Abstract
Background & objectives:
Among cell surface proteins, biofilm-associated protein (Bap) promotes biofilm development in Staphylococcus aureus strains. The aim of this study was to investigate proteinase-mediated biofilm dispersion in different isolates of S. aureus.
Methods:
Biofilm assay was done in 96-well microtitre plate to evaluate the effect of proteinase K on biofilms of bovine mastitis S. Aureus isolates. Extracellular polymeric substances were extracted and evaluated for their composition (protein, polysaccharides and extracellular DNA), before and after the proteinase K treatment.
Results:
Biofilm assay showed that 2 µg/ml proteinase K significantly inhibited biofilm development in bap-positive S. aureus V329 as well as other S. aureus isolates (SA7, SA10, SA33, SA352), but not in bap-mutant M556 and SA392 (a weak biofilm-producing strain). Proteinase K treatment on S. aureus planktonic cells showed that there was no inhibition of planktonic growth up to 32 µg/ml of proteinase K. Proteinase K treatment on 24 h old preformed biofilms showed an enhanced dispersion of bap-positive V329 and SA7, SA10, SA33 and SA352 biofilms; however, proteinase K did not affect the bap-mutant S. aureus M556 and SA392 biofilms. Biofilm compositions study before and after proteinase K treatment indicated that Bap might also be involved in eDNA retention in the biofilm matrix that aids in biofilm stability. When proteinase K was used in combination with antibiotics, a synergistic effect in antibiotic efficacy was observed against all biofilm-forming S. aureus isolates.
Interpretation & conclusions:
Proteinase K inhibited biofilms growth in S. aureus bovine mastitis isolates but did not affect their planktonic growth. An enhanced dispersion of preformed S. aureus biofilms was observed on proteinase K treatment. Proteinase K treatment with antibiotics showed a synergistic effect against S. aureus biofilms. The study suggests that dispersing S. aureus by protease can be of use while devising strategies against S. aureus biofilms.
Keywords: Antibiotic treatment, bacterial surface proteins, biofilm-associated protein, biofilm dispersal, extracellular DNA, microbial biofilms, proteinase K, Staphylococcus aureus
Most bacteria in nature execute definite development stages in biofilm formation; (i) adherence of cells to a substratum, (ii) development of microcolonies, (iii) maturation of microcolonies into biofilms, and (iv) detachment of bacteria and acquisition of motile phase, known as biofilm dispersal1. Biofilm disassembly/dispersion is believed to play an important role in pathogenicity, in environmental distribution and also in phase transition2. The dispersal phenomenon can also be triggered by several environmental signals or unfavourable condition3. Biofilms aid many advantages to microorganisms such as higher resistance to adverse environmental conditions, higher resistance to antimicrobial agents and enhanced protection from immune response in case of persistent infections4.
Staphylococcus aureus is a universal pathogen which causes mild to severely life-threatening diseases. This bacterium also constitutes a major cause of hospital-acquired/healthcare-associated infections (HAIs). According to Center for Disease Control and Prevention, S. aureus strains are associated with 15.6 per cent of the total HAIs reported between 2009 and 20105 and 12.3 per cent between 2011 and 2012 in Europe6. Commonly, a mature biofilm consists of polysaccharides, proteins, extracellular DNA (eDNA)7 and amyloid fibres as matrix. There are several reports on the role of surface proteins in S. aureus biofilm formation and its stability8,9. Among various surface proteins, biofilm-associated protein (Bap) was first reported as a large, multi-domain, cell surface-anchored protein, which plays a crucial role in S. aureus biofilm development, architecture and in the pathogenesis of bovine mastitis10,11,12,13. A study carried out in Brazil showed the presence of bap gene in all the coagulase-negative Staphylococcus spp. strains isolated from the nosocomial infections14. Another report showed a higher frequency of occurrence of bap gene (56.6%) in Staphylococcus spp. (189 samples) isolated from bovine subclinical mastitis. Apart from this, frequency of bap gene occurrence was significantly higher in coagulase-negative strains as compared with coagulase-positive15. The involvement of polysaccharide intercellular adhesin (ica-dependent) component of the S. aureus biofilm matrix has been studied comprehensively16. However, role of ica-independent mechanisms, which is predominantly mediated by biofilm-associated surface proteins [Bap, accumulation-associated protein (Aap), fibronectin-binding protein, etc.] in the stability of Staphylococci biofilm matrix, is poorly understood9,17,18. Previous reports on ica-independent biofilm formation in staphylococci showed a strong link between biofilm formation and cell wall-associated proteins, in particular, Bap11, Aap18 and Bap-homologue protein19. Repeated domains contain an amyloidogenic peptide motif (-STVTVTF- derived from the C-repeat of the Bap), which is responsible for cell-cell interaction20. Therefore, Bap and Bap-like surface proteins could be an important target for biofilm dispersal studies.
