Abstract
Background & objectives:
The emergence and rapid spread of carbapenem resistance mediated by metallo-beta-lactamase (MBL) in Pseudomonas aeruginosa is of major concern due to limited therapeutic options. This study was aimed at detecting the presence of MBL and its association with integrons in imipenem-resistant P. aeruginosa isolates and to determine their genetic relatedness.
Methods:
A total of 213 P. aeruginosa isolates were collected from two tertiary care centres and tested against anti-pseudomonal antibiotics by antimicrobial susceptibility testing, followed by the detection of MBL production by combined disk method. Minimum inhibitory concentration (MIC) of meropenem was determined by E-test. Multiplex polymerase chain reaction (PCR) was performed for the detection of blaSPM, blaIMP, blaVIM, blaNDM, blaGIM and blaSIM. PCR was carried out to characterize the variable region of class 1 integron. Transcongujation assay was carried out for the confirmation of plasmid-mediated resistance. Enterobacterial repetitive intergenic consensus sequence (ERIC)-PCR was performed for determining the genetic relatedness among P. aeruginosa isolates.
Results:
Of the 213 P. aeruginosa isolates, 22 (10%) were found to be carbapenem resistant and these were from pus 18 (82%), urine 2 (9%), sputum 1 (5%) and tracheal wash 1 (5%). Among 22 isolates, 18 (81.8%) were found to be MBL producers by phenotypic method and MIC range of meropenem was 8 to >32 µg/ml. PCR amplification showed that 20 (91%) isolates carried any one of the MBL genes tested: blaVIM and blaNDM in seven (32%) and six (27%) isolates, respectively; blaVIM and blaNDM in three (14%); blaIMP and blaNDM in two (9%); blaVIM and blaIMP in one (5%) isolate. The blaVIM, blaIMP and blaNDM were found to co-exist in one isolate. None of the isolates were positive for blaSPM, blaSIM and blaGIM. All 22 isolates carried class I integron. Of the 20 MBL-positive isolates, transconjugants were obtained for 15 isolates. ERIC-PCR analysis showed all isolates to be clonally independent.
Interpretation & conclusions:
Our results showed 10.3 per cent of carbapenem resistance among P. aeruginosa isolates, and the coexistence of MBL-encoding genes among P. aeruginosa mediated by class I integron.
Keywords: Class I integron, ERIC-PCR, metallo-beta-lactamase, Pseudomonas aeruginosa
Pseudomonas aeruginosa is an important cause of multidrug-resistant (MDR) nosocomial pneumonia, urinary tract infections, surgical site infections and bloodstream infections. It is also the fifth-most common etiological agent of infections, for which carbapenems serve as the last line of drug therapy1. The data of the Carbapenem Antimicrobials Pseudomonas Isolate Testing at regional Locations (CAPITAL) surveillance programme in 2010 reported that the overall rates of carbapenem-resistant P. aeruginosa ranged from 7.4 to 35.4 per cent2. In 2015, a meta-analysis study showed that the rates of carbapenem resistance ranged from 8.7 to 50.4 per cent among P. aeruginosa3. Carbapenem resistance is mainly due to upregulation of efflux pumps, decreased outer membrane permeability and acquired metallo-beta-lactamases (MBL)4. MBL belong to Amber class B, require zinc for its catalytic activity and are inhibited by metal chelators, such as ethylenediaminetetraacetic acid and thiol-based compounds. Various numbers of MBLs are identified worldwide including IMP, VIM, NDM, SPM, GIM and SIM, whereas in India, blaNDM, blaVIM and blaIMP genes are frequently encountered in P. aeruginosa5.
