency of blaVIM and blaIMP genes producing P. aeruginosa isolates based on demographic characteristics of patients is determined in Table 3. Prevalence of blaIMP and blaVIM genes in male and female patients was 32.3%, 16.1% and 29.4%, 11.8%, respectively. Comparing the sex group and presence of these genes by Chi-square analysis did not show any statistically significant difference (P > 0.05). Although the mean age of patients with blaIMP gene was lower than other patients, this difference was not statistically significant (P = 0.84). Nearly 52.6% (10 of 19) of the urine samples included blaIMP. There was a significant difference between the source of specimen and the presence of blaIMP gene by Fisher's exact test (P = 0.002). We could not find any statistically significant difference among the presence of blaVIM gene and MDR positive (P = 0.11), mean age (P = 0.64), or source of the specimen (P = 0.15). Furthermore, both multiplex and simplex PCR amplifications showed that all of the isolates were negative for blaSME and blaAIM genes.
Figure 2.
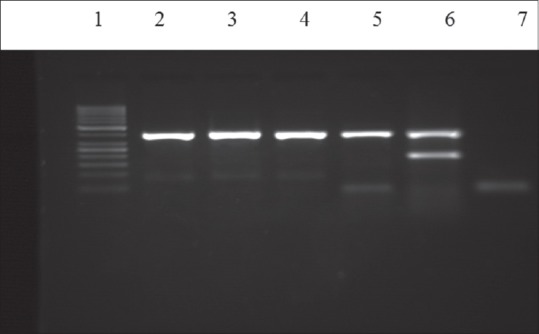
Gel electrophoresis of multiplex polymerase chain reaction products following amplification with specific primers for blaVIM gene (391bp). Lanes: (2) the 50bp ladder; (3-5) clinical isolates for blaVIM gene, (6) blaVIM gene blaIMP (232bp) positive control, (7) negative control
Figure 4.
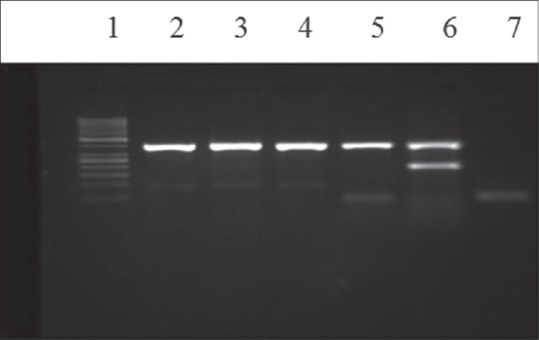
Gel electrophoresis of multiplex polymerase chain reaction products following amplification with specific primers for blaVIM gene (391bp). Lanes: (1) the 50bp ladder, (2-5) clinical isolates for blaVIM gene, (6) blaVIM gene blaIMP (232bp) positive control, (7) negative control
Table 3.
Results for β-lactamase genes in aeruginosa isolates according to demographic characteristics of patients
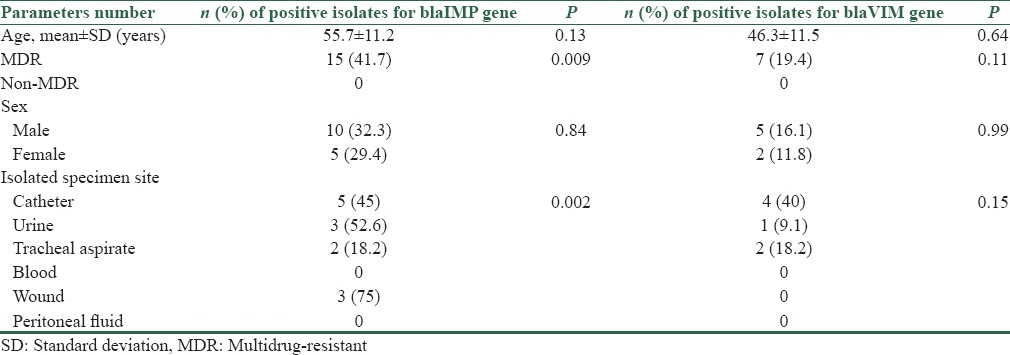
Figure 3.