Dispersal mechanisms vary in different bacteria and this event is considered as a novel approach to treat drug-resistant S. aureus which is common in body implants and catheter-related infections21. Among the natural ways of S. aureus biofilm dispersal, Agr-mediated biofilm dispersal and secretion of major extracellular proteases, SspA, SspB, Aur and Scp as pro-enzymes, were reported22. Theoretically, these enzymes may contribute to biofilm detachment, but very little is known about their role with regard to Staphylococci. An extracellular serine protease, Esp secreted by a subset of S. epidermidis, has shown to inhibit biofilm formation and nasal colonization by S. aureus23. Esp has proteolytic activity specifically towards biofilm-specific proteins that are associated with S. aureus biofilm formation and host-pathogen interaction24. Studies on Staphylococci chronic infections and biofilms as well as discovery of major dispersal mechanisms shifted the focus on the development of dispersal-mediated treatment options for S. aureus biofilm infections25. In an earlier report, we have shown that proteinase K can emulate the naturally produced proteases and can be used to enhance the biofilm dispersal through cleavage of surface proteins i.e. Bap-dependent S. aureus biofilm establishment17. In this study, we investigated whether this approach would be useful in general and has wider applicability using five other S. aureus mastitis isolates.
Material & Methods
Microorganisms and culture conditions: A bap-positive S. aureus V329 and its isogenic-mutant S. aureus M556 were used along with five other mastitis isolates of S. aureus viz., SA7, SA10 SA33, SA252 and SA392. Bovine mastitis S. aureus strains SA7, SA10 and SA33 were procured from Karnataka Veterinary College, Bengaluru, whereas SA352 and SA392 were procured from Madras Veterinary College, Chennai. All bovine mastitis S. aureus strains used in the study were isolated from infected site of bovine mastitis. M556 was generated by transposon insertion in the downstream part of bap gene of S. aureus V329 in such a way that Bap is synthesized but remains non-functional as cell wall anchoring region is truncated11. For each experiment, single colonies were picked from Tryptic Soy Agar (TSA) culture plates and inoculated in Tryptic Soy Broth (TSB) medium supplemented with 0.25 per cent glucose (TSB-G) and incubated at 37°C at 150 rpm. Overnight grown cultures were used for all experiments after checking for culture purity. All experiments were performed at Biofouling and Thermal Ecology Section, Water and Steam Chemistry Division, BARC Facilities, Kalpakkam, from January 2013 to December 2014.
Quantitative biofilm assay: Biofilm assay was performed in 96-well microtitre plates to estimate the inhibitory/dispersion action of proteinase K. The working concentration of proteinase K was chosen as 2 µg/ml in all the experiments. The overnight grown cultures of the S. aureus cells in TSB-G were diluted 1:40 in sterile TSB-G medium and added to the pre-sterilized 96-well flat bottom polystyrene microtitre plates. To estimate the inhibitory action, S. aureus biofilms were grown in the presence of 2 µg/ml of proteinase K. To study dispersion, biofilms were grown on microtitre plates and washed after prescribed time and 200 µl of fresh TSB-G amended with 2 µg/ml of proteinase K was added to the wells and the plates were incubated at 37°C for 24 h. To study the effect of Ca2+ on proteolytic cleavage of Bap in terms of biofilm formation, V329 biofilms were grown at 37°C for 24 h in the presence of 2 µg/ml of proteinase K along with increasing concentration of Ca2+ in the range of 1.56-50 mM. V329 biofilm grown in the presence of only Ca2+ in the similar concentration range acted as control for Ca2+. After 24 h of incubation, biofilm growth was quantified. Biofilm quantification was done by classical crystal violet assay as described previously2. To dissolve the bound crystal violet, 33 per cent acetic acid was used26. Biofilm growth was monitored in terms of absorbance at 570 nm using a multimode microplate reader (BioTek, USA).