Integrons are mobile genetic elements, usually located in plasmids or transposons. Class I integron are genetic structures capable of capturing gene cassettes, which possess 5’-CS and 3’-CS on either side of the integrated structures. The 5’-CS consists of integrase gene (Int1) and attachment site (attI), and 3’-CS contains antisepsis resistance qacED1 gene; the sul1 gene, which confers resistance to sulphonamides and an open reading frame (ORF) of unknown function, ORF56. MBL-encoding genes are found to be located within the integron structure along with other resistant determinants and rapidly spread among different species by horizontal gene transfer mechanism7. Various typing methods such as multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE)8, ribotying, BOX polymerase chain reaction (PCR), repetitive extragenic palindromic sequence PCR and enterobacterial repetitive intergenic consensus sequence (ERIC)-PCR are available. ERIC-PCR is an easy, inexpensive and reproducible method for DNA typing. Thus, this study was undertaken to detect the presence of MBL and its association with integrons in imipenem-resistant strains of P. aeruginosa and to investigate their genetic relatedness using ERIC-PCR.
Material & Methods
A total of 213 non-duplicate isolates of P. aeruginosa were collected during a period of November 2013- December 2014 from two tertiary care hospitals in southern India. One hundred and eighteen isolates from the department of Microbiology, Sri Manakula Vinayagar Medical College and Hospital, Puducherry, and 95 isolates from the department of Microbiology, ESIC Hospital, Chennai, were collected. Antimicrobial susceptibility testing was done by Kirby–Bauer disk diffusion method9 using the following antibiotics (µg): piperacillin (100), piperacillin-tazobactam (100/10), amikacin (30), gentamicin (10), tobramycin (10), netilmicin (30), colistin (10), polymyxin B (300 units), ciprofloxacin (5), ofloxacin (5), levofloxacin (10), ceftazidime (30), cefepime (30), aztreonam (30), ceftazidime-clavulanic acid (30/10), imipenem (10) and meropenem (10) and results were interpreted as per the Clinical and Laboratory Standards Institute (CLSI) guidelines 201310. P. aeruginosa ATCC 27853 was included as a standard strain.
Phenotypic detection of carbapenem resistance: MBL production was screened by combined disk method11. Briefly, a liquid culture of the test isolate was adjusted to a turbidity of 0.5 McFarland standard and spread on the surface of a Mueller-Hinton agar plate (HiMedia, Mumbai). Two disks of 10 µg imipenem were placed on the agar at a distance of 25 mm apart; 10 µl of 0.5 M EDTA was added to one of the disks. After overnight incubation at 37°C, an increase of 7 mm or more in zone diameter in the presence of imipenem-EDTA as compared to imipenem alone was considered as positive for the presence of an MBL. Minimum inhibitory concentration (MIC) was determined for meropenem using E-strips (HiMedia).
PCR amplification protocols: DNA extraction was done by boiling lysis method12. The cell suspension from an overnight culture was boiled at 100°C for 10 min and immediately kept at -20°C for at least 6 h. The supernatant was used as a template for PCR amplification and was stored at -20°C.
The presence of MBL-encoding genes was detected by multiplex PCR using primers specific for blaSPM, blaIMP, blaVIM, blaNDM, blaGIM and blaSIM (Table I)13,14. The reaction mixture consisted of 2.5 µl of 10x Taq buffer with MgCl2, 0.5 µl of 10 mM dNTP (2.5 mM each), 0.5 µl of Taq DNA polymerase (1.5 unit), 10 µM of each primer (forward and reverse) in a total volume of 23 µl with 2 µl of DNA. For detection of class 1 integron, integrase gene PCR was performed15. The thermal cycling conditions included initial denaturation at 94°C for five minutes followed by thirty cycles of DNA denaturation at 94°C for 30 sec; annealing at 52°C for 40 sec, primer extension at 72°C for 50 sec with a final extension step of 72°C for five minutes (Eppendorf Mastercycler Personal, Hamburg, Germany). BAA 2146 (Klebsiella pneumoniae) was used as positive control for blaNDM; NCTC 13476 (Escherichia coli) for blaIMP and NCTC 13439 (Klebsiella pneumonia) for blaVIM.
Table I.