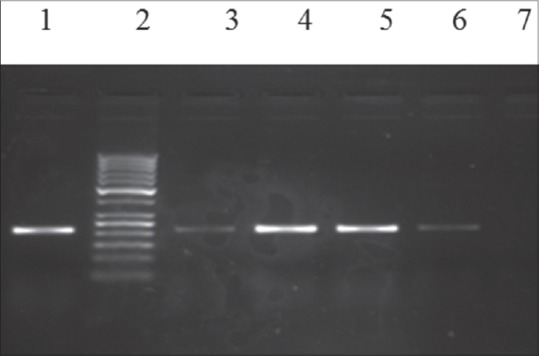
Gel electrophoresis of multiplex polymerase chain reaction products following amplification with specific primers for blaIMP gene (232bp). Lanes (1) blaIMP (232bp) positive control, (2) the 50bp ladder, (3-6) clinical isolates for blaIMP gene, (7) negative control
Discussion
MBLs have been recognized in clinical isolates throughout the world. These strains are responsible for extended nosocomial outbreaks and accompanied by severe infections.[19] In recent decades, the carbapenem-resistant P. aeruginosa (CRPA) has emerged as an important pathogen.
In a similar study that was performed in the USA, the resistant rate ranged from 7.4% to 35.4%.[20] According to the results of another study in Taiwan, 18.1% of all P. aeruginosa isolates in the ICUs of medical centers were CRPA. In the present study, the prevalence of blaVIM gene was assessed 14.6%. These different results implied the prevalence of CRPA was varied by geographic areas, specimen source, patient age, clinical setting, and broad-spectrum antibiotics consumption.[14]
Previous researches exhibited that IMP and VIM types of MBLs are also prevalent in Asian countries.[21] VIM type is also the most frequent enzyme in Turkey.[22] In a previous study in Iran, like our findings, blaVIM gene was detected among the isolates by PCR. MBL genes are placed on mobile genetic cassettes with other resistance determinants inserted into integrons, the isolates of P. aeruginosa producing these enzymes often show resistance to additional classes of drugs and act as MDR. Our data also demonstrated that all of the VIM-producing isolates are resistant to aminoglycosides and to fluoroquinolones that is in accordance with results of the similar study that was performed by Shahcheraghi et al. in Tehran.[21]
Prevalence of VIM-type MBL-producing P. aeruginosa strains in the recent similar study that was performed in Zanjan, Iran was 56% that also significantly was different (P < 0. 05) from other districts of Iran and also higher than our results (14.6%). Most of MBL-producing isolates carried Class 1 integron gene, which can easily spread the resistance encoding genes among these isolates. Several studies have also reported different frequencies of MBL-positive isolates carrying Class 1 integrons.[2,23]
Other carbapenemase enzymes such as blaDIM, blaSME, blaAIM, blaNMC, and blaCcrA are comparably infrequent and rarely in both their geographic occurrence and in the strains that they can be detected.[9] The blaSME genes are assumed to be chromosomal and nonmobile. The SME-1 β-lactamase, along with the nearly identical SME-2 and SME-3, has been found sporadically in different geographical locations such as London and the United States.[24] Infections caused by SME were small clusters of up to 19 isolates.[25] In this study, all isolates were negative for blaSME or blaAIM genes by PCR screening that is in accordance with another similar study in 2009.[1]
Due to these different results, every organization should usually check the resistant rate of pathogens and the antibiotics consumption and report the information to infection control programs.[26]
The prevalence of antibiotic resistance to imipenem, meropenem, and ceftazidime in our P. aeruginosa isolates was 97.9%, 97.9%, and 79.2%, respectively, that showed higher resistance in comparison to other regions of our country such as Tehran, Ahvaz, and Hamedan.[6,11] Several studies from other countries also showed the similar finding in recent years.[27]
Some researchers reported the emergence of colistin-resistant organisms in their study. Our results showed that about 50% of isolates were resistant to colistin; therefore, selection of antibiotics to treat such patients should be carried out. In our study, 36 (75%) isolates were MDR, but we could not find any significance relation between the MDR-positive and demographic characteristics such as age, sex, and source of infection. Several studies have found CRPA were more resistant to multiple drugs than carbapenem susceptible P. aeruginosa isolates and higher mortality rates of P. aeruginosa infection to be related to the site of infection, age, MDR, and improperness of empirical treatment.[14]