Planktonic growth studies: Overnight grown bacterial cultures were harvested and washed twice with phosphate buffered saline (PBS) and OD600 of each culture was set to 0.1. One hundred microlitres of each re-suspended culture was inoculated in 1900 µl of TSB-G and different concentrations of proteinase K. Cultures were incubated at 37°C and 150 rpm. Absorbance of each culture was recorded at different time intervals after vortexing for five seconds, to re-suspend the settled cells.
Extracellular polymeric substance (EPS) extraction and quantification of biofilm matrix components: S. aureus biofilm was grown for 48 h on glass slides immersed in 20 ml TSB-G. After 48 h, planktonic cells were aspirated and biofilm was gently washed twice with PBS. S. aureus biofilms were treated with 2 µg/ml proteinase K in TBS-G for 4 h at 37°C. After four hours, biofilm was gently washed with PBS and then remaining biofilm was scrapped and collected in 5 ml of PBS. Biofilm was disintegrated by gentle vortexing using glass beads. Five millilitres of the biofilm sample was centrifuged at 8000 rpm and 4°C for 30 min. Supernatant was collected and mixed with double volume of 90 per cent chilled ethanol and kept at 4°C for overnight. EPS was collected by centrifugation at 10,000 rpm and 4°C for 10 min. The supernatant was discarded and pellet was collected and dried at 60°C to remove ethanol. Pellet was re-suspended in 100 µl PBS buffer. The protein and eDNA content in the re-suspension was quantified using Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA); the quantization protocol was followed as published by the vendor27,28. Glucose concentration as a measure of polysaccharide content was quantified by the method as described elsewhere29.
Anti-biofilm activity of antibiotic-proteinase K treatment: To investigate the effect of proteinase K on the efficacy of antibiotics, proteinase K treatment was given a combination of gentamicin against biofilm forming S. aureus isolates viz., SA7, SA10, SA33 and SA352. The antibiotic concentrations were chosen as 10 and 50 times of the minimum inhibitory concentration (indicated as X and 5X, respectively) against S. aureus planktonic cells. Proteinase K treatment was given in combination with X concentration of antibiotics. S. aureus biofilms were grown in microtitre plates at 37°C and 150 rpm. After 24 h, planktonic cells were aspirated by pipette and the biofilms were gently rinsed twice with sterile PBS. After rinsing, the biofilms were treated with antibiotics alone and antibiotic-proteinase K combinations. After 24 h, planktonic cells were aspirated and biofilms were gently rinsed twice with PBS. Two hundred microlitres of PBS was added to each well, and the biofilm cells were dislodged by ultra-sonication for five minutes. Cells released from the biofilms were harvested, and the viable cell count was obtained by plating on TSA media and incubated at 37°C overnight.
Statistical analysis: Two-tailed Student’s t test was used to determine the differences in biofilm formation between the groups.
Results
Effect of proteinase K on biofilm development and planktonic growth of S. aureus: Proteinase K treatment hampered the biofilm development of most S. aureus isolates viz., SA7, SA10, SA33, SA352 and bap-positive V329. All S. aureus isolates, except SA392 (weak biofilm-producing strain), showed significant inhibition in biofilm growth when treated with 2 µg/ml proteinase K (Fig. 1). SA7, SA10, SA33, SA352 biofilms showed 84, 71, 83 and 68 per cent reduction in biofilm growth in the presence of 2 µg/ml proteinase K. On the contrary, strains M556 and SA392 were found to be weak biofilm producers and there was no significant inhibition of biofilm formation in the presence of proteinase K. Planktonic growth studies of bap-positive V329 and bap-mutant M556 and other S. aureus strains were carried out in the presence of different concentration of proteinase K. There was no effect of proteinase K on the planktonic growth of S. aureus isolates when tested up to 32 µg/ml (Fig. 2).
Fig. 1.
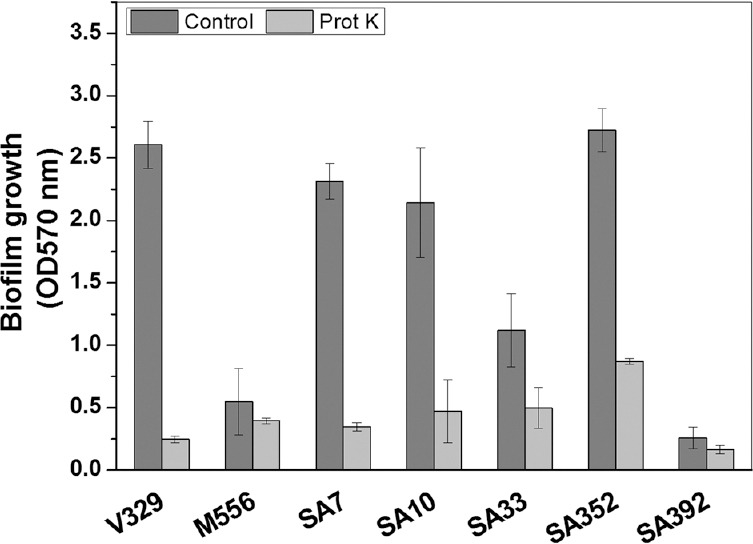
Effect of proteinase K on the growth of Staphylococcus aureus biofilms. Values are mean±standard deviation (n=3).