List of primers used in this study
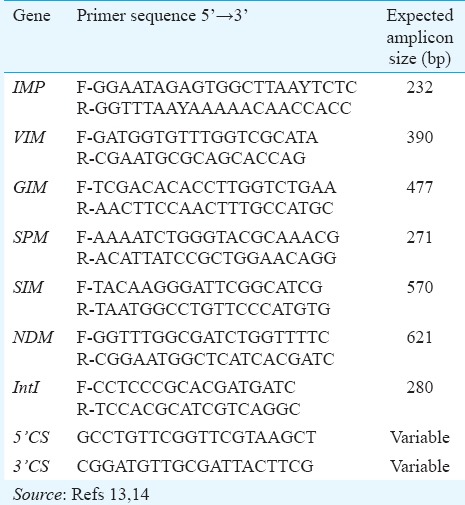
MBL-encoding genes are located in self- transferable plasmids within integrons; thus, PCR was performed for the variable region in class I integron using 5’-CS and 3’-CS specific primers16. The regions located upstream of the bla gene were amplified with forward primer of Int (5’-CS) gene of class 1 integron together with reverse primer of blaVIM or blaIMP or blaNDM (VIM-R/5’-CS, IMP-R/5’-CS, NDM-R/5’-CS). The sequences located downstream of the bla gene were amplified with 3’-CS with reverse-primer of blaVIM or blaIMP or blaNDM (VIM-F/3’-CS, IMP-F/3’-CS, NDM-R/3’-CS).
Conjugation assay: Transfer of resistant genes was evaluated through conjugation assay using a rifampin-resistant mutant of P. aeruginosa PU2117,18. All MBL-producing isolates were used as donors and P. aeruginosa PU21 as the recipient. An overnight culture of donor (0.1 ml) and recipient cells (0.4 ml) were incubated for 18-24 h, and transconjugants grown in trypticase soy agar plates containing rifampin (100 µg/ml) and ceftazidime (4 µg/ml) after 48 h of incubation were selected. Plasmid extraction of donor and transconjugants was done19 and screened for the presence of carbapenemase-encoding genes by PCR as described above.
Enterobacterial repetitive intergenic consensus- polymerase chain reaction (ERIC-PCR): Genomic bacterial DNA was used for the ERIC-PCR reactions using the sequences ERIC 1 (5’-ATGTAAGCTCCTGGGGATTCAC -3’) and ERIC 2 (5’-AAGTAAGTGACTGGGGTGAGCG -3’). The reaction mix contained 2.5 µl of 10x Taq buffer with MgCl2, 0.5 µl of 10 mM dNTP (2.5 mM each), 0.5 µl of Taq DNA polymerase (1.5 units), 10 µM of forward and reverse primer each in a total volume of 23 µl with 2 µl of DNA template. PCR amplification was carried out with an initial denaturation of 95°C for seven minutes followed by thirty cycles of DNA denaturation at 94°C for one minute, annealing at 52°C for one minute, primer extension at 65°C for eight minutes and final extension at 65°C for 16 min20. ATCC 27853 P. aeruginosa was used as control. ERIC-PCR analysis was performed using GelQuest version 3.2.1 (Digital DNA processing, Klein Raden, Germany) and SequentiX (Digital DNA processing) and ClusterVis version 1.8.2 (Digital DNA processing). Pearson Phi coefficient was calculated and compared to evaluate similarity among strains. Clustering was performed by neighbour-joining method.21
PCR products were analyzed by electrophoresis with 2% agarose gel in 1x Tris Borate EDTA buffer. The gel was stained with ethidium bromide, and the PCR products were visualized using the gel documentation system (Carestream Gel Logic 212 PRO, New York, USA).
The study protocol was cleared by the Institutional Ethics Committee.
Results
Two hundred and thirteen isolates of P. aeruginosa were obtained from various clinical specimens, such as pus (66%), urine (10%), sputum (15%), blood (4%), bronchoalveolar lavage (2%), semen (2%) and tracheal wash (1%).