Phenotypic tests such as the MHT can be used to identify strains that need molecular testing to reduce the costs. The manufacturers declare that the test can also be performed for direct detection in clinical specimens.[15] In the present study, carbapenemase gene was detected only in colonies because several studies have shown that EDTA can give false-positive results in some resistant P. aeruginosa isolates due to altered OprD levels and PCR confirmation is an important step.[19]
Out of seven MHT positive cases for VIM in our study, all of them were declared positive by PCR technique. Result of a study was carried out in 2007 to estimate different laboratory tests for identification of MBLs in Enterobacteriaceae showed that MBL test detected 98% cases keeping PCR as the gold standard, whereas only 0.03% was recognized as false positive.[28]
Our study has several limitations. First of all, this was a retrospective cross-sectional study and we could not investigate many independent factors. Second, although we detected the presence of blaVIM and blaIMP genes, we could not recognize AIM and SME genes because of the small sample size. Further studies should be performed to clarify if the presence of carbapenem-resistant genes affects the clinical outcome.
Conclusion
The multiplex PCR investigated in this study showed excellent performance and could supplement phenotypic tests in carbapenemase detection. The VIM prevalence in the present study was relatively high, and this MBL enzyme could emerge among clinical isolates of P. aeruginosa. Because of broad-spectrum antibiotic therapy and the presence of invasive devices, ICU patients are at increased risk for carbapenem resistance. As patients with infections caused by MBL-producing bacteria are at an intensified risk of treatment failure, fast determination of these organisms is necessary. Because of high resistant rate of imipenem or meropenem, so it is suggested that a simple test based on CLSI recommendation such as MHT should be adopted to confirm MBL-producing bacteria in clinical laboratories.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Wolter DJ, Khalaf N, Robledo IE, Vázquez GJ, Santé MI, Aquino EE, et al. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican Medical Center Hospitals: Dissemination of KPC and IMP-18 beta-lactamases. Antimicrob Agents Chemother. 2009;53:1660–4. doi: 10.1128/AAC.01172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doosti M, Ramazani A, Garshasbi M. Identification and characterization of metallo-ß-lactamases producing Pseudomonas aeruginosa clinical isolates in University Hospital from Zanjan Province, Iran. Iran Biomed J. 2013;17:129–33. doi: 10.6091/ibj.1107.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mataseje LF, Bryce E, Roscoe D, Boyd DA, Embree J, Gravel D, et al. Carbapenem-resistant gram-negative bacilli in Canada 2009-10: Results from the Canadian Nosocomial Infection Surveillance Program (CNISP) J Antimicrob Chemother. 2012;67:1359–67. doi: 10.1093/jac/dks046. [DOI] [PubMed] [Google Scholar]
- 4.Baumgart AM, Molinari MA, Silveira AC. Prevalence of carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii in high complexity hospital. Braz J Infect Dis. 2010;14:433–6. doi: 10.1590/s1413-86702010000500002. [DOI] [PubMed] [Google Scholar]
- 5.Dortet L, Boulanger A, Poirel L, Nordmann P. Bloodstream infections caused by Pseudomonas spp.: How to detect carbapenemase producers directly from blood cultures. J Clin Microbiol. 2014;52:1269–73. doi: 10.1128/JCM.03346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saderi H, Karimi Z, Owlia P, Bahar MA, Akhavi Rad SM. Phenotypic detection of metallo-beta-lactamase producing pseudomonas aeruginosa strains isolated from burned patients. Iran J Pathol. 2008;3:20–4. [Google Scholar]