Fig. 2.
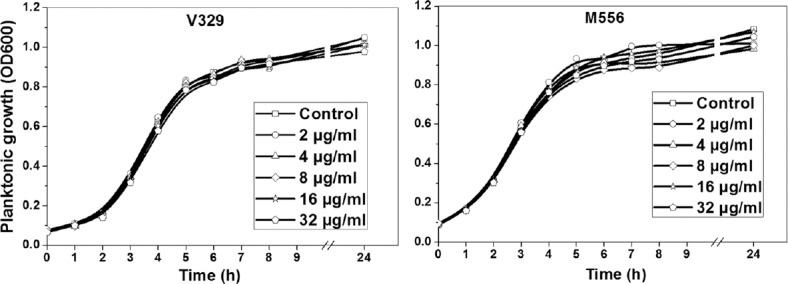
Effect of different concentrations of proteinase K on planktonic growth of Staphylococcus aureus V329 and M556.
Effect of different concentrations of Ca2+ on proteolytic degradation of biofilm-associated protein (Bap) and biofilm development of V329: Biofilm assay in the presence of proteinase K with increasing concentrations of Ca2+ was carried out. Addition of increasing concentrations of Ca2+ had no effect on proteinase K-mediated inhibition of biofilm development (Fig. 3). In other words, Ca2+ did not affect the proteolytic degradation of surface protein Bap by proteinase K. On the other hand, lower concentration of Ca2+ (up to 6.25 mM) had no significant effect on V329 biofilm formation, but higher concentrations showed an inhibitory effect.
Fig. 3.
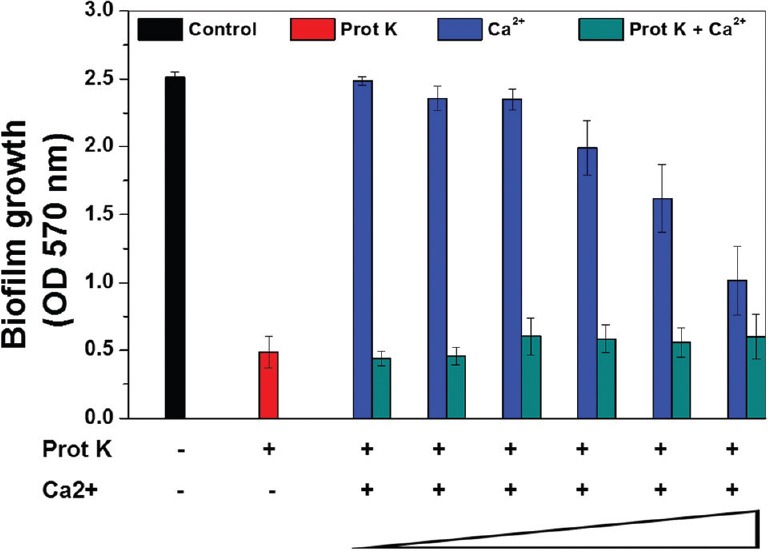
Effect of Ca2+ on proteinase K-mediated biofilm inhibition in bap-positive Staphylococcus aureus biofilm. Results are shown as mean±standard deviation (n=5).
Proteinase K enhances dispersal in S. aureus biofilms: To investigate the biofilm dispersal activity of proteinase K against S. aureus biofilms, proteinase K treatment was given to 24 h old S. aureus biofilms. Proteinase K treatment of S. aureus biofilms caused a significant disruption of all S. aureus biofilms except M556 and SA392 (Fig. 4). Upon 2 µg/ml proteinase K treatment for 24 h, approximately 92.5, 23.4, 90.1, 95.8, 81.9 and 60 per cent biofilm dispersal was observed in V329, M556, SA7, SA10, SA33 and SA352, respectively. Although proteinase K enhanced the biofilm dispersal, 100 per cent biofilm removal could not be achieved in any case. Moreover, SA392 formed a weak biofilm as compared to V329 after 24 h; hence, estimating SA392 biofilm dispersal was difficult and inappropriate, and therefore, SA392 was not involved in further experiments.