Among these, 22/213 (10.3%) isolates were resistant to all the antibiotics tested, except colistin and polymyxin B, and were chosen for the present study. Of the 22 carbapenem-resistant P. aeruginosa (CRPA) isolates, 18 (82%) were considered as positive and four (18%) isolates were negative for MBL production by combined disk method. These isolates had MIC values to meropenem which ranged from 8 to >32 µg/ml by E-test.
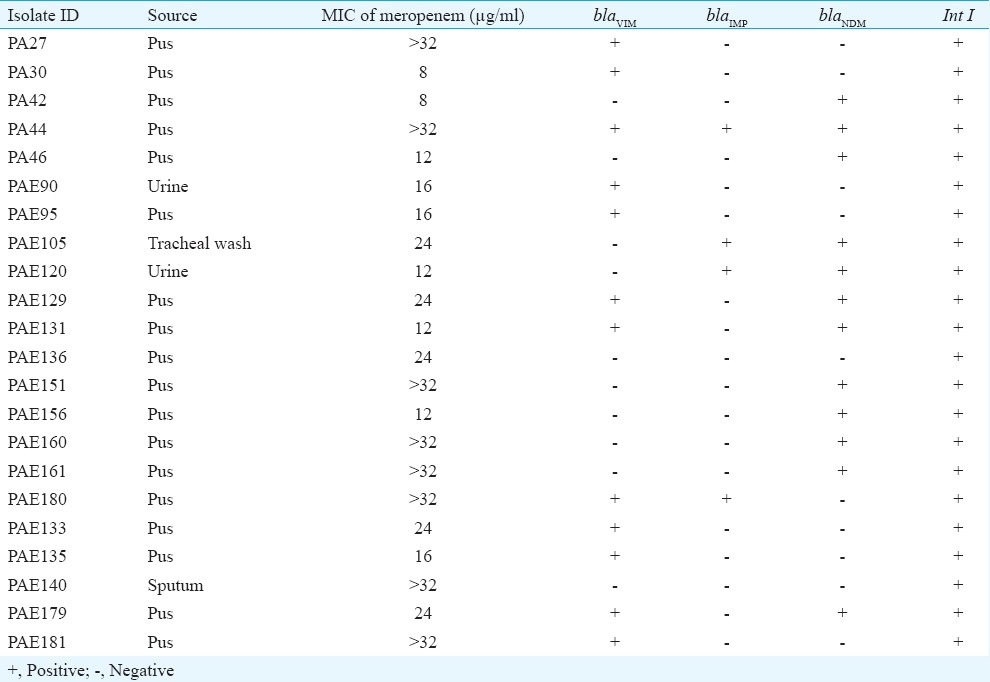
Genotypic detection of MBLs revealed that 91 per cent (20/22) of CRPA isolates harboured at least one MBL gene. Combined disk test was positive for isolates that harboured blaIMP, blaVIM and blaNDM genes. Of the 22 isolates, blaVIM and blaNDM genes were detected in seven (32%) and six (27%) isolates, respectively; three (14%) isolates had both blaVIM and blaNDM; two (9%) isolates had blaIMP and blaNDM and one (5%) isolate carried blaVIM and blaIMP in combination. Three MBLs (blaIMP, blaVIM and blaNDM) were found to co-exist in one isolate (Fig. 1). Two (9%) isolates positive for phenotypic MBL production were found to be negative for all the MBL genes tested. None of the isolates carried blaSPM, blaGIM and blaSIM. The distribution of MBL-encoding genes and their source is given in Table II.
Fig. 1.