- 7.Dortet L, Poirel L, Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clin Microbiol. 2012;50:3773–6. doi: 10.1128/JCM.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasteran F, Veliz O, Rapoport M, Guerriero L, Corso A. Sensitive and specific modified Hodge test for KPC and metallo-beta- lactamase detection in Pseudomonas aeruginosa by use of a novel indicator strain, Klebsiella pneumoniae ATCC 700603. J Clin Microbiol. 2011;49:4301–3. doi: 10.1128/JCM.05602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun SD, Monecke S, Thürmer A, Ruppelt A, Makarewicz O, Pletz M, et al. Rapid identification of carbapenemase genes in Gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS One. 2014;9:e102232. doi: 10.1371/journal.pone.0102232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dally S, Lemuth K, Kaase M, Rupp S, Knabbe C, Weile J. DNA microarray for genotyping antibiotic resistance determinants in Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. 2013;57:4761–8. doi: 10.1128/AAC.00863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safari M, Alikhani MY, Arabestani MR, Kamali Kakhki R, Jafari R. Prevalence of metallo-ß-lactamases encoding genes among aeruginosa strains isolated from the bedridden patients in the Intensive Care Units. Avicenna J Clin Microbiol Infect. 2014;1:e19216. [Google Scholar]
- 12.Bashir D, Thokar MA, Fomda BA, Bashir G, Zahoor D, Ahmad S, et al. Detection of metallo-beta-lactamase (MBL) producing Pseudomonas aeruginosa at a tertiary care hospital in Kashmir. Afr J Microbiol Res. 2011;5:164–72. [Google Scholar]
- 13.Cockerill FR. USA: Clinical and Laboratory Standards Institute; 2012. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty Second International Supplement M100-S22. [Google Scholar]
- 14.Lin KY, Lauderdale TL, Wang JT, Chang SC. Carbapenem-resistant Pseudomonas aeruginosa in Taiwan: Prevalence, risk factors, and impact on outcome of infections. J Microbiol Immunol Infect. 2016;49:52–9. doi: 10.1016/j.jmii.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Kaase M, Szabados F, Wassill L, Gatermann SG. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol. 2012;50:3115–8. doi: 10.1128/JCM.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noyal MJ, Menezes GA, Harish BN, Sujatha S, Parija SC. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res. 2009;129:707–12. [PubMed] [Google Scholar]
- 17.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–82. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–35. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrow BJ, Pillar CM, Deane J, Sahm DF, Lynch AS, Flamm RK, et al. Activities of carbapenem and comparator agents against contemporary US Pseudomonas aeruginosa isolates from the CAPITAL surveillance program. Diagn Microbiol Infect Dis. 2013;75:412–6. doi: 10.1016/j.diagmicrobio.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Shahcheraghi F, Nikbin VS, Feizabadi MM. Identification and genetic characterization of metallo-beta-lactamase-producing strains of Pseudomonas aeruginosa in Tehran, Iran. New Microbiol. 2010;33:243–8. [PubMed] [Google Scholar]
- 22.Strateva T, Yordanov D. Pseudomonas aeruginosa – A phenomenon of bacterial resistance. J Med Microbiol. 2009;58(Pt 9):1133–48. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 23.Khosravi Y, Tay ST, Vadivelu J. Analysis of integrons and associated gene cassettes of metallo-ß-lactamase-positive Pseudomonas aeruginosa in Malaysia. J Med Microbiol. 2011;60(Pt 7):988–94. doi: 10.1099/jmm.0.029868-0. [DOI] [PubMed] [Google Scholar]
- 24.Walther-Rasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57:373–83. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 25.Queenan AM, Bush K. Carbapenemases: The versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen IL, Lee CH, Su LH, Tang YF, Chang SJ, Liu JW. Antibiotic consumption and healthcare-associated infections caused by multidrug-resistant Gram-negative bacilli at a large medical center in Taiwan from 2002 to 2009: Implicating the importance of antibiotic stewardship. PLoS One. 2013;8:e65621. doi: 10.1371/journal.pone.0065621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal VD, Maki DG, Jamulitrat S, Medeiros EA, Todi SK, Gomez DY, et al. International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003-2008, issued June 2009. Am J Infect Control. 2010;38:95–104.e2. doi: 10.1016/j.ajic.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Amjad A, Mirza IA, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3:189–93. [PMC free article] [PubMed] [Google Scholar]