Fig. 4.
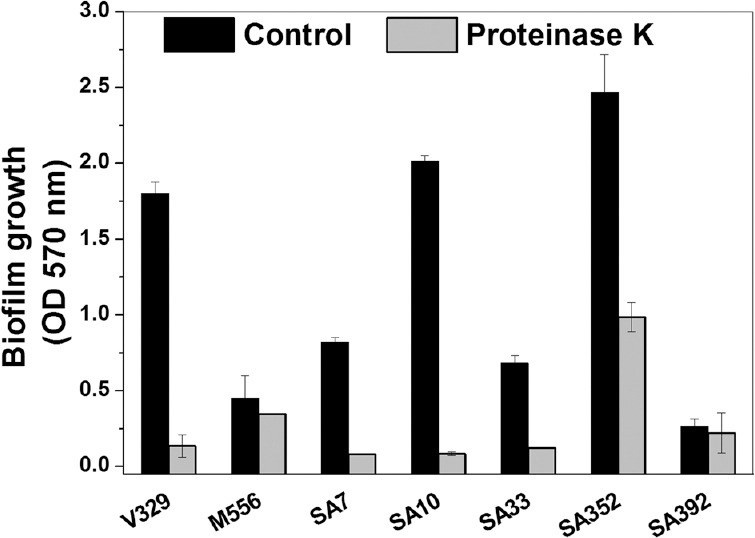
Dispersal of pre-grown 24 h-old Staphylococcus aureus biofilms by proteinase K. Results are shown as mean±standard deviation (n=5).
S. aureus V329 biofilm had significantly higher amount of carbohydrate as well as eDNA (P<0.05, n=3) as compared to Bap-mutant M556 biofilm. On the other hand, M556 biofilm was comprised significantly higher amount of biofilm matrix proteins (P<0.05, n=3), (Fig. 5). There was significant decrease in the protein as well as eDNA content in V329 and M556 biofilms after proteinase K treatment; however, there was no significant decrease in the carbohydrate content of biofilm matrix in either case.
Fig. 5.
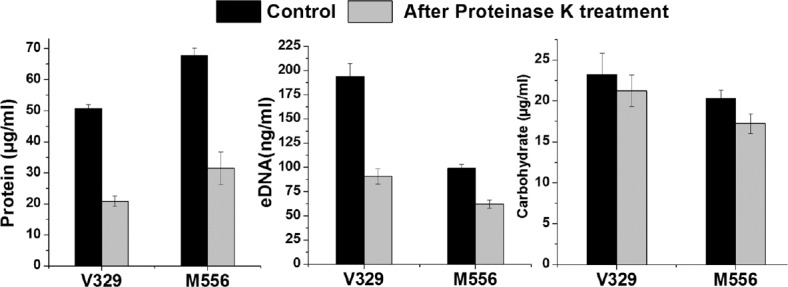
Constituents of Staphylococcus aureus extracellular polymeric substance before and after treatment of proteinase K. Results are shown as mean ± standard deviation (n=3).
Proteinase K enhances antibiotic efficacy against S. aureus biofilms: Fig. 6 shows the reduction in colony forming unit (cfu) count in S. aureus biofilms (SA7, SA10, SA33 and SA352) upon the treatment of gentamicin in various combinations. It was observed that addition of proteinase K in combination with gentamicin had more impact against S. aureus biofilm cells when compared to gentamicin alone. When the antibiotic concentration was increased by five times, there was no significant increase in log reduction of cfu count in any case. On the other hand, the addition of 2 µg/ml proteinase K in combination with antibiotics resulted in significant log reductions in each case. Proteinase K treatment caused an increase in the reduction in cfu in biofilms by 3.85, 5.0, 3.03 and 3.76 logs unit in the case of SA7, SA10, SA33 and SA352, respectively. Only 0.45, 1.27, 0.88 and 0.79 log units enhancement was noticed when gentamicin concentration was increased five times.
Fig. 6.