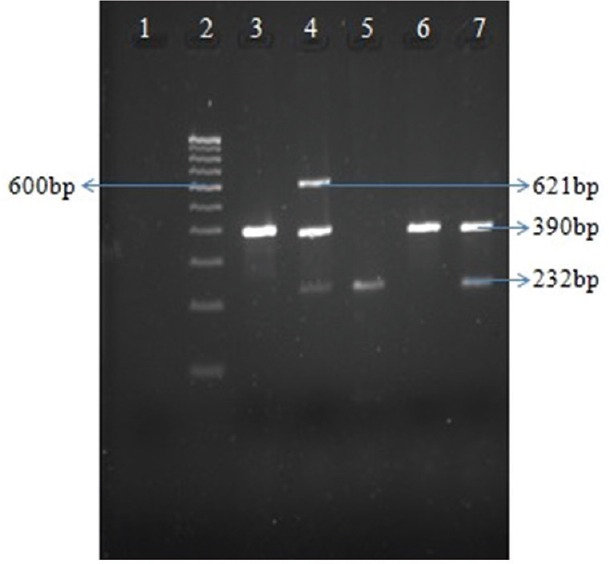
Multiplex polymerase chain reaction for detection of metallo-beta-lactamase-encoding genes. Lane 1, negative control; lane 2, 100 bp ladder; lane 3, PA32 (VIM positive); lane 4, PA44 (VIM, IMP, NDM positive); lane 5, PAE181 (IMP positive); lane 6, PA27 (VIM positive); lane 7, PAE180 (IMP, VIM positive).
Table II.
Distribution of bla genes and their source and minimum inhibitory concentration (MIC) value of carbapenem resistant isolates
Screening of class I integron revealed that all 22 isolates carried Int I gene. Using the 59-CS and 39-CS primers, the variable regions of the integrons were amplified. Further, PCR with class I integron (5’ & 3’) conserved sequence was amplified in 12 (55%) isolates which showed approximately 1700 and 2250 bp in size (Fig. 2) and were not amplified in 10 (45%) isolates. Among 12 VIM-positive isolates, 5’-CS/VIM-R was amplified in five isolates (amplicon size ~1500 bp) whereas 3’-CS/VIM-F was amplified in four isolates (~1000 bp). Of the four IMP-positive isolates, amplification was observed in two isolates with 5’-CS/IMP-R (~500 bp) and no amplification was observed in 3’-CS/IMP-F. Amplification was observed in all 12 NDM-positive isolates using 5’-CS/NDM-R (~500 bp) and 3’-CS/NDM-F (~400 bp). Transconjugants of PU21 P. aeruginosa were obtained for 15 isolates and repeated attempts failed to produce the transconjugants for the remaining isolates. The multiplex PCR which was done on the PU21 transconjugants revealed resistance to the same carbapenems as the donor strain.
Fig. 2.
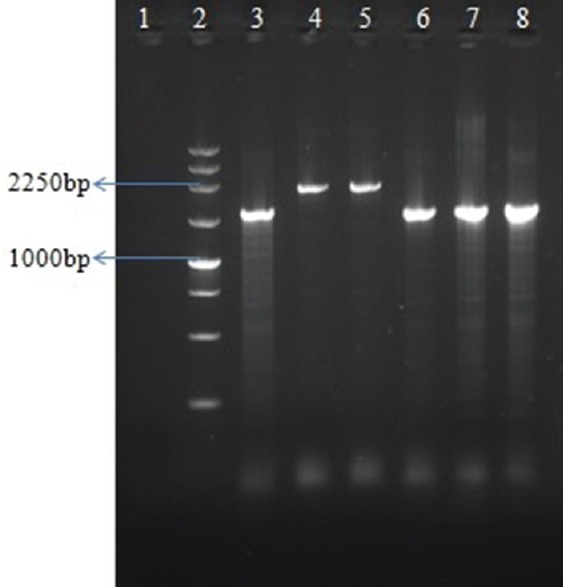
Polymerase chain reaction for detection of gene cassettes (5’-CS with 3’-CS). Lane 1, negative control; lane 2, 250 bp Ladder; lane 3-8, PA27, PA30, PA42, PA44, PAE120, PAE131.