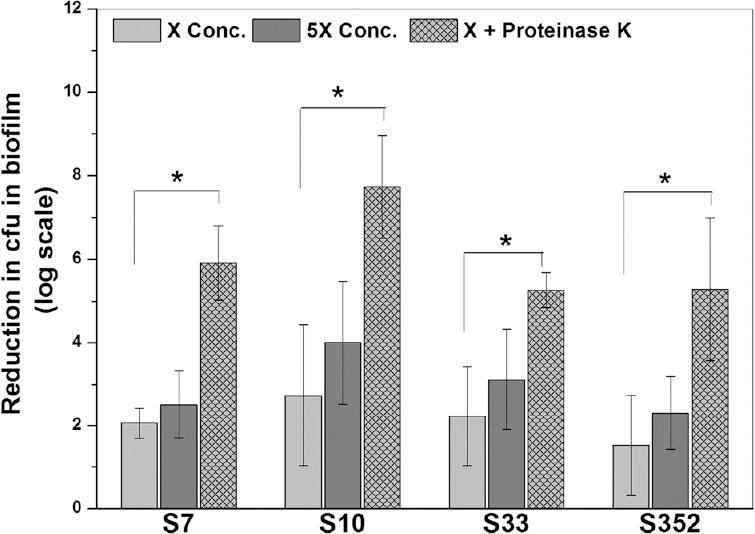
Antimicrobial efficacy of antibiotics in combination with proteinase K against 24 h old Staphylococcus aureus biofilms. Proteinase K was used at 2 µg/ml. Two concentrations of gentamicin were used; X=150 µg/ml and 5X=750 µg/ml. *P<0.05.
Discussion
In clinical settings, S. aureus biofilms impose resistance to host immune/defence mechanisms and antimicrobial therapy, thus enabling the bacterium to persist30. It was anticipated that proteolytic cleavage of these biofilms would hamper the initial adhesion process and in turn progression of biofilm. Proteinase K treatment significantly impacted the biofilm development of most S. aureus isolates. To investigate if the biofilm inhibition was due to the hampered growth or due to the switch-over of lifestyle of the cells to planktonic form, the effect of different concentrations of proteinase K on planktonic cells was tested. There was no impact of proteinase K on cell viability; hence, biofilm inhibition was due to proteolytic cleavage of surface proteins. These observations also reemphasized the important role played by Bap-like surface proteins in biofilm development as similar effect was not observed in bap-mutant M556.
Bap contains four putative Ca2+ binding EF-hand motifs, thus, it was evaluated whether binding of Ca2+ to Bap would confer any resistance against proteinase K-mediated degradation and in turn biofilm dispersion. The result showed that there was no apparent difference between biofilms grown in the presence of proteinase K alone and proteinase K along with increasing concentrations of Ca2+, whereas another set of increasing concentrations of Ca2+ alone showed inhibition of V329 biofilm at higher concentrations. This result demonstrated that binding of Ca2+ ions to Bap had no effect on proteinase K-mediated inhibition of V329 biofilm. In other words, biofilm assay using Ca2+ alone and Ca2+ with proteinase K showed that Ca2+ did not confer any immunity against proteolytic degradation of Bap.
Proteinase K treatment caused a significant dispersal of all pre-grown (24 h) S. aureus biofilms, except M556 and SA392. Since, in the case of M556, Bap does not remain attached to the cell wall, it remains non-functional and does not contribute to biofilm stability. M556 harbours functional ica-operon and hence could produce significant amount of biofilm after 48 h as shown in earlier report11,17. Therefore, it can also be speculated that in weak biofilms by M556 and SA392, polysaccharide intercellular adhesin (PIA) might play a predominant role as their biofilms were not affected by proteinase K treatment.
After proteinase K treatment, a significant decrease in the protein and eDNA but not in the carbohydrate content in EPS was observed. eDNA is also known to play very important role in S. aureus biofilm stability31. As there was a significant decrease in eDNA content along with the biofilm matrix protein content, it was speculated that matrix proteins might also be involved in eDNA retention in the biofilm. Since Ca2+ binds with Bap12 as well as eDNA32, it is speculated that Ca2+ might act as a cross-linking agent between Bap and eDNA, thereby the presence of Bap can assist in retention of eDNA. Therefore, upon proteinase K treatment that degraded Bap, a significant amount of eDNA was also lost along with the proteins. M556 biofilm comprised significant amount of eDNA and carbohydrate, which suggested that in the absence of functional Bap, eDNA and carbohydrate i.e. polysaccharide polymers in matrix might play a crucial role in S. aureus biofilms. In M556 biofilm, matrix proteins do not contribute to biofilm stability despite having higher amount of protein content. The results obtained also indicate that matrix proteins were neither protected by sugars and DNA nor resistant to proteinase K and hence degraded by proteinase K. It is speculated that carbohydrate polymers retain the biofilm structure and do not allow the M556 biofilm to get dispersed upon proteinase K treatment.
It is known that biofilm cells are extremely (1000 times or more) resistant to antibiotics as compared to planktonic cells due to physical as well as genetic reasons33,34. The proteinase K-mediated dispersal of S. aureus biofilms suggested its potential use in enhancing the susceptibility of bacterial cells towards antibiotic treatment. Our earlier report showed that proteinase K treatment in combination with different antibiotics had a synergistic effect on efficacy of antibiotics17. On similar lines, gentamicin efficacy in combination with proteinase K was investigated against four other biofilm-forming S. aureus isolates in this study. The result showed that proteinase K treatment significantly enhanced the efficacy of gentamicin against all S. aureus biofilm tested i.e. SA7, SA10 SA33 and SA352. As shown in our previous report17, proteinase K treatment increases the surface to volume ratio and roughness coefficient of biofilm. Thus, the enhanced values of surface to bio-volume ratio and roughness coefficient after proteinase K treatments leave more biofilm surface available for antibiotics action. Moreover, it was also shown that proteinase K treatment significantly decreased the average diffusion distance as well as maximum diffusion distance17. This enhances the antibiotics penetration in biofilms and in turn its efficacy. Thus, proteinase K shows synergistic effect when associated with antibiotics for biofilm removal.
Enzyme-based S. aureus biofilm disruption has emerged as a promising strategy to combat biofilm-related persistent infections as enzyme-based antibiotic treatment enhances the antibiotic sensitivity of microbial biofilm25,35. Among such enzymes, DNase I, dispersin B and proteinase K are commercially produced. Dispersal mechanisms using such enzymes could be utilized in the prevention of biofilm formation associated with implanted medical devices25. Several studies have found that pre-treatment of polymeric surfaces36 or local delivery of dispersal agents from the implanted device7 should prevent biofilm development. Studies done with recombinant human DNase I to reduce the antigenicity of DNase I enzyme have shown promising results in terms of anti-biofilm activity against S. aureus biofilm37. While these approaches sound promising, there are several concerns that need to be thoroughly addressed before clinical trials. For example, if the antibiotic treatment along with the proteinase K fails to fully eradicate the dispersed microbial cells, it might result in acute infections. Thus, more studies need to be performed to confirm dispersal mechanisms in relevant animal models of infection before treating S. aureus biofilm infections in clinical set-ups.
The present study showed a wider applicability of proteinase K treatment against S. aureus biofilms, and that antibiotics in combination with proteinase K could be more effective in controlling S. aureus biofilm-mediated infection.
Footnotes
Conflicts of Interest: None.
References
- 1.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Mangwani N, Kumari S, Shukla SK, Rao TS, Das S. Phenotypic switching in biofilm-forming marine bacterium Paenibacillus lautus NE3B01. Curr Microbiol. 2014;68:648–56. doi: 10.1007/s00284-014-0525-8. [DOI] [PubMed] [Google Scholar]
- 3.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–47. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention 2009-2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 6.Gravel D, Taylor G, Ofner M, Johnston L, Loeb M, Roth VR, et al. Point prevalence survey for healthcare-associated infections within Canadian adult acute-care hospitals. J Hosp Infect. 2007;66:243–8. doi: 10.1016/j.jhin.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez CJ, Jr, Prieto EM, Krueger CA, Zienkiewicz KJ, Romano DR, Ward CL, et al. Effects of local delivery of D-amino acids from biofilm-dispersive scaffolds on infection in contaminated rat segmental defects. Biomaterials. 2013;34:7533–43. doi: 10.1016/j.biomaterials.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon BP, Geoghegan JA, Waters EM, McCarthy H, Rowe SE, Davies JR, et al. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J Bacteriol. 2014;196:4268–75. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cucarella C, Tormo MA, Ubeda C, Trotonda MP, Monzón M, Peris C, et al. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun. 2004;72:2177–85. doi: 10.1128/IAI.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–96. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla SK, Rao TS. Effect of calcium on Staphylococcus aureus biofilm architecture: A confocal laser scanning microscopic study. Colloids Surf B Biointerfaces. 2013;103:448–54. doi: 10.1016/j.colsurfb.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Snel GG, Monecke S, Ehricht R, Piccinini R. Molecular characteristics of bap-positive Staphylococcus aureus strains from dairy cow mastitis. J Dairy Res. 2015;82:312–6. doi: 10.1017/S0022029915000199. [DOI] [PubMed] [Google Scholar]