Analysis of ERIC-PCR showed four main clusters (A-D). Cluster A and B contained the majority (n=17, 77%) of the isolates. Four isolates (18%) and one isolate (5%) formed cluster C and cluster D, respectively (Fig. 3).
Fig. 3.
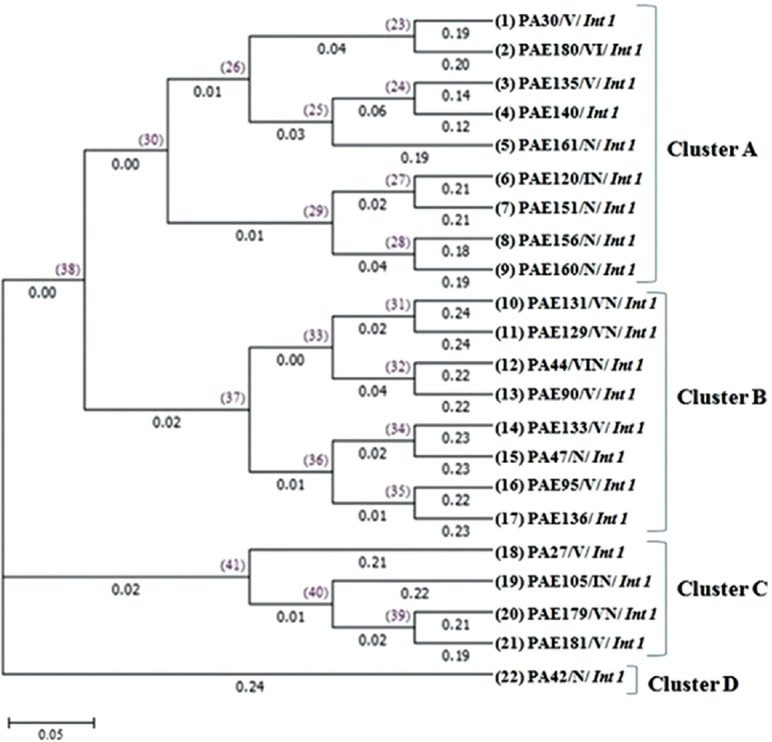
Dendrogram tree of unweighted pair group method with arithmetic mean method of enterobacterial repetitive intergenic consensus sequence-polymerase chain reaction (ERIC-PCR). The values on top of the horizontal lines represent the node identity and the values given below the horizontal lines represent the branch length. The bar at the bottom of the figure represents an amount of genetic change (0.05).
Discussion
Treatment of infections caused by P. aeruginosa is challenging due to increase in the prevalence of MDR strains. Carbapenems are the most effective drugs used for the treatment of MDR infections; however, their inappropriate usage has led to the emergence of resistance. In south India, 12 per cent of carbapenem resistance was reported in Karnataka and Vellore22,23. Others have reported 10.9 and 16 per cent of carbapenem resistance in Puducherry24 and Chennai25, respectively. Mendiratta et al26 have shown 8.6 per cent of imipenem-resistance amongst P. aeruginosa in a hospital setup. In the present study, 10.3 per cent of isolates were resistant to imipenem and meropenem.