- 14.Potter A, Ceotto H, Giambiagi-Demarval M, dos Santos KR, Nes IF, Bastos Mdo C. The gene bap, involved in biofilm production, is present in Staphylococcus spp. strains from nosocomial infections. J Microbiol. 2009;47:319–26. doi: 10.1007/s12275-009-0008-y. [DOI] [PubMed] [Google Scholar]
- 15.Zuniga E, Melville PA, Saidenberg AB, Laes MA, Gonsales FF, Salaberry SR, et al. Occurrence of genes coding for MSCRAMM and biofilm-associated protein Bap in Staphylococcus spp. isolated from bovine subclinical mastitis and relationship with somatic cell counts. Microb Pathog. 2015;89:1–6. doi: 10.1016/j.micpath.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693–8. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar Shukla S, Rao TS. Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J Antibiot (Tokyo) 2013;66:55–60. doi: 10.1038/ja.2012.98. [DOI] [PubMed] [Google Scholar]
- 18.Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Büttner H, et al. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun. 2015;83:214–26. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tormo MA, Knecht E, Götz F, Lasa I, Penadés JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: Evidence of horizontal gene transfer? Microbiology. 2005;151(Pt 7):2465–75. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 20.Lembré P, Vendrely C, Martino PD. Identification of an amyloidogenic peptide from the Bap protein of Staphylococcus epidermidis. Protein Pept Lett. 2014;21:75–9. doi: 10.2174/09298665113209990072. [DOI] [PubMed] [Google Scholar]
- 21.Götz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–78. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 22.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–9. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto S, Iwamoto T, Takada K, Okuda K, Tajima A, Iwase T, et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol. 2013;195:1645–55. doi: 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister JL, Horswill AR. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front Cell Infect Microbiol. 2014;4:178. doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stepanovic S, Vukovic D, Jezek P, Pavlovic M, Svabic-Vlahovic M. Influence of dynamic conditions on biofilm formation by staphylococci. Eur J Clin Microbiol Infect Dis. 2001;20:502–4. doi: 10.1007/s100960100534. [DOI] [PubMed] [Google Scholar]
- 27.Bajrami B, Shi Y, Lapierre P, Yao X. Shifting unoccupied spectral space in mass spectrum of peptide fragment ions. J Am Soc Mass Spectrom. 2009;20:2124–34. doi: 10.1016/j.jasms.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Beaudet MP, Ahnert N, Dallwig JA, Goodman T, Thomas JA. A fast, easy, accurate method for protein quantitation. Faseb J. 2007;21:A1006–7. [Google Scholar]
- 29.Paraneeiswaran A, Shukla SK, Prashanth K, Rao TS. Microbial reduction of [Co(III)-EDTA]- by Bacillus licheniformis SPB-2 strain isolated from a solar salt pan. J Hazard Mater. 2015;283:582–90. doi: 10.1016/j.jhazmat.2014.09.058. [DOI] [PubMed] [Google Scholar]
- 30.del Pozo JL, Patel R. The challenge of treating biofilm-associated bacterial infections. Clin Pharmacol Ther. 2007;82:204–9. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 31.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das T, Sehar S, Koop L, Wong YK, Ahmed S, Siddiqui KS, et al. Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS One. 2014;9:e91935. doi: 10.1371/journal.pone.0091935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyle BD, Costerton JW. Bacterial resistance to antibiotics: The role of biofilms. Prog Drug Res. 1991;37:91–105. doi: 10.1007/978-3-0348-7139-6_2. [DOI] [PubMed] [Google Scholar]
- 34.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–10. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen UT, Burrows LL. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int J Food Microbiol. 2014;187:26–32. doi: 10.1016/j.ijfoodmicro.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 36.Donelli G, Francolini I, Romoli D, Guaglianone E, Piozzi A, Ragunath C, et al. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob Agents Chemother. 2007;51:2733–40. doi: 10.1128/AAC.01249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan JB, LoVetri K, Cardona ST, Madhyastha S, Sadovskaya I, Jabbouri S, et al. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J Antibiot (Tokyo) 2012;65:73–7. doi: 10.1038/ja.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]