Various phenotypic methods have been described for the detection of MBL. Lee et al27 have shown 100 and 88 per cent sensitivity and specificity in modified Hodge test and 100 per cent sensitivity and specificity for EDTA disk synergy test. A similar finding has been described by Noyal et al28. In this study, 18 isolates were found to be MBL producers by combined disk method and four isolates which were negative by phenotypic method harboured IMP and VIM genes. Amudhan et al29 have reported 51.4 per cent of carbapenem resistance due to blaVIM /blaIMP in P. aeruginosa. Another study reported five types of VIM enzymes in Pseudomonas spp30. Further, a prospective study conducted in Puducherry showed 13.3 per cent of VIM-2 and 1.3 per cent of IMP-1 type MBL producing isolates of P. aeruginosa31. In the present study, VIM and NDM genes were detected in 55 per cent and IMP in 18 per cent of CRPA isolates. Two isolates which were positive by phenotypic method were negative with PCR. This may be due to the contribution of integron-mediated ESBLs with carbapenem hydrolyzing activity, increased expression of efflux systems, reduced porin expression and increased chromosomal cephalosporinase activity32. Class I integron is the most frequently observed type of integrons. In our study all imipenem resistant isolates carried class I integron. The class I integron conserved sequence was not observed in 10 of 22 isolates and subsequent amplification of the 3’-CS with combination of primers also failed. This showed the lack of 3’CS in class I integron and this may be due to the recombination of 5’-CS of class 1 integron with the tns module of class 2. This module is similar to Tn402 (TN5090), progenitor of class I integron16. Epidemiological typing by ERIC-PCR demonstrated the presence of four clusters among MDR isolates with majority of isolates falling into cluster A and B. ERIC-PCR profile did not show any genetic relatedness and it indicated individual origins of dissemination. The study has certain lacunae such as lack of clinical details and patient follow up for assessment of clinical outcomes.
In conclusion, our findings showed 10.3 per cent of carbapenem resistance among P. aeruginosa isolates. Combined disk method was found to be less reliable than genotypic methods for presumptive identification of MBLs. All resistant isolates were associated with class I integron. Our study also documented the coexistence of bla MBL genes.
Acknowledgment
Authors thank Dr George A. Jacoby, Infectious Disease Specialist, Lahey Clinic Medical Center, Burlington, for providing PU21 P. aeruginosa, Dr Sunil Santaji Shivekar, Sri Manakula Vinayagar Medical College and Hospital, Puducherry, and Drs Lakshmi Priya and Esther Mary, ESIC Hospital, Chennai, for providing clinical isolates.
Footnotes
Conflicts of Interest: None.
References
- 1.El Solh AA, Alhajhusain A. Update on the treatment of Pseudomonas aeruginosa pneumonia. J Antimicrob Chemother. 2009;64:229–38. doi: 10.1093/jac/dkp201. [DOI] [PubMed] [Google Scholar]
- 2.Morrow BJ, Pillar CM, Deane J, Sahm DF, Lynch AS, Flamm RK, et al. Activities of carbapenem and comparator agents against contemporary US Pseudomonas aeruginosa isolates from the CAPITAL surveillance program. Diagn Microbiol Infect Dis. 2013;75:412–6. doi: 10.1016/j.diagmicrobio.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Li X, Li W, Du X, He JQ, Tao C, et al. Influence of carbapenem resistance on mortality of patients with Pseudomonas aeruginosa infection: A meta-analysis. Sci Rep. 2015;5:11715. doi: 10.1038/srep11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:4783–8. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanthi M, Sekar U, Kamalanathan A, Sekar B. Detection of New Delhi metallo beta lactamase-1 (NDM-1) carbapenemase in Pseudomonas aeruginosa in a single centre in Southern India. Indian J Med Res. 2014;140:546–50. [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Lambert T, Türkoglü S, Ronco E, Gaillard J, Nordmann P. Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla(VIM-2) carbapenem-hydrolyzing beta-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother. 2001;45:546–52. doi: 10.1128/AAC.45.2.546-552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khosravi Y, Tay ST, Vadivelu J. Analysis of integrons and associated gene cassettes of metallo-β-lactamase-positive Pseudomonas aeruginosa in Malaysia. J Med Microbiol. 2011;60(Pt 7):988–94. doi: 10.1099/jmm.0.029868-0. [DOI] [PubMed] [Google Scholar]
- 8.Maâtallah M, Bakhrouf A, Habeeb MA, Turlej-Rogacka A, Iversen A, Pourcel C, et al. Four genotyping schemes for phylogenetic analysis of Pseudomonas aeruginosa: Comparison of their congruence with multi-locus sequence typing. PLoS One. 2013;8:e82069. doi: 10.1371/journal.pone.0082069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement M100-S23. Wayne, PA: CLSI; 2013. [Google Scholar]
- 11.Behera B, Mathur P, Das A, Kapil A, Sharma V. An evaluation of four different phenotypic techniques for detection of metallo-beta-lactamase producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26:233–7. doi: 10.4103/0255-0857.39587. [DOI] [PubMed] [Google Scholar]
- 12.Pitout JDD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–35. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59:321–2. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother. 2011;55:934–6. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, et al. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother. 2001;45:723–6. doi: 10.1128/AAC.45.3.723-726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toleman MA, Vinodh H, Sekar U, Kamat V, Walsh TR. blaVIM-2-harboring integrons isolated in India, Russia, and the United States arise from an ancestral class 1 integron predating the formation of the 3’conserved sequence. Antimicrob Agents Chemother. 2007;51:2636–8. doi: 10.1128/AAC.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001;45:2598–603. doi: 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby GA. Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1974;6:239–52. doi: 10.1128/aac.6.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–23. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stehling EG, Leite DS, Silveira WD. Molecular typing and biological characteristics of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Brazil. Braz J Infect Dis. 2010;14:462–7. [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Navaneeth BV, Sridaran D, Sahay D, Belwadi MR. A preliminary study on metallo-beta-lactamase producing Pseudomonas aeruginosa in hospitalized patients. Indian J Med Res. 2002;116:264–7. [PubMed] [Google Scholar]
- 23.Gladstone P, Rajendran P, Brahmadathan KN. Incidence of carbapenem resistant nonfermenting Gram negative bacilli from patients with respiratory infections in the Intensive Care Units. Indian J Med Microbiol. 2005;23:189–91. doi: 10.4103/0255-0857.16593. [DOI] [PubMed] [Google Scholar]
- 24.Shashikala Kanungo R, Srinivasan S, Devi S. Emerging resistance to carbapenems in hospital acquired Pseudomonas infection: A concern for concern. Indian J Pharmacol. 2006;38:287–8. [Google Scholar]
- 25.Hemalatha V, Sekar U, Kamat V. Detection of metallo betalactamase producing Pseudomonas aeruginosa in hospitalized patients. Indian J Med Res. 2005;122:148–52. [PubMed] [Google Scholar]
- 26.Mendiratta DK, Deotale V, Narang P. Metallo-beta-lactamase producing Pseudomonas aeruginosa in a hospital from a rural area. Indian J Med Res. 2005;121:701–3. [PubMed] [Google Scholar]
- 27.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 28.Noyal MJ, Menezes GA, Harish BN, Sujatha S, Parija SC. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res. 2009;129:707–12. [PubMed] [Google Scholar]
- 29.Amudhan MS, Sekar U, Kamalanathan A, Balaraman S. bla(IMP) and bla(VIM) mediated carbapenem resistance in Pseudomonas and Acinetobacter species in India. J Infect Dev Ctries. 2012;6:757–62. doi: 10.3855/jidc.2268. [DOI] [PubMed] [Google Scholar]
- 30.Castanheira M, Bell JM, Turnidge JD, Mathai D, Jones RN. Carbapenem resistance among Pseudomonas aeruginosa strains from India: Evidence for nationwide endemicity of multiple metallo-beta-lactamase clones (VIM-2, -5, -6, and -11 and the newly characterized VIM-18) Antimicrob Agents Chemother. 2009;53:1225–7. doi: 10.1128/AAC.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramakrishnan K, Rajagopalan S, Nair S, Kenchappa P, Chandrakesan SD. Molecular characterization of metallo β-lactamase producing multidrug resistant Pseudomonas aeruginosa from various clinical samples. Indian J Pathol Microbiol. 2014;57:579–82. doi: 10.4103/0377-4929.142670. [DOI] [PubMed] [Google Scholar]
- 32.Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16:303–7. [PMC free article] [PubMed] [Google Scholar]


